Back to Journals » Drug Design, Development and Therapy » Volume 19
Alisol Triterpenoids Exhibit Triple Modulatory Mechanisms on the Cell Membrane to Overcome Cancer Multidrug Resistance
Authors Chen JY, Chang YT , Ho YC, Teng YN
Received 17 February 2025
Accepted for publication 21 June 2025
Published 26 June 2025 Volume 2025:19 Pages 5471—5488
DOI https://doi.org/10.2147/DDDT.S521116
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Manfred Ogris
Jing-Yi Chen,1,* Ying-Tzu Chang,2,* Yu-Cheng Ho,3 Yu-Ning Teng2– 4
1Department of Medical Laboratory Science, College of Medical Science and Technology, I-Shou University, Kaohsiung, Taiwan, Republic of China; 2Department of Pharmacy, College of Pharmacy, China Medical University, Taichung City, Taiwan, Republic of China; 3School of Medicine, College of Medicine, I-Shou University, Kaohsiung, Taiwan, Republic of China; 4Department of Pharmacy, E-Da Cancer Hospital, Kaohsiung, Taiwan, Republic of China
*These authors contributed equally to this work
Correspondence: Yu-Ning Teng, Department of Pharmacy, College of Pharmacy, China Medical University, 100, Sec. 1, Jingmao Road, Beitun Dist, Taichung City, 406040, Taiwan, Republic of China, Tel +886-4-22053366 ext. 5158, Fax +886-4-22078083, Email [email protected]; [email protected]
Purpose and Study Design: Multi-drug resistance (MDR) in cancer significantly hinders effective treatment, leading to poor patient outcomes. The study investigates the potential of natural compounds, Alisol B 23-acetate (B23) and Alisol A 24-acetate (A24), to reverse MDR through various mechanisms on cancer cell membranes.
Results: Cytotoxicity assays established non-toxic concentrations of B23 and A24, which were then tested in drug-sensitive and drug-resistant cancer cell lines with or without chemotherapeutic drugs. Both compounds significantly enhanced reactive oxygen species (ROS) production and apoptosis in HepG2/VIN MDR cells while preserving cell membrane integrity. They also improved membrane fluidity and inhibited the function of P-glycoprotein (P-gp) efflux transporters in both HepG2/VIN and ABCB1/Flp-InTM-293, leading to increased drug accumulation. Molecular docking studies revealed that B23 and A24 interact with distinct binding sites on P-gp, demonstrating allosteric and competitive inhibition.
Conclusion: B23 and A24 effectively reverse cancer MDR by (1) modulating ROS levels and inducing apoptosis, (2) maintaining membrane integrity but improving membrane fluidity, and (3) inhibiting drug efflux by membrane transporters. These findings provide a promising basis for developing new therapeutic strategies to combat MDR in cancer, highlighting the potential use of these natural product derivatives in adjunctive cancer therapy.
Keywords: alisol triterpenoids, multi-drug resistance, cell membrane fluidity, efflux transporter inhibitor, reactive oxygen species
Introduction
Multi-drug resistance (MDR) is a significant barrier to the effective treatment of cancer, contributing to therapeutic failure and poor prognosis for patients. It is defined as the phenomenon in which cancer cells develop resistance to various drugs with differing mechanisms of action, ultimately making standard chemotherapy ineffective.1 MDR is responsible for over 90% of cancer-related deaths, primarily due to the ability of cancer cells to evade drugs designed to combat them. This challenge has propelled researchers to investigate the underlying mechanisms of MDR to identify novel therapeutic strategies and to develop drugs that either evade these resistance mechanisms or reverse them.2 As the overactivity of drug-efflux transporters plays a major role in contributing to the MDR phenomenon, there are several strategies targeting this dilemma. One approach involves the co-administration of MDR transporter inhibitors alongside cytotoxic drugs, a strategy known as “engagement or inhibition”. This method aims to block the action of MDR transporters, allowing for higher concentrations of chemotherapy agents within the cancer cells. Various techniques and substances are involved in this strategy, such as the application of nanotechnology or natural products. However, it is not easy to achieve the balance between efficacy and safety, and the combinatorial ratio challenge when applied from preclinical research to clinical study.3–5 Another strategy focuses on using cytotoxic agents designed to evade MDR transporter-mediated efflux, thus ensuring that these drugs can enter and accumulate in the cancer cells without being expelled. This tactic is referred to as “evasion or bypass”. Nevertheless, the design of the structure escaping from the recognition of MDR transporters is challenging, given the nature of the diversified structure recognition of MDR transporters.6 Additionally, a third strategy capitalizes on the collateral sensitivity exhibited by multidrug-resistant cells, termed “exploitation or target”. This approach identifies and utilizes drugs to which MDR cells have developed an unexpected sensitivity. The challenge of this strategy is the diversity of cancer cells, especially the resistant ones. Finding a specific compound that can target all stages and phenotypes of MDR cells is still tricky at present.7 Therefore, the discovery of novel mechanisms behind MDR and the development of promising combinatorial strategies might have an opportunity to unveil and combat the MDR cancer issue.
The mechanisms contributing to MDR in cancer are multifaceted and involve various cellular processes, particularly those related to the cancer cell membrane. One of the prominent mechanisms includes the action of ATP-binding cassette (ABC) transporters, such as P-glycoprotein (P-gp), which actively pump out chemotherapeutic agents from the cells, thereby reducing drug accumulation.8 In addition, alterations in reactive oxygen species (ROS) levels play a crucial role, as increased ROS can lead to cellular damage that either promotes apoptosis or allows cells to adapt and survive despite treatment.9 Changes in apoptosis pathways, such as those resulting in necrosis or autophagy, further complicate the response to chemotherapy.10 Additionally, modifications in the fluidity and integrity of cancer cell membranes can influence drug uptake and resistance. Membrane fluidity is strongly linked to cholesterol levels, with many studies indicating a relationship between the activity of P-gp ATPase and both cholesterol content and membrane fluidity.11 Cholesterol acts as a transcriptional regulator for MDR efflux transporters, influencing both the expression and function of key multidrug transporters that contribute to MDR.12 Substrates of P-gp may accumulate in cholesterol-rich membrane areas, thereby increasing P-gp activity.13 Additionally, interactions between lipids and transporters facilitate structural changes in P-gp, which in turn regulate its catalytic function.14 Collectively, these mechanisms highlight the adaptability of cancer cells, reinforcing the need for innovative approaches to combat MDR.
Among the various approaches, natural products emerged as potential resources and attracted researchers’ attention.3 Alisol B 23-acetate (B23)15 and Alisol A 24-acetate (A24)16(Supplementary Figure 1), two natural compounds derived from Alismatis Rhizoma, offer promising therapeutic potential in both clinical applications and basic research, particularly in the context of cancer17 B23 has demonstrated efficacy in inhibiting various coronaviruses, including SARS-CoV-2, by obstructing viral entry and mitigating proinflammatory responses, positioning it as a candidate for COVID-19 treatment.18 Furthermore, it has been found to promote the browning of white adipose tissue, making it a viable option for managing obesity.19 In cancer research, B23 has shown anti-tumor properties, inducing apoptosis in several cancer cell lines, including those of gastric and lung origins, highlighting their multifaceted biological activities and therapeutic implications.20–22 Nevertheless, the potential and detailed modulatory mechanisms of B23 and A24 in resolving MDR cancer warrant further investigations.
This study explores the ability of B23 and A24 to combat MDR in cancer cells through a series of mechanisms. Results suggest these compounds significantly increase ROS levels and enhance apoptosis in MDR cancer cells. However, the secondary necrosis is not initiated while cell membrane fluidity is enhanced, preserving membrane integrity. Notably, both compounds inhibit the function of ABC drug efflux transporters, thereby enhancing the accumulation of chemotherapeutic agents within the cells. These findings indicate that B23 and A24 may serve as effective agents in overcoming MDR, providing substantial support for their further investigation as potential adjuncts in cancer chemotherapy.
Materials and Methods
Chemicals and Reagents
B23 and A24 were purchased from TargetMol Chemicals Inc. (Boston, MA, USA). Acetic acid, calcein-AM, dimethyl sulfoxide (DMSO), doxorubicin, menadione, methyl-β-cyclodextrin (MBCD), paclitaxel, rhodamine 123, sulforhodamine B (SRB), trichloroacetic acid, verapamil, vinblastine, and vincristine were purchased from Sigma-Aldrich, Corp. (St. Louis, Missouri, USA). Dulbecco’s Modified Eagle Media (DMEM), fetal bovine serum (FBS), hygromycin B, phosphate-buffered saline (PBS), Roswell Park Memorial Institute-1640 (RPMI-1640) medium, trypsin-EDTA, and zeocin were purchased from Invitrogen, Corp. (Carlsbad, California, USA).
Cell Lines and Cell Culture
The human Flp-InTM-293 cell line (Thermo Fisher Scientific; #R75007) was used to construct a stable P-glycoprotein (P-gp) over-expressed cell line (ABCB1/Flp-InTM-293) as described previously.23 Flp-InTM-293 and ABCB1/Flp-InTM-293 were selected based on 100 μg/mL zeocin and 100 μg/mL hygromycin B resistance, respectively. The human cervical epithelioid carcinoma (HeLaS3) was sourced from the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan), while Dr. Kuo-Hsiung Lee generously provided the corresponding multi-drug resistant KB/VIN cells from the University of North Carolina, Chapel Hill, USA. The use of KB/VIN cells was approved by the research ethics committee at I-Shou University on 13 Dec 2022 (approval for the application and conduct of NSTC proposal: NSTC 112–2320-B-214-003-MY3 and NSTC 112–2320-B-039-063-MY3). The human breast cancer cell line (MCF-7) was obtained from BCRC. Doxorubicin-resistant MCF-7 cells (MCF-7/DOX) were developed by exposing MCF-7 cells to doxorubicin, resulting in a drug-resistant clone. The human hepatocellular carcinoma (HepG2) was purchased from BCRC. The corresponding multi-drug resistant HepG2/VIN was constructed by constant vincristine exposure. To preserve the MDR characteristics, KB/VIN, MCF-7/DOX, and HepG2/VIN were maintained in RPMI-1640 containing 100 nM vincristine, 500 nM doxorubicin, and 2 μM vincristine, respectively. All cells were cultured in DMEM or RPMI-1640 media containing 10% FBS at 37°C under a humidified atmosphere of 5% CO2.
Cytotoxic SRB Assay
72-h cytotoxicity was assessed using the SRB colorimetric assay, as previously described.24 Cells were incubated with the testing compounds and subsequently fixed with 50% trichloroacetic acid for 30 min. After washing them with water and allowing them to air dry, the cells were stained with 0.04% SRB for 30 min. Unbound dye was removed by rinsing with 1% acetic acid. Once air-dried, the bound stain was dissolved in 10 mM tris-base, and the absorbance was measured at 515 nm using a SpectraMax® ABS plus reader (Molecular Devices, LLC).
ROS Production Evaluation
The ROS generation after testing compounds treatment was performed using ROS-Glo™ H2O2 Assay (Promega). Cells were incubated with testing compounds for 24-h in a 96-well white plate. Then, the H2O2 Substrate Solution was added to the plate and incubated for an additional 2-h. Finally, the ROS-Glo™ Detection Solution was added and incubated for 20 min at RT. The luminescent signals were detected using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC).
Apoptosis and Secondary Necrosis Evaluation
The RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay (Promega) was used for determination. Briefly, Detection Reagent (containing Annexin V NanoBiT™ Substrate, CaCl2, Necrosis Detection Reagent, Annexin V-SmBiT, Annexin V-LgBiT), testing compounds, and cells were placed into a 96-well white plate at a total volume 200 μL. Then, the plate was incubated for the indicated period. The 48-h apoptotic events were evaluated by luminescence detection with the interaction between NanoBiT® Luciferase, Annexin V-LgBiT, and Annexin V-SmBiT. The 72-h secondary necrosis and membrane integrity were evaluated by fluorescence detection (excitation/emission: 485/528nm) with the Necrosis Detection Reagent. SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC) was used.
Membrane Fluidity Assessment
Cell membrane fluidity was evaluated using a Membrane Fluidity Kit (Abcam, #ab189819), as previously described.24 Cells cultured in a 96-well black plate were treated with testing compounds or 2.5 mM methyl-β-cyclodextrin (MBCD) (positive control) for 72-h. The cells were incubated with the labeling solution for 1-h at 25°C in the dark. After discarding the labeling solution and replacing it with fresh culture medium (DMEM for Flp-InTM-293 and ABCB1/Flp-InTM-293, RPMI-1640 for HepG2 and HepG2/VIN), the fluorescence intensity of the pyrene excimer and monomer was measured using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC)., with excitation/emission wavelengths set at 350/460 nm for the excimer and 350/400 nm for the monomer. Membrane fluidity was calculated based on the excimer-to-monomer fluorescence ratio.
Calcein Retention Assay
As previously described, the P-gp efflux function was assessed by performing an intracellular calcein retention assay.24 Cells were plated in a 96-well black plate and incubated for 24-h. After a 30-minute pre-treatment with the specified concentration of testing compounds, the culture medium was removed and replaced with fresh media (DMEM for Flp-InTM-293 and ABCB1/Flp-InTM-293, RPMI-1640 for HepG2 and HepG2/VIN) containing 1 μM of calcein-AM. The cells’ real-time fluorescence signals of calcein were monitored every three minutes for 30 min using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC)., with an excitation wavelength set at 485 nm and an emission wavelength at 535 nm. The fluorescent pictures of this assay were performed with a 30-minute pre-treatment with verapamil, B23, or A24, followed by a 15-minute treatment with 1 μM of calcein-AM. The cells were then washed with PBS, collected by trypsinization, centrifuged, rewashed, and resuspended in ice-cold PBS. Samples of each treatment were put into the Countess Cell Counting Chamber Slides and read the fluorescence on the Countess 3 FL Automated Cell Counter (Thermo Fisher Scientific Inc). The green, fluorescent picture was generated with auto-focus and saved as images.
Rhodamine 123 Accumulation Assay
The inhibitory effects of testing compounds on P-gp efflux function were confirmed with rhodamine 123 accumulation assay. Briefly, cells were cultured in a 6-well plate overnight. After a 30-minute pre-treatment with the specified concentration of testing compounds, the culture medium was removed and replaced with fresh DMEM media containing 10 μM of rhodamine 123 for 30 min. Then, cells were washed with PBS and harvested into 1.5 mL centrifuge tubes. The cells were resuspended in PBS, and the intracellular rhodamine 123 signals were detected using the NovoCyte Flow Cytometer (Agilent Technologies, Inc).
ABCB1 Gene Expression Assessment
Cells were treated with testing compounds for 72-h. Then, cells were harvested, and the total RNA of cell lines was extracted by PureLink® RNA Mini Kit (Thermo Fisher Scientific Inc) and was converted to cDNA using Applied BiosystemsTM High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific Inc). The concentrations and quality of RNA and cDNA were measured with a Nano-300 Micro-Spectrophotometer. Quantitative real-time PCR was performed with 2x master mix (TaqMan™ Universal Master Mix II, with UNG), 20x probe, and cDNA. TaqMan Assay-On-DemandTM reagents of primers and probes for ABCB1 and GAPDH genes were Hs00184500_m1 and Hs02758991_g1, respectively. The relative ABCB1 expression levels were normalized to the amount of GAPDH in the same cDNA and evaluated by CFX Duet Real-Time PCR System (Bio-Rad Laboratories, Inc).
P-gp Protein Expression Assessment
Cells were treated with testing compounds for 72-h. Then, cells were harvested, and the extracts were collected. Protein concentrations were determined by BCA method. After that, samples were separated with 4–20% or 8% GenScript SurePAGETM Bis-Tris gels (using GenBox Mini Electrophoresis Tank, GenScript) and transferred to PVDF membrane (using eBlot® L1-Fast Wet Protein Transfer System, GenScript). Then, the antibody incubation was performed with eZwestTM Diluent Kit, first primary monoclonal antibody (P-gp_ab170904 or GAPDH_ab181602), followed by a secondary antibody ab97051 (using eZwest Lite, Automated Western Device, GenScript). Finally, the protein signals and chemiluminescent images were detected with ECL reagents using ChemiDoc XRS+ (Bio-Rad Laboratories, Inc). Protein standard was Broad Multi Color Pre-Stained Protein Standard (M00624-250, GenScript).
P-gp Conformation Determination
The influences of testing compounds on P-gp conformation were evaluated using MDR1 Shift assay.25 Cells were harvested and treated with testing compounds for 30 min at 37°C. Then, cells were incubated with IgG2a (background control) or UIC2 antibody (anti-P-gp antibody, detecting conformational extracellular epitope) for 15 min at 37°C. After that, the reaction was stopped and washed by adding iced UIC2 binding buffer, continuing with secondary antibody incubation for 15 min at 4°C. Finally, the cells were washed and resuspended with UIC2 binding buffer, and the signals were detected using the NovoCyte Flow Cytometer (Agilent Technologies, Inc).
Rhodamine 123 and Doxorubicin Efflux Quantification Assay
The molecular interaction mechanisms between testing compounds and P-gp at the transmembrane domain (TMD) were analyzed by efflux kinetics quantitation and Lineweaver–Burk plot.26 Cells were incubated in 96-well plates overnight. After that, the cells were pre-treated with testing compounds at the indicated concentration for 30 min. Then, the medium was replaced with fresh DMEM culture medium containing rhodamine 123 or doxorubicin for 30 min or 3-h, respectively. Subsequently, the cells were washed with warm PBS and then further incubated in PBS to facilitate the rhodamine 123 or doxorubicin efflux for 10 minutes or 2-h, respectively. Finally, the supernatant was transferred to a 96-well black plate, and fluorescence signals were determined using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC). The excitation/emission wavelengths for rhodamine 123 and doxorubicin were 485/535 nm and 485/590 nm, respectively. The nonlinear regression was employed to analyze efflux kinetics by the following equation: V = (Vmax × C) / (Km + C). Here, V represents the efflux rate, Vmax is the maximum efflux rate, Km is the Michaelis-Menten constant, and C is the substrate concentration. The analysis used Scientist version 2.01 (MicroMath Scientific Software, Salt Lake City, UT, USA).
Molecular Docking
Molecular docking at TMD or nucleotide-binding domain (NBD) of P-gp was conducted using AutoDock 4.2.6.26 The three-dimensional P-gp structure was obtained from the RCSB Protein Data Bank (PDB: 7O9W).27 The structure-data files for testing compounds were retrieved from PubChem or ChEBI. All files were converted to PDBQT format using Open Babel version 2.4.1. The docking grid box was set at the drug-binding site (TMD) or ATP-binding site (NBD). The dimensions in the x/y/z axes were set at 60 points, with a spacing of 0.5. The grid centers (x, y, z) for TMD and NBD were (144.585, 143.275, 145) and (110.2, 110.2, 200.2), respectively. Docking parameters were kept at default settings, except for the number of genetic algorithm runs (set to 50) and population size (set to 300). The lowest binding energy and the optimal docking pose were determined by analyzing docking log files and post-docking evaluations.
ATPase Activity Assay
The effects of testing compounds at NBD ATP-binding sites were evaluated using the luminescent Pgp-Glo™ assay (Promega).26 P-gp inside-out plasma membrane vesicles were incubated with ATP detection buffer containing standard ATPase stimulator verapamil with or without various concentrations of testing compounds in a 96-well untreated white plate. Sodium orthovanadate acted as an ATPase inhibitor. The reaction was initiated by adding 5 mM MgATP and incubating at 37°C. Then, the reaction was stopped after 120 min by adding 50 μL ATPase Detection Reagent and incubated for an additional 20 min. Luminescence was measured using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices, LLC).
Statistical Analysis
Data are presented as mean ± standard error of the mean (SEM). Experiments were performed on different days and repeated at least nine times. Differences between the two groups were assessed using a two-tailed Student’s t-test. * Indicated p-value < 0.05 compared to the corresponding control group. All statistical analyses were performed using SigmaPlot v.10.0, Microsoft Excel 2016, or GraphPad Prism v.9.0.
Results
B23 and A24 Reversed MDR in Various Types of Cancer
The non-cytotoxic concentrations of B23 and A24 were obtained by cytotoxic IC50 examination and used in the combinatorial experiments (Table 1). The paired cell lines (drug-sensitive and drug-resistant) were adopted to evaluate the reversal potentials of B23 and A24 on various types of cancer. As shown in Table 2, B23 and A24 exhibited prominent re-sensitizing effects on the cytotoxicity of doxorubicin, especially in the drug-resistant cell lines. For instance, at the concentration of 10 μM, B23 and A24 showed a 7.95- and 8.14- fold reversion when co-treated with doxorubicin on HepG2/VIN cell line, whereas the reversing effects on HepG2 cell line were 1.22- and 1.25- fold, respectively. This resistance overcome phenomenon was also observed in KB/VIN, MCF-7/DOX, and P-gp stably over-expressing ABCB1/Flp-InTM-293 cell lines. Furthermore, when B23 and A24 were co-treated with other chemotherapeutic drugs, paclitaxel and vincristine, on KB/VIN and MCF-7/DOX cell lines, the cytotoxic IC50 were significantly decreased (Figure 1A–D). In other words, B23 and A24 exhibited great potential on MDR reversion in a variety of cancer categories.
 |
Table 1 The Cytotoxic IC50 of B23 and A24 on the Investigated Cell Lines |
 |
Table 2 The Resistance Reversal Effects of B23 and A24 on Doxorubicin Cytotoxicity |
Influences on Cell Membrane: 1. Induced ROS Production and Apoptosis
B23 and A24 demonstrated a 24-h enhanced ROS production at the concentration of 10 μM (Figure 2A). Although drug-sensitive HepG2 cell line was also influenced, the drug-resistance HepG2/VIN showed a more prominent ROS induction effect. Furthermore, this oxidative stress progressed to cell apoptosis induction. After treated HepG2 and HepG2/VIN with B23 and A24 (10 μM) for 48-h, they triggered apoptosis like the positive control paclitaxel (Figure 2B). Again, this effect was more significant in the resistant cell line HepG2/VIN. Therefore, B23 and A24 exhibited an oxidative stress induced apoptosis on the MDR cancer cell membrane.
Influences on Cell Membrane: 2. Cell Retained Membrane Integrity but Enhanced Membrane Fluidity
When B23 and A24 were treated in the HepG2 and HepG2/VIN for 72-h, they did not cause secondary necrosis like the positive control paclitaxel (Figure 2C), indicating the cell membrane integrity was intact. Nevertheless, B23 and A24 showed a significant influence on the cell membrane fluidity. As demonstrated in Figure 3A and B, B23 and A24 (10 μM) enhanced the membrane fluidity in HepG2/VIN and ABCB1/Flp-InTM-293 cell lines. In other words, B23 and A24 showed influences on the cancer MDR cell membrane via modulating the fluidity, but not the integrity.
Influences on Cell Membrane: 3. The Efflux Function of Membrane Transporter Was Inhibited
As the resistance phenomenon was largely caused by the over-expression of efflux pumps on the MDR cancer cell membrane, the efflux inhibitory ability of B23 and A24 was also examined. As results in Figure 3C and D showed, B23 and A24 significantly increased the intracellular calcein retention, indicating the P-gp efflux function was prohibited, like the effects of positive control verapamil, a first-generation P-gp inhibitor. The results of the fluorescent picture also showed that drug-sensitive cell line Flp-InTM-293 retained more calcein inside the cells, regardless of compound treatment. Nevertheless, the drug-resistant cell line ABCB1/Flp-InTM-293 exhibited less calcein intracellular fluorescence. After compound (verapamil, B23, or A24) pre-treatment, the intracellular calcein retention significantly increased in ABCB1/Flp-InTM-293, indicating inhibitory effects of B23 and A24 on the P-gp efflux function (Supplementary Figure 2). Besides, this P-gp inhibitory potential of B23 and A24 was as dose-dependent event (Figure 4A). When the P-gp substrate calcein-AM was replaced with rhodamine 123, similar dose-dependent trend was observed (Figure 4B and C). Therefore, B23 and A24 influenced the MDR cell membrane by inhibiting the pump-out effect of transmembrane transporters.
Underlying Modulatory Mechanisms of B23 and A24 on Human P-gp
The overall effects of B23 and A24 on P-gp efflux transporters were ensuing investigated. B23 and A24 showed a decreasing trend on the ABCB1 gene expression and P-gp protein expression in the MDR HepG2/VIN cell line (Figure 4D–F, Supplementary Figure 3). However, the conformation of P-gp was not influenced by B23 and A24 treatment, not like the positive control vinblastine, which exhibited a rightward shift on the flow cytometry result (Figure 4G).
The molecular interactions of B23 and A24 on the P-gp protein were studied on the TMD drug-binding sites and NBD ATP-binding sites. As shown in Figure 5A, B23 exhibited an allosteric modulation on the transport of rhodamine 123 and doxorubicin, the LB plot demonstrated an uncompetitive and non-competitive inhibition, respectively. The quantitative analyses of maximum efflux rate (Vm) and affinity (Km) supported this result. B23 showed dual influences on both Vm and Km on the rhodamine 123 transport while decreased Vm only and retained Km the constant on the doxorubicin transport (Table 3). Nevertheless, A24 exhibited competitive inhibition on both the rhodamine 123 and doxorubicin transport. The Km was significantly increased with A24 treatment, but Vm remained constant (Figure 5B and Table 3). This differential interactions between B23 and A24 were further supported via molecular docking. As Figure 5C and D showed, the binding of A24 on the P-gp drug-binding site was similar to the binding site of doxorubicin (R site), while B23 bonded to P-gp at the H site, like the Hoechst 33342, another P-gp fluorescent substrate.
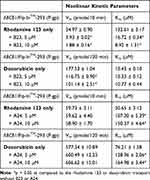 |
Table 3 The Effects of B23 and A24 on the Transport of Rhodamine 123 or Doxorubicin in Human P-gp Over-Expressed Cell Line |
In terms of the ATP-binding site, B23 and A24 alone stimulated the basal P-gp ATPase activity (Figure 6A and B). When co-treated with standard stimulator verapamil, B23 and A24 showed a contrary result. B23 inhibited the verapamil-stimulated ATPase activity, and a different binding site with verapamil on the NBD pocket. On the contrary, A24 further stimulated the verapamil-stimulated ATPase activity, and showed a similar binding behavior as verapamil at the NBD pocket (Figure 6A–C). Above results demonstrated the differential molecular interactions of Alisol Triterpenoids on human P-gp modulation.
Discussion
This study demonstrated that B23 and A24 effectively reverse MDR in various cancer cell lines, specifically showing significant re-sensitization effects to doxorubicin in drug-resistant cells compared to sensitive counterparts. Non-cytotoxic concentrations of these compounds, established through IC50 assessments, were used in combination treatments, leading to notable increases in drug cytotoxicity, particularly in resistant lines such as HepG2/VIN. As the results of cytotoxicity showed that among the cancer cell lines, B23 and A24 exhibited the greatest reversal effects in the HepG2/VIN; therefore, in the investigation of the following mechanisms, the hepatocellular counterpart cell lines (HepG2 and HepG2/VIN), were selected as a research platform. Mechanistically, both B23 and A24 heightened ROS levels, resulting in enhanced apoptosis while maintaining cell membrane integrity without inducing secondary necrosis. These compounds also increased membrane fluidity and inhibited the function of P-gp efflux transporters, confirming their role in combating MDR by reducing drug efflux and enhancing drug accumulation within cells. Their interaction with P-gp suggests intricate allosteric and competitive modulation, thereby offering potential as therapeutic agents in overcoming cancer drug resistance.
B23 and A24 are potential molecules derived from natural products, and the results of the present study are consistent with several previous reports. B23 demonstrated an apoptotic effect in gastric cancer by modulating cell cycle arrest and mitochondrial-related pathways, as well as increasing ROS production. The apoptotic pathway was primarily intrinsic, involving increased caspase-3 and caspase-9 activity; also, the mitogen-activated protein kinases (MAPK) pathway was activated.20 Another study used the SGC7901 cell line as a gastric cancer research platform and reported that B23 induced apoptosis via mitochondria-caspase, PI3K/Akt-dependent pathways, and caspase-3 and caspase-9 activation.28 In terms of hepatocellular carcinoma, the study of Xia et al showed that B23 initiated hepatoma cell death by regulating cell cycle arrest and apoptosis. The FAS/caspase-related apoptotic proteins, caspase-3 and PARP cleavage, and mitochondrial dysfunction played a major role in contributing to apoptosis.29 Li et al showed that the effects of B23 on hepatocellular carcinoma were associated with the up-regulation of Bax, Caspase-3, and Caspase-9, down-regulation of Bcl-2, and the PI3K/Akt signaling pathway.30 Additionally, B23 was found to generate ROS and activate JNK phosphorylation to induce autophagic-dependent apoptosis in colon cancer.31 In terms of lung cancer, Liu et al showed that B23 induces apoptosis in A549 non-small cell lung cancer (NSCLC) cells by upregulating the ratio of Bax/Bcl-2, PI3K/AKT/mTOR inhibition, and arresting the cell cycle in the G1 phase.22 Another study performed on A549 and NCI-H292 cells reported that B23 activated the intrinsic apoptotic pathway, increased the Bax/Bcl-2 ratio, and increased the release of cytochrome c from mitochondria and the translocation of apoptotic inducing factor (AIF) into nuclei.21 Unlike the aforementioned results, the present research found that B23 and A24 exhibited significant effects in MDR cancer cell lines, including enhanced ROS production and apoptosis. This finding is promising, as MDR remains a significant unresolved issue in clinical practice. Furthermore, our study revealed that B23 and A24 regulated membrane fluidity without compromising the integrity of MDR cancer cell membranes. Lipids, particularly phospholipids and cholesterol, play a crucial role in modulating the expression and activity of efflux pumps, which contribute to the development of MDR in cancer cells. The altered lipid composition in MDR cells not only decreases membrane fluidity and enhances drug-binding efficiency but also protects against oxidative stress and promotes resistance, presenting opportunities to reverse MDR through the regulation of this pathway.11 Overall, B23 and A24 demonstrate great potential in cell membrane regulation, including modulation of membrane fluidity and ROS-initiated apoptosis to re-sensitize MDR cancer cells.
Another important factor contributing to the manifestation of MDR in cancer cells is the overexpression of ABC drug efflux pumps on the cell membrane. Previous research has dedicated significant effort to developing P-gp inhibitors from various sources, including small molecule modification, nano-delivery technology, natural products, and siRNA development.8,32,33 In the present study, B23 and A24 were found to potentially modulate P-gp through distinct molecular interacting mechanisms. Using molecular docking to further confirm the results of in vitro cell models, B23 and A24 exhibited binding at the H site and R site on the P-gp TMD drug-binding domain, demonstrating allosteric and competitive inhibition of doxorubicin efflux, respectively. These results provide a novel perspective, as the standard P-gp inhibitor, verapamil, binds at a different M site compared to B23 and A24,34 thereby offering detailed information on the development of P-gp inhibitors at three distinct binding sites. Regarding the NBD site, a previous study revealed that B23 exhibited partially non-competitive inhibition when treated with verapamil.35 Our results corroborate this finding, as B23 demonstrated a distinct binding site at the NBD in the presence of verapamil. Consequently, the verapamil-stimulated P-gp ATPase activity was inhibited in a dose-dependent manner by B23. In contrast, A24 demonstrated a similar binding mode to verapamil and further enhanced the stimulation of ATPase activity. These findings reveal the underlying differences between B23 and A24 and provide in-depth information on their structure-activity relationships.
The novelty of the present study lies in our revelation that B23 and A24 can reverse cancer MDR through three cell membrane-related mechanisms. These independent events represent a promising target for addressing the clinical issue of MDR. By using paired drug-sensitive and drug-resistant cell lines, the differences in the effects of natural compounds on these two cancer conditions can be studied in detail. With this information, it may be possible to develop specific drugs tailored to distinct cancer scenarios. Furthermore, the use of molecular docking strategies enhances our understanding of the interactions between protein targets and compound ligands, which will also aid in future drug design.36 The clinical potential of the present research includes that the distinct molecular mechanisms of B23 and A24 might provide the opportunity to overcome different MDR phenotypes based on the tumor characteristics of patients. Furthermore, the optimistic cytotoxic results of the combination of B23 and A24 with various chemotherapeutic drugs give the potential for these alisol triterpenoids to be applied to the current diverse chemotherapeutic regimens. However, as MDR cancer is an ever-changing disease and includes diverse stages and phenotypes, the application and translation of these findings to clinical practice will face obstacles and challenges, such as the interference of therapeutic efficacy because of drug-drug interactions, the more complicated drug regimens and stable formulation design are also important factors to consider for successful clinical translation. Besides, the underlying genetic differences among patients will influence the treatment outcome as well. Therefore, more detailed mechanistic investigations such as apoptosis pathway analysis, cell cycle array, the interplay between distinct pathways, or the impacts of genetic polymorphisms on the effects are warranted. Additionally, preclinical in vivo models are also an important platform that can reveal the efficacy and safety of these alisol triterpenoids. Hence, future studies of B23 and A24 or their structural derivatives should include a further in-depth mechanism investigation and in vivo efficacy confirmation.
Conclusion
In conclusion, this research highlights the promising effects of two alisol triterpenoids in reversing cancer MDR. Compared to Alisol A and Alisol B, these two acetate derivatives (B23 and A24) show significant potential for future applications. Nevertheless, as this study was preclinical research focused on mechanisms exploration, more efficacy confirmation studies, such as the in vivo xenograft model, are warranted to strengthen the potential further. Besides, based on the molecular mechanistic results of B23 and A24, the modifications of chemical structures on these alisol triterpenoids might provide another promising application of this research.
Abbreviations
ABC, ATP-binding cassette; A24, Alisol A 24-acetate; BCRC, Bioresource Collection and Research Center; B23, Alisol B 23-acetate; DMSO, dimethyl sulfoxide; DMEM, Dulbecco’s Modified Eagle Media; FBS, fetal bovine serum; MBCD, methyl-β-cyclodextrin; MDR, Multi-drug resistance; NBD, nucleotide-binding domain; PBS, phosphate-buffered saline; PDB, Protein Data Bank; P-gp, P-glycoprotein; ROS, reactive oxygen species; RPMI-1640, Roswell Park Memorial Institute-1640; SRB, sulforhodamine B; TMD, transmembrane domain.
Acknowledgments
We gratefully acknowledge Dr. Chin-Chuan Hung (China Medical University, Taiwan) for the assistance with the construction of ABCB1/Flp-InTM-293 cell line. We extend our thanks to Dr. Kuo-Hsiung Lee (University of North Carolina, Chapel Hill) for providing the multidrug-resistant human cervical cancer cell line KB/VIN. We also acknowledge the Basic Medical Core Laboratory, I-Shou University College of Medicine for their technical support.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the China Medical University [CMU113-MF-53]; and the National Science and Technology Council, Taiwan [NSTC 112-2320-B-214-003-MY3 and NSTC 112-2320-B-039-063-MY3]. The funders had no role in study design, data collection, and analysis; decision to publish; or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bukowski K, Kciuk M, Kontek R. Mechanisms of multidrug resistance in cancer chemotherapy. Int J Mol Sci. 2020;21(9):3233. doi:10.3390/ijms21093233
2. Vaidya FU, Sufiyan Chhipa A, Mishra V, et al. Molecular and cellular paradigms of multidrug resistance in cancer. Cancer Rep. 2022;5(12):e1291. doi:10.1002/cnr2.1291
3. Chen T, Xiao Z, Liu X, et al. Natural products for combating multidrug resistance in cancer. Pharmacol Res. 2024;202:107099. doi:10.1016/j.phrs.2024.107099
4. Bhusare N, Gade A, Kumar MS. Using nanotechnology to progress the utilization of marine natural products in combating multidrug resistance in cancer: a prospective strategy. J Biochem Mol Toxicol. 2024;38(6):e23732. doi:10.1002/jbt.23732
5. Yadav P, Ambudkar SV, Rajendra Prasad N. Emerging nanotechnology-based therapeutics to combat multidrug-resistant cancer. J Nanobiotechnology. 2022;20(1):423. doi:10.1186/s12951-022-01626-z
6. Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi:10.1038/nrd1984
7. Pluchino KM, Hall MD, Goldsborough AS, Callaghan R, Gottesman MM. Collateral sensitivity as a strategy against cancer multidrug resistance. Drug Resist Updat. 2012;15(1–2):98–105. doi:10.1016/j.drup.2012.03.002
8. Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18(7):452–464. doi:10.1038/s41568-018-0005-8
9. Cui Q, Wang JQ, Assaraf YG, et al. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist Updat. 2018;41:1–25. doi:10.1016/j.drup.2018.11.001
10. Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36(1):52. doi:10.1186/s40880-017-0219-2
11. Kopecka J, Trouillas P, Gašparović A, Gazzano E, Assaraf YG, Riganti C. Phospholipids and cholesterol: inducers of cancer multidrug resistance and therapeutic targets. Drug Resist Updat. 2020;49:100670. doi:10.1016/j.drup.2019.100670
12. Kim S, Lee M, Dhanasekaran DN, Song YS. Activation of LXRɑ/β by cholesterol in malignant ascites promotes chemoresistance in ovarian cancer. BMC Cancer. 2018;18(1):1232. doi:10.1186/s12885-018-5152-5
13. Subramanian N, Schumann-Gillett A, Mark AE, O’Mara ML. Understanding the accumulation of P-glycoprotein substrates within cells: the effect of cholesterol on membrane partitioning. Biochim Biophys Acta. 2016;1858(4):776–782. doi:10.1016/j.bbamem.2015.12.025
14. Alam A, Kowal J, Broude E, Roninson I, Locher KP. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science. 2019;363(6428):753–756. doi:10.1126/science.aav7102
15. PubChem. PubChem Compound Summary for CID 14036811, alisol B 23-acetate. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/alisol-B-23-acetate.
16. PubChem. PubChem compound summary for CID 76336194, alisol A 24-acetate. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/76336194.
17. Bailly C. Pharmacological properties and molecular targets of alisol triterpenoids from alismatis rhizoma. Biomedicines. 2022;10(8):1945. doi:10.3390/biomedicines10081945
18. Du Q, Liang R, Wu M, et al. Alisol B 23-acetate broadly inhibits coronavirus through blocking virus entry and suppresses proinflammatory T cells responses for the treatment of COVID-19. J Adv Res. 2024;62:273–290. doi:10.1016/j.jare.2023.10.002
19. Han LL, Zhang X, Zhang H, et al. Alisol B 23-acetate promotes white adipose tissue browning to mitigate high-fat diet-induced obesity by regulating mTOR-SREBP1 signaling. J Integr Med. 2024;22(1):83–92. doi:10.1016/j.joim.2024.01.003
20. Kwon MJ, Kim JN, Lee MJ, Kim WK, Nam JH, Kim BJ. Apoptotic effects of alisol B 23‑acetate on gastric cancer cells. Mol Med Rep. 2021;23(4). doi:10.3892/mmr.2021.11887
21. Wang J, Li H, Wang X, Shen T, Wang S, Ren D. Alisol B-23-acetate, a tetracyclic triterpenoid isolated from Alisma orientale, induces apoptosis in human lung cancer cells via the mitochondrial pathway. Biochem Biophys Res Commun. 2018;505(4):1015–1021. doi:10.1016/j.bbrc.2018.10.022
22. Liu Y, Xia XC, Meng LY, Wang Y, Li YM. Alisol B 23‑acetate inhibits the viability and induces apoptosis of non‑small cell lung cancer cells via PI3K/AKT/mTOR signal pathway. Mol Med Rep. 2019;20(2):1187–1195. doi:10.3892/mmr.2019.10355
23. Teng YN, Sheu MJ, Hsieh YW, Wang RY, Chiang YC, Hung CC. β-carotene reverses multidrug resistant cancer cells by selectively modulating human P-glycoprotein function. Phytomedicine. 2016;23(3):316–323. doi:10.1016/j.phymed.2016.01.008
24. Chen JY, Sung CJ, Chen SC, Hsiang YP, Hsu YC, Teng YN. Redefine the role of d-α-Tocopheryl polyethylene glycol 1000 succinate on P-glycoprotein, multidrug resistance protein 1, and breast cancer resistance protein mediated cancer multidrug resistance. Eur J Pharm Sci. 2023;190:106579. doi:10.1016/j.ejps.2023.106579
25. Teng YN, Hung CC, Kao PH, Chang YT, Lan YH. Reversal of multidrug resistance by Fissistigma latifolium-derived chalconoid 2-hydroxy-4,5,6-trimethoxydihydrochalcone in cancer cell lines overexpressing human P-glycoprotein. Biomed Pharmacother. 2022;156:113832. doi:10.1016/j.biopha.2022.113832
26. Ho YC, Chiu WC, Chen JY, Huang YH, Teng YN. Reversal potentials of Tween 20 in ABC transporter-mediated multidrug-resistant cancer and treatment-resistant depression through interacting with both drug-binding and ATP-binding areas on MDR proteins. J Drug Target. 2024;1–14. doi:10.1080/1061186x.2024.2429006
27. Urgaonkar S, Nosol K, Said AM, et al. Discovery and characterization of potent dual P-glycoprotein and CYP3A4 inhibitors: design, synthesis, Cryo-EM analysis, and biological evaluations. J Med Chem. 2022;65(1):191–216. doi:10.1021/acs.jmedchem.1c01272
28. Xu YH, Zhao LJ, Li Y. Alisol B acetate induces apoptosis of SGC7901 cells via mitochondrial and phosphatidylinositol 3-kinases/Akt signaling pathways. World J Gastroenterol. 2009;15(23):2870–2877. doi:10.3748/wjg.15.2870
29. Xia J, Luo Q, Huang S, et al. Alisol B 23-acetate-induced HepG2 hepatoma cell death through mTOR signaling-initiated G(1) cell cycle arrest and apoptosis: a quantitative proteomic study. Chin J Cancer Res. 2019;31(2):375–388. doi:10.21147/j.issn.1000-9604.2019.02.12
30. Li L, Cheng J, Zhu D, et al. The effects of Alisol B 23-acetate in hepatocellular carcinoma via inducing cell apoptosis and inhibiting cell migration and invasion. Gen Physiol Biophys. 2020;39(3):219–228. doi:10.4149/gpb_2020005
31. Zhao Y, Li ETS, Wang M. Alisol B 23-acetate induces autophagic-dependent apoptosis in human colon cancer cells via ROS generation and JNK activation. Oncotarget. 2017;8(41):70239–70249. doi:10.18632/oncotarget.19605
32. Engle K, Kumar G. Cancer multidrug-resistance reversal by ABCB1 inhibition: a recent update. Eur J Med Chem. 2022;239:114542. doi:10.1016/j.ejmech.2022.114542
33. Hour MJ, Tsai FJ, Lai IL, et al. Efficacy of HMJ-38, a new quinazolinone analogue, against the gemcitabine-resistant MIA-PaCa-2 pancreatic cancer cells. Biomedicine. 2023;13(4):20–31. doi:10.37796/2211-8039.1423
34. Ferreira RJ, Ferreira MJ, Dos Santos DJ. Molecular docking characterizes substrate-binding sites and efflux modulation mechanisms within P-glycoprotein. J Chem Inf Model. 2013;53(7):1747–1760. doi:10.1021/ci400195v
35. Wang C, Zhang JX, Shen XL, Wan CK, Tse AK, Fong WF. Reversal of P-glycoprotein-mediated multidrug resistance by Alisol B 23-acetate. Biochem Pharmacol. 2004;68(5):843–855. doi:10.1016/j.bcp.2004.05.021
36. Yalcin-Ozkat G. Molecular modeling strategies of cancer multidrug resistance. Drug Resist Updat. 2021;59:100789. doi:10.1016/j.drup.2021.100789
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







