Back to Journals » International Journal of Nanomedicine » Volume 20
Au@Pt@HP1-HP2@Fe3O4 Nanoenzymatic Complexes Based on CHA Signal Amplification Strategy for Ultrasensitive SERS Detection of ctDNA in Liver Cancer
Authors Wang X, Sheng J, Yang H, Shen K, Yao J, Qian Y , Chen G
Received 31 March 2025
Accepted for publication 28 June 2025
Published 8 July 2025 Volume 2025:20 Pages 8891—8905
DOI https://doi.org/10.2147/IJN.S531541
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Eng San Thian
Xiaoyong Wang,1,* Jinxin Sheng,1,* Haifan Yang,2,3 Kang Shen,2,3 Jie Yao,1 Yayun Qian,2,3 Gaoyang Chen4
1Department of General Surgery, Nantong Haimen People’s Hospital, Nantong, Jiangsu, People’s Republic of China; 2Institute of Translational Medicine, Medical College, Yangzhou University, Yangzhou, Jiangsu, People’s Republic of China; 3The Key Laboratory of Syndrome Differentiation and Treatment of Gastric Cancer of the State Administration of Traditional Chinese Medicine, Yangzhou, Jiangsu, People’s Republic of China; 4Department of Oncology, The Affiliated Taizhou Second People’s Hospital of Yangzhou University, Taizhou, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Gaoyang Chen, Department of Oncology, The Affiliated Taizhou Second People’s Hospital of Yangzhou University, Taizhou, Jiangsu, People’s Republic of China, Email [email protected]
Purpose: Early diagnosis of liver cancer requires highly sensitive detection of biomarkers. This study aims to develop a novel method for detecting circulating tumor DNA (ctDNA) in the serum of liver cancer patients, leveraging a catalytic hairpin self-assembly (CHA) signal amplification strategy combined with surface-enhanced Raman scattering (SERS) technology and nano-enzyme catalysis.
Methods: We synthesized Au@Pt@HP1-HP2@Fe3O4 nano-enzyme complexes, utilizing the SERS-enhancing properties of Pt-coated Au nanoparticles (Au@Pt) and the separation-enrichment capability of Fe3O4 magnetic beads. The complexes catalyzed the oxidation of colorless TMB by H2O2 to produce blue ox-TMB, enabling quantitative detection of PIK3CA E542K mutant ctDNA. The assay’s performance was validated using gold standard qRT-PCR.
Results: Under optimized conditions, the method achieved a detection limit for PIK3CA E542K as low as 4.12 aM. The assay demonstrated high sensitivity, specificity, and efficient magnetic separation, making it a robust tool for ctDNA detection.
Conclusion: This study presents a highly sensitive and specific detection platform for liver cancer early diagnosis, characterized by magnetic separation and nano-enzyme catalysis. The method holds significant clinical potential for the accurate and early detection of liver cancer biomarkers.
Keywords: surface-enhanced Raman scattering, nano-enzymes, circulating tumor DNA, liver cancer, catalytic hairpin self-assembly
Introduction
Liver cancer is a highly aggressive malignancy with a poor prognosis, primarily attributable to the absence of early symptoms and frequent late-stage diagnosis.1,2 As a real-time indicator of tumor genetic alterations and progression, circulating tumor DNA (ctDNA) has demonstrated significant value in early cancer detection. Studies reveal markedly elevated ctDNA levels in liver cancer patients, with its short half-life enabling dynamic tumor monitoring, making it an ideal diagnostic marker.3 Notably, PIK3CA gene mutations drive hepatocarcinogenesis by activating the PI3K/AKT/mTOR pathway, establishing this gene as a critical target for diagnosis and personalized therapy.4–6 However, conventional detection methods exhibit significant limitations. Real-time quantitative PCR (qPCR) has restricted applicability for minimal residual disease (MRD) monitoring. Droplet digital PCR (ddPCR) lacks standardized detection protocols and requires interpretation alongside multiparameter flow cytometry (MFC) data, and next-generation sequencing (NGS) demands sophisticated bioinformatics tools that are unavailable in most routine clinical laboratories.7,8 These constraints have accelerated the development of alternative diagnostic technologies, highlighting the urgent need for rapid and sensitive alternatives.9,10
Surface Enhanced Raman Scattering (SERS), a highly sensitive spectroscopic analysis technique, has been widely used in biomedical detection in recent years.11–13 SERS can dramatically enhance Raman signals by adsorbing target analytes onto nanostructured noble metal surfaces, enabling highly sensitive detection of biomolecules even at trace concentrations.14–16 The latest clinical validation study confirms that SERS technology has significant advantages in the early diagnosis of liver cancer.17,18 Multiple hepatocellular carcinoma biomarkers miRNA122 and miRNA233 based on asymmetric competitive CRISPR (acCRISPR) and surface-enhanced Raman spectroscopy coupled to PTS with LODs of 10.36 and 4.65 fM, respectively.19 In addition, the SERS platform has a detection limit of 952 aM translation for liver cancer-associated long chain non-coding RNA (lncRNA).20 Nevertheless, these approach still face limitations, including suboptimal sensitivity or inadequate target specificity, which may restrict its clinical applicability.
In recent years, the innovative design of nanomaterials has reinvigorated the development of biosensing technology.21–23 Nano-enzymes, as a kind of nanomaterials with enzyme-like catalytic activity, show great potential for application in the field of biosensing. Compared with natural enzymes, nano-enzymes not only have similar high catalytic activity, but also have the advantages of good stability, low cost and easy modification.24–27 Integrating nano-enzymes with SERS technology enables multiplexed signal amplification of target molecules. The catalytic activity of nano-enzymes synergistically enhances the signal intensity of the reaction system, substantially improving detection sensitivity and specificity.28,29 This combination not only extends the utility of SERS in analytical applications but also introduces an innovative paradigm for ultrasensitive bioanalysis in challenging sample environments. Pt-coated Au nanoparticles (Au@Pt) are ideal materials for SERS detection due to their excellent stability and enhancement effects. The Au@Pt not only inherits the excellent SERS enhancement properties of gold nanomaterials, but also further improves the catalytic activity and chemical stability of the material through the introduction of platinum.30–32 This unique structure enables the Au@Pt nanomaterials to simultaneously exert the surface plasmon resonance effect and the catalytic effect of the nano-enzymes in SERS detection,33,34 thus realizing dual signal amplification. In addition, magnetic beads, as an efficient separation and enrichment tool, can realize efficient capture and enrichment of low-abundance ctDNA in complex biological samples through specific binding to target molecules.35 The integration of magnetic beads with Au@Pt nanomaterials enhances both the capture efficiency of target molecules and the sensitivity and accuracy of SERS-based detection.
Furthermore, DNA hairpin-based self-assembly has emerged as a prominent nucleic acid amplification strategy due to its operational simplicity and mild reaction requirements.36,37 The catalytic hairpin self-assembly (CHA) reaction, as a non-enzymatic signal amplification method, is capable of signal amplification by constructing a target strand cycling loop at room temperature.38,39 Combining the CHA reaction with SERS technology and nano-enzymes is expected to construct a novel and highly sensitive ctDNA detection platform for liver cancer.40
In this study, we developed a CHA-based signal amplification system using synthesized Au@Pt@HP1-HP2@Fe3O4 nanozymes for ultrasensitive detection of ctDNA in liver cancer patient serum. As shown in Scheme 1, hairpin DNA1 (HP1) was modified on the surface of platinum-coated gold nanoparticles (Au@Pt) to prepare Au@Pt@HP1. Hairpin DNA2 (HP2) was modified on the surface of Fe3O4 to prepare HP2@ Fe3O4. PIK3CA E542K, as a target, can open its corresponding HP1 by complementary pairing, and HP2 replaces the target to form a large number of HP1-HP2 double-stranded structures, while the replaced ctDNA will continue to participate in the next round of CHA reaction. As the reaction cycle proceeds, more Au@Pt@HP1-HP2@Fe3O4 complex structures are formed with magnetic separation, enzyme-like catalytic activity and SERS-enhancing effect. The complex facilitated the oxidation of colorless TMB by H2O2, producing blue ox-TMB. Quantitative detection of PIK3CA E542K was accomplished by establishing a linear correlation between the SERS signal intensity at ox-TMB’s characteristic peak and the logarithmic concentration of the target ctDNA.
Materials and Methods
Materials
HAuCl4 (≥99.9%), H2PtCl6 (≥99.9%) were purchased from Sinopharm Chemical Reagent Co., Ltd. (China), trisodium citrate (≥99.0%), iron oxide (Fe3O4, ≥90%), acetic acid-sodium acetate buffer (ACS grade), phosphate buffered saline (PBS, molecular biology grade), ethanol (EtOH, ≥99.7%), and 3,3′,5,5′-tetramethylbenzidine (TMB, ≥99%) were purchased from Bioengineering Biotechnology (Shanghai) Co. and used without further purification. The nucleotide sequences were custom-synthesized by Suzhou GeneWise Biotechnology Co. as shown in Table 1, and all experiments were conducted using deionized water with a resistivity exceeding 18.3 MΩ·cm.
 |
Table 1 Nucleotide Sequences Used in the Experiment |
Samples Collection and Processing
Serum samples were collected from 30 healthy volunteers and 30 liver cancer patients at the Affiliated Taizhou Second People’s Hospital of Yangzhou University. The study protocol received ethical approval from the hospital’s Institutional Review Board, and all participants provided written informed consent in compliance with the Declaration of Helsinki guidelines. Following collection, blood samples were immediately centrifuged (12,000 rpm, 10 min, 4°C) to isolate serum, which was subsequently aliquoted and stored at −80 °C until analysis. Table 2 summarizes the demographic and clinical characteristics of all study participants.
 |
Table 2 Statistics of Sample Provider Information |
Synthesis of Platinum-Coated Gold (Au@Pt) and Preparation of Au@Pt@HP1
First, the Au@Pt core-shell NPs were prepared by following a typical procedure with minor modifications.41 In this strategy, Au NPs were synthesized as the core. The aqueous HAuCl4 solution (0.5 mL, 1.0 wt.%) and ultrapure water (50.0 mL) were mixed. Trisodium citrate solution (0.8 mL, 1.0 wt.%) was rapidly injected into the boiling mixture. After the mixture was stirred for 10 min under boiling, ascorbic acid (1.0 mL, 0.1 M) and aqueous H2PtCl6 solution (1.25 mL, 1.0 wt.%) were introduced successively and were boiled for 25 min. The final solution changed from wine-red to brownish-black. The cooled mixture was washed three times by centrifugation at 10000 rpm/min. The Au@Pt was redispersed into ultrapure water and stored at 4 °C until use.
To prepare Au@Pt@HP1, fresh TECP buffer (160 μL, 1 mM) was used to activate H1 (0.1 mM) on Au@Pt through a 12-hour reaction. The mixture was then dispersed in 80 μL of BSA solution (1 wt%) and incubated for 60 minutes, followed by purification at 9000 RPM for 25 minutes. This process yielded the final Au@Pt@HP1 complex.
Synthesis of HP2@Fe3O4
The capture probe was synthesized by modifying HP on the surface of Fe3O4. First, 500 mL of Fe3O4 (0.5 mg/mL) was measured in a test tube, and a magnet was placed at the bottom of the outer surface of the test tube after tilting the tube. 470 mL of PBS solution (10 mM) was added and the above steps were repeated several times after removing the supernatant. The carboxyl groups on the surface of MBs were activated with EDC (5 mL, 0.1 M) and NHS (5 mL, 0.1 M) at room temperature and incubated with shaking (500 rpm, 30 min). A drop of 10 mL of BSA solution (10 wt%) was added to seal the surface sites of Fe3O4. After rinsing with PBS, Fe3O4 was mixed with 470 mL of PBS solution, then TECP-activated HP2 was added and incubated for 12 hours. After repeated washing, the mixture was dispersed in PBS buffer to obtain HP2@Fe3O4.
Optimal Peroxidase-Like Activity of Au@Pt@HP1-HP2@Fe3O4 Under Various Reaction Conditions
To optimize the experimental conditions, the effects of reaction time, pH, TMB concentration, and H2O2 concentration on the SERS signal were systematically investigated. The influence of reaction time was studied by incubating a mixture of 40 mM TMB (40 μL), 10 M H2O2 (100 μL), and Au@Pt@HP1-HP2@Fe₃O₄ (100 μL) in a pH 4.0 buffer (1770 μL) for 0 to 20 minutes, followed by SERS spectra collection of the catalytic product oxTMB. The pH dependence was evaluated by adjusting the buffer pH from 3.0 to 8.0 while maintaining the same reactant concentrations and a 15-minute incubation period. For TMB concentration optimization, TMB solutions ranging from 0.5 to 1.0 mM were prepared in ethanol, mixed with 10 M H2O2 (100 μL) and Au@Pt@HP1@HP2@Fe₃O₄ (100 μL) in pH 4.0 buffer (1770 μL), and incubated for 15 minutes before SERS measurement. Similarly, the effect of H2O2 concentration was examined by varying its concentration from 0.1 to 0.8 mM in the reaction mixture, followed by a 15-minute incubation and SERS spectra acquisition. All SERS spectra of oxTMB were collected using a Raman spectrometer.
SERS Signal Acquisition
The SERS spectra of catalytic product oxTMB were collected using Raman spectrometer after incubation of 100 μL of freshly prepared H2O2 solution (10 M), 40 mM TMB solution (40 μL), and 100 μL of Au@Pt@HP1-HP2@Fe3O4 in acetic acid-sodium acetate buffer (1770 μL) at pH 4.0 for 15 min.
Measurements and Characterization Techniques
The main instruments used in the experiment included scanning electron microscopy (SEM, Hitachi S-4800), transmission electron microscopy (TEM, Philips Tecnai 12), field emission transmission electron microscopy (FE-TEM, FEI Tecnai G2 F30 S-TWIN), UV-Vis spectrophotometer (Cary 5000, Varian), and Raman spectrometer (Renishaw inVia Raman). Microscope). These instruments are used to characterize the morphology and structure of nanomaterials, as well as to perform SERS spectroscopy measurements and analysis. Raman spectra were obtained using a Renishaw inVia microscope with a 5 mW laser. SERS measurements were performed at 785 nm using a 50× objective, with a fixed exposure time of 10s for all experiments.
Results and Discussions
To systematically validate the proposed CHA-nanozyme-SERS integrated strategy (as illustrated in Scheme 1), the experimental results are presented through three hierarchical levels: (1) At the material characterization level, TEM, EDX and etc. analyses confirm the precise assembly of Au@Pt@HP1-HP2@Fe3O4; (2) At the molecular mechanism level, gel electrophoresis and enzyme kinetics verify the synergistic effects between CHA cycling and nanozyme catalysis; (3) At the clinical application level, the high concordance between serum tests from 30 liver cancer patients and qPCR results confirms the method’s reliability. This progressive demonstration directly addresses the two key challenges raised in the Introduction: the sensitivity (LOD=4.12 aM) and specificity of ctDNA detection.
 |
Scheme 1 Schematic representation of the detection principle and process. |
Characterization of Au@Pt@HP1-HP2@Fe3O4
Figure 1A demonstrates that the synthesized Au@Pt nanoparticles exhibit uniform spherical morphology with an average diameter of 55 nm. SEM characterization (Figure 1B) reveals the Fe3O4 microspheres display well-defined spherical structures (200 nm diameter) with excellent size uniformity. This morphological consistency enabled the successful preparation of stable Au@Pt@HP1-HP2@Fe3O4 nanocomposites with controlled particle size distribution. The composite structure Au@Pt@HP1 was observed. Au@Pt was homogeneously dispersed on the surface of HP2@Fe3O4 with uniform morphology, structural integrity, and good dispersion, which had an extremely strong SERS enhancement effect (Figure 1C–F). HRTEM images of the Au@Pt@HP1 surface showing clear lattice fringes with a layer spacing of 0.24 nm corresponding to the {111} facets of Au and Pt are shown in Figure 2G. The SAED patterns of Au@Pt@HP1-HP2@Fe3O4 are shown in Figure 2H. The characteristic peak intensity of TMB at 1607 cm−1 was selected to study the SERS enhancement effect of Au@Pt@HP1-HP2@Fe3O4. As shown in Figure 2I, free TMB (1 mM) exhibited only baseline Raman signals, whereas the TMB@Au@Pt@HP1-HP2@Fe3O4 complex at 1 nM concentration generated intense characteristic peaks with significant signal enhancement. This dramatic improvement in SERS response demonstrates the excellent plasmonic activity of our Au@Pt@HP1-HP2@Fe3O4 nanocomposite system.
 |
Figure 2 Elemental analysis diagram for Au@Pt@HP1@HP2@Fe3O4. (A) HAADF-STEM images of Au@Pt@HP1-HP2@Fe3O4. (B–E) elemental mappings of Au@Pt@HP1-HP2@Fe3O4. (F) EDX spectra of the Au@Pt@HP1-HP2@Fe3O4. |
The HAADF-STEM image in Figure 2A clearly shows the structure of Au@Pt@HP1-HP2@Fe3O4. Figure 2B–E forms composite Au (blue), Fe (orange), Pt (green) and O (red) elemental maps to further show the specific elemental arrangement of Au@Pt@HP1-HP2@Fe3O4 and its structure. Platinum is densely coated on the surface of the gold particles in the form of granules. Figure 2F shows the EDX spectrum of Au@Pt@HP1-HP2@Fe3O4, which reveals that the complex contains the elements Au, Pt, Fe, and O. Among them, Au and Pt are composed of Au@Pt, Fe and O are mainly from Fe3O4, while the Cu peak is caused by the copper mesh that carries the sample.
Evaluation of CHA Reaction
To validate the CHA reaction for ctDNA detection and assess its performance, we conducted gel electrophoresis analysis using PIK3CA E542K as the model target (Figure 3). In lane 5, the appearance of HP1-HP2 was observed in the presence of PIK3CA E542K along with HP1 and HP2. Formation of the HP1-PIK3CA E542K product and release of the target strand were clearly observed when PIK3CA E542K and HP1 were placed in lane 6. The above experimental results indicate that CHA reactions were successfully performed in this study.
 |
Figure 3 Validation of CHA reactions by gel electrophoresis. Lane 1: Marker; Lane 2: PIK3CA E542K; Lane 3: HP1; Lane 4: HP1+HP2; Lane 5: HP1+HP2+PIK3CA E542K; Lane 6: HP1+PIK3CA E542K. |
Experimental Optimization
Reaction conditions play a crucial role in the activity of Au@Pt@HP1-HP2@Fe3O4. Usually, the influencing factors of enzymatic reaction include temperature, pH and substrate concentration. Firstly, the effect of reaction time was investigated, as shown in Figure 4A. With the increase of time, the Raman characteristic peak intensity of ox-TMB at 1607 cm−1 gradually increased, and basically stopped changing after 15 min, Due to the instability of the oxTMB signal, the SERS signal gradually weakened after 15 min.42,43 Therefore, the optimal reaction time was set at 15 min. The effect of pH on the SERS signals was shown in Figure 4B. The peroxidase-like activity of Au@Pt@HP1-HP2@Fe₃O₄ nanocomposites exhibited strong pH dependence in the H2O2-TMB system. Quantitative SERS analysis revealed optimal catalytic performance at pH 4.0, with signal intensity increasing progressively from pH 3 to 4, then decreasing significantly across the pH 5–8 range. This pH-activity profile aligns with established literature reports demonstrating accelerated TMB oxidation kinetics in weakly acidic conditions (pH 3–5) compared to neutral or alkaline environments.44,45 By analyzing the effect of H2O2 concentration on the catalytic activity of Au@Pt@HP1-HP2@Fe3O4, the results are shown in Figure 4C. The relative activity of H2O2 concentration in the range of 0.1–0.5 mM showed an increasing trend, whereas it began to decrease at a concentration greater than 0.5 mM.46 It indicates that the catalytic activity has reached saturation at H2O2 concentration of 0.5 mM. In order to determine the effect of the concentration of TMB on the reaction, the concentration of TMB was investigated in the range of 0.5 to 1.0 mM. The results are shown in Figure 4D. The SERS signal reached its highest value at a TMB concentration of 0.8 mM. This is mainly due to the poor solubility of TMB in aqueous solution.47 Therefore, 0.8 mM was chosen as the optimal concentration. In summary, the optimal reaction time, pH, TMB concentration and H2O2 concentration were 15 min, 4, 0.8 mM and 0.5 mM, respectively.
 |
Figure 4 Optimization of H2O2 detection using Au@Pt@HP1-HP2@Fe3O4 in the presence of TMB. (A) Incubation time, (B) solution pH, (C) H2O2 concentration, (D) TMB concentration. |
Enzyme Catalytic Kinetics
The peroxidase-mimetic activity of Pt@Au@HP1-HP2@Fe3O4was evaluated using the H2O2 -TMB catalytic system. Upon simultaneous addition of both H2O2 and Pt@Au@HP1-HP2@Fe3O4 to the TMB solution, an immediate color transition from colorless to blue was observed (Figure 5A), indicating rapid TMB oxidation. UV-vis spectroscopy confirmed this activity through the appearance of a characteristic absorption peak at 651 nm (Figure 5B), corresponding to the oxidized TMB product (oxTMB). These results unequivocally demonstrate the intrinsic peroxidase-like catalytic capability of the Pt@Au@HP1-HP2@Fe3O4 nanocomposite. The effect of different TMB concentrations on the catalytic reaction in the presence of Au@Pt@HP1-HP2@Fe3O4 was investigated. As shown in the Figure 5C, the absorbance increases with time at different TMB concentrations, and it is clear that the reaction speed increases with increasing TMB concentration. In addition, we evaluated the Michaelis-Menten kinetics of Au@Pt@HP1-HP2@Fe3O4 to gain further insight into its catalytic performance (Figure 5D). Compared with other reported nanozymes,48 Au@Pt@HP1-HP2@Fe3O4 exhibited a relatively lower Km (0.4089 mM) and higher Vmax (0.9533 μM/s), indicating that Au@Pt@HP1-HP2@Fe3O4 has desirable catalytic properties, It has a good affinity for the substrate.
Performance Evaluation
The magnetic properties of the Au@Pt@HP1-HP2@Fe3O4 nanocomposites were systematically characterized to assess their applicability in SERS-based detection. When exposed to an external magnetic field, the composites were quickly drawn from the solution and aggregated, while the supernatant no longer catalyzed TMB (Figure 6A). This highlights the strong magnetic properties of Au@Pt@HP1-HP2@Fe3O4. The rapid magnetic separation capability simplifies washing and product isolation, enhancing assay efficiency and sensitivity. The system demonstrated excellent SERS signal reproducibility, with oxTMB exhibiting consistent Raman intensities (RSD = 4.1%) across multiple measurements (n=10) over 24 hours (Figure 6B and C). This remarkable stability highlights the robustness of the Au@Pt@HP1-HP2@Fe3O4 platform for quantitative analysis. To verify the homogeneity, SERS spectra of 10 randomly selected points on the same composite material were measured to evaluate uniformity (Figure S1). The results show that the peak intensities at all points are relatively consistent, and the relative standard deviation (RSD) at 1607 cm⁻¹ is 7.88% (Figure S2), indicating good homogeneity. Subsequently, five batches of composites were prepared at different times for the detection of PIK3CA E542K solution. The corresponding SERS spectra are shown in Figure 6D. It can be seen that the spectral waveforms are basically the same with almost no significant difference, and the RSD value of the SERS signal intensity of the characteristic peak at 1607 cm−1 is 4.26%, which demonstrates that the composite material has a good reproducibility and strengthens the confidence of the determination. In addition, the stability of the composite was explored. Over time, the SERS spectral profile showed no notable changes, with only a minor reduction in intensity. The signal stabilized after 6 days and maintained 91.72% of its original intensity even after 18 days (Figure 6E). This sustained signal demonstrates the composite’s long-term stability for assay applications. To evaluate the specificity, experiments included interference sequences such as a single-base mismatch (MT1), a three-base mismatch (MT3), and a random sequence. As shown in Figure 6F, the I1607/I1183 ratio of PIK3CA E542K is much larger than the signal intensity of the interfering sequences, which is the result of the specific binding of HP1 to PIK3CA E542K.
Quantitative Testing
The assay’s sensitivity is vital for detecting low-abundance biomarkers, particularly in early-stage cancer diagnosis. To evaluate this, varying concentrations of PIK3CA E542K were spiked into serum samples, and the SERS signals were analyzed (Figure 7A). The results revealed a gradual decline in SERS intensity as the concentration increased. A strong linear correlation was observed between the logarithm of PIK3CA E542K concentration and the peak intensity at 1607 cm⁻¹, described by the equation: y = 4985.19x - 2821.85 (R² = 0.9928) (Figure 7B). The LOD was calculated based on the characteristic peaks of the SERS spectra using the following equation: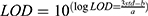 where a and b were the variables obtained with a linear regression of the signal-concentration curve, SD was the standard deviation and Cblank is the SERS intensity of the blank sample.49 The detection limit of PIK3CA E542K was calculated to be 4.12 aM. This sensitivity places the proposed SERS microfluidic chip among the most advanced methods currently available (Table 3).
where a and b were the variables obtained with a linear regression of the signal-concentration curve, SD was the standard deviation and Cblank is the SERS intensity of the blank sample.49 The detection limit of PIK3CA E542K was calculated to be 4.12 aM. This sensitivity places the proposed SERS microfluidic chip among the most advanced methods currently available (Table 3).
 |
Table 3 Comparison of the Proposed Method with Currently Reported Methods |
Characterization of Clinical Samples
Magnetic resonance imaging (MRI) can provide multi-parameter, multi-sequence, and multi-directional images to evaluate the extent of liver cancer lesions. Due to its excellent contrast resolution for liver tissue, MRI can accurately and meticulously display the anatomical structure of the liver and its pathological features. Axial plane images can clearly demonstrate the characteristics of liver cancer (Figure 8A–C and 8E–G). On non-contrast T1-weighted sequences, liver cancer typically appears as hypointense or isointense, while on post-contrast scans, it shows significant enhancement in the arterial phase and a “wash-in and wash-out” pattern in the portal venous phase. Coronal plane images can clearly reveal the size, boundaries of the tumor, and its invasion into surrounding tissues or blood vessels (Figure 8D and H). The diagnosis of liver cancer primarily relies on pathological examination. Figure 8I–K shows pathological images of liver cancer, where disordered arrangement of tumor cells can be observed, with large and irregular nuclei, reduced cytoplasm, prominent nucleoli, and areas of necrosis or hemorrhage in some regions. The markedly abnormal cells exhibit an increased nuclear-to-cytoplasmic ratio, unclear intercellular connections, and areas of fibrous tissue hyperplasia or inflammatory cell infiltration.
 |
Figure 8 Pathologic findings in patients with liver cancer. (A–H) MRI images of two patients with liver cancer. (I–K) Pathological HE staining of liver cancer tissues. |
Real Sample Analysis
To further test the reliability and accuracy of this SERS microfluidic chip in the analysis of clinical samples, it was utilized to detect the expression levels of PIK3CA E542K in serum samples from 30 healthy subjects and 30 liver cancer patients. The obtained spectra were processed to obtain the average spectra (Figure 9A), corresponding to the SERS signal intensities at 1607 cm−1 as shown in Figure 9B. It can be seen that the expression level of PIK3CA E542K was significantly elevated in the serum of liver cancer patients compared with that of healthy subjects. After that, the SERS signal intensity at 1607 cm−1 in the SERS spectrum was substituted into the linear regression equation to calculate the expression level of PIK3CA E542K. The accuracy of this assay was verified by comparing the results with those of the qRT-PCR assay (Table 4). The results showed that the assay was highly consistent with the results of qRT-PCR assay and had good detection accuracy.
 |
Table 4 The Results of SERS and qRT-PCR for Clinical Samples |
Conclusion
In this study, a highly sensitive method for the detection of ctDNA in the serum of liver cancer patients was successfully developed based on the CHA signal amplification strategy using the synthesized Au@Pt@HP1-HP2@Fe3O4 nano-enzyme complex. By modifying HP1 on the surface of Au@Pt and (HP2 with Fe3O4 magnetic beads, a multifunctional detection platform with magnetic separation, catalytic activity of the nano-enzymes, and SERS-enhanced effect was constructed. PIK3CA E542K was used as a target, and complementary pairing of HP1 and HP2 was triggered by CHA reaction. A large number of HP1-HP2 double-stranded structures were formed, while the released ctDNA continued to participate in the next round of cyclic reaction to achieve signal amplification. In comparison with existing detection technologies, we have for the first time organically integrated three techniques: catalytic hairpin assembly (CHA) cyclic amplification, gold-platinum nanozyme catalysis, and magnetic aggregation-based surface-enhanced Raman scattering (SERS), to construct a novel nanozyme-SERS detection platform that achieves multilevel signal amplification. The complex catalyzed the oxidation of colorless TMB by H2O2 to generate blue ox-TMB. A linear relationship between ctDNA concentration and signal intensity was established by detecting the signal intensity of the SERS characteristic peak of ox-TMB. The results showed that the detection limit of this method for PIK3CA E542K was as low as 4.12 aM, 2 orders of magnitude improvement over existing detection technologies. The assay can be completed in just 15 minutes, much faster than the hour-long cycle time of conventional sequencing methods. In addition, the platform exhibits excellent reproducibility, stability and specificity. Analysis of the clinical samples showed that the expression level of PIK3CA E542K in the serum of liver cancer patients was significantly higher than that of healthy subjects, and the detection results were highly consistent with qRT-PCR. In conclusion, the SERS microfluidic chip developed in this study combined with the CHA signal amplification strategy can efficiently and accurately determine the expression level of ctDNA, which provides a new technical means for the early diagnosis of liver cancer, and has an important potential for clinical application.
Acknowledgments
This study was financially supported by grants from the Social Development Foundation of Taizhou (TS202225); and the Key Research Institute of State Administration of Traditional Chinese Medicine (202259); General Program of Nantong Municipal Health Commission Research Project (MS2024111, MS2024112); Nantong University Special Research Fund for Clinical Medicine (2024LZ028).
Disclosure
There are no conflicts of interest in this study to declare.
References
1. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188314. doi:10.1016/j.bbcan.2019.188314
2. Yao Y, Hu Y, Meng X, et al. Dual-targeting liposomal delivery of sorafenib and docetaxel for enhanced synergistic therapy in advanced hepatocellular carcinoma. Pharm Sci Adv. 2024;2:100046. doi:10.1016/j.pscia.2024.100046
3. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. doi:10.1186/s12943-019-1043-x
4. Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31(2):173–187. doi:10.1055/s-0031-1276646
5. Yang S, Qian L, Li Z, et al. Integrated multi-omics landscape of liver metastases. Gastroenterology. 2023;164(3):407–423.e417. doi:10.1053/j.gastro.2022.11.029
6. Zhang A, Lakshmanan J, Motameni A, Harbrecht BG. MicroRNA-203 suppresses proliferation in liver cancer associated with PIK3CA, p38 MAPK, c-Jun, and GSK3 signaling. Mol Cell Biochem. 2018;441(1–2):89–98. doi:10.1007/s11010-017-3176-9
7. Kalil JA, Krzywon L, Petrillo SK, et al. Feasibility of ctDNA in detecting minimal residual disease and predicting recurrence for colorectal cancer liver metastases. Front Oncol. 2024;14:1418696. doi:10.3389/fonc.2024.1418696
8. Sun Y, Kong X, Yu J, et al. Characterization of genomic clones by targeted deep sequencing of ctDNA to monitor liver cancer. Transl Cancer Res. 2021;10(10):4387–4402. doi:10.21037/tcr-21-1005
9. Zhang L, Chen J, He M, Su X. Molecular dynamics simulation-guided toehold mediated strand displacement probe for single-nucleotide variants detection. Exploration. 2022;2(1):20210265. doi:10.1002/EXP.20210265
10. Li R, Liu X, Chen J. Opportunities and challenges of hole transport materials for high-performance inverted hybrid-perovskite solar cells. Exploration. 2023;3(3):20220027. doi:10.1002/EXP.20220027
11. Beeram R, Vepa KR, Soma VR. Recent trends in SERS-based plasmonic sensors for disease diagnostics, biomolecules detection, and machine learning techniques. Biosensors. 2023;13(3). doi:10.3390/bios13030328
12. Liu H, Gao X, Xu C, Liu D. SERS tags for biomedical detection and bioimaging. Theranostics. 2022;12(4):1870–1903. doi:10.7150/thno.66859
13. Lee S, Dang H, Moon JI, et al. SERS-based microdevices for use as in vitro diagnostic biosensors. Chem Soc Rev. 2024;53(11):5394–5427. doi:10.1039/d3cs01055d
14. Kukkar D, Chhillar M, Kim KH. Application of SERS-based nanobiosensors to metabolite biomarkers of CKD. Biosens Bioelectron. 2023;232:115311. doi:10.1016/j.bios.2023.115311
15. Huang Z, Peng J, Xu L, Liu P. Development and application of surface-enhanced Raman scattering (SERS). Nanomaterials. 2024;14(17):1417. doi:10.3390/nano14171417
16. Andreiuk B, Nicolson F, Clark LM, et al. Design and synthesis of gold nanostars-based SERS nanotags for bioimaging applications. Nanotheranostics. 2022;6(1):10–30. doi:10.7150/ntno.61244
17. Shin H, Choi BH, Shim O, et al. Single test-based diagnosis of multiple cancer types using exosome-SERS-AI for early stage cancers. Nat Commun. 2023;14(1):1644. doi:10.1038/s41467-023-37403-1
18. Sun T, Lin Y, Yu Y, et al. Low-abundance proteins-based label-free SERS approach for high precision detection of liver cancer with different stages. Anal Chim Acta. 2024;1304:342518. doi:10.1016/j.aca.2024.342518
19. Jiang H, Qian C, Deng Y, et al. Novel multimode assay based on asymmetrically competitive CRISPR and Raman barcode spectra for multiple hepatocellular carcinoma biomarkers detection. Anal Chem. 2024;96(50):20004–20014. doi:10.1021/acs.analchem.4c04593
20. Tao H, Weng S, Xu L, et al. Target-triggered assembly of plasmon resonance nanostructures for quantitative detection of lncRNA in liver cancer cells via surface enhanced Raman spectroscopy. Biosens Bioelectron. 2024;261:116488. doi:10.1016/j.bios.2024.116488
21. Ai Y, Tian Y, Qiao J, Wang C, Li H. “Yin-Yang philosophy” for the design of anticancer drug delivery nanoparticles. Biomater Transl. 2024;5(2):144–156. doi:10.12336/biomatertransl.2024.02.005
22. Qi L, Zhao T, Yan J, et al. Advances in magnesium-containing bioceramics for bone repair. Biomater Transl. 2024;5(1):3–20. doi:10.12336/biomatertransl.2024.01.002
23. Jia G, Jiang Y, Li X. Targeted drug conjugates in cancer therapy: challenges and opportunities. Pharm Sci Adv. 2024;2:100048. doi:10.1016/j.pscia.2024.100048
24. Ansari SA, Husain Q. Potential applications of enzymes immobilized on/in nano materials: a review. Biotechnol Adv. 2012;30(3):512–523. doi:10.1016/j.biotechadv.2011.09.005
25. Ramya RK, Theraka K, Ramprasadh SV, et al. Pragmatic treatment strategies for polyaromatic hydrocarbon remediation and anti-biofouling from surfaces using nano-enzymes: a review. Appl Biochem Biotechnol. 2023;195(9):5479–5496. doi:10.1007/s12010-022-03848-1
26. Jacob E, Mathew D, Benny L, Varghese A. Emerging nanomaterials as versatile nanozymes: a new dimension in biomedical research. Top Curr Chem. 2024;382(3):28. doi:10.1007/s41061-024-00473-w
27. Li J, Cai X, Jiang P, et al. Co-based nanozymatic profiling: advances spanning chemistry, biomedical, and environmental sciences. Adv Mater. 2024;36(8):e2307337. doi:10.1002/adma.202307337
28. Zhang T, Zhu S, Wang J, et al. Construction of a novel nano-enzyme for ultrasensitive glucose detection with surface-enhanced Raman scattering. Spectrochim Acta A Mol Biomol Spectrosc. 2023;291:122307. doi:10.1016/j.saa.2022.122307
29. Yao D, Wen G, Gong L, Li C, Liang A, Jiang Z. A Highly Sensitive SERS and RRS Coupled Di-Mode Method for CO Detection Using Nanogolds as Catalysts and Bifunctional Probes. Nanomaterials. 2020;10(3):450. doi:10.3390/nano10030450
30. Engelbrekt C, Šešelj N, Poreddy R, Riisager A, Ulstrup J, Zhang J. Atomically thin Pt shells on Au nanoparticle cores: facile synthesis and efficient synergetic catalysis. J Mater Chem A. 2016;4(9):3278–3286. doi:10.1039/C5TA08922K
31. Zhang Y, Janyasupab M, Liu C-W, et al. Improvement of amperometric biosensor performance for H2O2 detection based on bimetallic PtM (M = Ru, Au, and Ir) nanoparticles. Int J Electrochem. 2012;2012(1):410846. doi:10.1155/2012/410846
32. Lee J-W, Son J, Yoo K-M, Lo YM, Moon B. Characterization of the antioxidant activity of gold@platinum nanoparticles. RSC Adv. 2014;4(38):19824–19830. doi:10.1039/c4ra01765j
33. He W, Liu Y, Yuan J, et al. Au@Pt nanostructures as oxidase and peroxidase mimetics for use in immunoassays. Biomaterials. 2011;32(4):1139–1147. doi:10.1016/j.biomaterials.2010.09.040
34. Zhang B, Lv Y, Yu C, et al. Au-Pt nanozyme-based multifunctional hydrogel dressing for diabetic wound healing. Biomater Adv. 2022;137:212869. doi:10.1016/j.bioadv.2022.212869
35. Pathania H, Chauhan P, Chaudhary V, et al. Engineering core-shell mesoporous silica and Fe(3)O(4)@Au nanosystems for targeted cancer therapeutics: a review. Biotechnol Genet Eng Rev. 2024;40(4):3653–3681. doi:10.1080/02648725.2022.2147685
36. Abolhasan R, Mehdizadeh A, Rashidi MR, Aghebati-Maleki L, Yousefi M. Application of hairpin DNA-based biosensors with various signal amplification strategies in clinical diagnosis. Biosens Bioelectron. 2019;129:164–174. doi:10.1016/j.bios.2019.01.008
37. Hua Y, Ma J, Li D, Wang R. DNA-based biosensors for the biochemical analysis: a review. Biosensors. 2022;12(3):183. doi:10.3390/bios12030183
38. Xing C, Dai J, Huang Y, et al. Active self-assembly of train-shaped DNA nanostructures via catalytic hairpin assembly reactions. Small. 2019;15(27):e1901795. doi:10.1002/smll.201901795
39. Bai Y, Xu P, Li S, et al. Signal amplification strategy of DNA self-assembled biosensor and typical applications in pathogenic microorganism detection. Talanta. 2024;272:125759. doi:10.1016/j.talanta.2024.125759
40. Wang S, Guang J, Gao Y, et al. Fluorescent DNA tetrahedral probe with catalytic hairpin self-assembly reaction for imaging of miR-21 and miR-155 in living cells. Mikrochim Acta. 2024;191(8):462. doi:10.1007/s00604-024-06529-4
41. Zhang M, Bu T, Bai F, et al. Gold nanoparticles-functionalized three-dimensional flower-like manganese dioxide: a high-sensitivity thermal analysis immunochromatographic sensor. Food Chem. 2021;341:128231. doi:10.1016/j.foodchem.2020.128231
42. Josephy PD, Eling T, Mason RP. The horseradish peroxidase-catalyzed oxidation of 3,5,3’,5’-tetramethylbenzidine. Free radical and charge-transfer complex intermediates. J Biol Chem. 1982;257(7):3669–3675. doi:10.1016/S0021-9258(18)34832-4
43. Wan J, Liu Q, Tang P, et al. SERS-based error calibration of a TMB-H(2)O(2) colorimetric system. Analyst. 2023;148(4):869–875. doi:10.1039/D2AN01914K
44. Tang M, Ni J, Yue Z, et al. Polyoxometalate-nanozyme-integrated nanomotors (POMotors) for self-propulsion-promoted synergistic photothermal-catalytic tumor therapy. Angew Chem Int Ed Engl. 2024;63(6):e202315031. doi:10.1002/anie.202315031
45. Yue Z, Li J, Tang M, Sun T, Chen C, Wu Z. Nanozyme-based clusterphene for enhanced electrically catalytic cancer therapy. Adv Healthc Mater. 2024;13(9):e2303222. doi:10.1002/adhm.202303222
46. He W, Han X, Jia H, Cai J, Zhou Y, Zheng Z. AuPt alloy nanostructures with tunable composition and enzyme-like activities for colorimetric detection of bisulfide. Sci Rep. 2017;7:40103. doi:10.1038/srep40103
47. Pham XH, Seong B, Bock S, et al. Nonenzymatic hydrogen peroxide detection using surface-enhanced Raman scattering of gold-silver core-shell-assembled silica nanostructures. Nanomaterials. 2021;11(10).
48. Luo S, Liu Y, Rao H, Wang Y, Wang X. Fluorescence and magnetic nanocomposite Fe(3)O(4)@SiO(2)@Au MNPs as peroxidase mimetics for glucose detection. Anal Biochem. 2017;538:26–33. doi:10.1016/j.ab.2017.09.006
49. Mukanova Z, Gudun K, Elemessova Z, Khamkhash L, Ralchenko E, Bukasov R. Detection of paracetamol in water and urea in artificial urine with gold nanoparticle@Al foil cost-efficient SERS substrate. Anal Sci. 2018;34(2):183–187. doi:10.2116/analsci.34.183
50. Chen C, He R, Zhang Z, Chen Y. Dual-recognition-based determination of ctDNA via the clamping function of peptide nucleic acid and terminal protection of small-molecule-linked DNA. Analyst. 2020;145(23):7603–7608. doi:10.1039/D0AN01305F
51. Wang K, Lin X, Zhang M, et al. ACEK biosensor for the minute-scale quantification of breast cancer ctDNA. Sensors. 2024;24(2).
52. Sun Y, He S, Peng Y, Liu M, Xu D. A novel label-free capillary electrophoresis LED-induced fluorescence platform based on catalytic hairpin assembly for sensitive detection of multiple circulating tumor DNA. Analyst. 2024;149(5):1548–1556. doi:10.1039/D3AN01993D
53. Uygun ZO, Yeniay L, Gi Rgi NSF. CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs. Anal Chim Acta. 2020;1121:35–41. doi:10.1016/j.aca.2020.04.009
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Combined SERS Microfluidic Chip with Gold Nanocone Array for Effective Early Lung Cancer Prognosis in Mice Model
Qian Y, Gu Y, Deng J, Cai Z, Wang Y, Zhou R, Zhu D, Lu H, Wang Z
International Journal of Nanomedicine 2023, 18:3429-3442
Published Date: 23 June 2023






