Back to Journals » International Journal of Nanomedicine » Volume 20
Antibacterial Activity of CuO Nanoparticles, Ethanolic Extract of Amphipterygium adstringens, and Their Combination Against Multidrug-Resistant Bacteria
Authors Ruvalcaba-Ontiveros RI , Esparza-Ponce HE , Reyes-Martínez R, Manjarrez-Nevárez LA, Gómez-Benítez V, Piñon-Castillo HA
Received 24 January 2025
Accepted for publication 9 June 2025
Published 23 June 2025 Volume 2025:20 Pages 8003—8022
DOI https://doi.org/10.2147/IJN.S517465
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 9
Editor who approved publication: Professor Dong Wang
Rosa I Ruvalcaba-Ontiveros,1 Hilda E Esparza-Ponce,2 Reyna Reyes-Martínez,1 Laura A Manjarrez-Nevárez,1 Valente Gómez-Benítez,1 Hilda A Piñon-Castillo1
1Facultad de Ciencias Químicas, Universidad Autónoma de Chihuahua, Circuito Universitario S/N Campus II, Chihuahua, 31125, Mexico; 2Centro de Investigación en Materiales Avanzados, S.C., Complejo Industrial Chihuahua, Chihuahua, 31136, Mexico
Correspondence: Hilda A Piñon-Castillo, Facultad de Ciencias Químicas, Universidad Autónoma de Chihuahua, Circuito Universitario S/N Campus II, Chihuahua, 31125, Mexico, Tel +52-614-236-6000, Email [email protected]
Introduction: This study investigated the incorporation of Amphipterygium adstringens ethanolic extract into the synthesis of copper oxide nanoparticles (CuO+Aa) and evaluates their antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and carbapenem-resistant Acinetobacter baumannii (CRAB). To assess the impact of the extract, chemically synthesized CuO nanoparticles (CuO-NPs) and the extract alone were also tested. Both CuO-NPs and A. adstringens are known for their antimicrobial properties.
Methods: CuO+Aa nanoparticles were synthesized using A. adstringens extract and characterized through Transmission Electron Microscopy (TEM), Dynamic Light Scattering (DLS), Zeta Potential, Thermogravimetric Analysis (TGA), and X-ray Diffraction (XRD), and compared to CuO-NPs. The influence of the extract was analyzed using Inductively Coupled Plasma Mass Spectroscopy (ICP-MS), UV-Vis, FTIR, Raman, and Nuclear Magnetic Resonance Spectroscopy (NMR). Antibacterial effects were tested using the microdrop technique and biofilm inhibition. Bacterial structural changes were observed via Scanning Electron Microscopy (SEM), and cytotoxicity was measured through hemolysis assays.
Results: CuO+Aa nanoparticles were smaller (3.46 nm) than CuO-NPs (5.32 nm). TGA indicated improved thermal degradation in CuO+Aa, suggesting incorporation of organic compounds. XRD revealed a shift from CuO to a mixed CuO-Cu2O phase (75.15%– 24.84%) in CuO+Aa due to the functional groups present in the extract. Antibacterial assays showed that CuO+Aa inhibited MRSA and CRAB by 77% and 49%, respectively, using only 17.5 ppm of copper oxides—significantly lower than CuO-NPs, which required 150 ppm to achieve 96% and 78% inhibition. SEM revealed bacterial surface damage, including roughness, perforations, and cell wall collapse. All treatments showed low cytotoxicity (< 2% hemolysis). Biofilm formation increased by 180% in MRSA and 131% in CRAB.
Conclusion: A. adstringens ethanolic extract enhances CuO nanoparticle synthesis, reducing size and maintaining strong antibacterial activity with low toxicity. CuO+Aa represents a promising candidate for future biomedical applications against resistant pathogens.
Keywords: nanoparticles, A. adstringens ethanolic extract, MRSA, CRAB, bacterial structural damage
Introduction
The indiscriminate use of antibiotics in humans, livestock, and agriculture, along with self-medication and poor adherence to prescribed treatments, has significantly contributed to the emergence of antimicrobial resistance (AMR) in various bacterial pathogens.1 The Centers for Disease Control and Prevention (CDC) estimates that at least 2 million individuals in the United States experience serious infections caused by antibiotic-resistant bacteria annually, resulting in approximately 23,000 fatalities.2 Beyond its impact on public health, AMR imposes a substantial economic burden due to prolonged hospitalizations, extended and costlier treatments, and increased demands on medical resources. These factors result in higher healthcare expenditures and elevated mortality rates compared to infections that remain susceptible to antibiotic therapy. Notably, infections caused by Gram-negative bacteria have become particularly concerning due to their ability to develop resistance to nearly all available antibiotics.3
The increasing prevalence of antibiotic-resistant bacteria underscores the urgent need for alternative antimicrobial strategies. The World Health Organization (WHO) recently updated its Priority Pathogens List, identifying CRAB as a critical priority and MRSA as a high-priority pathogen.4 Acinetobacter baumannii is a Gram-negative pathogen responsible for an estimated 12,000 infections annually in the United States, with approximately 63% of cases classified as multidrug-resistant. These infections contribute to approximately 500 deaths per year.5 S. aureus is a Gram-positive bacterium in which methicillin-resistant strains represent a major cause of healthcare-associated infections. MRSA can cause a wide range of diseases, including skin and soft tissue infections, pneumonia, bloodstream infections, sepsis, and, in severe cases, death.6
To address these challenges, researchers have explored the potential of metallic nanoparticles, such as CuO-NPs, as alternatives to conventional antibiotics. CuO-NPs exhibit unique physicochemical properties, including a high surface-area-to-volume ratio, enhanced chemical reactivity, electrical conductivity, and tunable size and morphology (typically 1–100 nm). These characteristics contribute to their broad-spectrum biological activity, making them valuable for antimicrobial applications. The physicochemical properties of nanoparticles are highly dependent on the synthesis method employed, which influences their size, shape, and overall reactivity.
Green synthesis offers a sustainable and cost-effective approach to nanoparticle production by utilizing biological sources such as plant extracts and microbial metabolites from bacteria, algae, and fungi.7 For instance, CuO-NPs synthesized using Turbinaria species algae have demonstrated antibacterial activity against Streptococcus mutans, Klebsiella spp., and Staphylococcus mutans.8 Similarly, plant extracts from Lonicera japonica and Ocimum sanctum have exhibited antimicrobial effects against Aspergillus niger, S. aureus, Escherichia coli, Candida albicans, Enterobacter faecalis (MTCC-439), Agrobacterium tumefaciens (MTCC-609), and Klebsiella pneumoniae (MTCC-4030). Both plants are recognized for their medicinal properties. CuO-NPs also possess strong antioxidant and antibacterial effects, reducing bacterial viability and inducing oxidative stress, particularly in Gram-negative bacteria.9–11
Traditional medicine offers promising natural alternatives for combating antimicrobial resistance. A. adstringens, commonly known as Cuachalalate, is a medicinal plant traditionally used in Mexican medicine for its antimicrobial and antiproliferative properties.12 It has been shown to be effective in wound healing, particularly for burn injuries.13 The pharmacological effects of A. adstringens are attributed to its diverse bioactive compounds in its bark, including Triterpenoids (Friedelin, Friedelanol, and Epifriedelanol),14 Steroids (Stigmasterol and β-Sitosterol),15 Flavonoids (Rutin, Quercetin, and Kaempferol), Phenolic Compounds (Gallic Acid and Ellagic Acid), and Tannins (Catechins and Proanthocyanidins).16 These compounds exhibit anti-inflammatory, antibacterial, and anticancer properties.17,18 To date, the only reported nanoparticle synthesis involving A. adstringens utilized gold nanoparticles mediated by a shock-wave extract.
Given the significance of CuO-NPs in nanomedicine and nanobiology, previous studies evaluated the antimicrobial activity of chemically synthesized CuO-NPs.11 These nanoparticles exhibited significant antibacterial effects, including bacterial cell wall damage and increased reactive oxygen species (ROS) activity in S. aureus ATCC 24213 and Pseudomonas aeruginosa ATCC 27833. Building upon this work, the present study explores an innovative approach by combining CuO-NPs with plant extracts to enhance their antimicrobial efficacy. Studies have demonstrated that the toxicity of nanoparticles can be increased when combined with plant extracts.19 Furthermore, plant-derived compounds can serve as eco-friendly reducing agents, improving the antimicrobial properties of nanoparticles while minimizing environmental impact.20–23
This study investigates the synthesis, characterization, and antibacterial activity of CuO-NPs, A. adstringens ethanolic extract, and a combination of CuO-NPs with A. adstringens (CuO+Aa) against MRSA and CRAB. As of the submission of this manuscript, no prior studies have reported the synthesis of CuO-NPs using A. adstringens extract.
Material and Methods
Chemicals and Extract Preparation
All chemicals used were of analytical reagent grade, and distilled water was used throughout the experiment. A. adstringens dried bark was purchased from a commercial entity. Its extract was prepared by grinding the bark using a hammer mill. The powdered bark was subjected to extraction using a conventional Soxhlet apparatus, with ethanol as the solvent. The ethanol was subsequently evaporated to dryness under reduced pressure using a Büchi rotary evaporator R-215 connected to a V-100 vacuum pump.
Synthesis of CuO-NPs and CuO+Aa Nanoparticles
For the synthesis of CuO-NPs, 300 mL of 0.02 M Copper(II) acetate (Cu(CH₃COO)2, ≥98%, Sigma-Aldrich) was heated to 94°C under magnetic stirring. One milliliter of glacial acetic acid (CH₃COOH, JT Baker, ACS ≥ 99.7%) was added, followed by 0.4 g of sodium hydroxide (NaOH, ≥98% pellets, Sigma-Aldrich) as the reducing agent. The solution was maintained at this temperature with magnetic stirring for 30 min. During this time, the color of the solution changed from blue to black, indicating the formation of CuO-NPs. The supernatant was removed by centrifugation (10 min at 112 × g), and the CuO-NPs were washed three times with water. The resulting precipitate was air-dried at room temperature. The same procedure was followed for the synthesis of CuO+Aa, with the addition of 0.1 g of A. adstringens ethanolic extract to the 0.02M copper acetate solution.
Characterization
To determine the morphology of both nanoparticles, they were dispersed in distilled water by sonication, and a drop of each solution was placed on a carbon-coated copper grid for TEM analysis using a Hitachi 7700 microscope. The arithmetic mean size of the nanoparticles was calculated using ImageJ software24 by measuring 150 particles from each sample. XRD patterns of the dried nanoparticles powders were recorded on a PANalytical X’Pert PRO MPD diffractometer with an X’Celerator detector, using CuKα radiation at λ = 1.54 Å over a 2θ range of 20° to 80°. The structural patterns of CuO-NPs and CuO+Aa were analyzed by Rietveld refinement using the FullProf Suite Software25 and its interface WinPLOTR.26
CuO-NPs, CuO+Aa, and A. adstringens ethanolic extract were characterized using the following techniques. Their hydrodynamic diameter and surface charge were assessed at a concentration of 1000 ppm using DLS and zeta potential measurements on a Malvern ZetaSizer Nano ZS instrument. TGA was conducted using a TA Instruments Discovery TGA Series Model STD Q600 to evaluate their thermal behavior. UV-Vis analysis was performed using a Thermo Scientific Spectrophotometer Evolution 220. Raman spectroscopy was performed on a Micro Raman Horiba HR, and FTIR was conducted using a PerkinElmer infrared spectrometer over a range of 500 to 4000 cm⁻¹. Additionally, the ethanolic extract of A. adstringens was analyzed by NMR. The extract was dissolved in DMSO-d6, and its 1H NMR spectrum was recorded at room temperature using a Bruker Advance NMR spectrometer (400 MHz) using tetramethylsilane (TMS) as internal standard.
The A. adstringens content in the CuO+Aa samples was quantified using ICP-MS on a Thermo Scientific iCAP 6500 emission spectrometer.
Impact of Treatments on Antibiotic-Resistant Bacteria
MRSA and CRAB were isolated from patients and identified as antibiotic-resistant during routine hospital procedures at the Central Universitario de Chihuahua Hospital. The strains were cultured in Tryptic Soy Broth (TSB) for 12 h and adjusted to a concentration of 1 × 108 CFU/mL in 0.4X broth, using a Spectronic Genesys 8 UV-visible spectrophotometer. Both bacterial strains were exposed to 50, 100, and 150 ppm of CuO-NPs, CuO+Aa, and A. adstringens. All concentrations were dispersed in water and sonicated for 30 min. The cultures exposed to the treatments and the positive controls were incubated at 37°C for 24 h with orbital shaking at 120 rpm using a Thermo Scientific MaxQ 8000 Incubator Shaker. After incubation, a 100 μL aliquot was serially diluted 1:10 in broth. The samples were plated on TSB agar Petri dishes using the microdrop technique.27 Aliquots (10 μL) were plated in triplicate for each dilution. After 24 h of incubation at 37°C, the inhibition percentages for each treatment were determined by comparing the bacterial CFU count in the treated samples to that in the untreated controls.28
To evaluate the dissociative effect of MRSA, CRAB, water, and TSB on CuO-NPs, both bacteria, water, and TSB were exposed to 50, 100, and 150 ppm of CuO-NPs for 24 h. The samples were centrifuged at 112 × g for 10 min, and the supernatants were analyzed by ICP-MS to quantify the percentage of dissociated Cu ions.
The effect of the treatments on biofilm formation was evaluated using the method described by Ball et al29 was followed with minor modifications. Briefly, 24-hour pre-inocula of CRAB and MRSA in TSB supplemented with 0.5% glucose were adjusted to 1 × 108 and 1 × 107 CFU/mL, respectively. The cultures were exposed in triplicate to CuO-NPs, CuO+Aa, and A. adstringens ethanolic extract at concentrations of 150, 100, and 50 ppm. A negative control (10 mm phosphate-buffered saline [PBS], pH 7.4) and a positive control (bacteria without any treatment) were included. Aliquots of 200 µL of each mixture were incubated in a 96-well plate at 37°C for 24 hours. Non-adherent cells were removed, and wells were washed with 200 µL of PBS and allowed to dry for 10 minutes. The cells were then fixed with 200 µL of absolute ethanol and left to dry. Biofilm staining was performed using 200 µL of 0.1% crystal violet solution prepared in distilled water by incubating the microplate at room temperature for 15 minutes. The wells were washed three times with distilled water and allowed to dry. Finally, 200 µL of an ethanol: acetone solution (80:20) was added to solubilize the dye. Absorbance was measured at 590 nm using microplate reader Bio-Rad Model 680.
Structural Analysis of MRSA and CRAB by SEM
To observe the effect of CuO-NPs, CuO+Aa, and A. adstringens ethanolic extract on the structure of MRSA and CRAB, both bacteria were exposed for 24 h to 150 ppm of each treatment. Sample preparation for SEM followed the protocol described by Ulloa-Ogaz.11 Briefly, bacterial cells were fixed in 2.5% glutaraldehyde for 4 hours, washed twice with 0.1 M PBS, and dehydrated through a graded ethanol series (30%, 50%, 70%, three times in 90%, and three times in 100%). The cells were then immersed in ethanol:hexamethyldisilazane (HMDS) mixtures (3:1, 1:1, 1:3, and 100% HMDS). Finally, a drop of the suspension was placed on a silica wafer and stored in a desiccator until analysis. Morphological changes were observed by field-emission scanning electron microscopy (FESEM, JEOL JSM-7401F).
Cytotoxicity Evaluation
Hemolytic activity was assessed using human red blood cells donated by a clinical analysis class from the faculty. Erythrocytes were separated from heparinized blood by centrifugation at 500 × g for 10 minutes at 4°C. A 5% (v/v) red blood cell suspension was prepared in PBS. Solutions of CuO-NPs, CuO+Aa and A. adstringens ethanolic extract at 150, 100, and 50 ppm in PBS were added to the red blood cell suspension. The mixtures were incubated at 37°C for 2 h. PBS was used as the negative control, while erythrocytes resuspended in distilled water served as the positive control to induce osmotic hemolysis. After incubation, the absorbance of the supernatant was measured at 540 nm.30
Statistical Analysis
Each sample was analyzed in triplicate, and the results were statistically evaluated using one-way ANOVA followed by Tukey’s Honestly Significant Difference (HSD) test. In the results, “ns” indicates no significant difference; * denotes a statistically significant difference at the 0.05 level; and ** indicates a highly significant difference. All analyses and plots were performed using OriginPro 2024.
Results and Discussion
Nanoparticle Characterization
Previous studies have demonstrated the antimicrobial properties of CuO-NPs against standard strains of P. aeruginosa and S. aureus.11 In this study, CuO-NPs were synthesized using A. adstringens ethanolic extract to evaluate their efficacy against clinical strains with antimicrobial resistance. The morphologies of the synthesized CuO-NPs and CuO+Aa were analyzed by TEM, as shown in Figure 1a and b, respectively. The average diameter of CuO-NPs was 5.32 nm, while their hydrodynamic diameter measured 368.50 nm, with a Zeta Potential of −16 mV (Figure 2a and b, respectively). The discrepancy between particle size observed via TEM and hydrodynamic diameter measured by DLS can be attributed to nanoparticle agglomeration, as also indicated by the Zeta Potential value.31 Evidence of this agglomeration is visible in Figure 1a. This agglomeration, however, does not indicate that the CuO+Aa nanoparticles lose their nanoscopic properties; on the contrary, the organic layer actually prevents aggregation.
 |
Figure 1 TEM micrographs of: (a) CuO-NPs with a diameter of 5.32nm, and (b) CuO+Aa with a diameter of 3.46 nm. Their diameter distributions are presented in the inner boxes. |
 |
Figure 2 (a) Hydrodynamic diameter and (b) Zeta potential of CuO-NPs, CuO+Aa, and A. adstringens. |
For CuO+Aa, the average particle diameter was 3.46 nm, and the hydrodynamic diameter was significantly larger, at 1,951.3 nm (Figure 2a), with a Zeta Potential of −12.8 mV (Figure 2b). Nanoparticle formation and characteristics, including size and morphology, are influenced by various factors such as pH, reducing agents, and temperature.11,32–35 The observed reduction in particle size and increased agglomeration were likely influenced by the addition of A. adstringens ethanolic extract. This extract contains hydroxyl and carbonyl groups, which serve as reducing and stabilizing agents during nanoparticle synthesis.36 The reduction effect of the extract was evident in the smaller nanoparticle size, while the formation of an organic layer around the CuO+Aa complex contributed to nanoparticle stabilization.
Although the average particle diameter of CuO+Aa was 1.86 nm smaller than that of CuO-NPs, its hydrodynamic diameter was approximately five times larger. This substantial increase is attributed to the presence of the A. adstringens extract, which formed a visible organic coating around the nanoparticles (Figure 1b). Zeta Potential data further support this observation, indicating a shift in surface charge from −16 mV to −12.8 mV. This change suggests that molecules from A. adstringens, which have a Zeta Potential of −8.3 mV, interact with the diffuse ionic layer surrounding CuO-NPs, thereby altering their surface properties.31
Low Zeta Potential values promote nanoparticle agglomeration, which can directly influence interactions with bacterial cells. The reduction in Zeta Potential and resulting agglomeration reflect the complex interplay between CuO-NPs and A. adstringens ethanolic extract, potentially affecting the antimicrobial activity of the nanoparticles. These findings underscore the importance of the organic layer in modulating nanoparticle stability and behavior in biological environments.
Thermogravimetric Analysis
The TGA of CuO-NPs, CuO+Aa, and A. adstringens, shown in Figure 3 and detailed in Table 1, reveals distinct thermal degradation patterns corresponding to their compositions. A primary weight loss event occurred around 200°C across all samples, indicative of water loss. Specifically, CuO- NPs showed a 5% weight loss, whereas CuO+Aa and A. adstringens ethanolic extract exhibited an 11% reduction, attributable to their higher water content.
 |
Table 1 Data Obtained from the Thermogravimetric Analysis of CuO-NPs, A. adstringens, and CuO+Aa |
 |
Figure 3 Dynamic Thermogravimetric analysis of CuO-NPs (Black solid line), CuO+Aa (red dotted line) and A. adstringens (blue dashed line). |
For CuO-NPs, no significant weight change was observed beyond 258°C, with the weight stabilizing at 91% of the initial value, indicating high thermal stability. In contrast, CuO+Aa demonstrated continuous weight loss until approximately 386°C, resulting in a substantial 67% reduction in mass. Beyond this point, the weight remained constant at approximately 33%, suggesting that the organic components had mostly decomposed, leaving a stable residue, likely copper oxide. In the case of A. adstringens, the degradation persisted until 577°C, with almost complete conversion to CO2 at 835°C. This continuous weight loss is consistent with the decomposition of organic matter.37
The crystallinity index, often associated with copper residues, appears at approximately 600°C.38 Analysis of the TGA data revealed that CuO+Aa consisted of approximately 10% water, 65% A. adstringens, and 35% copper oxide, confirming the transformation of copper acetate to copper oxide nanoparticles during synthesis.
It has been reported that the decomposition of polymer chains and imidazole groups, such as those in polyvinyl imidazole, results in oxidation products, such as carbon, NO, NO2, and CO dioxide.37 Additionally, the thermal stability of green-synthesized copper nanoparticles up to approximately 450°C supports their potential utility in biomedical applications, where heat resistance can enhance performance.39 This suggests that the inclusion of plant extracts in nanoparticle synthesis not only contributes to size reduction and stabilization but also affects the thermal behavior of the resulting nanoparticles, enhancing their applicability in diverse fields, including medicine.
X-Ray Diffraction Analysis
The XRD patterns of CuO-NPs and CuO+Aa (Figure 4) highlight the impact of A. adstringens extract on nanoparticle synthesis. The addition of the extract caused compositional change in CuO-NPs, shifting from pure CuO (monoclinic structure with calculated lattice parameters: a = 6.38 Å, b = 3.42 Å, c = 5.13 Å, β = 133.32°) to a mixed-phase composition consisting of 75.15% CuO and 24.84% Cu2O (cubic structure with a lattice parameter of a = 4.26 Å). These lattice parameters were obtained through Rietveld refinement using a Pseudo-Voigt function,40 achieving a goodness-of-fit (χ²) of 1.53 for CuO-NPs and 0.83 for CuO+Aa. The refinement employed the Crystallography Open Database (COD) cards 96-900-8962 for CuO41 and 96-900-7498 for Cu2O.42
The peak broadening observed in the XRD patterns confirms the small grain size of the nanoparticles, which aligns with the TEM results, indicating a reduced particle size upon addition of the extract. The presence of A. adstringens appears to promote partial transformation from CuO to Cu2O, which is likely influenced by the glucose moieties found in the sarsasapogenin molecules present in the extract.43 Glucose can act as a reducing agent, facilitating the reduction of Cu²+ ions to Cu+ and ultimately forming Cu2O nanoparticles. This reaction pathway was associated with the formation of smaller nanoparticles.44
The reduction in particle size observed by TEM (Figure 1b), where the average diameter decreased from 5.32 nm for CuO-NPs to 3.46 nm for CuO+Aa, further supports the influence of the extract. This combination of XRD and TEM data demonstrates that the use of A. adstringens extract not only alters the phase composition of the nanoparticles but also significantly affects their size, potentially enhancing their antibacterial properties due to increased surface area and altered surface chemistry.
CuO+Aa Composition Evaluation
The ICP-MS analysis of CuO+Aa nanoparticles revealed a composition of 37.95% carbon, 4.02% hydrogen, and 31.24% copper oxides. Rietveld refinement further distinguished the copper oxide fraction as 24.09% CuO and 7.15% Cu2O. This result closely matches the 35% copper oxide content estimated by TGA, demonstrating strong agreement across characterization methods. Based on the combined data from XRD, TGA, and ICP-MS analyses, the final composition of the CuO+Aa nanoparticles is estimated to be approximately 65% A. adstringens extract, 26% CuO, and 9% Cu2O.
UV-Vis, FTIR, Raman and NMR Spectroscopy Characterization
The impact of A. adstringens ethanolic extract on the synthesis of CuO-NPs was further investigated using UV-Vis, FTIR, Raman, and NMR spectroscopy. The UV-Vis spectrum of CuO-NPs (Figure 5, solid black line) displays a characteristic absorption peak between 233 and 341 nm, consistent with copper oxide nanoparticles.45 The A. adstringens extract showed typical features of its ethanolic extract, including strong absorbance between 200 and 300 nm and a secondary hump between 300 and 500 nm.46 These spectral features, though with reduced intensity, were also present in the CuO+Aa spectrum, confirming the incorporation of both the extract and CuO in the hybrid nanoparticles.
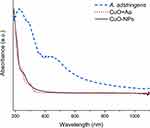 |
Figure 5 UV-Vis Spectrum of A. adstringens (blue dashed line), CuO+Aa (red dotted line), and CuO-NPs (black solid line). |
The FTIR spectrum of CuO-NPs (Figure 6a, solid black line) exhibited distinct peaks at 1399 and 672 cm⁻¹, characteristic of copper oxide nanoparticles.45 The FTIR and Raman spectra of A. adstringens (Figure 6a and b respectively) showed notable similarities, as summarized in Table 2. Spectral features such as hydroxyl and carbonyl functional groups, identified in the extract, played a significant role in the reduction and stabilization processes during the synthesis of CuO+Aa nanoparticles.47 The presence of these functional groups was further confirmed by the overlapping spectral patterns observed in both the extract and CuO+Aa spectra, highlighting their influence on nanoparticle formation.
 |
Table 2 Functional Groups Found in FTIR and Raman Spectra Analysis of CuO-NPs and CuO+Aa |
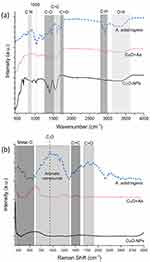 |
Figure 6 (a) FTIR, and (b) Raman spectrum of A. adstringens (blue dashed line), CuO+Aa (red dotted line), and CuO-NPs (black solid line). |
This spectroscopic evidence supports the successful incorporation of A. adstringens ethanolic extract into the synthesis of CuO nanoparticles, resulting in modified nanoparticle composition, size, and surface characteristics. These changes affect the physicochemical properties of the nanoparticles and may enhance their antibacterial activity against drug-resistant bacterial strains.
Further analysis of the CuO+Aa spectra revealed key insights into the interactions between the metal oxide and the plant extract. O–H bond vibrations—typically associated with phenols, alcohols, water, and carboxylic acids—were observed in the FTIR spectrum between 3100 and 3600 cm⁻¹ but were absent in the Raman spectrum due to a lack of detectable signals in that range. Conversely, metal–oxide bond vibrations, which are undetectable by FTIR, were clearly observed in the Raman spectrum between 180 and 600 cm⁻¹, underscoring the complementary nature of these two techniques. Additionally, a Raman peak between 640 and 1360 cm⁻¹ suggests the presence of aromatic compounds. Unique to the FTIR spectrum, a signal at 1050 cm⁻¹ indicates the presence of oxygenated compounds such as anhydrides, while bands between 800 and 900 cm⁻¹ correspond to C–O and C–N stretching vibrations. These features likely reflect the abundance of aromatic and cyclic compounds—including alkylphenols and terpenes—known for their antioxidant, antiproliferative, and anti-inflammatory properties.14,16,48,49
These findings are consistent with compounds identified in A. adstringens by Esquivel-Garcia et al.50 This species contains several bioactive molecules, including sarsasapogenin, a steroidal spirostane-type sapogenin; schinol, a lanostane-type triterpenoid; and masticadienolic acid along with 3β-hydroxymasticadienolic acid, both ursane-type pentacyclic triterpenoids. These compounds are known for their gastroprotective and antimicrobial properties. The presence of their functional groups in the nanoparticles indicates a strong interaction between the CuO-NPs and the extract, likely enhancing the bioactivity of the synthesized CuO+Aa nanoparticles. This interaction is crucial, as it may modulate nanoparticle properties and improve their effectiveness against resistant bacterial strains.
No references were found reporting the use of NMR analysis on the A. adstringens plant. However, signals in the region of 0.5 to 3.00 ppm are typically associated with hydrogen atoms in CH₃ and CH2 groups (Figure 7) from methoxides and aliphatic chains. These findings are consistent with the analysis reported by Galor-Linaldi et al51 in which 6-pentadecyl salicylic acid was isolated from A. adstringens and signals corresponding to the hydrogens of the aliphatic chain carbons were observed. Additionally, signals in the range of 6.5 to 7.5 ppm correspond to hydrogens associated with double bonds, commonly reported in phenolic compounds. The same study identified these signals in the phenolic structure of 6-pentadecyl salicylic acid. In the NMR analysis of guava leaf extract, signals corresponding to quercetin compounds were observed.52 Peaks at 1.325 and 1.334 ppm were not detected due to the strong presence of water. However, signals were observed in the 3.0 to 4.0 ppm region, as well as at 4.775 and 4.776 ppm, which are attributed to hydrogen atoms in aromatic rings with double bonds. Further signals were identified at approximately 6.393 ppm, corresponding to benzo[a]pyrene protons at the C6 and C8 positions of the quercetin molecule. Ai-Ping et al53 reported signals at 0.34 ppm and 0.55 ppm corresponding to saponins; however, these signals were not detected in the current NMR spectrum. Despite their absence in this analysis, saponins have been reported as components of aqueous A. adstringens extracts, as described by Sotelo et al.43
 |
Figure 7 1H NMR spectrum of A. adstringens ethanolic extract. |
Biosafety Evaluation of the Treatments
Figure 8 presents the hemolysis results for CuO-NPs, A. adstringens, and CuO+Aa. Exposure of red blood cells to CuO-NPs and A. adstringens resulted in negative absorbance values, indicating lower absorbance than the control. In contrast, CuO+Aa induced hemolysis of less than 2%. Similarly, green-synthesized copper and silver nanoparticles mediated by Cassia occidentalis have been reported to cause less than 2% hemolysis.54 However, hemolytic responses can vary with different nanoparticle formulations. For example, Ibne et al55 reported up to 10% hemolysis for copper nanoparticles coated with polyethylene glycol and loaded with ciprofloxacin. The results of this study indicate that all treatments, across all tested concentrations, are biocompatible by presenting less than 2% hemolysis.56
 |
Figure 8 Hemolysis produced by different concentrations of CuO-NPs, A. adstringens, and CuO+Aa. |
Antibacterial Properties of CuO-NPs, CuO+Aa, and A. adstringens Extract
CuO-NPs demonstrated significant antibacterial activity, inhibiting CRAB by 68%, 70%, and 78%, and MRSA by 86%, 96%, and 96% at concentrations of 50, 100, and 150 ppm, respectively (Figure 9a and b). These results indicate that increasing the concentration of CuO-NPs enhances their antibacterial effectiveness. In contrast, the ethanolic extract of A. adstringens exhibited substantially lower inhibition of CRAB (13%, −13%, and 7%) and MRSA (22%, 22%, and 47%) at the same concentrations.
 |
Figure 9 Antibacterial activity of CuO-NPs, A. adstringens, and CuO+Aa against (a) CRAB and (b) MRSA. |
However, the hybrid CuO+Aa nanoparticles—comprising A. adstringens (65%), CuO (26%), and Cu2O (9%)—showed improved antibacterial activity. At a total concentration of 50 ppm, they achieved 46% inhibition of CRAB, despite containing only 8% copper oxides. A similar trend was observed against MRSA, where CuO+Aa achieved 50% inhibition—which proportionally corresponds to a 13% inhibition from CuO, 4% inhibition from Cu2O, and 33% inhibition from A. adstringens— using only 17.5 ppm of copper oxides (35% of 50 ppm). In comparison, 50 ppm of CuO-NPs alone was required to reach 86% inhibition of MRSA. This suggests a synergistic interaction between the CuO, Cu2O and A. adstringens ethanolic extract, enhancing the overall antibacterial effect. The limited antibacterial activity of A. adstringens alone (13% for CRAB and 22% for MRSA at 50 ppm) supports the notion that the extract enhances the bioactivity of the hybrid nanoparticles rather than acting as a strong inhibitor independently. Previous studies have also noted that plant extracts alone may not significantly inhibit microbial growth.57
These results highlight the potential of green synthesis approaches in combating both Gram-positive and Gram-negative drug-resistant bacteria. The antibacterial activity of CuO+Aa nanoparticles may be facilitated by electrostatic interactions between their negative surface charge (−12.8 mV) and the positively charged regions of bacterial cell walls.58 Additionally, the reduced efficacy observed against CRAB may be due to efflux pump mechanisms—such as the Resistance-Nodulation-Division (RND) family pumps AdeABC, AdeIJK, and AdeFGH—which actively expel a wide range of compounds, including plant-derived substances like tannins, flavonoids, and polyphenols.59,60 These broad-specificity pumps contribute to the high level of drug resistance of CRAB.
The results also suggest that Cu-based nanoparticles are more effective against MRSA (Gram-positive) than CRAB (Gram-negative), consistent with findings by Du et al,61 who reported that nanoparticles with negative zeta potential exhibit stronger activity against Gram-positive strains.
The antibacterial effect of CuO-NPs is primarily attributed to the generation of reactive oxygen species (ROS), facilitated by surface defects and the electron-donating properties of CuO. Notably, the hydrodynamic diameter plays a more significant role than the actual particle size in enabling membrane penetration.62 Consequently, CuO-NPs, which possess a smaller hydrodynamic diameter, demonstrated enhanced antibacterial activity. In contrast, the antibacterial mechanism of Cu2O is associated with its affinity for intracellular proteins and its ability to disrupt bacterial homeostasis, independent of ROS production. Meghana et al63 emphasized the importance of the higher affinity of Cu2O to the bacterial cells, which explains the efficacy of CuO+Aa nanoparticles despite their larger hydrodynamic diameter. It is hypothesized that, upon contact with bacterial cells, the organic coating of CuO+Aa nanoparticles is partially lost, reducing the hydrodynamic diameter and facilitating a more effective interaction with the bacterial membrane.
Dissociation Effect of Bacteria and TSB on CuO-NPs
The dissociative effects of MRSA, CRAB, water, and TSB on CuO-NPs were evaluated to understand the interactions between the nanoparticles, media, and bacteria. CuO-NPs typically precipitate when dispersed in water, but when dissolved in TSB, CRAB, or MRSA, there is no visible evidence of the precipitation of the nanoparticles, suggesting that potential interactions affect their stability. To quantify the dissociation of Cu ions, the nanoparticles were exposed to TSB, both bacterial strains, and water for 24 h. The supernatant was analyzed using ICP-MS to quantify the Cu content. Figure 10 shows the percentage of dissociated Cu ions at three different concentrations of CuO-NPs (50, 100, and 150 ppm). The average dissociation percentage for each medium is listed in Table 3.
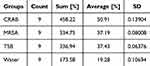 |
Table 3 Summary of the Data Used to Perform the One-Way ANOVA and Tukey’s HSD Test |
The results indicated that the CuO-NPs exposed to water showed minimal dissociation of Cu ions (19.28%), reflecting the stability of the nanoparticles in this medium. In contrast, exposure to TSB and MRSA resulted in significantly higher dissociation (37.43% and 37.19%, respectively), indicating that the nanoparticles interacted more extensively in these environments. Notably, the most substantial dissociation occurred when CuO-NPs were exposed to CRAB, with a dissociation rate of 50.91%.
These findings highlight that the dissociative behavior of CuO-NPs is strongly influenced by the presence of bacteria and TSB, suggesting that the biochemical environment plays a critical role in the stability and dissolution of CuO-NPs. The increased dissociation observed with CRAB compared to water and other media indicates specific interactions that could enhance the antibacterial activity of CuO-NPs, potentially owing to changes in the local microenvironment. Numerous studies have detailed the effects of factors, such as pH, ionic strength, proteins, and other substances, on the colloidal stability of nanoparticles.64,65 These factors also play a significant role in the biosynthesis of nanoparticles, which is mediated by secondary metabolites from microorganisms.66 Although much has been explored regarding nanoparticle synthesis via microbial pathways, there is a notable gap in the literature regarding the specific impact of these secondary metabolites on the solubilization and dissociation of nanoparticles. Understanding these interactions is essential to optimize the use of CuO-NPs as antibacterial agents, particularly against drug-resistant bacterial strains.
Structural Effects of the Treatments on CRAB and MRSA
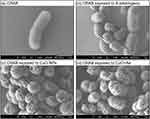
Microscopic analysis of the treated cells revealed significant changes in cell morphology for both CRAB (Figure 11) and MRSA (Figure 12). Upon exposure to 150 ppm A. adstringens, CRAB cells exhibited smoother surfaces; some cells appeared smaller than those in the control group, whereas others displayed elongated shapes, suggesting inhibition of cell division. When exposed to CuO- NPs or CuO+Aa, noticeable changes in the cell surface structure were observed, including roughened cell surfaces, potential perforations, and collapse of the cell wall (indicated by arrows in the figures).
 |
Figure 12 Microscopic analysis of: (a) MRSA control, (b) MRSA exposed to A. adstringens, (c) MRSA exposed to CuO-NPs, and (d) MRSA exposed to CuO-Aa. |
In the case of MRSA, control cells retained their typical cocci shape with smooth surfaces. However, upon exposure to A. adstringens, the cells developed a rough surface, with the formation of thread-like structures intertwining the cells. When exposed to CuO-NPs or CuO+Aa, rougher cell surfaces and biofilm formation were observed in both the treatments. These observations are consistent with the findings of Mohammed et al,67 where CuO-NPs used to decorate carbon nanoparticles led to similar alterations in the cellular structures of E. coli and S. aureus.
Evaluation of Biofilm Production
Biofilm production is one of the primary resistance mechanisms of CRAB and MRSA.29,68 Figure 13 shows the percentage of biofilm produced by CRAB (Figure 13a) and MRSA (Figure 13b) after exposure to 150, 100, and 50 ppm of CuO-NPs, A. adstringens, and CuO+Aa. An increase in biofilm production was observed with higher treatment concentrations. Notably, all values exceeded 100%, indicating that bacteria exposed to the treatments produced more biofilm than the untreated positive control. Yang et al69 reported that A. baumannii increases biofilm formation in response to antibiotic exposure. This response is attributed to stress induced by the treatment, which triggers gene regulation mechanisms that confer a fitness advantage to resistant strains.70
 |
Figure 13 Biofilm production of (a) CRAB and (b) MRSA when exposed to different concentrations of CuO-NPs, A. adstringens, and CuO+Aa. |
Conclusion
This study investigated the synthesis and characterization of CuO+Aa nanoparticles and their antibacterial activity against MRSA and CRAB, using CuO-NPs and A. adstringens ethanolic extract as controls. The incorporation of A. adstringens ethanolic extract led to the formation of smaller CuO-NPs, with the measured diameter decreasing from 5.32 nm to 3.46 nm. A. adstringens also enhanced the thermal stability and altered the structural composition of the nanoparticles, as demonstrated by TEM, XRD, and ICP-MS analyses. Notably, the CuO+Aa formulation contained only 35% copper oxide nanoparticles (26% CuO and 9% Cu2O). Spectroscopic analyses confirmed that functional groups such as hydroxyl and carbonyl in the extract played a key role in nanoparticle stabilization and size reduction.
All of the treatments used in this research, CuO-NPs, CuO+Aa, and A. adstringens ethanolic extract, showed good biocompatibility, as indicated by the very low red blood cell lysis rates.
The hybrid CuO+Aa nanoparticles, composed of A. adstringens extract (65%), CuO (26%), and Cu2O (9%), demonstrated enhanced antibacterial activity, supporting the presence of a synergistic effect. At a total concentration of 50 ppm, the formulation inhibited CRAB growth by 46%, despite containing only 8% copper oxides. Similarly, against MRSA, CuO+Aa achieved 50% inhibition with just 17.5 ppm of copper oxides (35% of the total concentration), a significantly lower amount compared to the 50 ppm of CuO-NPs required to achieve 86% inhibition. These findings highlight the potential of combining A. adstringens extract with copper oxides to improve antibacterial efficacy while reducing the required concentration of metallic components.
The results suggest that the bacterial cell wall structure influences the antibacterial response, with greater reactivity observed against Gram-positive bacteria than Gram-negative. However, the presence of efflux pump mechanisms in CRAB likely contributed to reduced treatment effectiveness.
Despite the larger hydrodynamic diameter of CuO+Aa nanoparticles, they retained strong antibacterial activity. This may be explained by the loss of the organic coating upon contact with bacterial cells, which facilitates membrane penetration. Structural damage to bacterial cells, including surface roughening, perforations, and cell wall collapse, further supported the antibacterial effect of the nanoparticles.
These findings highlight the potential of CuO+Aa nanoparticles for biomedical applications, particularly in combating drug-resistant bacterial strains. The results demonstrate that incorporating A. adstringens extract into nanoparticle synthesis can enhance antimicrobial activity, offering a promising green approach. However, further research is needed to better understand how A. adstringens influences the surface chemistry and reactivity of the nanoparticles, as well as the implications of their thermal stability. Additionally, in vivo studies are essential to assess the systemic effects and safety of these treatments before clinical application.
Acknowledgments
We extend our gratitude to César Leyva-Porras, Raúl Ochoa-Gamboa, and Jesús Uribe-Chavira for their invaluable contributions in obtaining scanning and transmission electron microscopy micrographs as well as conducting the Rietveld refinement analysis. We also acknowledge the postdoctoral scholarship provided by the Secretaría de Ciencia, Humanidades, Tecnología e Innovación (SECIHTI) and the Facultad de Ciencias Químicas.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ho CS, Wong CTH, Aung TT, et al. Antimicrobial resistance: a concise update. Lancet Microbe. 2024;6(1):100947. doi:10.1016/j.lanmic.2024.07.010
2. Centers for Disease Control and Prevention (U.S.), National Center for Emerging Zoonotic and Infectious Diseases (U.S.). Division of Healthcare, Quality Promotion. Antibiotic Resistance Coordination and Strategy Unit. Antibiotic Resistance Threats in the United States, 2019. 2019. doi:10.15620/cdc:82532
3. Naghavi M, Vollset SE, Ikuta KS, et al. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404(10459):1199–1226. doi:10.1016/S0140-6736(24)01867-1
4. World Health Organization. WHO Bacterial Priority Pathogens List 2024. 2024. Available from: https://www.who.int/publications/i/item/9789240093461.
5. Marino A, Augello E, Stracquadanio S, et al. Unveiling the Secrets of Acinetobacter baumannii: resistance, Current Treatments, and Future Innovations. Int J Mol Sci. 2024;25(13):6814. doi:10.3390/ijms25136814
6. Carrel M, Smith M, Shi Q, et al. Antimicrobial Resistance Patterns of Outpatient Staphylococcus aureus Isolates. JAMA Network Open. 2024;7(6):e2417199. doi:10.1001/jamanetworkopen.2024.17199
7. Huston M, Debella M, Dibella M, Gupta A. Green synthesis of nanomaterials. Nanomaterials. 2021;11(8):2130. doi:10.3390/nano11082130
8. Raja Rajamanikkam SCR, Anbalagan G, Subramanian B, Suresh V, Sivaperumal P. Green Synthesis of Copper and Copper Oxide Nanoparticles From Brown Algae Turbinaria Species’ Aqueous Extract and Its Antibacterial Properties. Cureus. 2024. doi:10.7759/cureus.57366
9. Das D, Nath BC, Phukon P, Dolui SK. Synthesis and evaluation of antioxidant and antibacterial behavior of CuO nanoparticles. Colloids Surf B Biointerfaces. 2013;101:430–433. doi:10.1016/j.colsurfb.2012.07.002
10. Grigore ME, Biscu ER, Holban AM, Gestal MC, Grumezescu AM. Methods of synthesis, properties and biomedical applications of CuO nanoparticles. Pharmaceuticals. 2016;9(4):75. doi:10.3390/ph9040075
11. Ulloa-Ogaz AL, Piñón-Castillo HA, Muñoz-Castellanos LN, et al. Oxidative damage to Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 induced by CuO-NPs. Environ Sci Pollut Res. 2017;24(27):22048–22060. doi:10.1007/s11356-017-9718-6
12. Rodriguez-Garcia A, Peixoto ITA, Verde-Star MJ, De La Torre-Zavala S, Aviles-Arnaut H, Ruiz ALTG. In Vitro Antimicrobial and Antiproliferative Activity of Amphipterygium adstringens. Evid Based Complement Alternat Med. 2015;2015:1–7. doi:10.1155/2015/175497
13. Gómez-Salgado MRH, Beltrán-Gómez JÁ, Díaz-Nuñez JL, et al. Efficacy of a Mexican folk remedy containing cuachalalate (Amphipterygium adstringens (Schltdl.) Schiede ex Standl) for the treatment of burn wounds infected with Pseudomonas aeruginosa. J Ethnopharmacol. 2024;319:117305. doi:10.1016/j.jep.2023.117305
14. Oviedo-Chávez I, Ramírez-Apan T, Soto-Hernández M, Martínez-Vázquez M. Principles of the bark of Amphipterygium adstringens (Julianaceae) with anti-inflammatory activity. Phytomedicine. 2004;11(5):436–445. doi:10.1016/j.phymed.2003.05.003
15. Arenas-Quevedo MG, Gracia-Fadrique J. Amphipterygium adstringens (cuachalalate) extract by supercritical CO2. Chem Thermodynamics Thermal Analysis. 2024;13:100128. doi:10.1016/j.ctta.2024.100128
16. Makino M, Motegi T, Fujimoto Y. Tirucallane-type triterpenes from Juliania adstringens. Phytochemistry. 2004;65(7):891–896. doi:10.1016/j.phytochem.2003.12.012
17. Naz R, Ayub H, Nawaz S, et al. Antimicrobial activity, toxicity and anti-inflammatory potential of methanolic extracts of four ethnomedicinal plant species from Punjab, Pakistan. BMC Complement Altern Med. 2017;17(1). doi:10.1186/s12906-017-1815-z
18. Bourgou S, Pichette A, Marzouk B, Legault J. Antioxidant, anti-inflammatory, anticancer and antibacterial activities of extracts from nigella sativa (black cumin) plant parts. J Food Biochem. 2012;36(5):539–546. doi:10.1111/j.1745-4514.2011.00567.x
19. Aziz WJ, Abid MA, Hussein EH. Biosynthesis of CuO nanoparticles and synergistic antibacterial activity using mint leaf extract. Mater Technol. 2020;35(8):447–451. doi:10.1080/10667857.2019.1692163
20. Sharma BK, Shah DV, Roy DR. Green synthesis of CuO nanoparticles using Azadirachta indica and its antibacterial activity for medicinal applications. Mater Res Express. 2018;5(9):095033. doi:10.1088/2053-1591/aad91d
21. Padil VVT, Černík M. Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int J Nanomed. 2013;8:889–898. doi:10.2147/IJN.S40599
22. Acharyulu NPS, Dubey RS, Swaminadham V, Kollu P, Kalyani RL, Pammi SVN. Green Synthesis of CuO Nanoparticles using Phyllanthus Amarus Leaf Extract and their Antibacterial Activity Against Multidrug Resistance Bacteria Green synthesis of CuO nanoparticles. Int J Eng Res Technol. 2014;3(4):639–641. doi:10.17577/IJERTV3IS040918
23. Das P, Ghosh S, Ghosh R, Dam S, Baskey M. Madhuca longifolia plant mediated green synthesis of cupric oxide nanoparticles: a promising environmentally sustainable material for waste water treatment and efficient antibacterial agent. J Photochem Photobiol B. 2018;189:66–73. doi:10.1016/j.jphotobiol.2018.09.023
24. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi:10.1038/nmeth.2089
25. Rodríguez-Carvajal J. Recent advances in magnetic structure determination by neutron powder diffraction. Phys B. 1993;192(1–2):55–69. doi:10.1016/0921-4526(93)90108-I
26. Roisnel T, Rodríguez-Carvajal J. WinPLOTR: a windows tool for powder diffraction pattern analysis. In: Materials Science Forum. Trans Tech Publications Ltd. 2001;378–381:118–123. doi:10.4028/www.scientific.net/msf.378-381.118
27. Miles AA, Misra SS, Irwin JO. The estimation of the bactericidal power of the blood. J Hygiene. 1938;38(6):732–749. doi:10.1017/S002217240001158X
28. Machado KN, dos SMF, Missio RF, Vendruscolo ECG. Bacterial Quantification in Different Plating Methodologies. J Adv Biol Biotechnol. 2024;27(8):964–973. doi:10.9734/jabb/2024/v27i81217
29. Ball AL, Augenstein ED, Wienclaw TM, et al. Characterization of Staphylococcus aureus biofilms via crystal violet binding and biochemical composition assays of isolates from hospitals, raw meat, and biofilm-associated gene mutants. Microb Pathog. 2022;167:105554. doi:10.1016/j.micpath.2022.105554
30. Elizondo-Luévano JH, Gomez-Flores R, Verde-Star MJ, et al. In Vitro Cytotoxic Activity of Methanol Extracts of Selected Medicinal Plants Traditionally Used in Mexico against Human Hepatocellular Carcinoma. Plants. 2022;11(21):2862. doi:10.3390/plants11212862
31. Clogston JD, Patri AK. Zeta Potential Measurement. In: Methods in Molecular Biology. Humana Press Inc.. 2011;Vol. 697:63–70. doi:10.1007/978-1-60327-198-1_6
32. Nahar B, Chaity SB, Gafur MA, Hossain MZ. Synthesis of Spherical Copper Oxide Nanoparticles by Chemical Precipitation Method and Investigation of Their Photocatalytic and Antibacterial Activities. J Nanomater. 2023;2023:1–10. doi:10.1155/2023/2892081
33. Lanje AS, Sharma SJ, Pode RB, Ningthoujam RS. Synthesis and optical characterization of copper oxide nanoparticles. Adv Appl Sci Res. 2010;1(2):36–40.
34. Matei A, Ţucureanu V, Popescu M, Romanitan C, Bita B, Cernica I. Synthesis and characterization of various surfactants for stabilized CuO powder. In: Powder Metallurgy and Advanced Materials. Vol 8. Materials Research Forum LLC. 2018:52–60. doi:10.21741/9781945291999-6
35. Ahamed M, Alhadlaq HA, Khan MAM, Karuppiah P, Al-Dhabi NA. Synthesis, Characterization, and Antimicrobial Activity of Copper Oxide Nanoparticles. J Nanomater. 2014;2014(1). doi:10.1155/2014/637858
36. Mohamad NAN, Arham NA, Jai J, Hadi A. Plant extract as reducing agent in synthesis of metallic nanoparticles: a review. Adv Mater Res. 2014;832:350–355. doi:10.4028/www.scientific.net/AMR.832.350
37. Pozdnyakov AS, Emel’yanov AI, Korzhova SA, et al. Green synthesis of stable nanocomposites containing copper nanoparticles incorporated in poly-n-vinylimidazole. Polymers. 2021;13(19):3212. doi:10.3390/polym13193212
38. Pérez-Alvarez M, Cadenas-Pliego G, Pérez-Camacho O, Comparán-Padilla VE, Cabello-Alvarado CJ, Saucedo-Salazar E. Green synthesis of copper nanoparticles using cotton. Polymers. 2021;13(12):1906. doi:10.3390/polym13121906
39. Mohamed HI, Mahmoud NMR, Ramadan A, Al-Subaie AM, Ahmed SB. Novel Biological-Based Strategy for Synthesis of Green Nanochitosan and Copper-Chitosan Nanocomposites: promising Antibacterial and Hematological Agents. Nanomaterials. 2024;14(13):1111. doi:10.3390/nano14131111
40. Uribe-Chavira JS, Herrera-Pérez G, Santillán-Rodríguez CR, Sáenz-Hernández RJ, Matutes-Aquino JA, Grijalva-Castillo MC. X-ray diffraction analysis by Rietveld refinement of FeAl alloys doped with terbium and its correlation with magnetostriction. J Rare Earths. 2023;41(8):1217–1224. doi:10.1016/j.jre.2022.07.006
41. Wyckoff RWG. Crystal Structures 1.
42. Kirfel A, Eichhorn K. Accurate structure analysis with synchrotron radiation. The electron density in Al2O3 and Cu2O. Acta Crystallogr A. 1990;46(4):271–284. doi:10.1107/S0108767389012596
43. Sotelo-Barrera M, Cília-García M, Luna-Cavazos M, et al. Amphipterygium adstringens (Schltdl.) Schiede ex Standl (Anacardiaceae): an Endemic Plant with Relevant Pharmacological Properties. Plants. 2022;11(13):1766. doi:10.3390/plants11131766
44. Carrasco-Hernández AR, Ruvalcaba-Ontiveros RI, Martínez-Guerra E, Duarte-Moller JA, Esparza-Ponce HE. Evolution of Structural and Optical Properties of Cuprous Oxide Particles for Visible Light Absorption. J Nanomater. 2022;2022(1). doi:10.1155/2022/7964428
45. Dulta K, Koşarsoy Ağçeli G, Chauhan P, Jasrotia R, Chauhan PK, Ighalo JO. Multifunctional CuO nanoparticles with enhanced photocatalytic dye degradation and antibacterial activity. Sustainable Environ Res. 2022;32(1). doi:10.1186/s42834-021-00111-w
46. Torres-Ortiz D, García-Alcocer G, Loske AM, et al. Green Synthesis and Antiproliferative Activity of Gold Nanoparticles of a Controlled Size and Shape Obtained Using Shock Wave Extracts from Amphipterygium adstringens. Bioengineering. 2023;10(4):437. doi:10.3390/bioengineering10040437
47. Munandar N, Kasim S, Arfah R, et al. Green synthesis of copper oxide (CuO) nanoparticles using Anredera cordifolia leaf extract and their antioxidant activity. Commun Scie Technol. 2022;7(2):127–134. doi:10.21924/cst.7.2.2022.1004
48. Rodríguez-Luis OE, Hernandez-Delgadillo R, Sánchez-Nájera RI, et al. Green synthesis of silver nanoparticles and their bactericidal and antimycotic activities against oral microbes. J Nanomater. 2016;2016:1–10. doi:10.1155/2016/9204573
49. Saitta M, Giuffrida D, La Torre GL, Potortì AG, Dugo G. Characterisation of alkylphenols in pistachio (Pistacia vera L.) kernels. Food Chem. 2009;117(3):451–455. doi:10.1016/j.foodchem.2009.04.043
50. Esquivel-García R, Ayiania M, Abu-Lail N, et al. Pyrolytic oils from Amphipterygium adstringens bark inhibit IL-8 production of IL-17-stimulated HaCaT keratinocytes. J Anal Appl Pyrolysis. 2020;145:104749. doi:10.1016/j.jaap.2019.104749
51. Galot-Linaldi J, Hernández-Sánchez KM, Estrada-Muñiz E, Vega L. Anacardic acids from amphipterygium adstringens confer cytoprotection against 5-fluorouracil and carboplatin induced blood cell toxicity while increasing antitumoral activity and survival in an animal model of breast cancer. Molecules. 2021;26(11):3241. doi:10.3390/molecules26113241
52. Huynh HD, Nargotra P, Wang HMD, Shieh CJ, Liu YC, Kuo CH. Bioactive Compounds from Guava Leaves (Psidium guajava L.): characterization, Biological Activity, Synergistic Effects, and Technological Applications. Molecules. 2025;30(6):1278. doi:10.3390/molecules30061278
53. Li AP, Li ZY, Qu TL, Qin XM, Du GH. Nuclear magnetic resonance based metabolomic differentiation of different Astragali Radix. Chinese J Nat Med. 2017;15(5):363–374. doi:10.1016/S1875-5364(17)30057-2
54. Gondwal M, Joshi Nee Pant G. Synthesis and Catalytic and Biological Activities of Silver and Copper Nanoparticles Using Cassia occidentalis. Int J Biomater. 2018;2018:1–10. doi:10.1155/2018/6735426
55. Ibne Shoukani H, Nisa S, Bibi Y, et al. Ciprofloxacin loaded PEG coated ZnO nanoparticles with enhanced antibacterial and wound healing effects. Sci Rep. 2024;14(1). doi:10.1038/s41598-024-55306-z
56. Witika BA, Makoni PA, Matafwali SK, et al. Biocompatibility of biomaterials for nanoencapsulation: current approaches. Nanomaterials. 2020;10(9):1–40. doi:10.3390/nano10091649
57. Yu W, Tang J, Gao C, Zheng X, Zhu P. Green Synthesis of Copper Nanoparticles from the Aqueous Extract of Lonicera japonica Thunb and Evaluation of Its Catalytic Property and Cytotoxicity and Antimicrobial Activity. Nanomaterials. 2025;15(2):91. doi:10.3390/nano15020091
58. Ashokkumar M, Palanisamy K, Ganesh Kumar A, Muthusamy C, Senthil Kumar KJ. Green synthesis of silver and copper nanoparticles and their composites using Ocimum sanctum leaf extract displayed enhanced antibacterial, antioxidant and anticancer potentials. Artif Cells Nanomed Biotechnol. 2024;52(1):438–448. doi:10.1080/21691401.2024.2399938
59. Gautam D, Dolma KG, Khandelwal B, et al. Acinetobacter baumannii: an overview of emerging multidrug-resistant pathogen. Med J Malaysia. 2022;77(3):357–370.
60. Esterly JS, Richardson CL, Eltoukhy NS, Qi C, Scheetz MH. Genetic mechanisms of antimicrobial resistance of Acinetobacter baumannii. Ann Pharmacother. 2011;45(2):218–228. doi:10.1345/aph.1P084
61. Du WL, Niu SS, Xu YL, Xu ZR, Fan CL. Antibacterial activity of chitosan tripolyphosphate nanoparticles loaded with various metal ions. Carbohydr Polym. 2009;75(3):385–389. doi:10.1016/j.carbpol.2008.07.039
62. Khurana C, Vala AK, Andhariya N, Pandey OP, Chudasama B. Antibacterial activity of silver: the role of hydrodynamic particle size at nanoscale. J Biomed Mater Res A. 2014;102(10):3361–3368. doi:10.1002/jbm.a.35005
63. Meghana S, Kabra P, Chakraborty S, Padmavathy N. Understanding the pathway of antibacterial activity of copper oxide nanoparticles. RSC Adv. 2015;5(16):12293–12299. doi:10.1039/c4ra12163e
64. Moore TL, Rodriguez-Lorenzo L, Hirsch V, et al. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem Soc Rev. 2015;44(17):6287–6305. doi:10.1039/c4cs00487f
65. Jabor Z, Sutton SC. Effects of Digestion, Cell Culture Media, and Mucous on the Physical Properties, Cellular Effects, and Translocation of Polystyrene and Polymethacrylate Nanoparticles. Toxics. 2023;11(8):708. doi:10.3390/toxics11080708
66. Schwan J, Markert S, Rosenfeldt S, Schüler D, Mickoleit F, Schenk AS. Comparing the Colloidal Stabilities of Commercial and Biogenic Iron Oxide Nanoparticles That Have Potential In Vitro/In Vivo Applications. Molecules. 2023;28(13):4895. doi:10.3390/molecules28134895
67. Mohammed SAA, Khashan KS, Jabir MS, et al. Copper Oxide Nanoparticle-Decorated Carbon Nanoparticle Composite Colloidal Preparation through Laser Ablation for Antimicrobial and Antiproliferative Actions against Breast Cancer Cell Line, MCF-7. Biomed Res Int. 2022;2022(1). doi:10.1155/2022/9863616
68. Gedefie A, Demsis W, Ashagrie M, et al. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: a review. Infect Drug Resist. 2021;14:3711–3719. doi:10.2147/IDR.S332051
69. Yang CH, Su PW, Moi SH, Chuang LY. Biofilm formation in Acinetobacter baumannii: genotype-phenotype correlation. Molecules. 2019;24(10). doi:10.3390/molecules24101849
70. Balderrama-González AS, Piñón-Castillo HA, Ramírez-Valdespino CA, Reyes-Martínez R, Esparza-Ponce HE. Effect of the AuNPs@amox system on antibiotic-resistant bacteria. J Nanopart Res. 2024;26(6). doi:10.1007/s11051-024-06048-6
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.