Back to Journals » Infection and Drug Resistance » Volume 18
Anti-Virulence Efficacy of Chinese Dragon’s Blood Against Pseudomonas aeruginosa Isolated from Wound Infections
Authors Shen Y, Gu Z, Li Y, Zhou H, Zhu S, Sheng L, Qiang X, Zheng X
Received 5 April 2025
Accepted for publication 17 June 2025
Published 27 June 2025 Volume 2025:18 Pages 3157—3167
DOI https://doi.org/10.2147/IDR.S532537
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yingjie Shen,1 Zengyue Gu,2 Yujie Li,2 Hanmeng Zhou,2 Shengzhe Zhu,2 Lingtao Sheng,2 Xinhua Qiang,3 Xiangkuo Zheng2
1School of Life Sciences, Huzhou University, Huzhou, 313000, People’s Republic of China; 2School of Medicine, Huzhou University; Key Laboratory of Vector Biology and Pathogen Control of Zhejiang Province, Huzhou, 313000, People’s Republic of China; 3Department of Clinical Laboratory, The First Affiliated Hospital of Huzhou University, Huzhou, 313000, People’s Republic of China
Correspondence: Xiangkuo Zheng, School of Medicine, Huzhou University; Key Laboratory of Vector Biology and Pathogen Control of Zhejiang Province, 759 East Second Ring Road, Wuxing District, Huzhou, 313000, People’s Republic of China, Tel +86-0572-2321531, Email [email protected] Xinhua Qiang, Department of Clinical Laboratory, The First Affiliated Hospital of Huzhou University, 158 Back Square Road, Wuxing District, Huzhou, 313000, People’s Republic of China, Tel +86-0572-2039408, Email [email protected]
Background: Pseudomonas aeruginosa (P. aeruginosa) wound infections are an emerging global health threat. Empiric therapy of infected wounds with Chinese dragon’s blood (CDB) is one of the most precious traditional Chinese medicines used in clinical settings. We investigated the anti-virulence efficacy of CDB against P. aeruginosa isolated from wound infections.
Methods: We collected six P. aeruginosa clinical isolates obtained from wound specimens. Antimicrobial susceptibility profiles were determined using the agar dilution method. Biofilm formation and eradication assays, quantitative real-time PCR (RT-qPCR), and bacterial motility assays were performed to evaluate the efficacy of CDB on biofilm formation, mature biofilm eradication, and motility ability of P. aeruginosa isolates.
Results: Minimal inhibitory concentration (MIC) values of CDB against P. aeruginosa isolates were ≥ 1024 μg/mL. The differences in biofilm formation ability between the CDB-containing LB broth and LB broth groups were statistically significant (P < 0.05). The results of mature biofilm-eradicating assays indicated that CDB had excellent efficacy on eradicating the biofilm formed by all experimental strains (P < 0.05). The mRNA relative expression of lasR, pslA, pelA, algD, and algU genes in P. aeruginosa strains was significantly downregulated after exposure to CDB at a concentration of 128 μg/mL (P < 0.05). CDB could inhibit the motility ability of P. aeruginosa isolates through swimming, swarming, and twitching motilities.
Conclusion: CDB exerts a positive anti-virulence efficacy on P. aeruginosa. CDB significantly reduced the biofilm formation by downregulating the mRNA relative expression of the biofilm-associated genes lasR, pslA, pelA, algD, and algU. In addition, CDB efficiently inhibited the motility ability of P. aeruginosa isolates by swimming, swarming, and twitching motilities in a concentration-dependent manner. Therefore, these findings position CDB as an alternative for P. aeruginosa wound infections management in clinical settings.
Keywords: Chinese dragon’s blood, wound infections, Pseudomonas aeruginosa, anti-virulence efficacy
Introduction
Wound infections are emerging as a major global health challenge, and studies have demonstrated that approximately 52–70% of trauma patients succumb to wound infections.1,2 While acute wounds generally exhibit self-healing capacity without requiring substantial clinical intervention, chronic wounds frequently fail to undergo spontaneous repair, which necessitates therapeutic management.3 Clinically, the treatment of chronically infected wounds poses significant difficulties owing to microbial colonization of the wound beds. This pathological colonization disrupts the physiological healing cascade, leading to impaired tissue regeneration and, ultimately, non-healing wound states.4,5 Notably, Pseudomonas aeruginosa (P. aeruginosa) and Staphylococcus aureus (S. aureus) are the most common pathogenic microorganisms isolated from infected wounds, and their ability to express virulence determinants and establish biofilm structures may adversely affect the wound healing process.6 Biofilms constitute structured microbial communities encased in a self-synthesized matrix composed of extracellular polymeric substances (EPS) derived from microbial and host sources.7,8 These biofilm-forming bacteria demonstrate distinct metabolic profiles embedded within this protective macromolecular architecture. Biofilm facilitates environmental adaptation through multifaceted mechanisms, thereby conferring intrinsic tolerance to conventional antimicrobial interventions via physical exclusion and altered microbial physiology.9,10 Some studies have revealed that quorum sensing system contributes to biofilm formation in P. aeruginosa wound isolates. Therefore, biofilm formation by P. aeruginosa critically contributes to the pathogenesis of non-healing wound infections. In addition, the incorporation of antibiotics into therapeutic regimens has proven to be effective in eradicating diverse pathogens. However, the widespread and often indiscriminate use of broad-spectrum antimicrobial agents has driven the progressive emergence of antimicrobial resistance (AMR).11,12 This escalating prevalence of AMR has precipitated formidable challenges in clinical infection management, compounding the therapeutic complexities of combating persistent infections.4
Within the framework of integrated medical approaches, traditional Chinese medicine (TCM) formulations have emerged as viable adjunctive treatment modalities for wound infection management, with Chinese dragon’s blood (CDB) being a particularly notable botanical agent.13,14 Pharmacopeial specifications (National Drug Standard WS3-082 (Z-016)-99(Z)) define CDB as a crimson-hued resinous exudate derived from the ligneous tissues of Dracaena cochinchinensis (Liliaceae family).15 There are some studies identify proanthocyanidins as the predominant phytochemical group in terms of relative quantity. Additionally, other significant phytochemical groups include alkaloids, diterpenoids, phytosteroids, saponins, phenolics, and polyphenolics.16,17 The pharmacological studies have demonstrated that this phytotherapeutic substance contains multiple bioactive compounds with anti-inflammatory, antimicrobial, antifungal, and cytotoxic properties, rendering it applicable to diverse pathological conditions.18 Results from an in vivo study on burn wounds showed that scaffolds containing 20% CDB exhibited excellent wound healing ability with 80.3% wound closure after 21 days,19 and the novel research illustrated the potential protective mechanism of CDB on intestinal inflammatory-related diseases and might be useful for further clinical application of CDB.20 Particularly relevant to wound care, CDB has been employed therapeutically in clinical practice to address suppurative lesions, diabetic foot complications, and traumatic tissue damage, among other conditions.14 The present investigation was designed to systematically evaluate the capacity of CDB to attenuate virulence determinants in P. aeruginosa isolates, while concurrently establishing preclinical evidence to optimize its clinical application in wound infection management.
Materials and Methods
Bacterial Isolates and Growth Conditions
Six P. aeruginosa clinical isolates were obtained from wound specimens of patients receiving treatment at the First Affiliated Hospital of Huzhou University (Zhejiang Province, China) in 2024. Bacterial identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the VITEK MS platform (bioMérieux, Lyon, France). The microbial cultures were cryopreserved at −80°C and subsequently subcultured on blood agar medium under standardized conditions (37°C for 18–24 h) prior to experimental analysis.
Determination of Antimicrobial Resistance Profiles
Antimicrobial susceptibility profiles were determined using the agar dilution technique in compliance with the latest Clinical and Laboratory Standards Institute guidelines (CLSI 2025).21 The experimental protocol involved resuspending overnight cultures in sterile saline solution (0.85% NaCl), with subsequent turbidity standardization to match the 0.5 McFarland reference (approximately 1.5 × 10⁸ CFU/mL). Ten-fold dilutions of the bacterial suspensions were then inoculated onto Mueller–Hinton agar plates containing antimicrobial agents. Following 16–18 h of aerobic incubation at 37°C, the inhibition zones were systematically evaluated. The investigational compound CDB (Lot Z25O9B84376, Yuanye Biotechnology, Shanghai, China) was dissolved in dimethyl sulfoxide to create serial dilutions spanning 1–512 μg/mL. Strain validation was performed using P. aeruginosa ATCC 27853 as the reference organism. Minimal inhibitory concentration (MIC) determinants were conducted through independent experiments in triplicate to ensure methodological reproducibility.
Biofilm Formation and Eradication Assays
Biofilm formation and eradication assays were performed according to the methods described by Bukhari, with some modifications.22,23 This study was carried out using seven P. aeruginosa strains (PA676, PA679, PA731, PA732, PA764, PA769, and PAO1). A single bacterial colony isolated from the blood agar plate was inoculated into 3 mL fresh LB medium and incubated overnight at 37°C with continuous agitation (180 rpm). The bacterial suspension was standardized to 0.5 McFarland units using sterile normal saline, followed by a 1:100 dilution in fresh LB broth. Aliquots (100 μL) were transferred to 96-well microplates containing different concentrations of CDB (0, 32, 64, 128, or 256 μg/mL). Following 24-hour incubation at 37°C, non-adherent cells were removed through two consecutive washes with 200 μL of phosphate-buffered saline (PBS, 1×, Sigma-Aldrich). The microplates were air-dried and inverted at ambient temperature. Subsequent staining was performed using 150 μL of 1% (w/v) crystal violet (CV; Solarbio Biotechnology, Beijing, China; Lot number: 20240846) for 15 min at room temperature. Excess stain was discarded and the wells were rinsed three times with PBS. Bound CV was eluted with 150 μL of destaining solution (95% ethanol: 5% glacial acetic acid, v/v), and biofilm quantification was achieved by measuring the optical density at 595 nm using a Multiskan FC microplate reader. All experimental conditions were tested in triplicate with three independent biological replicates. Biofilm formation and mature biofilm eradication experiments differed according to the timing of the drug addition. The drug was added before the biofilm formed in the biofilm formation experiment and after the biofilm had matured in the eradication experiments.
Quantitative Real-Time PCR (RT-qPCR)
All P. aeruginosa strains (PA676, PA679, PA731, PA732, PA764, PA769, and PAO1) were either exposed to CDB at a concentration of 128 μg/mL or left untreated. Total RNA was subsequently isolated from bacterial cultures using the Bacterial RNA Miniprep Kit (Biomiga, Shanghai, China). First-strand cDNA synthesis was performed with 1000 ng of RNA template per reaction using the RevertAid First Strand cDNA Synthesis System (Thermo Scientific, Waltham, MA, USA), following the manufacturer’s recommended protocols. Gene mRNA relative expression analysis was conducted via quantitative real-time PCR (RT-qPCR) targeting lasR, pslA, pelA, algD, and algU transcripts. The rpsL housekeeping gene served as an endogenous control for normalization. Gene-specific primers (listed in Table 1) were employed for amplification using SYBR Green chemistry with TB Green Premix Ex Taq II (Tli RNaseH Plus; Takara, Japan) on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, USA).24 Thermal cycling conditions consisted of initial denaturation at 95°C for 30s, followed by 40 cycles at 95°C for 5s and 60°C for 34s, with melt curve analysis for reaction specificity verification.
 |
Table 1 Primers Used for RT-qPCR in This Study |
Bacterial Motility Assays
Microbial motility assays were conducted using standardized agar-based platforms, fabricated according to established protocols.25,26 For swarming and swimming behavior assays, 2 μL aliquots of overnight bacterial cultures were centrally inoculated onto low-viscosity (0.3% agar) and high-viscosity (0.5% agar with 0.5% glucose) substrates, respectively. Twitching motility assay involved vertical inoculation of bacterial suspensions at the LB plate (1% agar) interface through stab inoculation. Following 24-hour aerobic incubation at 37°C, radial migration distances from the inoculation site were systematically quantified using digital caliper measurements. For twitching motility assay, removed the agar and subsequent staining was performed using 1 mL of 1% (w/v) crystal violet for 15 min at room temperature. Then, excess stain was discarded and the wells were rinsed three times with PBS. All experiments were performed in triplicate to ensure methodological reproducibility.
Statistical Analysis
All experimental procedures were performed in triplicate with independent biological repetitions. Quantitative data are presented as mean values ± standard deviation (SD), and intergroup comparisons were analyzed using the one-way ANOVA. Statistical computations were performed using GraphPad Prism, version 9.02 (GraphPad Software Inc., San Diego, CA, USA), with two-tailed analyses employed throughout the study. Statistical significance was set at P < 0.05.
Results
Determination of MICs of CDB and Antimicrobial Agents Against P. aeruginosa Isolates
Antimicrobial susceptibility testing using the agar dilution method revealed that the MIC values of CDB against P. aeruginosa isolates were ≥1024 µg/mL (Table 2).
 |
Table 2 Minimum Inhibitory Concentrations (MICs) of CDB and Clinical Routine Antimicrobial Agents Against Six P. aeruginosa Isolates |
Moreover, three P. aeruginosa isolates (PA676, PA764, and PA767) were resistant to routine clinical antimicrobial agents, including piperacillin/tazobactam (TZP), ceftazidime (CAZ), cefepime (FEP), imipenem (IPM), meropenem (MEM), ciprofloxacin (CIP), and levofloxacin (LVX) (Table 2).
Efficacy of CDB on Biofilm Formation and Mature Biofilm Eradication of P. aeruginosa Isolates
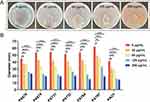
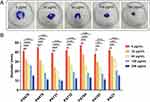
The effects of CDB on biofilm formation and mature biofilm eradication of P. aeruginosa isolates were investigated using crystal violet staining. Our results illustrated the biofilm structures of P. aeruginosa isolates in LB broth in the presence and absence of CDB and noted that the differences in biofilm formation ability between the drug-containing LB broth and LB broth groups were statistically significant (P < 0.05; Figure 1). Figure 2 shows the results of the mature biofilm-eradicating assays, which indicated that CDB had excellent efficacy on eradicating the biofilm formed by all experimental strains (P < 0.05).
 |
Figure 1 Effects of different concentrations of CDB on the biofilm formation ability of P. aeruginosa strains. N.S., P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. |
 |
Figure 2 Effects of different concentrations of CDB on the mature biofilm eradication of P. aeruginosa strains. N.S., P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. |
Expression of Genes Involved in Biofilm Formation of P. aeruginosa Strains
RT-qPCR results revealed that mRNA relative expression levels of lasR, pslA, pelA, algD, and algU genes in P. aeruginosa strains were significantly downregulated after exposure to CDB at a concentration of 128 μg/mL (P < 0.05; Figure 3A–G). These results indicated that CDB could decrease the capacity of biofilm formation of P. aeruginosa strains through the inhibition of the mRNA relative expression of lasR, pslA, pelA, algD, and algU genes.
Efficacy of CDB on the Motility of P. aeruginosa Isolates
To study the effect of CDB on the motility of P. aeruginosa isolates, motility assays were carried out. By measuring the diffusion diameter of P. aeruginosa on the surface of LB plate, the effects of CDB on the mobility of P. aeruginosa was analyzed. As shown in Figures 4–6, we concluded that CDB could inhibit the motility ability of P. aeruginosa isolates through swimming, swarming, and twitching motilities in a concentration-dependent manner.
Discussion
Clinically, infected wounds not only impair regenerative cascades but also precipitate life-threatening complications through systemic dissemination.4 Empirical data have identified P. aeruginosa and S. aureus, particularly multidrug-resistant strains, as the predominant etiological agents in refractory wound infections.1 Emerging evidence highlights CDB as a promising phytotherapeutic agent, demonstrating quorum-sensing attenuation and biofilm disruption capacities.14,18 However, CDB is currently used empirically in clinical practice for wound infection treatment, and there is no standardized dosage used in clinical practice in China. Our investigation delineates the mechanistic basis of the anti-pathogenic efficacy of CDB against P. aeruginosa clinical isolates obtained from wound specimens, providing preclinical validation for integrating CDB-derived compounds into combinatorial antimicrobial strategies targeting wound infection recalcitrance.
Our previous studies have documented how CDB exerts positive antibacterial efficacy against S. aureus and could also reduce biofilm formation and retard the virulence factor alpha-hemolysin of S. aureus by downregulating the expression levels of saeR, saeS, and hla genes.27 As a clinically comparable pathogen causing wound infections, P. aeruginosa also deserves further parallel investigation. Based on the results of the antimicrobial susceptibility testing, we found that CDB exhibits no significant antibacterial activity against P. aeruginosa at a concentration of 1024 μg/mL. Fortunately, CDB showed positive anti-virulence efficacy against P. aeruginosa isolated from wound infections. Sub-inhibitory concentrations of CDB could reduce the biofilm formation ability of P. aeruginosa, and CDB showed excellent efficacy on eradicating the biofilm formed by all experimental strains. These findings were further supported by the RT-qPCR results. We further found that CDB could downregulate the mRNA relative expression of lasR, pslA, pelA, algD, and algU genes and inhibit the motility of P. aeruginosa isolates.
Previous investigations have established that, within the microenvironment of chronically infected wounds, bacterial biofilm formation plays a pivotal role in microbial pathogenesis. Biofilms enhance microbial adhesion to wound beds, evade host immune defenses, and promote environmental adaptability.28–30 Consequently, the development of novel therapeutic compounds targeting biofilm formation has emerged as a critical imperative in wound management research. Experimental protocols for biofilm formation and mature biofilm eradication are distinguished by the temporal administration of therapeutic agents.22,23 In our study, biofilm formation and eradication assays revealed that sub-inhibitory concentrations of CDB could effectively reduce the biofilm formation ability and eradicating the mature biofilm formed by all experimental P. aeruginosa strains. P. aeruginosa biofilm formation is a critical determinant of wound healing trajectory and patient mortality in wound infections. Complementing this pathogenic mechanism, Ho et al demonstrated that CDB stimulates angiogenesis, while enhancing keratinocyte proliferation (1.8-fold) and fibroblast migration rates (42% increase vs control).31 We hypothesized that the dual modulatory effects of CDB, which simultaneously counteract biofilm-mediated healing impairment and activate tissue regenerative pathways, collectively contribute to its therapeutic efficacy in recalcitrant wound management.
Moreover, RT-qPCR was performed to detect the expression of genes involved in biofilm formation of P. aeruginosa strains. Interestingly, we found that the mRNA relative expression levels of lasR gene in P. aeruginosa strains were significantly downregulated after exposure to CDB at a concentration of 128 μg/mL. The quorum sensing (QS) system enables bacterial consortia to synchronize virulence factor secretion, biofilm maturation, metabolic reprogramming, and antimicrobial resistance phenotypes.32 Previous studies have shown that the lasRI gene in the las system is upstream of other subsystems and is the main control factor of the quorum sensing system network, and the expression of other subsystem genes can be upregulated after activation of the lasRI gene.33,34 Our findings reveal that CDB may be a potential quorum sensing inhibitor, which warrants further investigation.
P. aeruginosa exhibits niche-specific motility, which is a critical determinant of its pathogenic potential.35,36 The locomotive behavior of bacteria is intrinsically linked to virulence, demonstrating phenotypic plasticity dependent on physicochemical cues: (i) flagellum-driven swimming in aqueous matrices, (ii) surfactant-mediated swarming across viscous interfaces, and (iii) type IV pilus-dependent twitching on solid substrates.37 Mechanistically, the rotary flagellar apparatus propels aqueous-phase navigation, whereas surface-associated motility requires the coordinated action of flagella and retractile type IV pili for biofilm expansion and tissue colonization.38 Bacterial motility assays revealed that CDB could inhibit the motility ability of P. aeruginosa isolates through swimming, swarming, and twitching motilities in a concentration-dependent manner.
Meanwhile, we found that CDB could downregulate the mRNA relative expression of lasR, pslA, pelA, algD, and algU genes at a concentration of 128 μg/mL CDB. While higher concentrations (eg, 512 µg/mL) could theoretically be tested, our goal was to prioritize clinically translatable doses. Due to the formulation barriers and metabolic clearance, bioavailable concentrations in tissues are substantially lower, thus results from in vitro testing may not fully replicate the pharmacological effects observed in vivo. Building on these findings, our subsequent investigation will employ a murine cutaneous wound infection model to systematically evaluate the in vivo anti-virulence efficacy of CDB against P. aeruginosa.39 Crucially, elucidating the anti-virulence mechanisms of CDB, particularly its capacity to disrupt P. aeruginosa biofilm ontogeny, suppress virulence determinants (eg, elastase and pyocyanin), and interfere with quorum-sensing circuitry, constitutes the pivotal first step in deconstructing its multimodal antimicrobial action. This mechanistic blueprint will inform the development of phytotherapeutic strategies targeting pathogenicity rather than bacterial lethality.
Conclusion
In conclusion, CDB, which is one of the most precious traditional Chinese medicines, exerts the positive anti-virulence efficacy on P. aeruginosa. CDB could significantly reduce the biofilm formation by downregulating the mRNA relative expression of biofilm-associated genes lasR, pslA, pelA, algD, and algU. In addition, CDB could efficiently inhibit the motility ability of P. aeruginosa isolates through swimming, swarming, and twitching motilities in a concentration-dependent manner. Our findings elucidate novel therapeutic paradigms for P. aeruginosa wound infection management through mechanism-guided clinical deployment of CDB. The collective evidence suggests that CDB is a promising phytotherapeutic candidate capable of suppressing P. aeruginosa virulence determinants, thereby addressing the critical limitations of conventional antimicrobial approaches for P. aeruginosa wound infection management in clinical settings.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article, and further inquiries can be directed to the corresponding authors.
Ethical Statement
This study complies with the Declaration of Helsinki. All study protocols were approved by the Ethics Committee of the School of Medicine, Huzhou University. There are no studies with humans or animals performed by any of the authors in this article. Informed consent was waived because this study with observational nature mainly focused on bacteria and did no interventions to patients.
Acknowledgments
The authors acknowledge the financial support of the National Natural Science Foundation of China (no. 82304004), Huzhou Municipal Natural Science Foundation (no. 2024YZ47), and Ningbo Key Laboratory of Skin Science, Ningbo College of Health Sciences (no. PFKX2024002).
Author Contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas, took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Xiangkuo Zheng and Xinhua Qiang are joint corresponding authors.
Funding
This work was supported by the National Natural Science Foundation of China (no. 82304004), Huzhou Municipal Natural Science Foundation (no. 2024YZ47), and Ningbo Key Laboratory of Skin Science, Ningbo College of Health Sciences (no. PFKX2024002).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Maitz J, Merlino J, Rizzo S, et al. Burn wound infections microbiome and novel approaches using therapeutic microorganisms in burn wound infection control. Adv Drug Deliv Rev. 2023;196:114769. doi:10.1016/j.addr.2023.114769
2. Cefalu JE, Barrier KM, Davis AH. Wound infections in critical care. Crit Care Nurs Clin North Am. 2017;29(1):81–96. doi:10.1016/j.cnc.2016.09.009
3. Hurlow J, Bowler PG. Acute and chronic wound infections: microbiological, immunological, clinical and therapeutic distinctions. J Wound Care. 2022;31(5):436–445. doi:10.12968/jowc.2022.31.5.436
4. Hurlow J, Wolcott RD, Bowler PG. Clinical management of chronic wound infections: the battle against biofilm. Wound Repair Regen. 2025;33(1):e13241. doi:10.1111/wrr.13241
5. Falcone M, De Angelis B, Pea F, et al. Challenges in the management of chronic wound infections. J Glob Antimicrob Resist. 2021;26:140–147. doi:10.1016/j.jgar.2021.05.010
6. Serra R, Grande R, Butrico L, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. 2015;13(5):605–613. doi:10.1586/14787210.2015.1023291
7. Karygianni L, Ren Z, Koo H, Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28(8):668–681. doi:10.1016/j.tim.2020.03.016
8. Kim SK, Lee JH. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol. 2016;54(2):71–85. doi:10.1007/s12275-016-5528-7
9. Diban F, Di Lodovico S, Di Fermo P, et al. Biofilms in chronic wound infections: innovative antimicrobial approaches using the in vitro Lubbock chronic wound biofilm model. Int J Mol Sci. 2023;24(2):1004. doi:10.3390/ijms24021004
10. Wang C, Su Y, Shahriar SMS, et al. Emerging strategies for treating medical device and wound-associated biofilm infections. Microb Biotechnol. 2024;17(10):e70035. doi:10.1111/1751-7915.70035
11. Qin S, Xiao W, Zhou C, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7(1):199. doi:10.1038/s41392-022-01056-1
12. Cosentino F, Viale P, Giannella M. MDR/XDR/PDR or DTR? Which definition best fits the resistance profile of Pseudomonas aeruginosa? Curr Opin Infect Dis. 2023;36(6):564–571. doi:10.1097/QCO.0000000000000966
13. Zhao Y, Li H, Wei S, et al. Antimicrobial effects of chemical compounds isolated from Traditional Chinese Herbal Medicine (TCHM) against drug-resistant bacteria: a review paper. Mini Rev Med Chem. 2019;19(2):125–137. doi:10.2174/1389557518666181017143141
14. Liu Y, Zhao X, Yao R, et al. Dragon’s blood from Dracaena Worldwide: species, traditional uses, phytochemistry and pharmacology. Am J Chin Med. 2021;49(6):1315–1367. doi:10.1142/S0192415X21500634
15. Lin Y, Xiong W, Xiao S, et al. Pharmacoproteomics reveals the mechanism of Chinese dragon’s blood in regulating the RSK/TSC2/mTOR/ribosome pathway in alleviation of DSS-induced acute ulcerative colitis. J Ethnopharmacol. 2020;263:113221. doi:10.1016/j.jep.2020.113221
16. de Albuquerque R, Leon-Vargas FR, Carrasco-Montanez DD, et al. A review on phytochemistry and recent pharmacology of Dragon’s Blood (Croton lechleri), a multifunctional ethnomedicinal resource from the Amazon Forest. Planta Med. 2025. doi:10.1055/a-2551-5681
17. Sun X, Huang Q, Wu M, et al. Metabolomics and quantitative analysis to determine differences in the geographical origins and species of Chinese dragon’s blood. Front Plant Sci. 2024;15:1427731. doi:10.3389/fpls.2024.1427731
18. Pona A, Cline A, Kolli SS, et al. Review of future insights of Dragon’s Blood in dermatology. Dermatol Ther. 2019;32(2):e12786. doi:10.1111/dth.12786
19. Seyedi D, Salehi M, Zamani S, et al. Evaluation of burn wound healing and skin regeneration in animal model using Alginate/PVA nanofibrous wound dressings containing Dragon’s blood. J Biomed Mater Res B Appl Biomater. 2025;113(3):e35553. doi:10.1002/jbm.b.35553
20. Liu H, Yan R, Li Y, et al. Dragon’s blood attenuates LPS-induced intestinal epithelial barrier dysfunction via upregulation of FAK-DOCK180-Rac1-WAVE2-Arp3 and downregulation of TLR4/NF-kappaB signaling pathways. J Nat Med. 2024;78(4):1013–1028. doi:10.1007/s11418-024-01824-z
21. CLSI. Performance Standard for Antimicrobial Susceptibility Testing. 35th Ed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2025.
22. Bukhari SI, Aleanizy FS. Association of OprF mutant and disturbance of biofilm and pyocyanin virulence in Pseudomonas aeruginosa. Saudi Pharm J. 2020;28(2):196–200. doi:10.1016/j.jsps.2019.11.021
23. Han Y, Zhang Y, Zeng W, et al. Synergy with farnesol rejuvenates colistin activity against Colistin-resistant Gram-negative bacteria in vitro and in vivo. Int J Antimicrob Agents. 2023;62(3):106899. doi:10.1016/j.ijantimicag.2023.106899
24. Zheng X, Zhang X, Zhou B, et al. Clinical characteristics, tolerance mechanisms, and molecular epidemiology of reduced susceptibility to chlorhexidine among Pseudomonas aeruginosa isolated from a teaching hospital in China. Int J Antimicrob Agents. 2022;60(1):106605. doi:10.1016/j.ijantimicag.2022.106605
25. Yang R, Guan Y, Zhou J, et al. Phytochemicals from camellia nitidissima chi flowers reduce the pyocyanin production and motility of Pseudomonas aeruginosa PAO1. Front Microbiol. 2017;8:2640. doi:10.3389/fmicb.2017.02640
26. Grace A, Sahu R, Owen DR, Dennis VA. Host-mimicking conditions promote Pseudomonas aeruginosa PA14 virulence gene expression. Front Microbiol. 2025;16:1557664. doi:10.3389/fmicb.2025.1557664
27. Zheng X, Chen L, Zeng W, et al. Antibacterial and anti-biofilm efficacy of Chinese Dragon’s blood against Staphylococcus aureus isolated from infected wounds. Front Microbiol. 2021;12:672943. doi:10.3389/fmicb.2021.672943
28. Wu YK, Cheng NC, Cheng CM. Biofilms in chronic wounds: pathogenesis and diagnosis. Trends Biotechnol. 2019;37(5):505–517. doi:10.1016/j.tibtech.2018.10.011
29. Uberoi A, McCready-Vangi A, Grice EA. The wound microbiota: microbial mechanisms of impaired wound healing and infection. Nat Rev Microbiol. 2024;22(8):507–521. doi:10.1038/s41579-024-01035-z
30. Garg SS, Dubey R, Sharma S, et al. Biological macromolecules-based nanoformulation in improving wound healing and bacterial biofilm-associated infection: a review. Int J Biol Macromol. 2023;247:125636. doi:10.1016/j.ijbiomac.2023.125636
31. Ho TJ, Jiang SJ, Lin GH, et al. The in vitro and in vivo wound healing properties of the Chinese herbal medicine “Jinchuang Ointment”. Evid Based Complement Alternat Med. 2016;2016:1654056. doi:10.1155/2016/1654056
32. Guo L, Ruan Q, Ma D, Wen J. Revealing quorum-sensing networks in Pseudomonas aeruginosa infections through internal and external signals to prevent new resistance trends. Microbiol Res. 2024;289:127915. doi:10.1016/j.micres.2024.127915
33. Rather MA, Saha D, Bhuyan S, et al. Quorum Quenching: a drug discovery approach against Pseudomonas aeruginosa. Microbiol Res. 2022;264:127173. doi:10.1016/j.micres.2022.127173
34. Mohan MS, Salim SA, Ranganathan S, et al. Attenuation of Las/Rhl quorum sensing regulated virulence and biofilm formation in Pseudomonas aeruginosa PAO1 by Artocarpesin. Microb Pathog. 2024;189:106609. doi:10.1016/j.micpath.2024.106609
35. Sanchez-Pena A, Winans JB, Nadell CD, Limoli DH. Pseudomonas aeruginosa surface motility and invasion into competing communities enhance interspecies antagonism. mBio. 2024;15(9):e0095624. doi:10.1128/mbio.00956-24
36. Khan F, Pham DTN, Oloketuyi SF, Kim YM. Regulation and controlling the motility properties of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2020;104(1):33–49. doi:10.1007/s00253-019-10201-w
37. Tang D, Lin Y, Yao H, et al. Effect of L-HSL on biofilm and motility of Pseudomonas aeruginosa and its mechanism. Appl Microbiol Biotechnol. 2024;108(1):418. doi:10.1007/s00253-024-13247-7
38. Lewis KA, Baker AE, Chen AI, et al. Ethanol decreases Pseudomonas aeruginosa flagellar motility through the regulation of flagellar stators. J Bacteriol. 2019;201(18):10–128. doi:10.1128/JB.00285-19
39. Hou J, Wu Q, Xiong R, et al. A standardized mouse model for wound infection with Pseudomonas aeruginosa. Int J Mol Sci. 2024;25(21):11773. doi:10.3390/ijms252111773
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.