Back to Journals » International Journal of Nanomedicine » Volume 20
Advancing Nanomedicine Through Electron Microscopy: Insights Into Nanoparticle Cellular Interactions and Biomedical Applications
Received 17 October 2024
Accepted for publication 16 January 2025
Published 8 March 2025 Volume 2025:20 Pages 2847—2878
DOI https://doi.org/10.2147/IJN.S500978
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sachin Mali
Sultan Akhtar,1 Fatimah Zuhair2
1Department of Biophysics Research, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, 31441, Saudi Arabia; 2Department of Infection Control, Alzahra General Hospital, Qatif, 31911, Saudi Arabia
Correspondence: Sultan Akhtar, Department of Biophysics Research, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, 31441, Saudi Arabia, Tel +966133330876, Email [email protected]
Abstract: Nanomedicine has revolutionized cancer treatment by the development of nanoparticles (NPs) that offer targeted therapeutic delivery and reduced side effects. NPs research in nanomedicine significantly focuses on understanding their cellular interactions and intracellular mechanisms. A precise understanding of nanoparticle interactions at the subcellular level is crucial for their effective application in cancer therapy. Electron microscopy has proven essential, offering high-resolution insights into nanoparticle behavior within biological systems. This article reviews the role of electron microscopy in elucidating the cellular uptake and intracellular interactions of NPs. Transmission electron microscopy (TEM) provides imaging capabilities, such as cryo three-dimensional tomography, which offer in-depth insights into nanoparticle localization, endocytosis pathways, and subcellular interactions, while high resolution-TEM is primarily used for studying the atomic structure of isolated NPs rather than nanoparticles within cells or tissues. On the other hand, scanning electron microscopy (SEM) is ideal for examining larger surface areas and provides a broader perspective on the morphology and topography of the samples. The review highlights the advantages of electron microscopy in visualizing nanoparticle interactions with cellular structures and tracking their mechanisms of action. It also addresses the challenges associated with electron microscopy characterization, such as tedious sample preparation, static imaging limitations, and a restricted field of view. By examining various nanoparticle uptake pathways, and cellular destination of NPs with examples, the article emphasizes the importance of these pathways to optimize nanoparticle design and enhance therapeutic efficacy. This review underscores the need for continued advancement in electron microscopy techniques to improve the effectiveness of nanomedicine and address existing challenges. In summary, electron microscopy is a key tool for advancing our understanding of nanoparticle behavior in biological contexts, aiding in the design and optimization of nanomedicines by providing insights into nanoparticle cellular dynamics, uptake mechanisms, and therapeutic applications.
Plain Language Summary: Cancer remains one of the leading causes of death worldwide, with incidence and mortality rates projected to rise significantly in the coming years. In recent years, tiny-objects known as nanoparticles are widely utilized in therapeutic applications to improve treatment outcomes and effectiveness in combating cancer. This article explores how electron microscopy is transforming cancer treatment using nanoparticles. Nanoparticles are designed to deliver drugs directly to cancer cells, improving treatment options while reducing side effects. A detailed information is crucial in order to understand how these nanoparticles interact with cells. Electron microscopy techniques such as transmission electron microscopy and scanning electron microscopy play a key role in obtaining such information. Electron microscopes enables scientists to visualize nanoparticles and cells with enhanced clarity and magnification. This capability is helping to understand how nanoparticles enter the cells, transport in specific locations, and interact with various cellular components. Moreover, it allows the examination of the larger surface areas of the treated cells, revealing the overall morphology and topography of the nanoparticles and their interactions with cell structures.
The objective of this study is to investigate the role of electron microscopy in understanding the cellular uptake, intracellular interactions, and subcellular localization of nanoparticles used in cancer nanomedicine. It aims to highlight how advanced electron microscopy techniques like transmission electron microscopy and scanning electron microscopy provide critical insights into nanoparticle behavior within biological systems, while addressing challenges such as sample preparation and imaging limitations. The study underscores the need for continued innovation in electron microscopy technologies to optimize nanoparticle-based therapeutic design and improve cancer treatment outcomes.
This review outlines studies evaluating the use of electron microscopy techniques in nanoparticle applications for cancer research, highlighting their advantages and limitations. It focuses on nanoparticle uptake pathways, cellular destinations, and recent examples of nanoparticle interactions with cancer cells. Electron microscopy, particularly transmission electron microscopy, plays a vital role in visualizing nanoparticle behavior within cells, revealing ultrastructural changes such as nanoparticle aggregation, shape, and interactions with cellular membranes and organelles. Transmission electron microscopy offers precise insights into nanoparticle uptake, transport, and exit, while three-dimensional (3D) tomography provides detailed reconstructions of nanoparticle localization. However, challenges like sample preparation and artifacts persist, with rapid freezing methods, such as plunge-freezing particularly, high-pressure freezing, proving effective in preserving native cellular structures. Combining electron microscopy with techniques like flow cytometry improves the study of nanoparticle interactions by offering both ultrastructural and quantitative data, enabling accurate analysis of nanoparticle uptake and behavior in cancer cells.
Keywords: cancer cells, ultrastructural analysis, uptake pathways, challenges, nanomedicine
Graphical Abstract:

Introduction
Cancer remains one of the leading global causes of death, with an estimated 19 million new cases and 10 million deaths in 2020 alone, representing about one death in six worldwide. This trend is expected to rise due to environmental pollutants, increased risk factors, and demographic changes.1 The American Cancer Society predicts over 600,000 cancer deaths and about 2 million new cases in 2024. Though breast cancer has a higher incidence rate, Lung cancer, in particular, is the deadliest form among both males and females.2,3 Traditional cancer treatments, including surgery, chemotherapy, and radiotherapy, have been effective for some cancers but they have limitations, especially with metastatic cancers. These treatments often result in severe side effects by damaging normal cells.4,5 Consequently, there has been significant interest in developing drug delivery systems that specifically target cancer cells to minimize side effects and improve therapeutic outcomes.6
NPs have emerged as promising tools in cancer therapy due to their ability to deliver drugs more precisely.7 Nanotechnology-based diagnostics and therapies are rapidly advancing, offering potential improvements in cancer treatment.8 NPs are categorized based on their properties, shape, and size, including fullerene, metallic, ceramic, and polymeric NPs.9,10 Targeted therapies using NPs are often more effective than traditional chemotherapeutics because they can deliver drugs more accurately, reducing side effects.11 Various types of NPs have been developed for drug delivery, including lipid-based NPs,12 polymeric NPs,13 semiconductor-based NPs,14 inorganic NPs15 nucleic acid therapeutic,16 chemotherapy,17 and immunotherapy.18,19
As the field of cancer nanomedicine grows, understanding of NP interaction with biological systems becomes increasingly important.20 NPs are used in various applications, such as drug delivery,21 biosensing,22 imaging,23 sort systems,24 and scaffolding.25,26 In vivo and in vitro imaging techniques are crucial for confirming NP targeting, distribution, and elimination.27–30 Given the diverse properties of NPs, it is essential to assess their toxic potential and side effects through rigorous microscopic monitoring. This includes observing undesirable reactions like necrosis, inflammation, fibrosis, or tumor progression.31
No imaging technique is ideal; each has its limitations. Optical imaging (OM), magnetic resonance imaging (MRI), and positron emission tomography, for instance, offer lower resolution (hundreds of micrometres), making subcellular visualization challenging. Techniques such as fluorescence microscopy provide better cellular detail but still struggle with individual NPs detection due to resolution limits.32–34 Electron microscopy (EM), such as TEM and SEM, offers high-resolution analysis, making it suitable for detailed NP studies.35–39
TEM, with its high spatial resolution (down to 0.1 nm), is beneficial for visualizing single NPs and their localization within cellular structures.40 It also aids in toxicological assessments by revealing structural changes in organelles affected by NPs.41 However, TEM has limitations, including tedious sample preparation and time-consuming imaging processes.31,42 Nonetheless, Ostrowski et al, 2015 claimed that TEM is a practical addition to other microscopic techniques, instead of being the preferred imaging technique for studying NPs in a cellular environment.31 Moreover, TEM is limited by the thickness of samples, typically around 300 nm, as thicker samples result in fewer transmitted electrons and noisier images due to overlapping elastic and inelastic electron scattering. Scanning transmission electron microscopy (STEM) offers an alternative, allowing imaging of thicker samples (up to 1 μm) by spatially separating elastic and inelastic electron, making it less sensitive to sample thickness and improving composition analysis. However, both methods often involve embedding samples in resin and heavy metal staining, which can denature structures and hinder high-resolution studies of delicate cellular components.43 Recent advancements, such as TEM/STEM electron tomography on plastic-embedded cells and tissues, have significantly improved 3D visualization of NP localization and cellular ultrastructure, providing high-quality imaging data. Additionally, energy-dispersive X-ray spectroscopy (EDX)-based chemical mapping in TEM projection images enables the identification of NP composition and their distribution within cellular environments.44
On the other hand, SEM provides high-resolution images (3–20 nm) of the specimen’s surface and is valuable for studying NP interactions with cell surfaces and morphological changes45–48 Despite its surface sensitivity, SEM’s capability to visualize surface morphology helps in understanding NP distribution. Furthermore, advanced SEM techniques, such as serial block-face scanning electron microscopy (SBFSEM), allow for the imaging of large tissue volumes at nanoscale resolution in an automated manner. By coupling SEM with a built-in ultramicrotome or focused ion beam (FIB-SEM), researchers can achieve 3D reconstructions of cellular structures and tissue morphology, providing comprehensive insights into NP interactions within complex biological environments.49–51
Correlative Light Electron Microscopy (CLEM) is an advanced imaging approach that combines the strengths of light microscopy (LM) and EM to study biological specimens at multiple scales. CLEM allows researchers to first locate and analyze regions of interest with fluorescence microscopy, such as proteins or specific cellular structures, and then examine those same areas at high resolution using EM to reveal ultrastructural details. This method is particularly valuable for studying dynamic cellular processes, such as nanoparticle uptake or protein localization.52,53
While cryo-TEM offers significant advantages, including the ability to determine two-dimensional (2D) structures, it also faces challenges in obtaining high-information 2D images.54 Room temperature electron tomography of thick cells or tissue slices (200–500 nm) is a powerful technique for obtaining detailed three-dimensional (3D) structural information. This method involves collecting a series of 2D projections at various tilt angles, which are then computationally reconstructed into a 3D model, providing insights into cellular architecture and the spatial relationships of subcellular components.55 Electron tomography, particularly cryo-electron tomography (cryo-ET), enables the determination of 3D structures at nanometer resolution and provides nearly native shape visualization of biological specimens, such as proteins and cellular components. However, the achievable resolution for biological samples in cryo-ET is typically limited to a range of 1–2 nm due to constraints like beam damage and sample quality. These techniques, which involve samples preserved in a frozen state with vitreous ice, enhance our understanding of protein structures and cellular interactions, including ribosomes and chemoreceptor arrays.56 Cryo-ET thus remains an indispensable tool for elucidating the spatial organization of complex biological specimens while maintaining their near-native states.57 This review aims to outline studies evaluating the use of electron microscopy techniques in various NP applications for cancer research. It will highlight the advantages and limitations of these techniques, focusing on NP uptake pathways and cellular destinations, and providing examples from recent literature on NP interactions with cancer cells.
Significance of Subcellular Morphology in Advancing Nanomedicine Research
In nanomedicine, precise delivery of NPs to specific subcellular targets is crucial for effective treatment and minimal side effects.58 The successful nanomedicine relies on the ability to control and track NP transport within the body while addressing safety concerns related to cellular interactions.59–61 Achieving such precision remains a significant challenge in the field.62,63 Understanding the interactions between NPs and the biological systems is essential for overcoming these challenges and improving therapeutic outcomes.64 While conventional optical microscopy is widely used,65 it has limitations in distinguishing the exact localization of NPs within cells and detecting subcellular changes associated with NP interactions.66 Optical microscopy cannot analyze cell stress such as nuclear fragmentation, cell membrane blebbing, membrane protrusions, reformation of the endoplasmic reticulum, chromatin condensation, mitochondrial deterioration, generating apoptotic bodies, or necrotic alterations.67–69 EM plays a crucial role in addressing these limitations. TEM provides high-resolution images that allow detailed visualization of NP interactions with cellular structures and mechanisms of action. It can track NP localization, distribution, and intracellular degradation with exceptional clarity, making it a powerful tool for understanding how NPs affect cellular environments and contribute to their therapeutic effects.
Nanoparticle Cellular Interaction Mechanisms
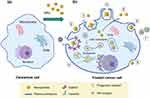
TEM offers substantial advantages for studying NPs’ interactions with biological systems, thereby enhancing our understanding of their mechanisms of action. TEM enables detailed examination of NP uptake and intracellular fate, revealing critical insights into how NPs interact with and affect cells.70 The physiochemical properties of NPs—such as charge, size, shape, and hydrophobicity—play a significant role in their cellular uptake via active or passive uptake.71 Achieving safe cellular entry is crucial for the efficacy of nanomedicines, dictating a thorough understanding of uptake mechanisms to design effective and targeted therapies.72 Cellular uptake of NPs involves complex processes and biomolecular interactions that facilitate crossing the plasma membrane.73 The plasma membrane, a thin layer surrounding cells, separates intracellular components from the external environment and is composed of lipids and proteins.74 Lipids form a negatively charged bilayer that serves as a barrier, while proteins mediate substance exchange and cellular signaling.75 Understanding how NPs traverse the plasma membrane is essential for elucidating their role, intracellular fate, and responses within the cell.76 NPs can enter cells through several pathways, which are generally classified into two categories: (1) endocytosis-based uptake (active uptake) and (2) direct cellular entry (passive uptake). These pathways are crucial for optimizing NP design and functionality. The endocytosis uptake further includes various pathways (Figure 1):
- Clathrin-mediated Endocytosis (CDE).
- Caveolae-mediated Endocytosis (CADE).
- Clathrin- and Caveolae-independent Endocytosis (CCIE).
- Phagocytosis (PGC).
- Macropinocytosis (MPC).
Each pathway is regulated by specific lipids and transport proteins, and NPs are encapsulated into vesicles such as endosomes, phagosomes, or macropinosomes after endocytosis. These vesicles do not directly fuse with the cytoplasm or organelles.77 Endocytosis routes are categorized based on the size of NPs: larger particles (>500 nm) are typically internalized via phagocytosis or macropinocytosis, while smaller particles utilize CDE, CADE, or CCIE.78 Figure 1 illustrates the various uptake mechanisms emphasizing the characteristics of the membrane-related specific uptake mechanism.
Clathrin-Dependent Endocytosis (CDE)
CDE is a widely studied cellular uptake mechanism characterized by membrane invaginations about 100 nm in size, coated with the protein clathrin. This pathway facilitates the internalization of NPs and the uptake of essential nutrients and membrane components like cholesterol and iron. CDE begins when NP surface ligands bind to cell membrane receptors, leading to the formation of a clathrin-coated pit. This pit is visible across various cell types and is crucial for receptor-mediated endocytosis.79 Recent studies have shown that pathogens such as SARS-CoV-2 utilize CDE for cell entry, highlighting its role in viral infectivity.80 The process involves several stages: (a) nucleation of cytosolic proteins to form a clathrin-coated pit, (b) membrane invagination and budding, (c) vesicle formation and separation from the plasma membrane, and (d) vesicle decapsulation and protein restoration.81 Research indicates that the surface charge of NPs influences their uptake pathway. For instance, positively charged NPs have been observed to enter HeLa cervical cancer cells via CDE, leading to higher internalization and accumulation compared to negatively charged NPs.82
Caveolin-Dependent Endocytosis (CADE)
CADE is another critical uptake pathway involving caveolae, small membrane invaginations coated with caveolin. This pathway plays a key role in various biological processes, including cell signaling, transcytosis, and regulation of lipids and proteins. Caveolin-coated vesicles are typically transported to intracellular locations such as the Golgi apparatus (GA) and endoplasmic reticulum (ER).83 CADE is significant for NP research, especially for those targeting intracellular organelles. Engineered NPs often utilize surface ligands such as cholesterol, albumin, and folate to exploit this pathway for cellular internalization.84
Clathrin/Caveolin Independent Endocytosis (CCIE)
CCIE describes an uptake mechanism that operates in the absence of clathrin and caveolin. This pathway allows viruses and NPs to cross the plasma membrane through a specific lipid composition, often involving cholesterol. CCIE is responsible for internalizing substances such as interleukin-2, growth hormones, and folic acid.78 Given that cancer cells have a heightened demand for folic acid, NPs modified with folic acid are particularly effective in targeting folate receptors on tumor cells. Folate-functionalized NPs can thus achieve successful delivery into the cytoplasm via this pathway.85
Macropinocytosis (MPC)
MPC is a cellular uptake mechanism characterized by the extension of the plasma membrane to enclose extracellular fluids, forming large vesicles known as macropinosomes. This process is facilitated by actin-driven membrane ruffling and does not rely on lipid rafts or specific membrane proteins.86 NPs and other particles are internalized within these large vesicles.87 Macropinosomes are generally larger than vesicles formed through other endocytic pathways and have been observed to be leaky vesicles, which can lead to premature escape of NPs before reaching lysosomal degradation compartments.62
Phagocytosis
The plasma membrane extends around the particle to form a phagosome, which subsequently fuses with lysosomes for degradation. Phagocytosis of smaller particles, including NPs, is less well understood but does occur. The efficiency of phagocytosis can be influenced by the size and morphology of NPs, as well as the specific type of phagocytic receptor involved. For instance, Fc receptor-mediated phagocytosis can initiate pro-inflammatory responses, whereas complement receptor-mediated phagocytosis is typically noninflammatory.88–91
Phagocytosis is an uptake mechanism that occurs by bending the plasma membrane and swallowing the particles. It is used primarily by professional phagocytes—such as neutrophils, macrophages, dendritic cells, and monocytes—as well as some nonprofessional phagocytes. This process involves the engulfing of larger particles, generally greater than 0.5 micrometers in diameter, including microorganisms, apoptotic cells, and foreign materials.84,92,93 The plasma membrane extends around the particle to form a phagosome, which subsequently fuses with lysosomes for degradation. Phagocytosis of smaller particles is less understood. The efficiency of phagocytosis can be influenced by the size and morphology of NPs, as well as the specific type of phagocytic receptor involved. For instance, Fc receptor-mediated phagocytosis can initiate pro-inflammatory responses, whereas complement receptor-mediated phagocytosis is typically noninflammatory.68,80,82,83,85,88,89,93
Intracellular Trafficking and Action of Nanoparticles
Several studies have employed electron microscopy to investigate the cellular uptake and intracellular interaction of various types of nanoparticles, revealing important insights into their applications and mechanisms, as detailed in Table 1. This overview highlights the diverse roles and imaging approaches used to discuss NPs’ interactions with cancer cells.
 |
Table 1 Summary of Various Nanoparticle Applications in Cancer Therapy and Their Characterization Using Electron Microscopy Techniques |
Hui et al (2024) studied the effect of silica nanocapsules (SNCs) with varying elasticity on macrophage cells (RAW264.7) and ovarian adenocarcinoma cells (SKOV3). They used TEM to show that the elasticity of NPs significantly affects their cellular uptake. Stiff NPs retained their shape, whereas soft NPs deformed during receptor-dependent binding, reducing their uptake efficiency. RAW264.7 cells internalized NPs via phagocytosis and macropinocytosis, while SKOV3 cells used receptor-mediated endocytosis (see Figures 2 and 3).94
 |
Figure 2 Endocytosis paths and distortion of silica nanocapsules (SNCs). (a) Ratios for cellular uptake of SNCs in (ovarian adenocarcinoma) SKOV3 cells. (b) TEM images illustrating the changes in the morphology of the SNCs with the greatest stiffness (top) and softness (bottom) during receptor-dependent binding to SKOV3 cells. (c) Scheme demonstrating the alteration of the soft SNCs along with receptor-dependent uptake. (d) Substantial extent of SKOV3 cell surface can be surrounded with numerous stuck SNCs. (e) Ratios for cellular uptake of SNCs in macrophage (RAW264.7) cells. (f) TEM images illustrating the changes in the morphology of the soft SNCs while interacting with RAW264.7 cells. All scale bars ae 200 nm. The uptake ratio refers to the uptake of SNCs in cells exposed to endocytic inhibitors (cytochalasin D (CytD), Nystatin, and chlorpromazine (CPZ)), expressed relative to the uptake in untreated cells. Results are reported as mean ± standard deviation (n = 3), with statistical significance denoted as *P < 0.05, **P < 0.01, #P < 0.001, and N.S. indicating no significance. Reproduced from Hui Y, Yi X, Wibowo D, Yang G, Middelberg APJ, Gao H, Zhao CX. Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Sci Adv. 2020 Apr 17;6(16):eaaz4316. Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original US Government Works. Distributed under a Creative Commons Attribution Non-Commercial License 4.0 (CC BY-NC).94 |
 |
Figure 3 (a) Ratios for cellular uptake of PEG-modified SNCs within SKOV3 cells. (b) TEM images illustrating changes in the morphology of the soft PEG-SNCs while interacting with SKOV3 cells; scale bars are 200 nm. (c) A scheme demonstrating the proteins involved in the different designs of SNCs-cell interactions. Reproduced from Hui Y, Yi X, Wibowo D, Yang G, Middelberg APJ, Gao H, Zhao CX. Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Sci Adv. 2020 Apr 17;6(16):eaaz4316. Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original US Government Works. Distributed under a Creative Commons Attribution Non-Commercial License 4.0 (CC BY-NC).94 |
Gunn et al (2011) developed a nanoparticle system for targeted cancer cell labeling using fusion proteins and biotinylated magnetic nanoparticles (FeO-NPs). TEM analysis revealed effective endocytosis of large NPs into Ramos cells via anti-CD20 fusion proteins, demonstrating successful targeting of lymphoma cells (see Figure 4).95
 |
Figure 4 The internalization of NPs by cells. (a) TEM image of the Ramos cell line after the treatment of anti-CD20 fusion protein as well as exposure to NPB-AF647. The image showed a great number of NPs (black spots) within endosomes (rounded white zones with black spots). The white box indicates the area in the endosome illustrating the distribution of NPs within endosome. (b) The analysis of TEM with higher magnification illustrating the feature lattice planes of NP crystal (rounded zones) obtained from endosomes encompassing NPs. (c) Ramos cells without anti-CD20 fusion protein pretreatment, indicating no NP uptake post-treatment with NPB-AF647. Reproduced from Gunn J, Park SI, Veiseh O, Press OW, Zhang M. A pretargeted nanoparticle system for tumor cell labeling. Mol BioSyst. 2011;7(3):742–748. Copyright 2011 ROYAL SOCIETY OF CHEMISTRY.95 |
M. S. Pudovkin et al (2021) investigated the uptake of Pr3+: LaF3 nanoplates and nanospheres by A-549 lung carcinoma cells using TEM and flow cytometry. They found that nanomaterials were internalized via macropinocytosis and were not found in the nucleus or other organelles. Macropinocytosis was shown to be dynamic, with macropinosomes shifting within the cytoplasm. TEM and flow cytometry analysis revealed that both nanospheres and nanoplates have the same action against lung carcinoma cells.68
Guo et al (2010) investigated the anticancer potential of resveratrol-bovine serum albumin nanoparticles (BSA-NPs) on SKOV3 cancer cells in a murine model. The study focused on the in vivo effects of these NPs by measuring tumor volume and examining tissue morphology using TEM. The results indicated that the BSA-NPs exhibited enhanced dispersity and solubility in water, which notably influenced their distribution within tissues and organs. Treatment with the NPs led to a significant reduction in tumor growth. TEM analysis of the tumor tissues revealed morphological changes, including apoptotic and necrotic bodies, suggesting the NPs induced cell death. Additionally, protein expression analysis pointed to the activation of the mitochondrial apoptotic pathway, highlighting the potential of these NPs in cancer therapy.69
Cheng et al (2014) explored the in vitro anticancer activity of round and rod-shaped iron oxide magnetic nanoparticles (MNPs) on human cervical cancer cells (HeLa). The study compared the effects of these differently shaped NPs on cell viability and observed their interactions with cancer cells using optical microscopy, SEM, and TEM. The findings revealed that exposure to the NPs led to significant damage to cell membranes and cytoplasm, resulting in decreased cell viability. The study concluded that rod-shaped NPs, influenced by an oscillating magnetic field, were more effective at damaging cancer cells compared to spherical NPs. SEM images showed NP accumulation on cell membranes, while TEM revealed their distribution within the cell cytoplasm, demonstrating the superior efficiency of rod-shaped NPs in cancer treatment as illustrated in Figure 5.96
 |
Figure 5 Anticancer activity of iron oxide magnetic nanoparticles (MNPs) on human cervical cancer cells (HeLa). (a and b) SEM micrographs displaying MNPs-loaded HeLa cell membranes. The nanoparticles either spherical or rod-shaped are marked with yellow arrows. (c and d) Optical microscopy images of semi-thin slices of HeLa cells. The cells are indicated with black arrows. (e and f) TEM images demonstrating ultra-thin slices of HeLa cells; arrows in (f) indicate rod-like NPs that have been cut during ultramicrotomy preparation. The cell membrane is marked by a black arrow (panel e) while mitochondria with a white arrow and possibly lysosome or endosome with a yellow arrow (panel f). Reproduced from Cheng D, Li X, Zhang G, Shi H. Morphological effect of oscillating magnetic nanoparticles in killing tumor cells. Nanoscale Res Lett. 2014 Apr 28;9(1):195. Copyright © 2014 Cheng et al; licensee Springer. Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0.96 |
Richa Panwar et al (2016) investigated the use of chitosan-tripolyphosphate pentasodium (CS-TPP) NPs as carriers for drug delivery, focusing on the encapsulation of ferulic acid to enhance its therapeutic efficacy. The study specifically examined the anticancer activity of spherical ferulic acid-loaded NPs, which had an average size of 125 nm, against the human cervical cancer cell line ME-180. SEM images revealed significant morphological changes in the cancer cells, including apoptotic bodies, cytoplasmic fragments, and wrinkled, destructed cells as illustrated in Figure 6. A notable reduction in cancer cell proliferation was observed. Fluorescence microscopy further distinguished dead and viable cells and confirmed the induction of apoptosis.66
 |
Figure 6 Chitosan-tripolyphosphate pentasodium (CS-TPP) NPs as carriers for drug delivery, ME-180 cell line demonstrating changes in the morphology as observed by SEM: (a) untreated control cells, (b) cells exposed to 40 µM native ferulic acid, (c) cells exposed to CS-TPP NPs, (d) cells exposed to 40 µM ferulic acid-loaded CS-TPP NPs after 24 hours of incubation. The cell death features are highlighted with white arrow. (SEM magnification: ×10,000). Reproduced from Panwar, (R), Sharma, (A) K. Kaloti, (M) et al. Characterization and anticancer potential of ferulic acid-loaded chitosan nanoparticles against ME-180 human cervical cancer cell lines. Appl Nanosci 6, 803–813 (2016) Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).66 |
S. Akhtar et al (2019) investigated the cytotoxic effects of functionalized magnetic nanoparticles (f-MNPs) on colorectal cancer cells (HCT-116). The study employed TEM to analyze the morphology of cancer cells both before and after treatment with NPs. TEM micrographs revealed that spherical NPs, ranging from 100 to 400 nm in size, were successfully endocytosed by the cells and internalized within vesicles, as depicted in Figure 7. Within these vesicles, 3–12 NPs were typically observed, indicating that the morphology and distribution of NPs are crucial for understanding their cellular interactions. The study noted an irregular distribution of NPs within the cells, with some cells showing a high accumulation of NPs while others had little to none. Clusters of NPs were visible in the cytoplasm, and those within vesicles were distinguished by their light contrast. Notably, TEM images showed free NPs located near the nucleus, suggesting that these particles had escaped from lysosomes and reached the nucleus, where they may induce cell death.97
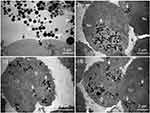 |
Figure 7 Localization of penetrated magnetic nanoparticles (MNPs) within cancer cells at varying magnifications. (a) MNPs are entering the cell in a streaming manner, with a black arrow indicating the cell and a white arrow pointing to the MNPs. (b) MNPs are observed within the cytoplasm and inside vesicles, marked by white arrows. (c) A lower number of MNPs are located inside vesicles, while (d) shows a higher number of MNPs clustered together within the vesicles, as indicated by white arrows. “Nu” denotes nucleus, “Cy” indicates cytoplasm, and “CW” refers to the cell wall. Reproduced from Akhtar S, Khan FA, Buhaimed A. Functionalized magnetic nanoparticles attenuate cancer cells proliferation: transmission electron microscopy analysis. Microsc Res Tech. 2019;82(7):983–992. © 2019 Wiley Periodicals, Inc.97 |
R. Vivek et al (2013) developed a pH-responsive drug delivery system (DDS) using chitosan nanoparticles (NPs) to enhance the efficacy of Tamoxifen, a common chemotherapeutic agent. In this research, they synthesized the Tamoxifen-loaded chitosan NPs by lowering the pH from 7.4 to 4.0, exploiting the acidic environment typical of tumor tissues. This pH-sensitive approach aims to improve drug release specifically within the tumor site, thereby potentially increasing the therapeutic impact. The effectiveness of this DDS was tested using MCF-7 cancer cells, a common breast cancer cell line. The results indicated that the inhibition rate of cancer cell growth increased with higher concentrations of Tamoxifen-loaded chitosan NPs, suggesting enhanced cytotoxicity. TEM analysis revealed that Tamoxifen-loaded chitosan NPs had a more effective cellular uptake compared to Tamoxifen alone, highlighting the potential of this pH-responsive system in improving drug delivery and therapeutic efficacy in cancer treatment.98
B. De Berardis et al (2010) conducted a study to investigate the effects of zinc oxide nanoparticles (ZnO-NPs) on colon cancer LoVo cells, focusing on cytotoxicity, oxidative stress, and apoptosis induction. Their findings revealed that exposure to ZnNPs significantly reduced cell viability, caused depolarization of mitochondrial membranes, and triggered apoptosis. Additionally, there was an increase in interleukin-8 (IL-8) release, which is often associated with inflammatory responses in cancer. The study showed that high concentrations of ZnO-NPs achieved up to 98% anticancer activity within 24 hours of treatment, indicating a potent effect. The authors suggested that oxidative stress is likely a key mechanism driving the cytotoxicity observed in the cancer cells. After 48 h of treatment with high concentrations of the NPs, the LoVo cells exhibited pronounced morphological changes, including shrinkage and severe damage, as depicted in Figure 8 of the study. This morphological damage further supports the potential of ZnO-NPs as a therapeutic agent in targeting cancer cells.99
 |
Figure 8 TEM images of untreated control cell (a) and cells exposed to 10 μg/cm2 ZnO-NPs (b–d). Treatment caused noticeable nuclear changes characterized by condensed chromatin (b) and alterations in the mitochondria, which showed signs of swelling and shrinkage (c and d). The scale bare are 2 µm (a and b) and 1 µm (c and d). Reproduced from De Berardis B, Civitelli G, Condello M, et al. Exposure to ZnO nanoparticles induces oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol Appl Pharmacol. 2010;246(3):116–127. Copyright © 2011 Elsevier Ltd. All rights reserved.99 |
Wang et al (2011) explored the use of acetylated dendrimer-entrapped gold nanoparticles (ADE@ AuNPs) for computed tomographic (CT) imaging of cancer cells. Their study involved both in vitro and in vivo applications of AuNPs using the SPC-A1 cell line (human lung cancer cell line), a model for lung cancer. The research demonstrated that after incubating SPC-A1 cells with acetylated dendrimer-entrapped AuNPs, the cells could be effectively visualized using micro-CT. This capability extended to visualizing tumor models through both intratumoral and intraperitoneal administration of the NPs. This observation suggests that AuNPs can serve as effective contrast agents for CT imaging in cancer diagnostics. The study also assessed the cytotoxicity of the AuNPs. MTT viability assays and cell cycle analyses using flow cytometry revealed that the NPs were non-cytotoxic, indicating their safety for imaging applications. TEM analysis showed a high accumulation of ADE@AuNPs in the cytoplasm, with a predominant presence in the lysosomes within the cells, as illustrated in Figure 9. This distribution supports their potential use in imaging applications while maintaining cellular safety.100
 |
Figure 9 TEM visualization of SPC-A1 cell: (a) cells in the untreated negative control group, (b) cells treated for 12 h with ADE@ AuNPs, and (c) high magnification image of the highlighted section in (b) to display the AuNPs in the cytoplasm (arrowhead) and cell nucleus (arrow) with clearness. The scale bare are 2.5 µm (a and b) and 1 µm (c). Reproduced from Wang H, Zheng L, Peng C, et al. Computed tomography imaging of cancer cells using acetylated dendrimer-entrapped gold nanoparticles. Biomaterials. 2011;32(11):2979–2988. Copyright © 2011 Elsevier Ltd. All rights reserved.100 |
Calero et al (2015) explored the anticancer potential of dimercaptosuccinic acid-coated superparamagnetic iron oxide nanoparticles (DMSA-SPION) with a diameter of 15 nm in breast cancer cells (MCF-7). The study demonstrated that these DMSA-SPION were successfully internalized by the cells through endocytosis, specifically via macropinocytosis and clathrin-dependent pathways, as confirmed by TEM analysis. Only a few nanoparticles were observed at short exposure time. The maximum accumulation of DMSA-SPION occurred after 24 h of incubation. During this time, the nanoparticles were predominantly localized in endosomes and accumulated near the nuclei as the vesicle count and size of nanoparticle aggregates increased. These results highlight the effective cellular uptake and prolonged intracellular retention of DMSA-SPION, suggesting their potential for targeted cancer therapy (Figure 10). Furthermore, TEM identified specific endocytosis pathways utilized by DMSA-SPION in MCF-7 breast cancer cells (see Figure 11). The strong contrast of the magnetic particles enabled clear visualization of their internalization and distribution. Smaller nanoparticle aggregates, less than 200 nm, were observed near clathrin-coated pits, indicating clathrin-mediated endocytosis (Figure 11a–d). In contrast, larger nanoparticle aggregates were found near the cell periphery, often engulfed by membrane extensions, suggesting micropinocytosis (Figure 11e–f). After a brief incubation, nanoparticles were detected near the plasma membrane within vesicles resembling early endosomes. Ultrastructurally, the early endosomes are characterized by complex and dynamic morphology, often appearing as tubulovesicular structures with a combination of vesicles and interconnected tubules. These compartments are typically about 500–1000 nanometers in diameter and possess a mildly acidic lumen.106 Despite the notable internalization and accumulation of nanoparticles, there was no significant impact on cell morphology, cytoskeleton structure, cell cycle, cell viability, or reactive oxygen species (ROS) production, suggesting that the DMSA-SPION did not adversely affect the overall cell health compared to untreated control cells.101
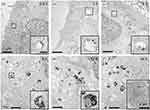 |
Figure 10 TEM analysis of uptake kinetics. Thin-section images of MCF-7 cell line treated with DMSA-SPION. Cells incubated for (a) 0.5 h, (b) 1 h, (c) 3 h, (d) 6 h, (e) 12 h and (f) 24 h. The overall cell morphology and shape are displayed in (f) inset. The small squares are the areas where the insets are taken and zoomed in. The scale bars are 1 μm for images (a–f); 200 nm insets (a–e) and 2 μm for the inset (f). Reproduced from Calero M, Chiappi M, Lazaro-Carrillo A, Rodríguez MJ, Chichón FJ, Crosbie-Staunton K, Prina-Mello A, Volkov Y, Villanueva A, Carrascosa JL. Characterization of interaction of magnetic nanoparticles with breast cancer cells. J Nanobiotechnology. 2015 Feb 26;13:16. © Calero et al; licensee BioMed Central. 2015. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0.101 |
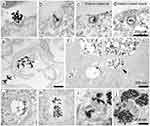 |
Figure 11 TEM analysis of SPION interaction and cellular uptake. (a–d) Thin sections of cells interacting with DMSA-SPION via clathrin-mediated endocytosis, resulting in aggregates less than 200 nm in diameter. (e and f) Two TEM images of thin cell sections demonstrating macropinocytosis associated with DMSA-SPION uptake, aggregates with size greater than 200 nm. (g–j) TEM images of various endosome types that contain SPION aggregates: Early endosome (g). (h) A multivesicular body with intraluminal vesicles inside of it. (i) A multilamellar morphology identifies a late endosome. (j) Late endosomes and lysosomes exhibit a multivesicular structure with extensive electron-dense zones. All scale bars correspond to 200 nm. Reproduced from Calero M, Chiappi M, Lazaro-Carrillo A, Rodríguez MJ, Chichón FJ, Crosbie-Staunton K, Prina-Mello A, Volkov Y, Villanueva A, Carrascosa JL. Characterization of interaction of magnetic nanoparticles with breast cancer cells. J Nanobiotechnology. 2015 Feb 26;13:16. © Calero et al; licensee BioMed Central. 2015. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0.101 |
Kasivelu Govindaraju et al (2015) investigated the anticancer effects of green synthesized silver nanoparticles (AgNPs) with a size of 10 nm on HL60 and HeLa cancer cell lines, as well as on normal peripheral blood mononuclear cells. In that study, they found that AgNPs induced cell death characterized by the formation of apoptotic bodies. The NPs were effective in preventing lipid peroxidation-induced reactive oxygen species (ROS) generation, thereby inhibiting carcinogenesis related to irradiation. TEM examination revealed that AgNPs were internalized by cells and localized within the nucleus and mitochondria as observed by Kasivelu Govindaraju et al (2015). The study observed several apoptotic indicators, including cellular shrinkage, chromatin compaction, nuclear damage, membrane leakage, and the breakdown of cells into apoptotic bodies. The structural damage to mitochondria caused by the accumulation of NPs was suggested to contribute to the observed mitochondrial destruction and subsequent cell death.102 Sato et al (2013) examined the effects of magnetic nanoparticles (MNPs) of Fe3O4, docetaxel (DTX), and their combination (DTX@ MNPs) on prostate cancer cells (DU145) in vitro. Their findings revealed that Fe3O4 NPs induced a dose-dependent increase in ROS and 8-hydroxydeoxyguanosine levels, which are markers of oxidative stress and DNA damage. The study suggested that combining MNPs with a low dose of DTX could be an effective strategy for treating prostate cancer, potentially reducing adverse effects compared to higher doses of DTX alone. TEM images demonstrated the uptake of Fe3O4 NPs by the cells and their localization within cellular vesicles (Figure 12), supporting the internalization and distribution of these NPs in the cancer cells.103
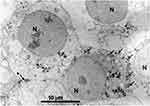 |
Figure 12 TEM visualization of DU145 cell line exposed to 10 μg/mL of MNPs of Fe3O4. Arrow: extracellular MNPs; arrowhead: intracellular MNPs. Abbreviation: N, nucleus. The scale bars correspond to 10 µm. Reproduced from Sato A, Itcho N, Ishiguro H, et al. Magnetic nanoparticles of Fe3O4 enhance docetaxel-induced prostate cancer cell death. Int J Nanomed. 2013;8:3151–3160. © 2013 The Author(s). This work is published and licensed by Dove Medical Press Limited.103 |
Wang et al (2019) investigated the cytotoxicity, DNA damage, and apoptosis induction of 5 nm titanium dioxide nanoparticles (TiO2 NPs) on the A549 cell line (isolated from the lung tissue). The MTT viability assay demonstrated that the NPs caused time- and concentration-dependent cytotoxicity. After 48 h of exposure, significant DNA damage was observed in the treated cells. Flow cytometry revealed a notable arrest in the G2/M phase of the cell cycle and a substantial increase in apoptotic cells. The NPs also impaired mitochondrial membrane potential. SEM showed distinct morphological changes: control cells exhibited a large, spherical shape with a smooth surface covered in dense microvilli and small protrusions. In contrast, cells exposed to 50 and 100 µg/mL of NPs became flattened, with roughened membranes and an increase in the extent and number of protrusions, along with thinner microvilli. At a 200 µg/mL concentration, cells showed wrinkling on their surface and pronounced apoptotic features, including cellular shrinkage. These results indicate that TiO2 NPs induce apoptosis in A549 cells, with the extent of damage correlating with the concentration of NPs.104
Preparation of Cellular Samples for Electron Microscopy Analysis
One of the challenges in visualizing cells using electron microscopy (EM) is managing the inherent water content of biological specimens. To address this, samples must be prepared before being placed in the vacuum of the EM column. For accurate preservation of the biological matrix at around 10 nm and the physicochemical properties of NPs within cellular compartments, standard room temperature preparation techniques can be applied to cells and tissues.107 Another major challenge is the intrinsic low contrast of biological samples under the electron beam, as they are primarily composed of light elements that scatter electrons poorly. To enhance contrast, heavy metal solutions such as osmium tetroxide, uranyl acetate, and lead citrate are commonly used during the TEM chemical preparation process. These staining agents bind to cellular structures, improving electron scattering and enabling detailed visualization of ultrastructural components of the biological specimens.97,108 The choice of specimen preparation method depends on the desired resolution and preservation of cellular ultrastructure, whether detailed localization of specific organelles or merely confirming NP uptake is required. Proper specimen preparation is crucial, as the mobility or dissolution of NPs can complicate uptake analysis.109 Conventional sample preparation techniques, commonly used for TEM, include chemical fixation to preserve cellular ultrastructure, producing thin sections (typically 50–150 nm) through embedding and slicing, and enhancing contrast through staining agent.110 The chemical fixation methods vary based on sample size and type and include the following: (1) in-situ fixation for cell cultures, (2) immersion fixation for tissue samples, (3) vascular perfusion for animals, and (4) vapor fixation for small samples like membranes. These methods ensure that the specimen maintains its nearly native structure, allowing for detailed visualization and accurate analysis of cellular and nanoparticle interactions. Notable, the chemical fixation method, using fixatives, specific buffer osmolarity, solvents, and contrasting agents, might alter the original structure of the sample to some extent.111
The standard fixation method using glutaraldehyde and osmium tetroxide (OsO4) is a widely adopted approach in conventional electron microscopy sample preparation (see Figure 13).69,97,112 This method begins with an initial fixation step using glutaraldehyde, which forms protein cross-links and deactivates enzymatic activity, thereby preserving the cellular structure. Subsequently, the second fixative, OsO4 is employed as a post-fixation agent. OsO4 interacts with unsaturated lipids in cellular membranes and other structures, undergoing reduction in the process. This step not only initiates staining by binding heavy osmium atoms but also enhances contrast by highlighting components with unsaturated carbon bonds, including NPs within the cellular matrix. The combined use of these fixatives: glutaraldehyde and OsO4 ensures the fixation and stabilization of both proteins and lipids, crucial for detailed ultrastructural analysis. However, it is important to note that this fixation process can affect the structural integrity and functionality of these biomolecules, potentially altering their morphology to some extent.
After chemical fixation, the crucial step is embedding samples in a polymeric resin network. The preparation process involves several key steps: first, the specimens undergo dehydration to remove water, followed by infiltration with reactive monomers. These monomers are then polymerized at temperatures typically 60–70 °C to form a solid resin with a three-dimensional network. This resin embedding preserves the sample’s structural integrity and allows for the creation of thin sections. Once embedded, the resin blocks (resin capsule) are sliced either into semithin (for light microscopy) or ultrathin sections (for TEM) using ultramicrotomy, and these slices are then mounted onto glass slides or electron microscopy copper grids. Thereafter, the staining is performed to further enhance the contrast of the sample and its constituents. OsO4 is commonly used to bind to unsaturated lipids, but other stains are employed to highlight different cellular structures. For instance, uranyl acetate and lead citrate are effective stains for accentuating various membrane structures and cellular components. Uranyl acetate binds to phosphates in DNA and RNA, as well as to amino groups in proteins, while lead citrate interacts with carboxyl groups and other functional groups. This differential staining enhances the contrast of ultrastructural features, such as membrane morphology and organelles, facilitating detailed visualization in TEM.
In addition to traditional chemical fixation methods, which can be harsh and may alter the sample’s original state, alternative approaches such as cryo-electron microscopy (cryo-EM) have been developed to better preserve the native state of hydrated, organic-rich samples (see Figure 14).104 Cryo-EM techniques involve fast-freezing methods to immobilize lipids and proteins without forming damaging ice crystals. One common technique is plunge-freezing where a small sample is rapidly immersed in liquid nitrogen or ethane to achieve vitrification, thereby preserving the sample’s original structure. However, plunge-freezing is generally limited to thin samples due to its slower cooling rates. An alternative approach involves freezing under high pressure, which combines ultra-low temperatures with increased pressures to prevent ice crystal formation in thicker samples. This method allows for the visualization of larger sample volumes under cryogenic conditions using cryo-EM; however, the signal-to-noise ratio remains low due to factors such as phase contrast, direct imaging of unstained samples, and the low electron dose required to minimize radiation damage. Alternatively, the frozen samples could be pre-stained in order to enhance the signal-to-noise ratio as followed by S. Rubino et al (2012).113 In cryo-FIB/SEM, the frozen samples are first protected by a thin layer of platinum coating, then prepared a small portion of the sample in a rectangular shape (known as a lamella), lamella is lifted-out from the holder, and welded on TEM copper grid. Finally, the lamella is thinned up to an electron transparent level using ions of the FIB. The samples are then examined under either SEM in a continued process or transferred to cryo-TEM for detailed investigation. To obtain detailed 3D structural information, multiple images are captured from various tilt angles to create a tilt series, which is then used to reconstruct the 3D structure. Tokuyasu cryo sectioning is another reliable and sensitive method for preparing biological samples. This technique involves initial chemical fixation with low concentrations of aldehydes, followed by infiltration with gelatin and sucrose instead of organic solvents. The gelatin- and sucrose-infiltrated sample is then rapidly frozen in liquid nitrogen and sliced at low temperatures using a dry knife. The ultra-thin sections are collected on Formvar-coated grids and stained with methylcellulose and uranyl acetate to enhance contrast for imaging.114 This approach helps maintain the integrity of the biological material while preparing it for detailed electron microscopy analysis as illustrated in Figure 15, showing different features of the vesicle from human adipocytes membrane and an area close to the cell’s periphery.105
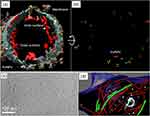 |
Figure 15 Cryo-electron microscopy of a vesicle from human adipocytes membrane preparations, treated with AuNP-FGF21 conjugate. (a) Isosurface rendering of tomographic reconstruction, with membrane density (blue-grey), density on the outer surface of the membrane (brown), density on the inner surface of the membrane (red), and AuNPs (yellow). (b) Same as (a), with all membrane and membrane-associated density removed, with different colours to distinguish pairs of AuNPs, and with rotation of 45° from the view in (a) for better visualization of AuNPs. (c) A section of the cryo-electron microscopy tomogram displaying an area close to the cell’s periphery. (d) 3D tomographic data with the membrane shown (blue), isosurface rendering showing the coated vesicle membrane (cyan), clathrin (magenta), actin (red), and microtubules (green), hexameric rings (emerald), and AuNPs (yellow). Reproduced from Azubel M, Carter SD, Weiszmann J, et al. FGF21 trafficking in intact human cells revealed by cryo-electron tomography with gold nanoparticles. Elife. 2019;8:e43146. © 2019, Azubel et al. Creative Commons Attribution License.105 |
Conclusion
The application of NPs in biomedicine, particularly for cellular and tissue imaging, is an expanding field with significant advancements driven by nanomedicine and nanotoxicology research. Imaging techniques, often integrated with various methods, have become essential for understanding NPs’ behavior within cellular environments. Each imaging method offers unique, providing specific types of data that contribute to a comprehensive understanding of NPs interactions with cells. EM has proven indispensable for studying NP-cell interactions due to its ability to reveal detailed ultrastructural changes, including NPs aggregation, shape, and their interaction with cellular membranes and organelles. TEM allows the precise observation of these interactions, which are challenging to discern with other techniques. By collecting specimens at various time points, TEM can offer dynamic insights into NPs uptake, transport, and exit. Additionally, computed tomography can provide 3D reconstructions of NPs localization within tissues and cells. The sample preparation process of the biological specimens is always challenging and can alter NPs and cellular ultrastructure. Traditional chemical fixation methods, while useful for simpler tasks, may cause artifacts to the NPs or cellular components. However, the electron dose required for imaging primarily leads to radiation damage in vitrified specimens within the cryo-EM workflow, whereas chemically fixed, plastic-embedded samples are only marginally affected by this issue. Rapid freezing techniques, such as plunge-freezing, are highly effective for preserving isolated NPs and thin biological samples close to their native state by producing vitreous ice. However, for vitrifying thicker specimens such as cells or tissues, high-pressure freezing is required to ensure uniform vitrification and minimize artifacts, providing a more accurate representation of the cellular environment. Conversely, conventional chemical fixation remains suitable for certain applications, such as identifying NPs within cells and tissues.
To enhance the study of NP interactions and improve the quantification of NP uptake, combining electron microscopy with techniques like flow cytometry can be beneficial. EM allows for detailed ultrastructural analysis, while flow cytometry provides quantitative data on NP uptake. Utilizing rapid freezing methods for specimen preparation ensures that cellular structures are preserved as much as possible, facilitating accurate and meaningful analysis of NP behavior in the cellular context.
Abbreviations
EM, Electron microscopy; SEM, Scanning electron microscopy; TEM, Transmission electron microscopy; HR-TEM, High-resolution transmission electron microscopy; STEM, Scanning transmission electron microscopy; EDX or EDS, Energy dispersive X-rays spectroscopy; Cryo-TEM, Cryo-transmission electron microscopy; Cryo-EM, Cryo-electron microscopy; Cryo-ET, Cryo-electron tomography; Cryo-FIB/SEM, Cryo-focused ion beam/scanning electron microscopy; FE-SEM, Field emission scanning electron microscopy; CLEM, Correlative Light Electron Microscopy; 3D view, Three-dimensional view; MRI, Magnetic resonance imaging; Ramos, lymphoma; Jurkat, leukemia; PM, Plasma membrane; CDE, clathrin-dependent endocytosis; CADE, Caveolin-dependent endocytosis; CCIE, Clathrin and caveolin independent endocytosis; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; HL60, Human myeloblastic leukemic cells; MCF- 7, Breast cancer cells; HCT – 116, Colon cancer cell line; HEK – 293, Human embryonic kidney cell line; HeLa, Cervical cancer cells; A549, Lung cancer cell line; SKOV3, Human ovarian adenocarcinoma cells; RAW264.7, Murine macrophage cells; DU145, Human prostate cancer cell line; GA, Golgi apparatus; ER, Endoplasmic reticulum; NPs, Nanoparticles; SNCs, silica nano-capsules; BSA-NPs, bovine serum albumin nanoparticles; Fe3O4, Iron Oxide; SPION, Superparamagnetic iron oxide nanoparticles; DMSA – SPION, Dimercaptosuccinic acid- coated superparamagnetic iron oxide nanoparticles; CS -TPP NPs, chitosan-tripolyphosphate pentasodium nanoparticles; Au- DENPs, Acetylated dendrimer-entrapped gold nanoparticles; AuNP- scFv, Gold-labelled single chain antibody fragment; AgNPs, Silver nanoparticles; AuNPs, Gold nanoparticles; MgNPs - Fe3O4, Magnetic nanoparticles of Fe3O4; TiO2 NPs, Titanium oxide nanoparticles; DTX, Docetaxel; MTT assays, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays; FGF21, Fibroblast growth factor 21 in human fat cells; CHO cells, Chinese hamster ovary cells; FA, Ferulic acid; FP, Fusion protein; DDS, Drug delivery system; ROS, Reactive oxygen species; PBS, phosphate-buffered saline; Glu, Glutaraldehyde; OsO4, Osmium tetroxide; (UO2(OOCCH3)2, Uranyl acetate; (Pb3(C6H5O7)2), lead citrate; PO43-, Phosphate groups.
Consent for Publication
This manuscript is approved by both authors for submission.
Acknowledgments
The authors are acknowledged Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia for providing the opportunity to complete this work.
Disclosure
The authors declare that they have no competing interests.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Cancer Facts & Figures. 2024. [cited February 18, 2024]. Available from: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2024-cancer-facts-figures.html.
3. CDCBreastCancer. Cancer Treatments [Internet]. Centers for Disease Control and Prevention. 2023 [cited November 4, 2023]. Available from: https://www.cdc.gov/cancer/survivors/patients/treatments.htm.
4. Nam J, Son S, Park KS, Zou W, Shea LD, Moon JJ. Cancer nanomedicine for combination cancer immunotherapy. Nat Rev Mater. 2019;4(6):398–414. doi:10.1038/s41578-019-0108-1
5. Gotwals P, Cameron S, Cipolletta D, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17(5):286–301. doi:10.1038/nrc.2017.17
6. Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18(3):175–196. doi:10.1038/s41573-018-0006-z
7. Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi:10.1038/nrc.2016.108
8. Nasir A, Khan A, Li J, et al. Nanotechnology, A tool for diagnostics and treatment of cancer. Curr Top Med Chem. 2021;21(15):1360–1376. doi:10.2174/1568026621666210701144124
9. Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arabian J Chem. 2019;12(7):908–931. doi:10.1016/j.arabjc.2017.05.011
10. Aghebati-Maleki A, Dolati S, Ahmadi M, et al. Nanoparticles and cancer therapy: perspectives for application of nanoparticles in the treatment of cancers. J Cell Physiol. 2020;235(3):1962–1972. doi:10.1002/jcp.29126
11. Targeted Therapies: immunologic Effects and Potential Applications Outside of Cancer - PMC. [cited February 19, 2024]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5972536/.
12. Mendonça MCP, Kont A, Kowalski PS, O’Driscoll CM. Design of lipid-based nanoparticles for delivery of therapeutic nucleic acids. Drug Discovery Today. 2023;28(3):103505. doi:10.1016/j.drudis.2023.103505
13. Dristant U, Mukherjee K, Saha S, Maity D. An overview of polymeric nanoparticles-based drug delivery system in cancer treatment. Technol Cancer Res Treatment. 2023;22:15330338231152083.
14. Terna AD, Elemike EE, Mbonu JI, Osafile OE, Ezeani RO. The future of semiconductors nanoparticles: synthesis, properties and applications. Mat Sci Eng. 2021;272:115363. doi:10.1016/j.mseb.2021.115363
15. Shi Z, Zhou Y, Fan T, Lin Y, Zhang H, Mei L. Inorganic nano-carriers based smart drug delivery systems for tumor therapy. Smart Mater Med. 2020;1:32–47. doi:10.1016/j.smaim.2020.05.002
16. Mendes BB, Conniot J, Avital A, et al. Nanodelivery of nucleic acids. Nat Rev Meth Primers. 2022;2;1:1–21.
17. Gholami A, Mousavi SM, Hashemi SA, Ghasemi Y, Chiang WH, Parvin N. Current trends in chemical modifications of magnetic nanoparticles for targeted drug delivery in cancer chemotherapy. Drug Metab Rev. 2020;52(1):205–224.
18. Nanomedicine in cancer therapy - PubMed. [cited February 19, 2024]. Available from: https://pubmed.ncbi.nlm.nih.gov/37544972/.
19. Anfray C, Mainini F, Andón FT. Chapter 11 - Nanoparticles for immunotherapy. In: Parak WJ, Feliu N, editors. Frontiers of Nanoscience. Vol. 16. Elsevier; 2020:265–306.
20. Haleem A, Javaid M, Singh RP, Rab S, Suman R. Applications of nanotechnology in medical field: a brief review. Global Health J. 2023;7(2):70–77. doi:10.1016/j.glohj.2023.02.008
21. Elumalai K, Srinivasan S, Shanmugam A. Review of the efficacy of nanoparticle-based drug delivery systems for cancer treatment. Biomed Technol. 2024;5:109–122. doi:10.1016/j.bmt.2023.09.001
22. Oliveira BB, Ferreira D, Fernandes AR, Baptista PV. Engineering gold nanoparticles for molecular diagnostics and biosensing. WIREs Nanomed Nanobiotechnol. 2023;15(1):e1836. doi:10.1002/wnan.1836
23. Nanomaterial-based CT contrast agents and their applications in image-guided therapy - PMC. [cited February 21, 2024]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9830442/.
24. Wang X, Liu S, Sun Y, et al. Preparation of selective organ-targeting (SORT) lipid nanoparticles (LNPs) using multiple technical methods for tissue-specific mRNA delivery. Nat Protoc. 2023;18(1):265–291. doi:10.1038/s41596-022-00755-x
25. Sadeghzadeh H, Dianat-Moghadam H, Del Bakhshayesh AR, Mohammadnejad D, Mehdipour A. A review on the effect of nanocomposite scaffolds reinforced with magnetic nanoparticles in osteogenesis and healing of bone injuries. Stem Cell Res Ther. 2023;14(1):194
26. Malatesta M. Transmission electron microscopy as a powerful tool to investigate the interaction of nanoparticles with subcellular structures. Int J mol Sci. 2021;22(23):12789. doi:10.3390/ijms222312789
27. Chen C, Ge J, Gao Y, Chen L, Cui J, Zeng J, Gao M. Ultrasmall superparamagnetic iron oxide nanoparticles: a next generation contrast agent for magnetic resonance imaging - Chen - 2022. WIREs Nanomed Nanobiotechnol. 2022;14(1):e1740
28. Magnetic nanoparticles in nanomedicine: a review of recent advances - IOPscience [Internet]. [cited February 21, 2024]. Available from: https://iopscience.iop.org/article/10.1088/1361-6528/ab4241/meta.
29. Karaman DŞ, Sarparanta MP, Rosenholm JM, Airaksinen AJ. Multimodality imaging of silica and silicon materials in vivo. Adv Mater. 2018;30(24):1703651. doi:10.1002/adma.201703651
30. Arami H, Khandhar A, Liggitt D, Krishnan KM. In vivo delivery, pharmacokinetics, biodistribution and toxicity of iron oxide nanoparticles. Chem Soc Rev. 2015;44(23):8576–8607. doi:10.1039/c5cs00541h
31. Ostrowski A, Nordmeyer D, Boreham A, et al. Overview about the localization of nanoparticles in tissue and cellular context by different imaging techniques. Beilstein J Nanotechnol. 2015;6:263–280. doi:10.3762/bjnano.6.25
32. Jeong D, Kim D. Recent developments in correlative super-resolution fluorescence microscopy and electron microscopy. Mol Cells. 2022;45(1):41–50. doi:10.14348/molcells.2021.5011
33. Tantra R, Knight A. Cellular uptake and intracellular fate of engineered nanoparticles: a review on the application of imaging techniques. Nanotoxicology. 2011;5(3):381–392. doi:10.3109/17435390.2010.512987
34. Balasubramanian H, Hobson CM, Chew TL, Aaron JS. Imagining the future of optical microscopy: everything, everywhere, all at once. Commun Biol. 2023;6(1):1–12. doi:10.1038/s42003-022-04154-6
35. Rennick JJ, Johnston APR, Parton RG. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat Nanotechnol. 2021;16(3):266–276. doi:10.1038/s41565-021-00858-8
36. FitzGerald LI, Johnston APR. It’s what’s on the inside that counts: techniques for investigating the uptake and recycling of nanoparticles and proteins in cells. J Colloid Interface Sci. 2021;587:64–78. doi:10.1016/j.jcis.2020.11.076
37. Calderan L, Malatesta M. Imaging techniques in nanomedical research. Eur J Histochem. 2020;64(3):3151. doi:10.4081/ejh.2020.3151
38. Abdulqadir SZ, Aziz FM. Internalization and effects on cellular ultrastructure of nickel nanoparticles in rat kidneys. Int J Nanomed. 2019;14:3995–4005. doi:10.2147/IJN.S200909
39. Franken LE, Grünewald K, Boekema EJ, Stuart MCA. A technical introduction to transmission electron microscopy for soft-matter: imaging, possibilities, choices, and technical developments. Small. 2020;16(14):1906198. doi:10.1002/smll.201906198
40. Holzhausen C, Gröger D, Mundhenk L, Welker P, Haag R, Gruber AD. Tissue and cellular localization of nanoparticles using 35S labeling and light microscopic autoradiography. Nanomedicine. 2013;9(4):465–468. doi:10.1016/j.nano.2013.02.003
41. Tremi I, Havaki S, Georgitsopoulou S, et al. A guide for using transmission electron microscopy for studying the radiosensitizing effects of gold nanoparticles in vitro. Nanomaterials. 2021;11(4):859. doi:10.3390/nano11040859
42. Fagerland JA, Wall HG, Pandher K, LeRoy BE, Gagne GD. Ultrastructural analysis in preclinical safety evaluation. Toxicol Pathol. 2012;40(2):391–402. doi:10.1177/0192623311430239
43. Trépout S, Sgarra ML, Marco S, Ramm G. An introduction to scanning transmission electron microscopy for the study of protozoans. Mol Microbiol. 2024;121(4):659–670. doi:10.1111/mmi.15213
44. Skorikov A, Heyvaert W, Albecht W, Pelt DM, Bals S. Deep learning-based denoising for improved dose efficiency in EDX tomography of nanoparticles. Nanoscale. 2021;13(28):12242–12249. doi:10.1039/D1NR03232A
45. Li L, Mak KY, Shi J, et al. Comparative in vitro cytotoxicity study on uncoated magnetic nanoparticles: effects on cell viability, cell morphology, and cellular uptake. J Nanosci Nanotechnol. 2012;12(12):9010–9017. doi:10.1166/jnn.2012.6755
46. Goldstein A, Soroka Y, Frušić‐Zlotkin M, Popov I, Kohen R. High resolution SEM imaging of gold nanoparticles in cells and tissues - GOLDSTEIN - 2014. J Microsc. 2014;256(3):237–247
47. Cambria MT, Villaggio G, Laudani S, et al. The Interplay between Fe3O4 Superparamagnetic Nanoparticles, Sodium Butyrate, and Folic Acid for Intracellular Transport. Int J Mol Sci. 2020;21(22):8473
48. Cellular uptake of polylactide particles induces size dependent cytoskeletal remodeling in antigen presenting cells - Biomaterials Science (RSC Publishing). [cited February 25, 2024]. Available from: https://pubs.rsc.org/en/content/articlelanding/2021/bm/d1bm01312b.
49. Enhancing Serial Block-Face Scanning Electron Microscopy to Enable High Resolution 3-D Nanohistology of Cells and Tissues | microscopy and Microanalysis | Cambridge Core. [cited December 16, 2024]. Available from: https://www.cambridge.org/core/journals/microscopy-and-microanalysis/article/enhancing-serial-blockface-scanning-electron-microscopy-to-enable-high-resolution-3d-nanohistology-of-cells-and-tissues/97C0B65A8639C351962D165DF1C77589.
50. Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2(11):e329. doi:10.1371/journal.pbio.0020329
51. Titze B, Genoud C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol Cell. 2016;108(11):307–323. doi:10.1111/boc.201600024
52. Cognigni F, Miraglia L, Contessi S, Biancardi F, Rossi M. Correlative light and electron microscopy (CLEM): a multifaceted tool for the study of geological specimens. J Exp Theoret Anal. 2023;1(2):74–85. doi:10.3390/jeta1020006
53. Tanida I, Yamaguchi J, Suzuki C, Kakuta S, Uchiyama Y. Recent advances in in-resin correlative light and electron microscopy of Epon-embedded cells. Microscopy. 2023;72(5):383–387. doi:10.1093/jmicro/dfad028
54. Thiberge S, Nechushtan A, Sprinzak D, et al. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc Natl Acad Sci USA. 2004;101:3346–3351. doi:10.1073/pnas.0400088101
55. Bárcena M, Koster AJ. Electron tomography in life science. Semin Cell Dev Biol. 2009;20(8):920–930. doi:10.1016/j.semcdb.2009.07.008
56. Moebel E, Martinez-Sanchez A, Lamm L, et al. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Springer Nat Exp. 2021;18(11):1386–1394.
57. De Yoreo JJ, A. J. M. S N. Investigating materials formation with liquid-phase and cryogenic TEM. Nat Rev Mater. 2016;1(8):1–18.
58. Bjornmalm M, Thurecht KJ, Michael M, Scott AM, Caruso F. Bridging bio–nano science and cancer nanomedicine. ACS Nano. 2017;11(10):9594–9613.
59. Luo YH, Chang LW, Lin P. Metal-based nanoparticles and the immune system: activation, inflammation, and potential applications. Biomed Res Int. 2015;2015:e143720. doi:10.1155/2015/143720
60. Boraschi D, Italiani P, Palomba R, et al. Nanoparticles and innate immunity: new perspectives on host defence. Semin Immunol. 2017;34:33–51. doi:10.1016/j.smim.2017.08.013
61. Rashid MM, Tavčer P F, Tomšič B. Influence of titanium dioxide nanoparticles on human health and the environment. Nanomaterials. 2024;11(9):2354. doi:10.3390/nano11092354
62. Donahue ND, Acar H, Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv Drug Delivery Rev. 2019;143:68–96. doi:10.1016/j.addr.2019.04.008
63. Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9(1):1410. doi:10.1038/s41467-018-03705-y
64. von RC, Jiang W, Chan CK, Weissman IL, Kim BYS. Breaking down the barriers to precision cancer nanomedicine. Trends Biotechnol. 2017;35(2):159–171. doi:10.1016/j.tibtech.2016.07.006
65. Thorn K. A quick guide to light microscopy in cell biology. Mol Biol Cell. 2016;27(2):219–222. doi:10.1091/mbc.e15-02-0088
66. Panwar R, Sharma AK, Kaloti M, Dutt D, Pruthi V. Characterization and anticancer potential of ferulic acid-loaded chitosan nanoparticles against ME-180 human cervical cancer cell lines. Appl Nanosci. 2016;6(6):803–813. doi:10.1007/s13204-015-0502-y
67. Kattner N, Dyson N, Bury Y, et al. Development and validation of a quantitative electron microscopy score to assess acute cellular stress in the human exocrine pancreas. J Pathol Clin Res. 2021;7(2):173–187. doi:10.1002/cjp2.185
68. Pudovkin MS, Shamsutdinov NI, Zelenikhin PV, Nizamutdinov AS. Transmission electron microscopy and flow cytometry study of cellular uptake of unmodified Pr3+:LaF3 nanoparticles in dynamic. J Nanopart Res. 2021;23(6):124. doi:10.1007/s11051-021-05249-7
69. Guo L, Peng Y, Yao J, Sui L, Gu A, Wang J. Anticancer activity and molecular mechanism of resveratrol-bovine serum albumin nanoparticles on subcutaneously implanted human primary ovarian carcinoma cells in nude mice. Cancer Biother Radiopharm. 2010;25(4):471–477. doi:10.1089/cbr.2009.0724
70. Zhang X, Ma G, Wei W. Simulation of nanoparticles interacting with a cell membrane: probing the structural basis and potential biomedical application. NPG Asia Mater. 2021;13(1):1–18. doi:10.1038/s41427-021-00320-0
71. Chen D, Wang J, Wang Y, et al. Promoting inter-/intra- cellular process of nanomedicine through its physicochemical properties optimization. Curr Drug Metab. 2018;19(1):75–82. doi:10.2174/1389200219666171221122119
72. Foroozandeh P, Aziz AA. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res Lett. 2018;13(1):339. doi:10.1186/s11671-018-2728-6
73. Zhang R, Qin X, Kong F, Chen P, Pan G. Improving cellular uptake of therapeutic entities through interaction with components of cell membrane. Drug Delivery. 2019;26(1):328–342. doi:10.1080/10717544.2019.1582730
74. Corradi V, Sejdiu BI, Mesa-Galloso H, et al. Emerging diversity in lipid–protein interactions. Chem Rev. 2019;119(9):5775–5848
75. Levental I, Lyman E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat Rev mol Cell Biol. 2023;24(2):107–122. doi:10.1038/s41580-022-00524-4
76. Behzadi S, Serpooshan V, Tao W, et al. Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev. 2017;46(14):4218–4244. doi:10.1039/C6CS00636A
77. de AMS, Susnik E, Drasler B, Taladriz-Blanco P, Petri-Fink A, Rothen-Rutishauser B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem Soc Rev. 2021;50(9):5397–5434. doi:10.1039/d0cs01127d
78. Sabourian P, Yazdani G, Ashraf SS, et al. Effect of physico-chemical properties of nanoparticles on their intracellular uptake. Int J Mol Sci. 2020;21(21):8019. doi:10.3390/ijms21218019
79. Francia V, Montizaan D, Salvati A. Interactions at the cell membrane and pathways of internalization of nano-sized materials for nanomedicine. Beilstein J Nanotechnol. 2020;11(1):338–353. doi:10.3762/bjnano.11.25
80. Bayati A, Kumar R, Francis V, McPherson PS. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296:100306. doi:10.1016/j.jbc.2021.100306
81. Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19(5):313–326. doi:10.1038/nrm.2017.132
82. Harush-Frenkel O, D N, B S, A Y. Targeting of nanoparticles to the clathrin-mediated endocytic pathway. Biochem Biophys Res Commun. 2007;353(1):26–32.
83. Wang X, Qiu Y, Wang M, et al. Endocytosis and organelle targeting of nanomedicines in cancer therapy. Int J Nanomed. 2020;15:9447–9467. doi:10.2147/IJN.S274289
84. Chatterjee M, Ben-Josef E, Robb R, et al. Caveolae-mediated endocytosis is critical for albumin cellular uptake and response to albumin-bound chemotherapy. Cancer Res. 2017;77(21):5925–5937. doi:10.1158/0008-5472.CAN-17-0604
85. Bellotti E, Cascone MG, Barbani N, et al. Targeting cancer cells overexpressing folate receptors with new terpolymer-based nanocapsules: toward a novel targeted DNA delivery system for cancer therapy. Biomedicines. 2021;9(9):1275. doi:10.3390/biomedicines9091275
86. Kay RR. Macropinocytosis: biology and mechanisms. Cells Dev. 2021;168:203713. doi:10.1016/j.cdev.2021.203713
87. Micropinocytosis - an overview | scienceDirect Topics [Internet]. [cited March 30, 2024]. Available from: https://www.sciencedirect.com/topics/medicine-and-dentistry/micropinocytosis.
88. How to build a phagosome: new concepts for an old process - ScienceDirect. [cited March 30, 2024]. Available from: https://www.sciencedirect.com/science/article/abs/pii/S0955067417301540.
89. Uribe-Querol E, Rosales C. Phagocytosis: our current understanding of a universal biological process. Front Immunol. 2020;11:1066. doi:10.3389/fimmu.2020.01066
90. Uribe-Querol E, Rosales C. Control of phagocytosis by microbial pathogens. Front Immunol. 2017;8:1368. doi:10.3389/fimmu.2017.01368
91. Varma S, Dey S, Palanisamy D. Cellular Uptake Pathways of Nanoparticles: process of Endocytosis and Factors Affecting their Fate – pubMed. Current Pharmaceut Biotechnol. 2022;23(5):679–706
92. Lee HJ, Woo Y, Hahn TW, Jung YM, Jung YJ. Formation and maturation of the phagosome: a key mechanism in innate immunity against intracellular bacterial infection. Microorganisms. 2020;8(9):1298. doi:10.3390/microorganisms8091298
93. Phagocytosis: a Fundamental Process in Immunity - PMC. [cited March 29, 2024]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5485277/.
94. Hui Y, Yi X, Wibowo D, et al. Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Sci Adv. 2020;6(16):eaaz4316.
95. Gunn J, Park SI, Veiseh O, Press OW, Zhang M. A pretargeted nanoparticle system for tumor cell labeling. Mol BioSyst. 2011;7(3):742–748. doi:10.1039/C005154C
96. Cheng D, Li X, Zhang G, Shi H. Morphological effect of oscillating magnetic nanoparticles in killing tumor cells. Nanoscale Res Lett. 2014;9(1):195. doi:10.1186/1556-276X-9-195
97. Akhtar S, Khan FA, Buhaimed A. Functionalized magnetic nanoparticles attenuate cancer cells proliferation: transmission electron microscopy analysis. Microsc Res Tech. 2019;82(7):983–992. doi:10.1002/jemt.23245
98. Vivek R, Nipun Babu V, Thangam R, Subramanian KS, Kannan S. pH-responsive drug delivery of chitosan nanoparticles as Tamoxifen carriers for effective anti-tumor activity in breast cancer cells. Colloids Surf B Biointerfaces. 2013;111:117–123. doi:10.1016/j.colsurfb.2013.05.018
99. De Berardis B, Civitelli G, Condello M, et al. Exposure to ZnO nanoparticles induces oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol Appl Pharmacol. 2010;246(3):116–127. doi:10.1016/j.taap.2010.04.012
100. Wang H, Zheng L, Peng C, et al. Computed tomography imaging of cancer cells using acetylated dendrimer-entrapped gold nanoparticles. Biomaterials. 2011;32(11):2979–2988. doi:10.1016/j.biomaterials.2011.01.001
101. Calero M, Chiappi M, Lazaro-Carrillo A, et al. Characterization of interaction of magnetic nanoparticles with breast cancer cells. J Nanobiotechnol. 2015;13:1–5
102. Govindaraju K, Krishnamoorthy K, Alsagaby SA, Singaravelu G, Premanathan M. Green synthesis of silver nanoparticles for selective toxicity towards cancer cells. IET Nanobiotechnol. 2015;9(6):325–330. doi:10.1049/iet-nbt.2015.0001
103. Sato A, Itcho N, Ishiguro H, et al. Magnetic nanoparticles of Fe3O4 enhance docetaxel-induced prostate cancer cell death. Int J Nanomed. 2013;8:3151–3160. doi:10.2147/IJN.S40766
104. Wang Y, Cui H, Zhou J, et al. Cytotoxicity, DNA damage, and apoptosis induced by titanium dioxide nanoparticles in human non-small cell lung cancer A549 cells. Environ Sci Pollut Res Int. 2015;22(7):5519–5530. doi:10.1007/s11356-014-3717-7
105. Azubel M, Carter SD, Weiszmann J, et al. FGF21 trafficking in intact human cells revealed by cryo-electron tomography with gold nanoparticles. Elife. 2019;8:e43146. doi:10.7554/eLife.43146
106. Klumperman J, Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol. 2014;6(10):a016857. doi:10.1101/cshperspect.a016857
107. Fischer ER, Hansen BT, Nair V, Hoyt FH, Dorward DW. Scanning electron microscopy. Curr Protoc Microbiol. 2012;25(1). doi:10.1002/9780471729259.mc02b02s25
108. Gray JL, Tian Y, Ning G, Cutler T, Zheng Y, Patterson AD. Optimization of Conventionally Processed Biological Samples for STEM-EDS Elemental Quantification. Microsc Microanal. 2024;30(Supplement_1):ozae044. doi:10.1093/mam/ozae044.428
109. Brown A, Hondow N. Chapter 4 - Electron Microscopy of Nanoparticles in Cells. In: Summers H, editor. Frontiers of Nanoscience. Vol. 5. Elsevier; 2013:95–120.
110. Kumar S, Filippi MD, An alternative approach for sample preparation with low cell number for TEM analysis. J Vis Exp. 2016;116:54724. doi:10.3791/54724
111. Dey P. Fixation of Histology Samples: principles, Methods and Types of Fixatives. In: Dey P, editor. Basic and Advanced Laboratory Techniques in Histopathology and Cytology. Singapore: Springer Nature; 2022:3–18.
112. Cisplatin delivery, anticancer and antibacterial properties of Fe/SBA-16/ZIF-8 nanocomposite - PubMed. [cited September 12, 2024]. Available from: https://pubmed.ncbi.nlm.nih.gov/35559226/.
113. Rubino S, Akhtar S, Melin P, Searle A, Spellward P, Leifer K. A site-specific focused-ion-beam lift-out method for cryo transmission electron microscopy. J Struct Biol. 2012;180(3):572–576. doi:10.1016/j.jsb.2012.08.012
114. Bos E, Hussaarts L, van Weering JRT, Ellisman MH, de Wit H, Koster AJ. Vitrification of Tokuyasu-style immuno-labelled sections for correlative cryo light microscopy and cryo electron tomography. J Struct Biol. 2014;186(2):273–282. doi:10.1016/j.jsb.2014.03.021
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.