Back to Journals » International Journal of Nanomedicine » Volume 19
Aptamer sgc8-Modified PAMAM Nanoparticles for Targeted siRNA Delivery to Inhibit BCL11B in T-Cell Acute Lymphoblastic Leukemia
Authors Zeng X, Nie D, Liu Z , Peng X, Wang X, Qiu K, Zhong S, Liao Z, Zha X, Li Y , Zeng C
Received 8 July 2024
Accepted for publication 6 November 2024
Published 20 November 2024 Volume 2024:19 Pages 12297—12309
DOI https://doi.org/10.2147/IJN.S477597
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Farooq A. Shiekh
Xiangbo Zeng,1,2,* Dingrui Nie,1,3,* Zhen Liu,2 Xueting Peng,1 Xianfeng Wang,1 Kangjie Qiu,1 Shuxin Zhong,1 Ziwei Liao,4 Xianfeng Zha,1,5 Yangqiu Li,1,2 Chengwu Zeng1
1Key Laboratory for Regenerative Medicine of Ministry of Education, Institute of Hematology, School of Medicine, Jinan University, Guangzhou, 510632, People’s Republic of China; 2Department of Hematology, First Affiliated Hospital, Jinan University, Guangzhou, 510632, People’s Republic of China; 3Department of Hematology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, People’s Republic of China; 4Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, 510180, People’s Republic of China; 5Department of Clinical Laboratory, First Affiliated Hospital, Jinan University, Guangzhou, 510632, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yangqiu Li; Chengwu Zeng, Jinan University, No. 601, West Huangpu Avenue, Guangzhou, Guangdong, People’s Republic of China, Email [email protected]; [email protected]
Introduction: T-cell acute lymphoblastic leukemia (T-ALL) is a malignant hematological disease with limited targeted therapy options. Overexpression of B-cell lymphoma/leukemia 11B is frequently observed in T-ALL and contributes to leukemogenesis. Knockdown of BCL11B inhibits T-ALL cell proliferation and induces apoptosis, making it a potential therapeutic target. However, the clinical application of siRNA therapies is hindered by challenges such as poor delivery efficiency and limited clinical outcomes.
Methods: We developed a targeted delivery system for BCL11B siRNA (siBCL11B) using generation 5 polyamidoamine (G5-PAMAM) dendrimers conjugated with the sgc8 aptamer, which specifically binds to the T-ALL cell membrane protein PTK7. This nanoparticle, designated G5-sgc8-siBCL11B, was designed to selectively deliver siRNA to T-ALL cells. In vitro and in vivo experiments were conducted to evaluate its therapeutic efficacy and safety.
Results: We demonstrate that sgc8-conjugated siBCL11B nanoparticles selectively and efficiently target BCL11B-overexpressing T-ALL cells, significantly inhibiting cell viability and promoting apoptosis while exhibiting minimal impact on the viability of normal T cells. In T-ALL mouse model studies, G5-sgc8-siBCL11B and G5-siBCL11B significantly inhibited the progression of T-ALL in vivo, extending the survival of mice compared to the control (CTR), G5, and G5-sgc8 groups. Although there was no significant difference in survival between the G5-sgc8-siBCL11B and G5-siBCL11B groups, a trend towards improved survival was observed (p = 0.0993).
Conclusion: The G5-sgc8-siBCL11B nanoparticle system demonstrated efficient delivery and significant therapeutic efficacy, highlighting its potential as a promising novel approach for the treatment of T-ALL.
Keywords: BCL11B, T-ALL, aptamer, sgc8, siBCL11B, PAMAM
Introduction
T-cell acute lymphoblastic leukemia is a highly aggressive leukemia with poor prognosis.1–5 Compared to B-cell acute lymphoblastic leukemia (B-ALL), T-ALL is characterized by high tumor heterogeneity and a lack of well-defined therapeutic targets.6–9 Due to the dysregulated differentiation and uncontrolled proliferation typical of T-ALL, our study focused on BCL11B, a critical regulator of T-cell differentiation involved in multiple pathways during T-cell maturation and differentiation.10–12 BCL11B is essential for T-cell lineage commitment and plays a crucial role in directing the differentiation of hematopoietic stem cells into T-cells instead of natural killer cells.13,14 However, mutations in BCL11B or its fusion with other genes have been implicated in the development of T-ALL and acute myeloid leukemia (AML).15 Our research, along with findings from other studies, indicates that BCL11B is expressed at higher levels in T-ALL cells compared to normal T-cells, potentially contributing to the inhibition of apoptotic signaling pathways.16,17 Moreover, reducing the BCL11B level has been demonstrated to promote apoptosis and inhibit the growth of T-ALL cells.18–21 Furthermore, research also demonstrated that demonstrated that knocking down BCL11B in normal T-cells had minimal impact on T-cell differentiation and proliferation.22 Based on this evidence, we hypothesize that targeting BCL11B may offer a promising therapeutic approach for treating T-ALL.
Aptamers are a class of targeted, small-molecule nucleic acids capable of forming specific spatial structures akin to the specificity observed in antigen-antibody binding with particular molecules.23–25 As a result, aptamers are recognized as molecules possessing definitive targeting capabilities and have been instrumental in targeted delivery in various studies.26–31 Previous studies have reported screening for the nucleic acid aptamer sgc8, a single-stranded DNA with a specific secondary structure that can bind specifically to the T-ALL cell membrane protein PTK7 and become internalized through PTK7 mediation.32–36 In previous studies, sgc8 served predominantly as a probe for detecting T-ALL, showcasing remarkable sensitivity and specificity. Nonetheless, contemporary research on aptamers in targeted drug delivery largely concentrates on optimizing the delivery efficacy of chemotherapy agents. The exploration of siRNA delivery for tumor treatment, particularly in the context of T-ALL, remains nascent, highlighting a domain ripe for deeper exploration.37–39
Polyamidoamine (PAMAM) is a dendrimer with primary amine groups in its structure, which interact with the phosphate groups in the nucleic acid backbone, forming a complex that is beneficial for nucleic acid delivery.40–42 PAMAM can block nuclease active sites, protecting nucleic acids from degradation and stabilizing their activity. In previous studies, PAMAM has shown favorable affinity towards various small molecule nucleic acids when controlled with appropriate pH, ratio, and concentration. This molecule can be taken up by cancer cells and subsequently release the small molecule nucleic acids within the acidic intracellular environment caused by heightened metabolic activity, making it a promising mediator for small molecule nucleic acid delivery.26–31 However, the delivery efficiency and inherent toxicity of PAMAM have limited its direct therapeutic application.
In this study, we employed generation 5 PAMAM (G5-PAMAM, henceforth referred to as G5) to encapsulate both sgc8 and siBCL11B, resulting in the formation of G5-sgc8-siBCL11B nanoparticles for targeted therapy of T-ALL. Enhanced by the sgc8 aptamer, these novel nanoparticles demonstrate improved cellular targeting and effectively deliver siBCL11B, underscoring their potential for T-ALL treatment.
Materials and Methods
Clinical Samples
Peripheral blood (PB) samples from healthy individuals (HIs) and bone marrow (BM) samples from T-ALL patients were obtained from the First Affiliated Hospital of Jinan University. Written informed consent was obtained from all participants before blood collection, and the study was approved by the Ethical Committee of the First Affiliated Hospital of Jinan University (Approval, No.KY-2019-041). All procedures were performed in accordance with guidelines in the Declaration of Helsinki. T-ALL patient-derived xenograft (PDX) cells were obtained from Prof. Peng Li at the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
The Cell Culture Studies
T-ALL cell lines, including Jurkat, Molt4, CEM, MT4 and KOPTK1, along with the chronic myeloid leukemia (CML) cell line K562 and 293T cells, were obtained from the American Type Culture Collection. All cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C with 5% CO₂, except for 293T cells, which were maintained in DMEM supplemented with 10% FBS.
The Formation of G5-sgc8-siBCL11B Nanoparticles
G5-PAMAM was purchased from Shandong Weihai Chenyuan Molecular New Materials Co., Ltd. (CYD-150A) and dissolved in dimethyl sulfoxide to prepare a 10 mmol/L stock solution. The working solution was diluted to 100 μmol/L. siBCL11B and siBCL11B-cy5 were purchased from Guangzhou Ruibo Biological Company and dissolved in diethylpyrocarbonate water to prepare a 100 μmol/L solution with sterile phosphate-buffered saline used as the solvent. To form stable nanoparticle complexes, G5 and nucleic acid were mixed at an N:P ratio of 30:1 and vortexed well followed by incubation at room temperature for 15 minutes. The final concentration of siBCL11B was adjusted to 100 nmol/L. The siBCL11B sequences are as follows: siBCL11B-935-sense: GCACAACAUGCAAGCAGCCCUUCAA, siBCL11B-935-antisense: UUGAAGGGCUGCUUGCAUGUUGUGC, siBCL11B-671-sense: GUCCCAAGCAGGAGAACAU, siBCL11B-671-antisense: AUGUUCUCCUGCUUGGGAC, siBCL11B-3’UTR-sense: GGACAACGCUGCUUAGAUA, siBCL11B-3’UTR-antisecse: UAUCUAAGCAGCGUUGUCC. sgc8 and sgc8-FAM were purchased from Guangzhou Ruibo Biological Company with the following sequence: ATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA.
Quantitative RT-PCR Analysis
Cells treated with the nanocomposites for 24 hours were harvested, and total RNA was isolated using TRIzol reagent. Afterward, cDNA was generated by reverse transcription. Alterations in the RNA expression levels were assessed using Real-time quantitative reverse transcription polymerase chain reaction kits (TIANGEN, FP215) utilizing the Bio-Rad fluorescence quantitative PCR system and BioRad CFX Manager software. Target gene expression levels were normalized by house-keeping gene GAPDH. The primer sequences used were as follows: BCL11B Forword: GCAGCTCTCCGGGCGAT; BCL11B Reverse: CACAGGTGAGCAGGTCAGG; GAPDH Forword: GTCTCCTCTGACTTCAACAGCG; GAPDH Reverse: ACCACCCTGTTGCTGTAGCCAA.
Western Blotting Analysis
Cells were harvested and lysed with SDS (FUDE, FD2040-100). Western blotting analysis was performed as previously describe.43,44 Primary antibodies used in this study include anti-BCL11B (Cell Signaling Technology, 12120) and anti-GAPDH (Beyotime Technology, AF0006). Secondary antibodies used in this study include goat anti-mouse IgG H&L (HRP) (Abcam, ab6789) and goat anti-rabbit IgG H&L (HRP) (Abcam, ab6721).
Isolation and Culture of PBMCs and T Cells
PB collected from HIs was utilized to isolate peripheral blood mononuclear cells (PBMCs) through density gradient centrifugation with Ficoll lymphocyte separation solution. Subsequently, T cells were isolated using CD3 magnetic beads (Miltenyi Biotec, 130–092-012) according to the manufacturer’s instructions. The isolated T cells were cultured in RPMI 1640 medium supplemented with 20 U/mL IL-2, 10% fetal bovine serum, 10mg/mL penicillin, 10mg/mL streptomycin. T cell activation was conducted using the T Cell TransAct kit (Miltenyi Biotec, 130–111-160) according to the manufacturer’s instructions.
Lentivirus Transduction
The coding sequence (CDS) sequence of BCL11B was amplified via PCR from cDNA reverse-transcribed from RNA of Molt4 cells and cloned into the pCDH-puroR vector. 293T cells were used as packaging cells for viral production, and transfections of pCDH-BCL11B-puroR, psPAX2, and pMD2.G were carried out using Polyethylenimine linear (PEI, YEASEN, 40816ES02). Harvested lentivirus mixed with Polybrene (YEASEN, 40804ES76, 1mg/mL) was added to CEM and Molt4 cells for infection. After 72 hours, the medium was replaced with puromycin-containing medium (YEASEN, 60209ES10, 1 ug/mL) for one week to select stable overexpression cell lines.
Cell Proliferation Assay
Molt4 and CEM cells were seeded in 12-well plates at a density of 1×10^5 cells per well and treated with different interventions in various groups. After 48 hours of incubation, the cells were transferred to 96-well plates at a density of 2×10^4 cells per well, and 10 μL of CCK-8 reagent was added to each well. Following 4 hours of incubation, the absorbance at 450 nm was measured using a microplate reader.
Flow Cytometry
After a 48-hour treatment with nanocomposites, cells were harvested and stained with an apoptosis detection kit (Multi Sciences, AP107) according to the manufacturer’s instructions. Flow cytometry analysis was performed using a NovoCyte flow cytometer to detect propidium iodide (PI) and annexin V. For the assessment of tumor burden in mice, CD45-FITC antibody (BD, 555482) was used to lable CD45-positive cells in T-ALL PDX mouse model. PTK7-APC antibody (Miltenyi, 130–091-366) was employed to measure PTK7 expression on the cell membrane. Flow cytometry data were analyzed using FlowJo 10.8 software.
Hematoxylin-Eosin Staining and Immunohistochemistry
After euthanizing mice, spleens and femurs were excised, and the specimens were fixed in a 4% paraformaldehyde solution for 48 hours. Hematoxylin-eosin staining (HE) and immunohistochemistry (IHC) staining, along with scanning, were conducted by Wuhan Borfu Biotechnology Co., Ltd.
Confocal Microscopy
For confocal imaging, siBCL11B-Cy5 and sgc8-FAM were used to label the nanoparticles. Molt4 and T-ALL cells were seeded at a density of 1×10^6 cells per well in a 12-well plate and treated with various intervention combinations for 20 hours. After treatment, the cells were incubated with Hoechst 33342 at a concentration of 1 mg/mL for 4 hours, collected, and deposited onto poly-L-lysine-coated slides. Confocal images were captured using a Leica confocal microscope (LEICA SP8).
Dynamic Light Scattering and Zeta Potential Measurements
The particle size distribution and polydispersity index (PDI) were assessed using dynamic light scattering (DLS), while the surface charge was evaluated through zeta potential analysis via laser-Doppler electrophoresis. Each sample was prepared in triplicate (n = 3), with three measurements conducted for each sample using the Zetasizer Nano ZS in Jinan University Analytical and Testing Center.
Transmission Electron Microscopy
The prepared nanoparticles were diluted in pure water to achieve a concentration of 1 mg/mL. The resulting nanoparticle solution was then applied to a copper grid for transmission electron microscopy (TEM) imaging and allowed to air dry at room temperature. Before TEM imaging, the samples were stained in uranyl acetate (Electron Microscopy Sciences, 22400) for 1 minute. The TEM images were acquired using an Thermo TECNAI G2 Spirit TWIN microscope in Jinan University Analytical and Testing Center.
Animal Experiments
Animal experiment was approved by the Laboratory Animal Ethics Committee of Jinan University (approval No. 20230320-06) and conducted in accordance with the Guidelines for the Ethical Review of Laboratory Animal Welfare (National Standard GB/T 35,892-2018) issued by the People’s Republic of China. The procedure for constructing the T-ALL PDX model is summarized as follows: Male B-NDG mice, aged 6 to 8 weeks, received total body irradiation at a dose of 1 Gy. Twenty-four hours later, 1×10^6 T-ALL leukemic cells were injected intravenously through the tail vein. Successful model establishment was confirmed by detecting human CD45 expression using flow cytometry 21 days post-injection. Following confirmation, treatment commenced immediately. After treatment, three mice from each group were randomly selected for histological analysis. HE staining and IHC were conducted to evaluate tumor infiltration, while CD45+ cell levels in PB were analyzed by flow cytometry. The remaining mice were monitored for survival outcomes.
Statistical Analysis
Statistical analyses between groups were conducted using the t-test, while comparisons among multiple groups were performed using analysis of variance (ANOVA). Survival analysis was performed using Kaplan-Meier methods. Graphs were generated using GraphPad Prism 9.0, with symbols indicating statistical significance: * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and **** for p < 0.0001.
Results
Development of G5-sgc8-siBCL11B Nanoparticles for Selective Delivery
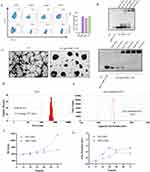
We developed sgc8-conjugated G5 nanoparticles for the efficient delivery of siBCL11B. Utilizing the nucleic acid affinity of G5, we conjugated the sgc8 aptamer with siBCL11B to enhance targeting specificity towards T-ALL cells. Flow cytometry analysis (Figure 1A) confirmed elevated PTK7 expression in T-ALL cell lines, including Molt4, Jurkat, and CEM, compared to the low expression observed in other leukemia cells such as K562. Based on these results, sgc8 were tailored to specifically target T-ALL cells. To optimize the G5 to siBCL11B ratio, we evaluated N:P ratios ranging from 1:1 to 30:1. Complexes were analyzed using Agarose gel electrophoresis, revealing that an optimal ratio of 30:1 effectively encapsulated siBCL11B within G5, resulting in nanoparticles that remained near the well due to increased charge and size (Figure 1B). Further characterization of the nanoparticles was conducted using TEM to visualize G5-sgc8-siBCL11B (Figure 1C), confirming that G5 successfully complexed sgc8 and siBCL11B into G5-sgc8-siBCL11B nanoparticles. Particle size analysis was performed using DLS (Figure 1D), while zeta potential measurements (Figure 1E) revealed a transition from the positive charge of G5 to a negative charge following the successful formation of G5-sgc8-siBCL11B. Supplementary Figure S1 provides the TEM, DLS, and zeta potential analyses for other formulations. These findings confirm the formation of G5 and nucleic acid nanoparticles. To evaluate its stability, G5-sgc8-siBCL11B was incubated in media containing 10% and 100% FBS for up to 72 hours, during which DLS and zeta potential measurements were monitored (Figure 1F–G). The results indicated that G5-sgc8-siBCL11B maintained stability for up to 72 hours in 10% FBS, mimicking the activity of serum nucleases. However, the presence of a higher concentration of FBS (100%) resulted in accelerated degradation of the nanoparticles.
G5-sgc8-siBCL11B Nanoparticles Selectively Suppressing Cell Viability and Enhancing Apoptosis in T-ALL Cells
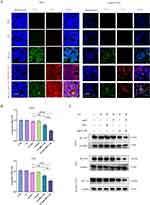
We subsequently investigated whether G5-sgc8-siBCL11B can efficiently and selectively target T-ALL cells. Following a 24-hour incubation with the cells, confocal microscopy was performed (Figure 2A). The FAM-labeled sgc8 exhibited green fluorescence, while the Cy5-labeled siBCL11B displayed red fluorescence. In both Molt4 and primary T-ALL cells, G5 effectively facilitated the intracellular delivery of sgc8 and siBCL11B. Notably, nanoparticles conjugated with sgc8 demonstrated superior delivery efficiency. Collectively, these findings validate the design of a nanoparticle system optimized for siBCL11B delivery.
Next, we investigated the anti-leukemia effects of G5-sgc8-siBCL11B nanoparticles on Molt4 and CEM cell lines. Cells were divided into five experimental groups: control (CTR), G5, G5-sgc8, G5-siBCL11B, and G5-sgc8-siBCL11B, and treated with the respective nanoparticle formulations. Following treatment, we assessed the knockdown efficiency of siBCL11B using qRT-PCR (Figure 2B) and Western blotting analyses (Figure 2C). Our data demonstrated successful delivery of siBCL11B by G5 nanoparticles, resulting in a significant reduction of BCL11B expression in both Molt4 and CEM cells, including de novo T-ALL leukemia cells. Notably, the G5-sgc8-siBCL11B group exhibited the most pronounced knockdown effect.
To ensure specificity and the observed inhibition was attributable to BCL11B suppression, two distinct siBCL11B sequences were utilized in subsequent experiments. Flow cytometric analysis (Figure 3A) revealed that the siBCL11B-935 and siBCL11B-671 sequences effectively induced apoptosis in Molt4 and CEM cells, with the G5-sgc8-siBCL11B group exhibiting the highest apoptosis rate. Consistent with these findings, CCK-8 assays (Figure 3B) further demonstrated the anti-leukemia effects of the nanoparticles, showing that the G5-sgc8-siBCL11B group achieved the most significant reduction in cell viability. Next, we designed an additional siRNA targeting the 3’ UTR of BCL11B, alongside the siBCL11B-671 and siBCL11B-935 sequences targeting the CDS region. Notably, siRNAs targeting the 3’ UTR were unable to inhibit the viability of cells overexpressing exogenous BCL11B that lacked the 3’ UTR (Supplementary Figure 2A). Furthermore, G5-sgc8-siBCL11B failed to suppress cell viability in T-ALL cell lines with low BCL11B expression (Supplementary Figure 2B), reinforcing that the observed inhibition was specifically attributable to BCL11B down-regulation. Additionally, the nanoparticles similarly induced apoptosis while inhibiting the cell viability of primary T-ALL cells (Figure 3C and D). To evaluate the safety profile of the nanoparticles, we assessed their cytotoxic effects on T cells isolated from healthy individuals. As shown in Figure 3E, G5-sgc8-siBCL11B exhibited minimal impact on the viability of normal T cells, underscoring their relative safety. Overall, our findings demonstrate that G5-sgc8-siBCL11B nanoparticles effectively deliver siBCL11B and downregulate BCL11B expression in T-ALL cells, thereby promoting apoptosis and inhibiting cell viability. These results highlight the therapeutic potential of G5-sgc8-siBCL11B for targeted T-ALL therapy.
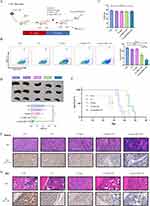
Anti-Leukemia Activity of G5-sgc8-siBCL11B Nanoparticles in T-ALL PDX Mice
We then investigated the potential anti-leukemia effect of nanoparticle formulation in T-ALL PDX mouse model (Figure 4A). Successful establishment of the T-ALL PDX mouse model was confirmed by detecting human CD45 expression in the PB of the recipient mice. Subsequently, the mice were divided into distinct experimental groups: CTR, G5, G5-sgc8, G5-siBCL11B, and G5-sgc8-siBCL11B. Each group received the corresponding nanoparticle formulation via intravenous injection through the tail vein. The nanoparticles encapsulating siBCL11B effectively reduced the tumor burden in PDX mice, with the G5-sgc8-siBCL11B group exhibiting the greatest reduction (Figure 4B). We measured the weight of PDX mice one week after treatment with nanoparticles. No significant weight loss was observed in the treated mice (Figure 4C), indicating the safety of G5-sgc8-siBCL11B in vivo. Mice treated with G5-sgc8-siBCL11B showed a significant reduction in spleen weight, whereas G5-siBCL11B exhibited a trend toward inhibition, though this was not statistically significant (Figure 4D). Survival analysis confirmed the therapeutic efficacy of G5-sgc8-siBCL11B and G5-siBCL11B, significantly extending lifespan in the treatment group compared to the CTR, G5 and G5-sgc8 group (Figure 4E). Although no statistically significant difference was observed between the G5-sgc8-siBCL11B and G5-siBCL11B groups, a trend was noted (p = 0.0993). Histological analysis of spleen and BM sections revealed a significant reduction in T-ALL cell infiltration in both G5-sgc8-siBCL11B and G5-siBCL11B group, particularly in the G5-sgc8-siBCL11B group, which exhibited the lowest BCL11B expression levels (Figure 4F–G). In conclusion, G5-sgc8-siBCL11B nanoparticles effectively reduce T-ALL tumor burden and prolong survival in vivo. The incorporation of sgc8 enhances the therapeutic potential of these nanoparticles, underscoring their promise for targeted T-ALL therapy.
Discussion
Hematologic malignancies have traditionally relied on chemotherapy for long-term management. Prolonged chemotherapy or allogeneic hematopoietic stem cell transplantation may be employed to sustain patient benefits.45,46 In the case of T-ALL, high-intensity chemotherapy regimens are commonly used. However, many patients encounter significant side effects during induction, experience relapse post-remission, and develop resistance to subsequent therapies, resulting in a poor prognosis and limited treatment options.45 Thus, targeted therapies for T-ALL must focus on identifying effective therapeutic targets or strategies to mitigate intolerance and provide salvage treatments after relapse during standard induction chemotherapy.4,45 While specific therapies, such as the CD19 and CD3 bispecific antibody blinatumomab, have been explored for B-ALL,47–49 there remains a notable absence of effective targeted or immunotherapeutic options for T-ALL.50
Our previous research identified BCL11B as a crucial therapeutic target in T-ALL due to its pivotal role in T cell development and its elevated expression in many T-ALL patients compared to HIs. Targeting BCL11B is essential because it regulates genes associated with proliferation and apoptosis, such as BCLxL and BCL2,51,52 which contribute to T-ALL progression. We demonstrated that silencing BCL11B in T-ALL cell lines reduces proliferation and resistance to apoptosis, with minimal impact on normal T cells. Furthermore, studies by others have shown that knocking out BCL11B not only does not inhibit immune function but can also induce normal T cells to adopt NK cell-like phenotypes,14 which could potentially promote tumor immunity. To achieve this, we employed the sgc8 aptamer to specifically target the T-ALL membrane protein PTK7, leading to the design of G5-sgc8-siBCL11B. This complex effectively targets T-ALL leukemia cells while minimizing non-specific side effects, and its straightforward assembly provides a robust foundation for advancing BCL11B-targeted drug development.
Delivering therapeutic nucleic acids to leukemia cells, including T-ALL, poses significant challenges due to reduced sensitivity to transfection methods and off-target effects.53,54 While small molecule nucleic acids, such as siRNA, have shown promise in tumor treatment, their application in non-tumor diseases and effective delivery to leukemia cells remain problematic due to low efficiency and off-target effects.55,56 Our prior research identified PTK7, a membrane protein highly expressed in T-ALL and correlated with BCL11B expression, as a viable target. Notably, PTK7 is recognized by the specific nucleic acid aptamer sgc8.34 By leveraging the synergistic advantages of BCL11B and sgc8, we developed nanoparticles that selectively target PTK7-expressing T-ALL cells, enhancing affinity while reducing necessary dosages and toxicity. In this study, we utilized PAMAM dendrimer, to construct high-affinity G5-sgc8-siBCL11B nanoparticles aimed at targeted downregulation of BCL11B in T-ALL cells. The nucleic acid affinity of G5 facilitated the formation of stable complexes with siBCL11B. Incorporating the sgc8 aptamer significantly enhanced the delivery efficiency and cell specificity of siBCL11B, leading to reduced proliferation and increased apoptosis in T-ALL cells. The G5-sgc8-siBCL11B nanoparticles reduced the required siBCL11B dosage,26,57 thereby enhancing the overall safety profile. Importantly, these nanoparticles demonstrated effective delivery of siBCL11B to both T-ALL cell lines and primary T-ALL cells. We further validated their delivery and therapeutic potential in a T-ALL PDX mouse model, where the nanoparticles, enhanced by the sgc8, exhibited improved delivery and therapeutic efficacy.
Conclusion
This study introduces a novel nanoparticle system utilizing G5 and the sgc8 aptamer for the targeted delivery of siBCL11B to T-ALL cells. Our findings demonstrate that these nanoparticles exhibit potent anti-leukemia activity both in vitro and in vivo, while maintaining a favorable safety profile. The unique combination of high specificity for T-ALL cells and effective gene silencing of BCL11B presents a promising therapeutic strategy for addressing T-ALL, particularly in cases resistant to conventional treatments. Furthermore, the safety and efficacy observed in our experiments highlight the potential for clinical applications of this system. Future research should investigate its long-term effects and explore its utility in combination therapies to further enhance its therapeutic impact.
Abbreviations
T-ALL, T-cell acute lymphoblastic leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CDS, coding sequence; CML, chronic myeloid leukemia; DLS, dynamic light scattering; HE, hematoxylin-eosin staining; HIs, healthy individuals; IHC, immunohistochemical; G5 PAMAM, generation 5 polyamidoamine; PBMCs, peripheral blood mononuclear cells; PDI, polydispersity index; PDX, patient-derived xenograft; TEM, transmission electron microscopy.
Acknowledgments
This work was supported by grants from the Intergovernmental International Cooperation on Scientific and Technological Innovation Project of Chinese Ministry of Science and Technology (No. 2017YFE0131600), National Natural Science Foundation of China (Nos. 82370167 and 32000802), Guangdong Basic and Applied Basic Research Foundation (No. 2023A1515012118), and Guangdong Innovation and Entrepreneurship Training Program for Undergraduate (No. S202310559078).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bardelli V, Arniani S, Pierini V, et al. T-cell acute lymphoblastic leukemia: biomarkers and their clinical usefulness. Genes. 2021;12(8):1118. doi:10.3390/genes12081118.
2. Brivio E, Baruchel A, Beishuizen A, et al. Targeted inhibitors and antibody immunotherapies: novel therapies for paediatric leukaemia and lymphoma. Eur J Cancer. 2022;164:1–17. doi:10.1016/j.ejca.2021.12.029
3. Duffield AS, Mullighan CG, Borowitz MJ. International consensus classification of acute lymphoblastic leukemia/lymphoma. Virchows Arch. 2023;482(1):11–26. doi:10.1007/s00428-022-03448-8
4. Fung SC, Man P, Hei M. Early T-cell precursor acute lymphoblastic leukemia: diagnosis, updates in molecular pathogenesis, management, and novel therapies. Front Oncol. 2021;11:750789. doi:10.3389/fonc.2021.750789
5. Wang P, Cai Q, Peng X et al. Increased co-expression of CTLA4/LAG3 predicted adverse clinical outcomes in patients with T-cell malignancies. Cell Investig. 2025;1:1 100004. doi:10.1016/j.clnves.2024.100004
6. Caracciolo D, Mancuso A, Polerà N, et al. The emerging scenario of immunotherapy for T-cell acute lymphoblastic leukemia: advances, challenges and future perspectives. Exp Hematol Oncol. 2023;12(1):5. doi:10.1186/s40164-022-00368-w
7. Lejman M, Chałupnik A, Chilimoniuk Z, Dobosz M. Genetic biomarkers and their clinical implications in b-cell acute lymphoblastic leukemia in children. IJMS. 2022;23(5):2755. doi:10.3390/ijms23052755
8. Xu J, Zhu HH. Targeted treatment of T-cell acute lymphoblastic leukemia: latest updates from the 2022 ASH annual meeting. Exp Hematol Oncol. 2023;12(1):30. doi:10.1186/s40164-023-00384-4
9. Zhu H, Dong B, Zhang Y, et al. Integrated genomic analyses identify high-risk factors and actionable targets in T-cell acute lymphoblastic leukemia. Blood Sci. 2022;4(1):16–28. doi:10.1097/BS9.0000000000000102
10. Li P, Leonard WJ. Chromatin accessibility and interactions in the transcriptional regulation of T cells. Front Immunol. 2018;9:2738. doi:10.3389/fimmu.2018.02738
11. Li P, Xiao Y, Liu Z, Liu P. Using mouse models to study function of transcriptional factors in T cell development. Cell Regener. 2012;1(1):8. doi:10.1186/2045-9769-1-8
12. Liao R, Wu Y, Qin L, et al.
13. Huang R, Wen Q, Zhang X. CAR-NK cell therapy for hematological malignancies: recent updates from ASH 2022. J Hematol Oncol. 2023;16(1):35. doi:10.1186/s13045-023-01435-3
14. Jiang Z, Qin L, Tang Y, et al. Human induced-T-to-natural killer cells have potent anti-tumour activities. Biomark Res. 2022;10(1):13. doi:10.1186/s40364-022-00358-4
15. Montefiori LE, Bendig S, Gu Z, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discovery. 2021;11(11):2846–2867. doi:10.1158/2159-8290.CD-21-0145
16. Huang X, Chen S, Shen Q, et al. Down regulation of BCL11B expression inhibits proliferation and induces apoptosis in malignant T cells by BCL11B −935-siRNA. Hematology. 2011;16(4):236–242. doi:10.1179/102453311X13025568941961
17. Li K, Chen C, Gao R, et al. Inhibition of BCL11B induces downregulation of PTK7 and results in growth retardation and apoptosis in T-cell acute lymphoblastic leukemia. Biomark Res. 2021;9(1):17. doi:10.1186/s40364-021-00270-3
18. Huang X, Du X, Li Y. The role of BCL11B in hematological malignancy. Exp Hematol Oncol. 2012;1(1):22. doi:10.1186/2162-3619-1-22
19. Huang X, Shen Q, Chen S, et al. Gene expression profiles in BCL11B-siRNA treated malignant T cells. J Hematol Oncol. 2011;4(1):23. doi:10.1186/1756-8722-4-23
20. Chen S, Huang X, Chen S, et al. The role of BCL11B in regulating the proliferation of human naive T cells. Hum Immunol. 2012;73(5):456–464. doi:10.1016/j.humimm.2012.02.018
21. Przybylski GK, Przybylska J, Li Y. Dual role of BCL11B in T-cell malignancies. Blood Sci. 2024;6(4):e00204. doi:10.1097/BS9.0000000000000204
22. Shen Q, Huang X, Chen S, et al. BCL11B suppression does not influence CD34 + cell differentiation and proliferation. Hematology. 2012;17(6):329–333. doi:10.1179/1024533212Z.000000000145
23. Kinghorn A, Fraser L, Liang S, Shiu S, Tanner J. Aptamer bioinformatics. IJMS. 2017;18(12):2516. doi:10.3390/ijms18122516
24. Wu L, Wang Y, Xu X, et al. Aptamer-based detection of circulating targets for precision medicine. Chem Rev. 2021;121(19):12035–12105. doi:10.1021/acs.chemrev.0c01140
25. Zhu G, Chen X. Aptamer-based targeted therapy. Adv Drug Delivery Rev. 2018;134:65–78. doi:10.1016/j.addr.2018.08.005
26. Alibolandi M, Taghdisi SM, Ramezani P, et al. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int J Pharm. 2017;519(1–2):352–364. doi:10.1016/j.ijpharm.2017.01.044
27. Chen Z, Peng Y, Li Y, et al. Aptamer-dendrimer functionalized magnetic nano-octahedrons: theranostic drug/gene delivery platform for near-infrared/magnetic resonance imaging-guided magnetochemotherapy. ACS Nano. 2021;15(10):16683–16696. doi:10.1021/acsnano.1c06667
28. Liu Y, Yang Y, Zhang Q, et al. Dynamics of delivering aptamer targeted nano-drugs into cells. J Mater Chem B. 2021;9(4):952–957. doi:10.1039/D0TB02527E
29. Mohammadi S, Salimi A, Hamd-Ghadareh S, Fathi F, Soleimani F. A FRET immunosensor for sensitive detection of CA 15-3 tumor marker in human serum sample and breast cancer cells using antibody functionalized luminescent carbon-dots and AuNPs-dendrimer aptamer as donor-acceptor pair. Anal Biochem. 2018;557:18–26. doi:10.1016/j.ab.2018.06.008
30. Ren M, Li Y, Zhang H, et al. An oligopeptide/aptamer-conjugated dendrimer-based nanocarrier for dual-targeting delivery to bone. J Mater Chem B. 2021;9(12):2831–2844. doi:10.1039/D0TB02926B
31. Zhang HJ, Zhao X, Chen LJ, Yang CX, Yan XP. Dendrimer grafted persistent luminescent nanoplatform for aptamer guided tumor imaging and acid-responsive drug delivery. Talanta. 2020;219:121209. doi:10.1016/j.talanta.2020.121209
32. Ding D, Yang C, Lv C, Li J, Tan W. Improving tumor accumulation of aptamers by prolonged blood circulation. Anal Chem. 2020;92(5):4108–4114. doi:10.1021/acs.analchem.9b05878
33. Xia F, He A, Zhao H, et al. Molecular engineering of aptamer self-assemblies increases in vivo stability and targeted recognition. ACS Nano. 2022;16(1):169–179. doi:10.1021/acsnano.1c05265
34. Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell‐specific internalization study of an aptamer from whole cell selection. Chem a Eur J. 2008;14(6):1769–1775. doi:10.1002/chem.200701330
35. Yang L, Liang M, Cui C, et al. Enhancing the nucleolytic resistance and bioactivity of functional nucleic acids by diverse nanostructures through in situ polymerization‐induced self‐assembly. ChemBioChem. 2021;22(4):754–759. doi:10.1002/cbic.202000712
36. Zhang L, Abdullah R, Hu X, et al. Engineering of bioinspired, size-controllable, self-degradable cancer-targeting DNA nanoflowers via the incorporation of an artificial sandwich base. J Am Chem Soc. 2019;141(10):4282–4290. doi:10.1021/jacs.8b10795
37. Taghdisi SM, Abnous K, Mosaffa F, Behravan J. Targeted delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer. J Drug Targeting. 2010;18(4):277–281. doi:10.3109/10611860903434050
38. Zhao N, Zeng Z, Zu Y. Self‐assembled aptamer‐nanomedicine for targeted chemotherapy and gene therapy. Small. 2018;14(4):1702103. doi:10.1002/smll.201702103
39. Qi J, Zeng Z, Chen Z, et al. Aptamer–gemcitabine conjugates with enzymatically cleavable linker for targeted delivery and intracellular drug release in cancer cells. Pharmaceuticals. 2022;15(5):558. doi:10.3390/ph15050558
40. Chauhan A. Dendrimers for drug delivery. Molecules. 2018;23(4):938. doi:10.3390/molecules23040938
41. Fox LJ, Richardson RM, Briscoe WH. PAMAM dendrimer - cell membrane interactions. Adv Colloid Interface Sci. 2018;257:1–18. doi:10.1016/j.cis.2018.06.005
42. Li X, Ta W, Hua R, Song J, Lu W. A review on increasing the targeting of PAMAM as carriers in glioma therapy. Biomedicines. 2022;10(10):2455. doi:10.3390/biomedicines10102455
43. Wang X, Chen Z, Nie D, et al. CASP1 is a target for combination therapy in pancreatic cancer. Eur J Pharmacol. 2023;961:176175. doi:10.1016/j.ejphar.2023.176175
44. Zeng C, Nie D, Wang X, et al. Combined targeting of GPX4 and BCR-ABL tyrosine kinase selectively compromises BCR-ABL+ leukemia stem cells. Mol Cancer. 2024;23(1):240. doi:10.1186/s12943-024-02162-0
45. McMahon CM, Luger SM. Relapsed T cell ALL: current approaches and new directions. Curr Hematol Malig Rep. 2019;14(2):83–93. doi:10.1007/s11899-019-00501-3
46. O’Dwyer KM. Optimal approach to T-cell ALL. Hematology. 2022;2022(1):197–205.
47. Nagorsen D, Kufer P, Baeuerle PA, Bargou R. Blinatumomab: a historical perspective. Pharmacol Ther. 2012;136(3):334–342. doi:10.1016/j.pharmthera.2012.07.013
48. Queudeville M, Ebinger M. Blinatumomab in pediatric acute lymphoblastic leukemia—from salvage to first line therapy (a systematic review). JCM. 2021;10(12):2544. doi:10.3390/jcm10122544
49. Zeng C. Advances in cancer treatment: the role of new technologies and research. Cell Investig. 2025;1(1):100001. doi:10.1016/j.clnves.2024.100001
50. Kimura S, Mullighan CG. Molecular markers in ALL: clinical implications. Best Pract Res Clin Haematol. 2020;33(3):101193. doi:10.1016/j.beha.2020.101193
51. Grabarczyk P, Przybylski GK, Depke M, et al. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26(26):3797–3810. doi:10.1038/sj.onc.1210152
52. Li W, Jiang Z, Li T, et al. Genome-wide analyses identify KLF4 as an important negative regulator in T-cell acute lymphoblastic leukemia through directly inhibiting T-cell associated genes. Mol Cancer. 2015;14(1):26. doi:10.1186/s12943-014-0285-x
53. Barati M, Mirzavi F, Atabaki M, Bibak B, Mohammadi M, Jaafari MR. A review of PD-1/PD-L1 siRNA delivery systems in immune T cells and cancer cells. Int Immunopharmacol. 2022;111:109022. doi:10.1016/j.intimp.2022.109022
54. Ramishetti S, Peer D. Engineering lymphocytes with RNAi. Adv Drug Delivery Rev. 2019;141:55–66. doi:10.1016/j.addr.2018.12.002
55. Tarab-Ravski D, Stotsky-Oterin L, Peer D. Delivery strategies of RNA therapeutics to leukocytes. J Control Release. 2022;342:362–371. doi:10.1016/j.jconrel.2022.01.016
56. Van Hoeck J, Braeckmans K, De Smedt SC, Raemdonck K. Non-viral siRNA delivery to T cells: challenges and opportunities in cancer immunotherapy. Biomaterials. 2022;286:121510. doi:10.1016/j.biomaterials.2022.121510
57. Ayatollahi S, Salmasi Z, Hashemi M, et al. Aptamer-targeted delivery of Bcl-xL shRNA using alkyl modified PAMAM dendrimers into lung cancer cells. Int J Biochem Cell Biol. 2017;92:210–217. doi:10.1016/j.biocel.2017.10.005
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.