Back to Journals » International Journal of Nanomedicine » Volume 20
A Super-Assembled Synergistically Nanoplatform AP@ZIF-8Pt for Hepatocarcinoma Therapy
Authors Luo Z, Wang D, Lin L, Zhou R, Su Y, Zhang Z, Hu J, Dai Y, Wu J , Huang X, Zhou Y, Gong L
Received 16 January 2025
Accepted for publication 16 April 2025
Published 1 May 2025 Volume 2025:20 Pages 5681—5692
DOI https://doi.org/10.2147/IJN.S516464
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Dong Wang
Zhenzhen Luo,2,* Dunhuang Wang,1,* Lie Lin,1 Rui Zhou,1 Yuanyuan Su,1 Zongkai Zhang,1 Jing Hu,1 Yaqing Dai,1 Jingjing Wu,1 Xiaoyan Huang,1 Yufei Zhou,1 Liuyun Gong1
1Department of Radiation Oncology, Xiamen Cancer Quality Control Center, Xiamen Cancer Center, Xiamen Key Laboratory of Radiation Oncology, The First Affiliated Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, Fujian, 361003, People’s Republic of China; 2Shenzhen Cancer Hospital, Chinese Academy of Medical Sciences, Shenzhen, Guangdong, 518000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liuyun Gong, Email [email protected] Yufei Zhou, Email [email protected]
Introduction: Intensive cancer treatment with nanoplatform is widely exploited in the clinic, the emerging nanomedicine offers an unparalleled opportunity for encapsulating potential antitumor drugs in a nano-carrier. Apoptin (AP), a coding protein of VP3 gene, stem from the chicken anemia virus (CAV), can be activated in malignant cells selectively and prevents the dividing cancer cells from repairing their DNA lesions, thereby forcing them to undergo apoptosis. Herein, a three-step intelligent biodegradable drug delivery nanoplatform was designed.
Methods: First, a hollow ZIF-8 was synthesized, embedded with platinum nanoparticle to form ZIF-8Pt, and then loaded with AP, and lastly formed AP@ZIF-8Pt, which possess pH-responsive drug release and cancer-targeted ability.
Results: As expected, both in vitro and in vivo experiment demonstrated that AP@ZIF-8Pt performed treatment effects in hepatocarcinoma through relieving tumor-hypoxic microenvironment, inhibiting cell proliferation and promoting cell apoptosis. Further transcriptomic analysis showed that the specific mechanism of the AP@ZIF-8Pt was thermogenesis, signaling pathways regulating pluripotency of stem cells, ribosome, prion disease and PI3K-Akt signaling pathway.
Discussion: This work highlights a new strategy for liver cancer treatment and provides a reference for treating malignant tumors.
Keywords: AP@ZIF-8Pt, tumor-hypoxic microenvironment, apoptosis, PI3K-Akt signaling pathway
Graphical Abstract:

Introduction
Liver cancer, specifically hepatocellular carcinoma (HCC), is one of the most frequent malignancies that causes cancer-related death.1–3 Although immune checkpoint blockade-based immunotherapies and targeted therapy of HCC have achieved remarkable success in the field of HCC treatment, few of them induce durable responses and provide prominent survival benefits in patients because of the HCC physiological complexity.4–6 To overcome it, effective strategies to optimizing HCC treatment are urgently needed,7,8 and there are two emerged strategies: finding new drugs according to specific mechanisms and developing novel drug delivery system to give full play to the new drugs.9
As for the new drugs finding, drug selectivity to cancer cells is the key issue of antitumor therapy.10 A group of proteins, killing cancer cells specifically without harming the normal cells, has attracted scientific interest.11 Apoptin (AP) is one of these proteins. In the normal cells, AP becomes filamentous to aggregated and then degraded through proteasomes. While in cancer cells, AP induces apoptosis.12–14 Our early study has proved that AP could induce apoptosis in hepatocarcinoma cells through targeting XPO1.15 So we focused on AP as the potential HCC therapy. As for the novel drug delivery system developing, the emerging nanoplatform provides a new direction, which delivering the drugs and regulating tumor-hypoxic microenvironment synergistically, therefore achieving remarkable therapeutic effect.16 Among them, zeolitic imidazolate framework-8 (ZIF-8) is a metal-organic framework (MOF) serving as a promising candidate for its stability in aqueous environments and decomposability in acidic environments.17–19 Moreover, ZIF-8 possesses good biocompatibility, can reduce immune reactions and increase drugs bioavailability in vivo.20–22 Platinum nanoparticles (Pt) can catalyze H2O2 (100 M-1 mm) to produce oxygen, thereby relieving tumor hypoxia in tumor tissues.23,24
Herein, we design a fire-new nanoplatform, using ZIF-8 implanted with Pt(ZIF-8Pt), after AP loading, the final AP@ZIF-8Pt was used to HCC therapy. First, AP@ZIF-8Pt had decomposability in the weakly acidic tumor microenvironment (TME), and then releases loaded AP. Second, the released AP can induce apoptosis in hepatocarcinoma cells. Third, Pt works as a catalyst, trigger endogenous H2O2 into O2, relieving hypoxia of TME and further improving the drug efficacy of AP. Expectedly, both in vitro and in vivo experiment results demonstrated that AP@ZIF-8Pt possessed the extraordinary biocompatibility and enhanced therapeutic effect through inducing apoptosis, promoting DNA damage of hepatocarcinoma cells. Further transcriptomic analysis showed that the specific mechanism of the AP@ZIF-8Pt was thermogenesis, signaling pathways regulating pluripotency of stem cells, ribosome, prion disease and PI3K-Akt signaling pathway. Collectively, this work highlights a new strategy for HCC therapy and can be a reference for developing the advanced antitumor therapy.
Materials and Methods
Materials
1,2-dimethylimidazole (2-MIL), zinc nitrate hexahydrate (Zn (NO3)2·6H2O), chloroplatinic acid hydrate (H2PtCl6·6H2O) and polyvinylpyrrolidone (PVP) were purchased from Aladdin. AP was synthesized by Shanghai Qiangyao Biotechnology Co., Ltd. 4ʹ6-diamidino-2-phenylindole (DAPI) and 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) were purchased from Beyotime Biotechnology. The γ-H2AX antibody, HIF-1α antibody and secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). All of the reagents used in the experiments were of analytical grade and used directly without treatment.
Characterizations
Malvern Zetasizer Nano ZS90 Apparatus, UV-2550 Ultraviolet-visible Spectrophotometer (Shimadzu, Japan), transmission electron microscopy system (JEOL/JEM-F200), XRD (Bruker/D8 ADVANCE), FTIR (Bruker/VERTEX70), FC500 flow cytometer (Beckman Coulter, USA) were used for material characterization same as our previous studies.25,26
Synthesis of AP@ZIF-8Pt
Zinc nitrate hexahydrate (25 mm) and 2-methyl imidazole (200 mm) were first dissolved in methanol. Then, 20 mL of methanol solution was stirred together with Pt nanoparticles. After continuous stirring for 3–4 h, the sample was washed three times with methanol and ZIF-8Pt was obtained. Then redispersed ZIF-8Pt in ethanol (10 mL) added AP and stirred for 6–8 h. The mixed solution was then washed with methanol (10000 rpm, 10 min) to remove excess AP then obtained AP@ZIF-8Pt.
In vitro Cytotoxicity Assay
The cytotoxicity against HCC cells was measured by CCK8 assay. The cells were cultured in a 96-well plate and incubated overnight. Next, gradient concentrations of AP@ZIF-8Pt (0, 20, 40, 60, 80 and 100 μg/mL) were added to each well and incubated for 24 h. Five parallel samples were set for each group, then 10 µL of CCK8 was added to each well and incubated for 4 h. The absorbance was quantified by a spectrophotometer at 450 nm.
Reactive Oxygen Species (ROS) Detection
ROS in cells was detected using the ROS probe, DCFH-DA. Briefly, HCC cells were seeded in a 6-well plate and cultured overnight. ZIF-8 and ZIF-8Pt were added to the cells and incubated in the dark for 2 h. Following incubation, the medium was removed and cells were washed with PBS three times. Following washing, the ROS probe, DCFH-DA, was added to the cells and incubated for a further 30 min, and then the fluorescence of cells generated by DCFH-DA was detected by fluorescence microscopy.
Cellular Uptake Analysis
HCC cells were seeded in confocal chambers and cultured overnight. The AP loaded Rhodamine B to track nanoparticles, were added to the chambers and incubated for 2 h. Next, the cells were washed with PBS three times and stained with phalloidin-FITC (30 min) and DAPI (10 min). Finally, the cells were observed by laser confocal microscopy (phalloidin-FITC, excitation: 490 nm; DAPI, excitation:400 nm and Rhodamine B, excitation:555 nm). We further investigated the cellular uptake of nanoparticles by FE-TEM. Briefly, HCC cells were cultured in a cell slide overnight. After incubating with AP@ZIF-8Pt for 2 h, the cell slides were washed with PBS. The cells were fixed with 3% glutaraldehyde overnight and then dehydrated in an ethanol series. Using a mini-gold sputter, cell samples were coated with gold-palladium for visual inspection by TEM.
Flow Cytometry Analysis
HCC cells at a concentration of 1×105 cells were seeded in six-well plates and incubated overnight. Cells were treated with AP@ZIF-8Pt, which were loaded with rose red B. After incubation for 3 h and 6 h, the cells were washed three times with cold PBS and the treated cells were harvested for flow cytometry analysis. The cells were seeded in 6 well plates at a density of 2×105 cells and cultured overnight. Apoptosis kit (BD Biosciences, CA) was used for apoptosis analysis and operated according to the instruction.
Immunofluorescence
HCC cells were fixed with 4% PFA at 37°C for 20 min, treated with 0.02% Triton X-100 for 10 min and then blocked at room temperature for 1 h. Cells were incubated with primary antibody overnight at 4°C followed by incubation with fluorescent-conjugated secondary antibody for 2 h at room temperature. The nuclei were stained with DAPI for 15 min. The images of the cells were obtained by laser confocal microscopy (γ-H2AX-FITC, excitation: 490 nm, HIF-1α-CY5, excitation: 650 nm and DAPI, excitation: 400 nm).
In vivo Therapeutic Effect
When the tumor reached approximately 100 mm3, tumor-bearing mice were randomly divided into 4 groups: (I) Control; (II) AP; (III) ZIF-8Pt, and (IV) AP@ZIF-8Pt. After the nanoparticles (50 mg∙kg−1) were intravenously injected into the tail veins every 2 d. For treatment, the solid tumor was dissected from one mouse in each group for H&E and TUNEL staining. The solid tumor was also used for HIF-1α and γ-H2AX IHC staining. The body weights of mice and the tumor volumes were recorded every other day.
Transcriptomics Analysis
When the tumor reached about 100 mm3, tumor-bearing mice were randomly divided into three groups: (I) Control; (II) ZIF-8Pt; (III) AP@ZIF-8Pt. Nanoparticles (50 mg/kg) were intravenously injected via tail vein every 2d. At the end of the experiment, tumors were dissected for transcriptomics analysis using an LC-ESI-MS/MS system.
Statistical Analyses
One-way and two-way analyses of variance (ANOVA) and t-tests were used to determine statistical significance (*p<0.05, **p<0.01, ***p<0.001), p-value less than 0.05 was considered statistically significant.
Results and Discussion
AP@ZIF-8Pt Synthesis and Characterization
First, zinc nitrate and 2-methylimidazole were used to synthesize zeolitic imidazolate framework 8 (ZIF-8).27 Second, polyvinylpyrrolidone (PVP)-modified Pt nanoparticles was added in to synthesize ZIF-8Pt.25,28,29 Third, loaded AP to form AP@ZIF-8Pt. The form of ZIF-8 was first scanned by scanning electron microscope (SEM) (Figure 1A), made sure the framework was correct. The particle size and morphology of AP@ZIF-8Pt was scanned by SEM (Figure 1B) and transmission electron microscopy (TEM) (Figure 1C), verified nanoparticles successful formation. The hydrated particle size of AP@ZIF-8Pt was measured by dynamic light scattering (DLS), which was about 150 nm (Figure 1D). Additionally, the X-ray diffraction (XRD) was performed to analyze the structure of AP@ZIF-8Pt. The result showed that AP@ZIF-8Pt XRD pattern was highly consistent with AP@ZIF-8 alone (Figure 1E), and the peaks (111), (200), (220), and (310) at high angles of 30°–90° corresponding to that of Pt (Figure 1F), suggesting that Pt were coated on ZIF-8 successfully. Further pH-responsive decomposition behavior of AP@ZIF-8Pt was investigated, as shown in Figure 1G, when the pH decreased to 6.0, the accumulative release of AP was significantly increased, suggesting that AP@ZIF-8Pt was easier to degrade in acidic tumor microenvironment. Furthermore, the surface potential of AP@ZIF-8Pt was +26.2 mV measured by Zeta potential (Figure 1H). Finally, size and polydispersity index (PDI) of AP@ZIF-8Pt proved the outstanding stability of AP@ZIF-8Pt, which contained stability in phosphate-buffered saline (PBS) with 10% fetal bovine serum (FBS) for a week (Figure 1I), suggesting that AP@ZIF-8Pt could be used in in vivo potentially.
Cellular Uptake and ROS Generation of AP@ZIF-8Pt
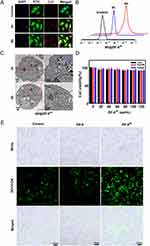
To verify the cellular uptake ability of AP@ZIF-8Pt in HCC cells, AP@ZIF-8Pt was added to Hep3B cells for 6 h. Confocal imaging showed that after 3 h, part of AP@ZIF-8Pt entered the cells and almost all AP@ZIF-8Pt entered the cells after 6 h (Figure 2A), consistently with the flow cytometry analysis for quantitative detection result (Figure 2B). Further field emission transmission electron microscopy (FE-TEM) was applied to verify the real-time absorption of AP@ZIF-8Pt to Huh7 cells. The versatility of FE-TEM allows detailed characterization of the endocytosis of nanoparticles. FE-TEM images showed much AP@ZIF-8Pt entered in HCT-116 cells (marked by blue circle) (Figure 2C). The biocompatibility of ZIF-8Pt was evaluated by Cell Counting Kit-8 (CCK-8) assay, when the concentration of ZIF-8Pt was up to 120 μg/mL, the survival rates of both normal liver cells (LO2) and HCC cells (Hep3B and Hepg2) were more than 95%, suggesting that the superior security of ZIF-8Pt (Figure 2D). The reactive oxygen species (ROS) generation ability of AP@ZIF-8Pt was evaluated by DCFH-DA, an ROS probe to detect H2O2 content and oxidative stress.30 As shown in Figure 2E, the green dots had no significant difference in the control and ZIF-8 treatment group, because the endogenous ROS producing in live cells. When treated with ZIF-8Pt, the green dots significantly decreased, as proved in our previous study, Pt nanoparticles could catalyze H2O2 to O2, elevating the source of ROS generation.25 The results showed that ZIF-8Pt presented significant concentration-dependent catalase-like activity.31,32
Therapeutic Efficacy and Mechanism of AP@ZIF-8Pt
Both the human HCC cell lines (Hepg2, Hep3B and Huh7) and the human liver cell line (LO2) were obtained from The American Type Culture Collection (Manassas, VA, USA). CCK-8 assay was first used to verify therapeutic efficacy of AP@ZIF-8Pt against three HCC cell lines (Hepg2, Hep3B and Huh7). The results showed that treatment with AP@ZIF-8Pt significantly inhibits the viability of HCC cells compared with single AP group (Figure 3A). Flow cytometry was then used to verify therapeutic efficacy of AP@ZIF-8Pt. The results showed that the apoptotic rate of AP@ZIF-8Pt treatment group was up to 33.42% for 6 h higher than 3 h (15.48%) (Figure 3B and C). Further therapeutic efficacy of AP@ZIF-8Pt was determined in vivo. Hepg2 tumor-bearing mice were randomly divided into control group, ZIF-8Pt group, AP group and AP@ZIF-8Pt group. The body weights, tumor volumes and tumor weights of the mice were recorded every 2 days in order to assess the effects of different treatments. The body weights of mice increased steadily, demonstrating the biosafety of AP@ZIF-8Pt (Figure 3D). The tumor weight and tumor volumes of mice treated with AP were smaller than those of the control group and ZIF-8Pt group, and the smallest tumors were detected in AP@ZIF-8Pt mice (Figure 3E and F).
Moreover, H&E and TUNEL staining analyses were performed to confirm the therapeutic efficacy of AP@ZIF-8Pt. The necrosis and green dots detected in the AP are more than those observed in the control group and ZIF-8Pt group, and the maximum necrosis and green dots were detected in AP@ZIF-8Pt group, indicates the remarkable therapeutic efficacy of AP@ZIF-8Pt (Figure 3G). The mechanism underlying the efficacy of AP@ZIF-8Pt was determined by immunofluorescence staining. The results showed that AP@ZIF-8Pt treatment up-regulated the expression of γ-H2AX, the DNA DSB marker33 (Figure 3H), indicated that AP@ZIF-8Pt promoted DNA damage by increasing the number of DSBs. In contrast, AP@ZIF-8Pt treatment down-regulated the expression of HIF-1α, a regulator of primary adaptive responses to hypoxia34,35 (Figure 3H), indicated that AP@ZIF-8Pt could alleviate hypoxia in tumor tissue to improve therapy sensitivity.
Transcriptomics Study
Transcriptomics is an extremely important tool for studying all RNA molecules in an organism.36 Cancer-associated protein expression can change levels of some RNA to promote cancer initiation and progression.37 After confirming the in vivo anticancer efficacy of AP@ZIF-8Pt, we further investigated the transcriptomics influence of it in Hepg2-tumor-bearing mice. The serum samples from mice treated with PBS, ZIF-8Pt and AP@ZIF-8Pt were used for transcriptomics analysis. The sample density result was shown in Figure 4A. All of the samples in each group were closely clustered, which demonstrated a good quality of samples with different groups. To focus on the differential genes influenced by AP@ZIF-8Pt, we performed analysis of differential genes between two of each of the groups (control and ZIF-8Pt, control and AP@ZIF-8Pt, ZIF-8Pt and AP@ZIF-8Pt) (Figure 4B). Differential transcriptomics analysis between the control and ZIF-8Pt groups and between the control and AP@ZIF-8Pt groups was carried out to eliminate the batch effect. A total of 173 genes were identified between the control and ZIF-8Pt groups, 2201 genes were identified between the control and AP@ZIF-8Pt groups, 1223 genes were identified between the ZIF-8Pt and AP@ZIF-8Pt groups. Finally, 1188 genes were identified as significant genes influenced by AP@ZIF-8Pt (Figure 4C).
The enrichment analysis was then performed for these significant genes. As shown in Figure 4D, the top 5 important Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways were thermogenesis, signaling pathways regulating pluripotency of stem cells, ribosome, rion disease and PI3K-Akt signaling pathway. As shown in Figure 4E, the top 3 important biological processes were purine nucleoside diphosphate metabolic process, purine ribonucleoside diphosphate metabolic process and ribonucleoside diphosphate metabolic process. The top 3 important cellular components were respiratory chain, cytosolic large ribosomal subunit and respiratory chain complex. And the 2 important molecular functions were extracellular mata structin consultent and Oxygen binding. Peixoto et al proved that HCC induced by hepatocyte Pten deletion reduced thermogenic capacity.38 As is known to all, the dysregulation of the PI3K-AKT-mTOR signaling pathway is common in HCC.39 Xue et al proved METTL16 promoted liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation.40 Read et al demonstrated that Soluble enzyme hydrolyzes purine nucleoside diphosphate resulted in HCC.41
Biocompatibility of AP@ZIF-8Pt
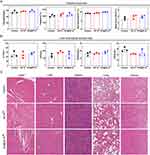
It is essential to evaluate the biosafety to determine whether AP@ZIF-8Pt can be applied in clinical treatment.42 Considering that changes in the behavior and body weights of mice are considered signs of acute toxicity and that changes in gross pathology reflect long-term toxicity,43 these parameters were assessed throughout the treatment period. Blood samples were collected from mice administered with i.p. injections of PBS, ZIF-8Pt and AP@ZIF-8Pt, and the behavior and body weights of these mice were monitored. The results showed that treatment with ZIF-8Pt and AP@ZIF-8Pt did not induce significant variations in mouse behavior and body weight compared to the control group (Figure 3D), which proved that the acute toxicity of ZIF-8Pt and AP@ZIF-8Pt was negligible. Similarly, routine blood tests proved that the ZIF-8Pt and AP@ZIF-8Pt groups did not cause hemolysis or myelosuppression even after 30 days of treatment (Figure 5A). Further liver and kidney function tests were performed to evaluate the liver or kidney damage induced by ZIF-8Pt and AP@ZIF-8Pt. ALT and AST were assessed as liver damage indexes, and BUN and CR were assessed as kidney damage indexes.44–46 The results demonstrated that both ZIF-8Pt and AP@ZIF-8Pt did not damage liver and kidney functions (Figure 5B). H&E staining analysis proved that no organ damage was induced by both ZIF-8Pt and AP@ZIF-8Pt (Figure 5C). All of these results proved that both ZIF-8Pt and AP@ZIF-8Pt possessed highly biocompatible and potential of clinical transformation.
Conclusion
In this study, a pH-responsive and catalase-like Pt nanoparticle-embedded metal-organic framework (ZIF-8Pt) loaded with AP (AP@ZIF-8Pt) was developed to treating HCC. Both in vitro and in vivo experiment results proved that AP@ZIF-8Pt had excellent biocompatibility and therapeutic efficacy. Further experiment verified that AP@ZIF-8Pt can promote DNA damage and relieving hypoxia. Transcriptomics analysis demonstrated that AP@ZIF-8Pt worked through thermogenesis, signaling pathways regulating pluripotency of stem cells, ribosome, rion disease, PI3K-Akt signaling pathway, purine nucleoside diphosphate metabolic process, purine ribonucleoside diphosphate metabolic process and ribonucleoside diphosphate metabolic process, respiratory chain, cytosolic large ribosomal subunit, respiratory chain complex, extracellular mata structin consultent and oxygen binding. Our study suggested that this nanoplatform AP@ZIF-8Pt offered a promising paradigm for HCC treatment and had clinical application potential.
Ethics and Consent to Participate Declarations
All animal experiments were approved by the Experimental Animal Management and Ethics Committee of Xiamen University. All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Funding
This work was supported by the National Natural Science Foundation of China (No.82403783), the Nature Science Foundation of Fujian Province (No.2024J08316) and National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (No. E010322014).
Disclosure
This paper has been uploaded to ResearchSquare as a preprint: https://www.researchgate.net/publication/387685173_A_super-assembled_synergistically_nanoplatform_APZIF-8_Pt_for_hepatocarcinoma_therapy. The authors declare no competing financial interest.
References
1. Cerreto M, Cardone F, Cerrito L, et al. The new era of systemic treatment for hepatocellular carcinoma: from the first line to the optimal sequence. Curr Oncol. 2023;30(10):8774–8792. doi:10.3390/curroncol30100633
2. Huang Z, Zhou J-K, Peng Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77. doi:10.1186/s12943-020-01188-4
3. Campani C, Zucman-Rossi J, Nault JC. Genetics of hepatocellular carcinoma: from tumor to circulating DNA. Cancers. 2023;15(3):817. doi:10.3390/cancers15030817
4. Oura K, Morishita A, Tani J, et al. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci. 2021;22(11):5801. doi:10.3390/ijms22115801
5. Xie P, Yu M, Zhang B, et al. CRKL dictates anti-PD-1 resistance by mediating tumor-associated neutrophil infiltration in hepatocellular carcinoma. J Hepatol. 2024;81(1):93–107. doi:10.1016/j.jhep.2024.02.009
6. Childs A, Aidoo-Micah G, Maini MK, et al. Immunotherapy for hepatocellular carcinoma. JHEP Rep. 2024;6(10):101130. doi:10.1016/j.jhepr.2024.101130
7. Luo X, He X, Zhang X, et al. Hepatocellular carcinoma: signaling pathways, targeted therapy, and immunotherapy. MedComm. 2024;5(2):e474. doi:10.1002/mco2.474
8. Brown ZJ, Tsilimigras DI, Ruff SM, et al. Management of hepatocellular carcinoma: a review. JAMA Surg. 2023;158(4):410–420. doi:10.1001/jamasurg.2022.7989
9. Yao C, Wu S, Kong J, et al. Angiogenesis in hepatocellular carcinoma: mechanisms and anti-angiogenic therapies. Cancer Biol Med. 2023;20(1):25–43. doi:10.20892/j.issn.2095-3941.2022.0449
10. Gao M, Yang T, Qin W, et al. Cell membrane-anchoring nano-photosensitizer for light-controlled calcium-overload and tumor-specific synergistic therapy. Small. 2022;18(48):e2204689. doi:10.1002/smll.202204689
11. Rohlena J, Dong LF, Neuzil J. Targeting the mitochondrial electron transport chain complexes for the induction of apoptosis and cancer treatment. Curr Pharm Biotechnol. 2013;14(3):377–389. doi:10.2174/1389201011314030011
12. Malla WA, Arora R, Khan RIN, et al. Apoptin as a tumor-specific therapeutic agent: current perspective on mechanism of action and delivery systems. Front Cell Dev Biol. 2020;8:524. doi:10.3389/fcell.2020.00524
13. Noteborn MH. Chicken anemia virus induced apoptosis: underlying molecular mechanisms. Vet Microbiol. 2004;98(2):89–94. doi:10.1016/j.vetmic.2003.10.003
14. Los M, Panigrahi S, Rashedi I, et al. Apoptin, a tumor-selective killer. Biochim Biophys Acta. 2009;1793(8):1335–1342. doi:10.1016/j.bbamcr.2009.04.002
15. Gong L, Lu Y, Wang J, et al. Cocktail hepatocarcinoma therapy by a super-assembled nano-pill targeting XPO1 and ATR synergistically. J Pharm Anal. 2023;13(6):603–615. doi:10.1016/j.jpha.2023.04.017
16. Dad HA, Gu T-W, Zhu A-Q, et al. Plant exosome-like nanovesicles: emerging therapeutics and drug delivery nanoplatforms. Mol Ther. 2021;29(1):13–31. doi:10.1016/j.ymthe.2020.11.030
17. Mi X, Hu M, Dong M, et al. Folic acid decorated zeolitic imidazolate framework (ZIF-8) loaded with baicalin as a nano-drug delivery system for breast cancer therapy. Int J Nanomed. 2021;16:8337–8352. doi:10.2147/IJN.S340764
18. Hoseinpour V, Shariatinia Z. Applications of zeolitic imidazolate framework-8 (ZIF-8) in bone tissue engineering: a review. Tissue Cell. 2021;72:101588. doi:10.1016/j.tice.2021.101588
19. Di Matteo V, Di Filippo MF, Ballarin B, et al. Cellulose/Zeolitic Imidazolate Framework (ZIF-8) composites with antibacterial properties for the management of wound infections. J Funct Biomater. 2023;14(9):472. doi:10.3390/jfb14090472
20. Xiang J, Zhu Y, Xie Y, et al. A Cu@ZIF-8 encapsulated antibacterial and angiogenic microneedle array for promoting wound healing. Nanoscale Adv. 2023;5(18):5102–5114. doi:10.1039/D3NA00291H
21. Wang Y, Zeng M, Fan T, et al. Biomimetic ZIF-8 nanoparticles: a novel approach for biomimetic drug delivery systems. Int J Nanomed. 2024;19:5523–5544. doi:10.2147/IJN.S462480
22. Zhang Y, Li -T-T, Shiu B-C, et al. Two methods for constructing ZIF-8 nanomaterials with good bio compatibility and robust antibacterial applied to biomedical. J Biomater Appl. 2022;36(6):1042–1054. doi:10.1177/08853282211033682
23. Yue S, He Y, Wang M, et al. Enhancement of sonodynamic treatment of ovarian cancer based on Pt-B-P ternary nanoparticles. Nanomedicine. 2023;51:102686. doi:10.1016/j.nano.2023.102686
24. Saha S, Banskota S, Liu J, et al. Genetically engineered nanoparticles of asymmetric triblock polypeptide with a Platinum(IV) cargo outperforms a Platinum(II) analog and free drug in a murine cancer model. Nano Lett. 2022;22(14):5898–5908. doi:10.1021/acs.nanolett.2c01850
25. Gong L, Zhang Y, Zhao J, et al. All-in-one biomimetic nanoplatform based on hollow polydopamine nanoparticles for synergistically enhanced radiotherapy of colon cancer. Small. 2022;18:e2107656. doi:10.1002/smll.202107656
26. Wang J, Zhao J, Ma F, et al. One stone, two birds: a Peptide-Au(I) infinite coordination supermolecule for the confederate physical and biological radiosensitization in cancer radiation therapy. Small. 2023;19(11):e2204238. doi:10.1002/smll.202204238
27. Ahmadi M, Asadian E, Mosayebnia M, et al. Optimization of the synthesis and radiolabeling of ZIF-8 nanoparticles. Iran J Pharm Res. 2024;23(1):e144928. doi:10.5812/ijpr-144928
28. Li C, Zhang M, Di X, et al. One-step synthesis of Pt@ZIF-8 catalyst for the selective hydrogenation of 1,4-butynediol to 1,4-butenediol. Chin J Catal. 2016;37(9):1555–1561. doi:10.1016/S1872-2067(16)62497-X
29. Whitford CL, Stephenson CJ, Gómez-Gualdrón DA, et al. Elucidating the Nanoparticle–Metal Organic Framework Interface of Pt@ZIF-8 Catalysts. J Phys Chem C. 2017;121(45):25079–25091. doi:10.1021/acs.jpcc.7b06773
30. Shi HT, Zhao X, Gao J, et al. Acid-resistant ROS-responsive hyperbranched polythioether micelles for ulcerative colitis therapy. Chin Chem Lett. 2020;31(12):3102–3106. doi:10.1016/j.cclet.2020.03.039
31. Fan Z, Wu S, Deng H, et al. Light-triggered nanozymes remodel the tumor hypoxic and immunosuppressive microenvironment for ferroptosis-enhanced antitumor immunity. ACS Nano. 2024;18(19):12261–12275. doi:10.1021/acsnano.4c00844
32. Luo T, Fan Z, Zeng A, et al. Biomimetic targeted co-delivery system engineered from genomic insights for precision treatment of osteosarcoma. Adv Sci. 2025;12(2):e2410427. doi:10.1002/advs.202410427
33. Dong Y, Zhang D, Cai M, et al. SPOP regulates the DNA damage response and lung adenocarcinoma cell response to radiation. Am J Cancer Res. 2019;9(7):1469–1483.
34. Martínez-Romero R, Cañuelo A, Siles E, et al. Nitric oxide modulates hypoxia-inducible factor-1 and poly(ADP-ribose) polymerase-1 cross talk in response to hypobaric hypoxia. J Appl Physiol. 2012;112(5):816–823. doi:10.1152/japplphysiol.00898.2011
35. Dubey R, Levin MD, Szabo LZ, et al. Suppression of tumor growth by designed dimeric epidithiodiketopiperazine targeting hypoxia-inducible transcription factor complex. J Am Chem Soc. 2013;135(11):4537–4549. doi:10.1021/ja400805b
36. Zheng Q, Lin R, Zheng C. Transcriptomics in the study of antiviral innate immunity. Methods Mol Biol. 2025;2854:83–91.
37. Aaltonen LA, Abascal F, Abeshouse A, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi:10.1038/s41586-020-1969-6
38. Peixoto ÁS, Moreno MF, Castro É, et al. Hepatocellular carcinoma induced by hepatocyte Pten deletion reduces BAT UCP-1 and thermogenic capacity in mice, despite increasing serum FGF-21 and iWAT browning. J Physiol Biochem. 2023;79(4):731–743. doi:10.1007/s13105-023-00970-4
39. Tian LY, Smit DJ, Jücker M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism. Int J Mol Sci. 2023;24(3):2652.
40. Xue M, Dong L, Zhang H, et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17(1):7. doi:10.1186/s13045-024-01526-9
41. Read R, Hansen G, Kramer J, et al. Ectonucleoside triphosphate diphosphohydrolase type 5 (Entpd5)-deficient mice develop progressive hepatopathy, hepatocellular tumors, and spermatogenic arrest. Vet Pathol. 2009;46(3):491–504. doi:10.1354/vp.08-VP-0201-R-AM
42. Zor F, Selek FN, Orlando G, et al. Biocompatibility in regenerative nanomedicine. Nanomedicine. 2019;14(20):2763–2775. doi:10.2217/nnm-2019-0140
43. Malayappan MS, Natarajan G, Mockaiyathevar L, et al. Acute and subacute toxicity assessment of Madhulai Manappagu (Siddha herbal syrup formulation) in animal model. J Complement Integr Med. 2021;19(1):9–18. doi:10.1515/jcim-2020-0327
44. Parker A, Kim Y. The effect of low glycemic index and glycemic load diets on hepatic fat mass, insulin resistance, and blood lipid panels in individuals with nonalcoholic fatty liver disease. Metab Syndr Relat Disord. 2019;17(8):389–396. doi:10.1089/met.2019.0038
45. Zhou J, He Z, Ma S, et al. AST/ALT ratio as a significant predictor of the incidence risk of prostate cancer. Cancer Med. 2020;9(15):5672–5677. doi:10.1002/cam4.3086
46. Cai A, Qi S, Su Z, et al. Method comparison and bias estimation of Blood Urea Nitrogen (BUN), Creatinine (Cr), and Uric Acid (UA) measurements between two analytical methods. Clin Lab. 2017;63(1):73–77. doi:10.7754/Clin.Lab.2016.160536
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.