Back to Journals » International Journal of Nanomedicine » Volume 19
Cinobufagin Suppresses Lipid Peroxidation and Inflammation in Osteoporotic Mice by Promoting the Delivery of miR-3102-5p by Macrophage-Derived Exosomes
Authors Geng Z , Sun T, Yu J, Wang N, Jiang Q, Wang P, Yang G, Li Y, Ding Y, Zhang J, Lin G, Zhao Y
Received 22 June 2024
Accepted for publication 10 October 2024
Published 18 October 2024 Volume 2024:19 Pages 10497—10512
DOI https://doi.org/10.2147/IJN.S483849
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. RDK Misra
Zixiang Geng,1,2,* Tiancheng Sun,1,2,* Jie Yu,1,2,* Ning Wang,1,2 Qiang Jiang,1,2 Peige Wang,1,2 Guangyue Yang,1,2 Yifei Li,3 Yue Ding,4 Jiange Zhang,5 Guoqiang Lin,5 Yongfang Zhao1,2
1Shi’s Center of Orthopedics and Traumatology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China; 2Institute of Traumatology and Orthopedics, Shanghai Academy of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China; 3Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China; 4School of Pharmacy, Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China; 5The Research Center of Chiral Drugs, Innovation Research Institute of Traditional, Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zixiang Geng, Institute of Traumatology and Orthopedics, Shanghai Academy of Traditional Chinese Medicine, Shanghai, People’s Republic of China, Email [email protected] Yongfang Zhao, Shi’s Center of Orthopedics and Traumatology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China, Email [email protected]
Background: Cinobufagin, the primary active compound in toad venom, is commonly used for anti-tumor, anti-inflammatory, and analgesic purposes. However, its specific bone-protective effects remain uncertain. This research aims to ascertain the bone-protective properties of cinobufagin and investigate underlying mechanisms.
Methods: Mice were ovariectomized to establish an osteoporosis model, followed by intraperitoneal injections of cinobufagin and cinobufagin-treated RAW.264.7-derived exosomes for therapy. MicroCT, HE staining, and TRAP staining were employed to evaluate bone mass and therapeutic outcomes, while mRNA sequencing and immunoblotting were utilized to assess markers of bone metabolism, inflammation, and lipid peroxidation. Osteoblast and osteoclast precursor cells were differentiated to observe the impact of cinobufagin-treated exosomes derived from RAW264.7 cells on bone metabolism. Exosomes characteristics were studied using transmission electron microscopy and particle size analysis, and miRNA binding targets in exosomes were determined by luciferase reporting.
Results: In ovariectomized mice, cinobufagin and cinobufagin-treated exosomes from RAW264.7 cells increased trabecular bone density and mass in the femur, while also decreasing inflammation and lipid peroxidation. The effect was reversed by an exosomes inhibitor. In vitro experiments revealed that cinobufagin-treated exosomes from RAW264.7 cells enhanced osteogenic and suppressed osteoclast differentiation, possibly linked to Upregulated miR-3102-5p in RAW-derived exosomes. MiR-3102-5p targets the 3′UTR region of alox15, thereby suppressing its expression and reducing the lipid peroxidation process in osteoblasts.
Conclusion: Overall, this study clarified cinobufagin’s bone-protective effects and revealed that cinobufagin can enhance the delivery of miR-3102-5p targeting alox15 through macrophage-derived exosomes, demonstrating anti-lipid peroxidation and anti-inflammatory effects.
Keywords: osteoporosis, cinobufagin, exosomes, miR-3102-5p
Graphical Abstract:

Introduction
Osteoporosis is a chronic metabolic bone disease characterized by chronic loss of trabecular bone as well as increased susceptibility to bone fracture and influenced by factors like menopause, aging, and tumors.1,2 Tumor-induced bone mass reduction is commonly linked to either the primary disease itself or treatments like bone metastasis, glucocorticoid therapy, hormone deprivation therapy, and chemotherapy-induced ovarian failure, all contributing to bone loss.3–5 However, the focus on tumor-targeting therapies often overlooks the significant impact of heightened fracture risks on patients’ quality of life. Therefore, the exploration of anti-tumor drugs with potential bone-protective properties is crucial for enhancing patients’ well-being, particularly those with hematologic systems or bone tumors. The underlying mechanism of osteoporosis stems from the imbalance between bone formation regulated by osteoblasts and bone resorption controlled by osteoclasts.6 The tumor-mediated microenvironment can directly or indirectly generate numerous inflammatory factors such as IL-6 and TNF, which not only facilitate tumor survival and metastasis but also stimulate osteoclast activity by engaging the RANKL/RANK signaling pathway.7 Prolonged chronic inflammation triggers reactive oxygen species build-up, prompting osteoblast apoptosis and subsequent bone loss.8 Consequently, mitigating chronic inflammation and oxidative stress is fundamental in disrupting the tumor microenvironment and managing bone loss.
Currently, natural products serve as a primary reservoir of active pharmaceutical ingredients and stand as pivotal avenues for drug development.9,10 Cinobufagin, an extract derived from the skin and parotid glands of toads, harbors a key active component extensively utilized in clinical settings for its anti-tumor, anti-inflammatory, and analgesic properties.11,12 Research indicates that multiple myeloma severity manifests in heightened osteoclast formation, escalated bone resorption, and diminished bone formation,13 whereas Cinobufagin exhibits the potential to induce apoptosis in multiple myeloma cells through the activation of the ERK, JNK, and p38MAPK pathways.14 Furthermore, Cinobufagin demonstrates the ability to curb inflammatory responses and bone cancer-induced pain sensitization by elevating the IL10/β-endorphin signaling pathway.12 Although the anti-tumor prowess of Cinobufagin is well-established, its specific impact on bone metabolism remains ambiguous. Thus, elucidating the bone-protective attributes of Cinobufagin for individuals battling cancer is imperative.
Exosomes, ranging in size from approximately 40–150nm, are secreted by diverse cells to facilitate intercellular communication through the carriage of bioactive substances such as RNA, DNA, proteins, lipids, and metabolites.15,16 These vesicles play crucial roles in the initiation and advancement of various cancers, orchestrating processes like bone metastasis, tumor microenvironment remodeling, angiogenesis, invasion, metastasis, and resistance to treatment.17–19 Specifically, exosomes originating from bone tumors can trigger osteoclast differentiation by targeting COL1A1 with transferred miRNA-92a-1-5p while concurrently impeding osteoblast differentiation.20 Furthermore, these vesicles are implicated in promoting estrogen receptor-positive breast cancer bone metastasis by cooperatively transferring miR-19a and IBSP.21 Cross-talk via exosomes among bone cells significantly impacts the regulation of bone metabolism. For instance, exosomes released from bone marrow mesenchymal stem cells modulate the Wnt pathway by targeting WIF1 and SMAD7 through delivered miR-424-5p and miR-21 to suppress bone formation.22,23 Additionally, exosomes transferred from bone marrow mesenchymal stem cells to osteoclasts release miR-143/145, targeting Cd226 and Srgap2, thereby enhancing osteoclast activity. Notably, osteoclasts derived from bone marrow monocytes secrete exosomes containing miR-324, which foster osteoblast differentiation in bone marrow mesenchymal stem cells.24 Recent investigations highlight the immunomodulatory impact of Cinobufagin on monocytes,25 indicating that macrophage-derived exosomes (RAW-Exos) could serve as effective carriers for immunomodulation,26 delivering active components to instigate anti-inflammatory responses and potentially elucidating the bone-protective properties of Cinobufagin.27,28
Upon our existing advancements,29,30 this study investigates the potential effects of cinobufagin on osteoporosis using the classical ovariectomy model, aiming to uncover its underlying mechanisms through exosomes exploration.
Methods
Animal Model Construction and Intervention
The animal model was established based on a previously published method.31,32 Briefly, 50 female C57BL/6 mice, aged 6–8 weeks, were procured from the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. These mice were randomly allocated into 5 groups, each group consisting of 10 mice. In the control group (Con), the periovarian fat tissue of the mice was excised, while in the other groups, both ovaries were surgically removed. Subsequently, two weeks post-ovariectomy, the model group (OVX) received intraperitoneal injections of PBS solution (200 µL) thrice a week for 8 weeks. The treatment group (Cinobufagin) was administered intraperitoneal injections of Cinobufagin (7.5 mg/kg) thrice a week for 8 weeks. The exosomes inhibitor group (Cinobufagin-GW4869) received intraperitoneal injections of Cinobufagin (7.5 mg/kg) thrice a week for 8 weeks, with a pre-treatment injection of the exosomes inhibitor GW4869 (2.5 µg/g). Lastly, the exosomes group (RAW-Exos) was intraperitoneally injected with RAW264.7 cell-derived exosomes treated with Cinobufagin (1 µM) (100 µg/mouse) thrice a week for 8 weeks. 4 mice were used for transcriptome sequencing, with 1 designated as a supplementary quality control measure. 3 mice for microCT and pathological examinations, and 3 mice for molecular biology analyses. All procedures were approved by the Ethics Committee of Shanghai University of Traditional Chinese Medicine (Approval No.: PZSHUTCM2305150001). All experimental procedures were strictly performed according to the Guidelines for Ethical Review of Laboratory Animal Welfare in China (GB/T3589-2018).
Bone Microarchitecture Analysis
Mouse femurs were fixed in 4% paraformaldehyde for 48 hours and scanned using a Quantum GX2 (PerkinElmer) micro-CT scanner at 90 kV, 88 μA, 18 FOV, and 36 VoxeSize for high-resolution scanning over 14 minutes. A total of 50 slices were selected from the distal to the proximal end of the femur for 3D reconstruction, visualized in coronal, sagittal, and transverse planes. The bone microarchitecture analysis comprised the evaluation of bone mineral density (BMD), bone volume (BV), bone volume fraction (BV/TV), bone surface area to bone volume ratio (BS/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), cortical bone thickness (Ct.Th), and cortical bone area fraction (Ct.Ar/Tt.Ar).
Enzyme-Linked Immunosorbent Assay (ELISA) for Serum P1NP and β-CTX Levels
Serum P1NP and β-CTX are sensitive indicators of bone metabolism, where P1NP signifies bone resorption and β-CTX denotes bone formation. Serum P1NP and β-CTX levels were measured using ELISA following the manufacturer’s instructions (YS-MY3126, Y-S Biotechnology). Briefly, serum samples were dispensed into enzyme-labeled plates and incubated at 37°C for 30 minutes, followed by rigorous washing steps. Then, the reaction reagent was added and incubated for another 30 minutes at 37°C, followed by thorough washing. Subsequently, the color development solution was added and left to incubate at 37°C for 10 minutes before measuring the optical density (OD) value using a microplate reader.
Exosomes Tracking
In accordance with the manufacturer’s instructions, Exosomes were labeled with DiD-Red, featuring an excitation wavelength of 644 nm and an emission wavelength of 665 nm. In vivo, after intraperitoneal injection of RAW-Exos for 24 hours, the main organs of the mice were imaged in fluorescent mode using an excitation wavelength of 640nm (IVIS Spectrum, PerkinElmer).
HE Staining
The femur sample was initially fixed in 4% paraformaldehyde for 48 hours, then decalcified in an EDTA decalcification solution (G1105, Servicebio) for a duration of three weeks. Following fixation and decalcification, the sample underwent dehydration, paraffin embedding, and sectioning. The sections were deparaffinized in xylene, dehydrated in an ethanol gradient, stained with hematoxylin at room temperature for 5 minutes, differentiated in 1% hydrochloric acid ethanol for 30 seconds, counterstained with alkaline ammonia solution for 1 minute, rinsed with distilled water for 5 minutes, stained with eosin at room temperature for 2 minutes, rinsed again with distilled water for 2 minutes, dehydrated in an ethanol gradient, permeabilized in xylene for 2 minutes, and ultimately sealed with neutral resin.
TRAP Staining
In summary, bone tissue or cells were fixed using 4% paraformaldehyde following the manufacturer’s protocol, thoroughly washed, and subsequently incubated in TRAP staining solution (G1050, Servicebio) at 37°C for a period of 30 minutes. Osteoclasts in bone tissue are identified as red cells, while in cell staining, the presence of three or more nuclei signifies mature osteoclasts.
MDA Content Detection
Following the extraction of total protein from bone tissue or cells using RIPA buffer, it is essential to adhere to the manufacturer’s guidelines for quantifying both protein concentration and MDA (S0131, Beyotime Biotechnology) levels.
Cell Culture and Exosomes Extraction
RAW264.7 cells (2×103 cells per well in a 96-well plate) were cultured in DMEM supplemented with 10% FBS and different concentrations of cinobufagin (0, 1, 10, 100, 150, 200 μM) for 24 hours. Cell viability was assessed using the CCK-8 assay (RM02823, abclonal). RAW264.7 cells were co-cultured with cinobufagin (1 μM), and exosomes extraction was carried out using the culture medium. The culture medium underwent initial centrifugation at 500×g for 10 minutes to eliminate cell debris and apoptotic bodies. Subsequent centrifugation at 16,500×g for 20 minutes was performed to harvest microvesicles. The resulting supernatant was filtered through a 0.22 μm filter (Merck Millipore) to remove proteins and debris. Exosomes were then isolated by ultracentrifugation at 118,000×g for 70 minutes. The exosomes, labeled with DiO-green (C1038, Beyotime Biotechnology) or DID-red (c1039, Beyotime Biotechnology), were resuspended in PBS and underwent final centrifugation at 118,000×g for 70 minutes. All cells originate from the Chinese Academy of Sciences Cell Center.
Transmission Electron Microscopy
A volume of 10 μL of exosomes solution was applied onto a copper grid and left to adsorb for 10 minutes. Following this, the grid was treated with a phosphotungstic acid staining solution for 2 minutes. Once air-dried, images were captured using an 80 kV high-contrast mode electron microscope.
Particle Size Analysis
Exosomes particle size was measured with a Nanoparticle Tracking Analysis (ZetaView, Particle Metrix) following the manufacturer’s guidelines.
Differentiation and Staining of RAW264.7 Cells
RAW264.7 cells were cultured in α-MEM supplemented with 10% FBS and 50 ng/mL RANKL for 7 days, treated with either PBS or RAW-Exos (100 μg/mL), and underwent medium changes every 2–3 days. Subsequently, the cells were fixed with 4% paraformaldehyde after being washed once with PBS. To identify osteoclasts, TRAP staining was performed following the manufacturer’s instructions, identifying cells with three or more nuclei and positive TRAP staining as osteoclasts. Levels of intracellular reactive oxygen species were quantified by measuring the intensity of DCF fluorescence according to the manufacturer’s guidelines.
Differentiation and ALP Activity Detection of MC3T3 Cells
MC3T3 cells were seeded at a density of 1×104 cells per well in a 96-well plate and cultured in α-MEM supplemented with 10% FBS, 0.05 mg/mL L-ascorbic acid, and 1.08 mg/mL Glycerol β phosphate disodium salt hydrate for 7 days, treated with either PBS or RAW-Exos (100 μg/mL), and subjected to medium changes every 2–3 days. Following the removal of the medium, the cells were fixed with 4% paraformaldehyde after being washed with PBS. ALP activity was assessed using the BCIP/NBT alkaline phosphatase assay kit and quantified using the alkaline phosphatase detection kit.
High-Throughput Transcriptome Sequencing and Analysis
The study was outsourced to Megagen Biosciences (China) for high-throughput transcriptome sequencing. Initially, total RNA extraction was conducted using QIAzol Lysis Reagent (Qiagen), followed by an assessment of RNA concentration and purity via Nanodrop 2000. RNA integrity was evaluated through agarose gel electrophoresis, and the RQN value was determined using Agilent 5300. Single library construction necessitated 1 μg of total RNA, a concentration of ≥ 30 ng/μL, RQN value > 6.5, and an OD260/280 ratio between 1.8 and 2.2. Oligo(dT)-coated magnetic beads were employed to pair with polyA for A-T base pairing to isolate mRNA from total RNA and subsequently fragment it. The mRNA underwent reverse transcription to cDNA, followed by adapter ligation and fragment selection for library construction (Illumina). Quality control of the sequenced data was executed using fastp software, and alignment of raw data to a reference gene allowed for the identification of differentially expressed genes, with selection criteria of False Discovery Rate (FDR) < 0.05 and |log2FC| ≥ 1. Subsequently, differential genes were annotated based on their involvement in Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) using Gene Ontology (GO) annotation. Further classification based on the pathways or functions was achieved through the KEGG database for the annotated differentially expressed genes.
Total RNA Isolation and Real-Time PCR
Total RNA was extracted from osteoblasts or osteoclasts utilizing a lysis buffer, and subsequent reverse transcription was performed following the manufacturer’s protocol. Similarly, miRNA extraction from exosomes was achieved using a lysis buffer, followed by reverse transcription according to the manufacturer’s guidelines. Real-time PCR analysis was conducted utilizing SYBR Green and specific primer pairs for the desired genes. The relative gene expression levels were quantified utilizing the comparative threshold method and normalized to β-actin or U6 reference genes. The primer sequences are provided below:
β-actin-f 5′GGCTGTATTCCCCTCCATCG3′, β-actin-r 5′CCAGTTGGTAACAATGCCATGT3′;
ocn-f 5′GAGGACCATCTTTCTGCTCACT3′, ocn-r 5′CGGAGTCTGTTCACTACCTTATTG3′;
opg-f 5′CCTTGCCCTGACCACTCTTAT3′, opg-r 5′CACACACTCGGTTGTGGGT3′;
runx2-f 5′GCCGGGAA TGA TGAGAACTA3′, runx2-r 5′GGTGAAACTCTTGCCTCGTC3′;
rankl-f 5′CGCTCTGTTCCTGTACTTTCG3′, rankl-r 5′GAGTCCTGCAAATCTGCGTT3′;
il6-f 5′CTGCAAGAGACTTCCATCCAG3′, il6-r 5′AGTGGTATAGACAGGTCTGTTGG3′;
Il4-f 5′ ATCATCGGCATTTTGAACGAGG3′, il4-r 5′ TGCAGCTCCATGAGAACACTA3′; tnfa-f 5′ CCTGTAGCCCACGTCGTAG3′, tnfa-r 5′GGGAGTAGACAAGGTACAACCC3′;
il1β-f 5′ GAAATGCCACCTTTTGACAGTG3′, tak1-r 5′ TGGATGCTCTCATCAGGACAG3′;
U6 5′GCGCTCGCTTCGGCAGCACA;
mmu-miR-124-5p 5′GCCGGCACAGCGGACCGA3′;
mmu-miR-298-5p 5′GGCAGAGGAGGGCTGTTCTTCC3′;
mmu-miR-23a-5p 5′GGGGTCCTGGGGGATGGGATTT3′;
mmu-miR-26b-3p 5′GCCCTGTTCTCCCATTACTTGGCTC3′;
mmu-miR-342-5p 5′GCCAGGGGTGCTATCTGTGATTGAG3′;
mmu-miR-379-5p 5′GCGCTGGTAGACTATGGAACGTAGG3′;
mmu-miR-470-5p 5′GCTTCTTGGACTGGCACTGGTGAGT3′;
mmu-miR-744-5p 5′GTGCGGGGCTAGGGCTAACAGCA3′;
mmu-miR-762 5′GGGGCTGGGGCCGGGACA3′;
mmu-miR-1193-3p 5′GCGCTAGGTCACCCGTTTTACTATC3′;
mmu-miR-1198-5p 5′GCTATGTGTTCCTGGCTGGCTTGG3′;
mmu-miR-3076-3p 5′CGCACTCTGGTCTTCCCTTGCAG3′;
mmu-miR-3090-5p 5′GTCTGGGTGGGGCCTGAGATC3′;
mmu-miR-3102-5p 5′GTGAGTGGCCAGGGTGGGG3′;
mmu-miR-5134-5p 5′GCTTGGCAGAAAGGGCAGCTGTG3′;
mmu-miR-5626-3p 5′GCCCAGCAGTTGAGTGATGTGACAC3′;
mmu-miR-6401 5′GCTTACACTCCAGTGGTGTCGGGT3′;
mmu-miR-6538 5′CGCGGGCTCCGGGGCGATA3′;
mmu-miR-6912-5p 5′GTACAGGGAGGGTGCTCAGGCA3′;
mmu-miR-6960-5p 5′GCCCAGGATGAGAAGAGTTTGGCTG3′;
mmu-miR-6983-5p 5′GCCTTGGAAGGGCATACTGATTCGG3′;
mmu-miR-6996-5p 5′GCTGCACAGGACAGAGCACAGTC3′;
mmu-miR-7003-3p 5′GCCCCGGGTTTTCCCCACAG3′;
mmu-miR-7018-3p 5′GTCACCCTGCTGCCGGCTTG3′;
mmu-miR-7027-5p 5′GCCTGGAAAGGAAGAAACAGCAGAGC3′;
mmu-miR-7028-5p 5′GTGGGCTGAGGCTTGGGTCAG3′;
mmu-miR-7083-5p 5′GTCGGGGCTGGACAAGCAGAGA3′;
mmu-miR-7683-3p 5′GCCTGGAAAGGTGGAACACGGAAC3′;
mmu-miR-7686-5p 5′CCTTCCACTGGACCTGGGGCT3′;
mmu-miR-7687-5p 5′GAGGCGGGGAACCTGAGGC3′;
mmu-miR-8100 5′CAGGAGGAAAGGGAGCAAGCAGGT3′;
mmu-miR-9768-5p 5′GTGGCACTCGGAGGACGGCA3′;
Western Blot Detection
Protein detection using Western blot analysis involved lysing cells or tissues with lysis buffer to extract total proteins, which were then separated by 12% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane. Subsequent procedures included blocking with skim milk, incubation with primary and secondary antibodies, and visualization of immunoreactivity using an enhanced chemiluminescence detection kit. The antibodies used in this study were β-ACTIN (4967, CST), OCN (59757, CST), RANKL (A13382, abclonal), CTSK (A5871, abclonal), ALOX15 (A22908, abclonal), CD9 (A19027, abclonal), CD63 (A19023, abclonal), TSG101 (A2216, abclonal), CALNEXIN (A4846, abclonal), anti-mouse IgG (7076, CST), anti-rabbit IgG (7074, CST).
Fluorescent Reporter Gene
The fluorescent reporter gene assay was conducted following a previously documented procedure (34180104). Briefly, mc3t3 cells were plated in a 96-well plate at a density of 1*104 cells per well and transfected with miR-3102-5p-mimics oligoribonucleotides (5′GTGAGTGGCCAGGGTGGGGCTG3′) or miR-3102-5p-mut oligoribonucleotides (5′GTGAGCACTCGCGGUGGGGCUG3′) in combination with either PGL6 or PGL6-alox15 plasmids using Lipofectamine 3000. The fluorescent reporter enzyme activity was assessed after 48 hours following the guidelines provided by the manufacturer (RG042S, Beyotime).
Statistical Analysis
Each experiment was conducted a minimum of three times. The data are presented as the mean ± standard deviation and analyzed using one-way analysis of variance (ANOVA) or Student’s t-test when suitable Significance was determined at a P-value of less than 0.05.
Results
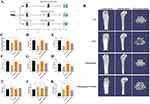
Cinobufagin Effectively Increases Bone Mass in Ovariectomized (OVX) Mice
An osteoporosis model was induced through ovariectomy in mice (Figure 1A), revealing uterine atrophy and reduced diameter in OVX mice upon macroscopic observation (Figure S1A and B). Histological examination using hematoxylin and eosin (HE) staining displayed features such as loose uterine tissue, increased cell spaces, nuclear condensation, and apoptosis in OVX mice (Figure S1C). Microcomputed tomography (MicroCT) scanning and reconstruction of the distal femur in mice exhibited a significant decrease in trabecular bone mineral density in OVX mice, with a moderate increase following Cinobufagin treatment (Figure 1B and C). Analysis of bone parameters indicated a marked reduction in bone volume (BV), bone volume fraction (BV/TV), bone surface density (BS/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), cortical thickness (Ct.Th), cortical area to total area ratio (Ct.Ar/Tt.Ar), and a significant increase in trabecular separation (Tb.Sp) in OVX mice, all of which were restored with Cinobufagin treatment (Figure 1D–I). Furthermore, Cinobufagin elevated the concentration of the serum bone formation marker P1NP and reduced the levels of the bone resorption marker β-CTX (Figure 1J and K). In conclusion, Cinobufagin effectively enhances bone mass in OVX mice.
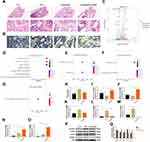
Cinobufagin Exhibits Anti-Inflammatory and Lipid Oxidation Inhibitory Effects in OVX Mice
HE staining revealed a significant increase in trabecular bone mass in OVX mice treated with Cinobufagin, accompanied by inhibited inflammatory infiltration (Figure 2A). Tartrate-resistant acid phosphatase (TRAP) staining illustrated the inhibition of osteoclast activity in trabecular bone by Cinobufagin (Figure 2B). Subsequently, RNA-sequencing transcriptome analysis was conducted on femoral tissues from the OVX and Cinobufagin group. The RNA-seq results identified 856 significantly altered genes in the Cinobufagin group, including 542 upregulated and 314 downregulated genes (Figure 2C). The top 200 upregulated and downregulated genes underwent Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. GO analysis revealed enrichment in biological processes are phagocytosis, plasma membrane invagination and membrane invagination, molecular functions are immunoglobulin receptor binding and antigen binding, and cellular components are immunoglobulin complex (Figure 2D–F). KEGG analysis highlighted linoleic acid metabolism as a uniquely enriched pathway, closely linked to ALOX15 (Figure 2G). Assessment of the lipid oxidation product malondialdehyde (MDA) content showed a significant decrease in the femurs of OVX mice treated with Cinobufagin (Figure 2H). Quantitative polymerase chain reaction (qPCR) analysis demonstrated that Cinobufagin upregulated the expression of bone formation genes Ocn and Opg (Figure 2I and J), downregulated bone resorption gene Rankl and pro-inflammatory genes il1β and il6 expression (Figure 2K–M), and increased the expression of anti-inflammatory genes il4 and il10 (Figure 2N and O). Western blot analysis indicated that Cinobufagin notably reduced the expression of key proteins involved in bone resorption (CTSK, Rankl) and lipid oxidation (ALOX15) while enhancing the expression of bone formation protein Ocn (Figure 2P and Q). Therefore, cinobufagin may regulate bone metabolism by inhibiting inflammation and lipid peroxidation.
The Therapeutic Efficacy of Cinobufagin is Closely Related to Exosomes (EXOs)
Given the regulatory role of exosomes in bone metabolism, we hypothesize that cinobufagin may inhibit bone loss by regulating exosomes. Hence prior to administering Cinobufagin, ovariectomized (OVX) mice were subjected to treatment with the exosomes inhibitor GW4869. Microcomputed tomography (MicroCT) imaging revealed that GW4869 reversed the therapeutic benefits of Cinobufagin (Figure 1B). Mice in the GW4869 group displayed a notable reduction in bone mass, with bone parameter assessments demonstrating significant decreases in BMD, BV/TV, Tb.N, Tb.Th (Figure 1C, E, G and H). HE staining and tartrate-resistant acid phosphatase (TRAP) results indicated that GW4869 hindered the anti-inflammatory, bone formation-enhancing, and anti-bone resorptive effects of Cinobufagin (Figure 2A and B). ELISA findings demonstrated a decrease in serum P1NP and an increase in β-CTX levels in the GW4869 group (Figure 1J and K), further supporting the notion that the exosomes inhibitor GW4869 countered the therapeutic properties of Cinobufagin. Given the potent anti-inflammatory properties of Cinobufagin, we postulated that Cinobufagin might influence the composition of exosomes released from monocytes to exert its effects. Intervention with various concentrations of Cinobufagin on RAW264.7 cells resulted in a notable increase in cell activity based on Cell Counting Kit-8 (CCK8) results (Figure 3A and B). Quantitative polymerase chain reaction (qPCR) analysis revealed that Cinobufagin significantly suppressed the expression of pro-inflammatory genes IL1, IL6 and TNFα (Figure 3C–E). Subsequently, exosomes derived from RAW264.7 cells were isolated, with protein quantification indicating a substantial elevation in protein concentration of RAW-Exos (Figure 3F). Electron microscopy imaging demonstrated that RAW-Exos exhibited a rounded shape, with particle sizes ranging from 50–200 nm (Figure 3G and H). Western blot analysis confirmed positive expression of exosomal marker proteins CD9, CD63 and TSG101 and negative expression of the exosomal control protein calnexin (Figure 3I). To investigate the impact of RAW-Exos on bone resorption, RAW-Exos were co-cultured with the osteoclast cell line RAW264.7, leading to significant internalization of a substantial quantity of RAW-Exos by RAW264.7 cells (Figure 3J). Functional assays showed that RAW-Exos notably inhibited the formation of TRAP+ osteoclasts and the production of reactive oxygen species (Figure 3K, Figure S2A and C).
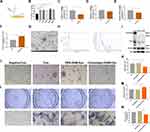
Cinobufagin Suppresses the Expression of ALOX15 in Osteoblasts by Upregulating miR-3102-5p in RAW-Exos
RAW-Exos was co-cultured with the osteoblast cell line MC3T3, revealing a significant uptake of RAW-Exos by MC3T3 cells (Figure S2B). The induced differentiation outcomes showed that RAW-Exos notably enhanced osteoblast differentiation, increased ALP activity, and reduced the production of the lipid oxidation product MDA (Figure 3L–N). Gene expression analysis via qPCR indicated that RAW-Exos notably upregulated the expression of Ocn and Runx2 while downregulating Rankl, TNFα and IL6 (Figure 4A–E). Western blot analysis demonstrated that RAW-Exos significantly elevated the levels of Ocn and antioxidant proteins SOD and GPX4 (Figure 4F). Notably, RAW-Exos exhibited the ability to downregulate the expression of the critical lipid oxidation protein ALOX15. To investigate the mechanism behind RAW-Exos’s suppression of lipid oxidation, bioinformatics tools were employed, predicting miRNAs regulating ALOX15. The findings suggested that 32 miRNAs could bind to ALOX15 to modulate its expression (Figure 4G). Validation through qPCR identified high levels of miR-3102-5p and miR-7683-3p in RAW-Exos, with miR-3102-5p displaying the most potential in regulating ALOX15 through an 8-mer binding site sequence 5′-CCACUCAA-3′ on the ALOX15 gene (Figure 4H–K). Luciferase reporter gene assays indicated a significant reduction in fluorescence intensity in plasmids containing ALOX15 when targeted by miR-3102-5p (Figure 4L). Hence, these results suggest that miR-3102-5p in RAW-Exos can enhance osteogenic differentiation by suppressing ALOX15 expression, thereby inhibiting lipid oxidation.
 |
Figure 4 RAW-exos suppress the expression of Arachidonate 15-lipoxygenase in osteoblasts by carrying miR-3102-5p. (A–E) mRNA expression level (**P<0.01, ***P<0.001, ****P<0.0001, N=3). mRNA expression level of Osteocalcin (Ocn) in osteoblasts (A). mRNA expression level of Runt-related transcription factor 2 (Runx2) in osteoblasts (B). mRNA expression level of Receptor activator of nuclear factor kappa-Β ligand (Rankl) in osteoblasts (C). mRNA expression level of Tumor necrosis factor-alpha (TNF-α) in osteoblasts (D). mRNA expression level of Interleukin-6 (IL-6) in osteoblasts (E). (F) Expression of ALOX15, SOD, GPX4, and OCN proteins in osteoblast. (G) Bioinformatics prediction of miRNAs binding to ALOX15 by TargetScan data(http://www.targetscan.org/) and miRWalk data (http://mirwalk.umm.uni-heidelberg.de/). (H–J) miRNA expression level of RAW-Exos (**P<0.01, N=3). (K) Binding site of miR-3102-5p, miR-7638-3p and Arachidonate 15-lipoxygenase (alox15). (L) Firefly luciferase reporter gene assay to detect the binding activity between miR-3102-5p and alox15 (*P<0.05, N=3). |
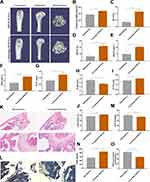
The Bone Protective Effect of RAW-Exos on OVX Mice
To further validate the bone-protective effect of RAW-Exos, OVX mice were treated with RAW-Exos. MicroCT analysis revealed a significant increase in BMD in OVX mice administered RAW-Exos (Figure 5A and B). Assessment of bone parameters showed notable improvements in BV, BV/TV, BS/TV, Tb.N, Tb.Th, Ct.Th, Ct.Ar/Tt.Ar, along with a decrease in Tb.Sp in OVX mice following RAW-Exos treatment (Figure 5C–J). The body interior tracking display shows that RAW-Exos can be enriched in the trabecular bone of the femur (Figure S3). Histological evaluation through HE staining and TRAP staining indicated a significant enhancement in trabecular bone mass and a reduction in the number of TRAP+ osteoclasts due to RAW-Exos treatment (Figure 5K and L). ELISA results exhibited that RAW-Exos led to an elevation in serum P1NP levels and a decrease in β-CTX in OVX mice (Figure 5M and N). Furthermore, RAW-Exos reduces MDA content in the femur of OVX mice (Figure 5O), suggesting that RAW-Exos serves as a key mediator for Cinobufagin in regulating bone metabolism and treating OVX mice.
Discussion
This study demonstrated the bone-protective effect of Cinobufagin and revealed its mechanism of action in altering bone metabolism by influencing the content of macrophage-derived exosomes. Toad venom, a natural product commonly used in clinical cancer treatment in East Asian countries,33 contains Cinobufagin as one of its primary active compounds with well-documented anti-tumor properties.11,34 Interestingly, while Cinobufagin exhibits a high IC50 in osteoblasts compared to tumor cells,35 indicating potential bone-protective qualities, its specific impact on bone metabolism has remained unclear. To investigate this, we induced simulated bone loss in ovariectomized mice and observed that Cinobufagin effectively restored trabecular bone mass in these mice and regulated abnormal markers of serum bone metabolism. Additionally, Cinobufagin was found to enhance the secretion of osteocalcin (OCN), decrease the secretion of receptor activator of nuclear factor-κB ligand (RANKL), and reduce the expression of cathepsin K (CTSK), a marker of bone-resorbing osteoclasts. Moreover, Cinobufagin demonstrated inhibitory effects on lipid peroxidation and the inflammatory state within the bones of ovariectomized mice, further supporting its bone-protective properties. Given that chronic inflammation and oxidative stress play crucial roles in bone loss pathology,36,37 further research is warranted to elucidate the precise molecular mechanisms through which Cinobufagin modulates these processes.
In the normal course of bone metabolism, various cell types such as osteoblasts, osteoclasts, adipocytes, immune cells, and muscle cells release exosomes that play a crucial role in regulating bone resorption and formation.24,38–40 These exosomes contain abundant genetic information and active substances, and any alterations in their contents can contribute to the development of osteoporosis.41–43 In individuals with osteoporosis, exosomes derived from bone marrow mesenchymal stem cells carrying microRNAs like miR-21, miR-424-5p, and miR-143/145 suppress osteogenic activity while enhancing osteoclastic activity.22,23,44 Consequently, modulations in exosomes composition may serve as a vital mechanism through which cinobufagin treats osteoporosis. GW4869, a non-competitive inhibitor of neutral sphingomyelinase (N-SMase) involved in exosomes synthesis and release, has been identified.45 Our research revealed that GW4869 can counteract the bone-protective effects of cinobufagin, validating the significant role of exosomes in bone metabolism, even though their origin remains uncertain. Monocytes, essential immune cells pivotal in inflammation regulation, are also the primary precursors of osteoclasts,46,47 differentiating into osteoclasts when exposed to M-CSF and RANKL cytokines. Therefore, monocytes stand as critical target cells for cinobufagin in both controlling inflammation and regulating bone metabolism. Our study demonstrated that cinobufagin inhibits the expression of inflammatory mediators in monocytes. Following intervention with cinobufagin, we isolated exosomes from monocytes, revealing disc-shaped structures with an average diameter ranging from 40 to 200 nm. Co-cultivation experiments confirmed that RAW-Exos, the isolated exosomes, are internalized by osteoblasts and osteoclasts, subsequently impeding osteoclast differentiation and fostering osteoblast differentiation. These results underscore the significance of RAW-exos as a primary target of cinobufagin in halting bone loss.
In ovariectomized (OVX) mice, inflammation is not independent, and the accumulation of reactive oxygen species (ROS) plays a role in the process of bone loss.48,49 During differentiation, osteoclasts produce ROS, which further enhances bone resorption activity.50 When OVX mice were treated with Cinobufagin, differentially expressed genes at the distal end of the femur showed enrichment in immune regulation and fatty acid metabolism pathways. Among these, Alox15, a key gene regulating lipid metabolism,51 was significantly downregulated. Alox15 encodes an enzyme involved in arachidonic acid metabolism, which in turn produces ROS in the lipid metabolism process, leading to the activation of inflammatory responses, inhibition of osteoblastic differentiation, and promotion of osteoclastic differentiation. Thus, it is hypothesized that the decreased expression of Alox15 may be influenced by changes in miRNAs within RAW-Exos. Through the prediction and validation of miRNAs that potentially regulate Alox15, it was discovered that miR-3102-5p is highly expressed in RAW-Exos and binds to the CCACUCAA sequence in the 3′UTR region of alox15, thereby suppressing Alox15 transcription. To confirm this finding, RAW-Exos was used as a therapeutic intervention in OVX mice, leading to a significant increase in bone mass in the femur of OVX mice. These results indicate that Cinobufagin may exert its bone-protective effect by modulating the expression of miRNAs in RAW-Exos.
Conclusion
In conclusion, our study has disclosed the bone-protective efficacy of Cinobufagin, a discovery of paramount importance for the potential clinical utilization of Cinobufagin, particularly for patients with bone tumors and bone metastases. Additionally, we have illustrated that RAW-Exos has the capability to deliver miR-3102-5p to osteoblasts, thus hindering the transcription of Alox15, consequently counteracting the inhibitory impact of lipid peroxidation on bone formation. Consequently, we postulate that Cinobufagin can mitigate inflammation and lipid peroxidation in OVX mice, thereby manifesting its bone-safeguarding influence by modulating the composition of RAW-Exos.
Abbreviations
OVX, Ovariectomy; RAW-Exos, macrophage (raw264.7 cells)-derived exosomes; BMD, bone mineral density; BV, bone volume; BV/TV, bone volume fraction; BS/TV, bone surface area to bone volume ratio; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation; Ct.Th, cortical bone thickness; Ct.Ar/Tt.Ar, cortical bone area fraction; ROS, reactive oxygen species.
Data Sharing Statement
Data will be made available on request.
Ethics Approval and Consent to Participate
All animal procedures were approved by the Animal Research Committee of Shanghai University of Traditional Chinese Medicine (PZSHUTCM2305150001).
Funding
This work was partially supported by Shanghai Science and Technology Development Fund from Central Leading Local Government (YDZX20223100001004), National Natural Science Foundation of China (82174404), Shanghai Chronic Bone and Joint Disease Clinical Research Center (20MC1920600), Shanghai Municipal Science and Technology Commission (23010505000).
Disclosure
All authors declare no conflict of interest.
References
1. Ensrud KE, Crandall CJ. Osteoporosis. Ann Intern Med. 2024;177(1):ITC1–ITC16. doi:10.7326/AITC202401160
2. Yu B, Gao Q, Sheng S, et al. Smart osteoclasts targeted nanomedicine based on amorphous CaCO(3) for effective osteoporosis reversal. J Nanobiotechnol. 2024;22(1):153.
3. Body JJ. Prevention and treatment of side-effects of systemic treatment: bone loss. Ann Oncol. 2010;21(7):vii180–5. doi:10.1093/annonc/mdq422
4. Eisen A, Somerfield MR, Accordino MK, et al. Use of Adjuvant Bisphosphonates and Other Bone-Modifying Agents in Breast Cancer: ASCO-OH (CCO) Guideline Update. J Clin Oncol. 2022;40(7):787–800. doi:10.1200/JCO.21.02647
5. Alibhai SMH, Zukotynski K, Walker-Dilks C, et al. Bone Health and Bone-Targeted Therapies for Nonmetastatic Prostate Cancer: a Systematic Review and Meta-analysis. Ann Intern Med. 2017;167(5):341–350. doi:10.7326/M16-2577
6. Yin JJ, Pollock CB, Kelly K. Mechanisms of cancer metastasis to the bone. Cell Res. 2005;15(1):57–62. doi:10.1038/sj.cr.7290266
7. Yang TL, Shen H, Liu A, et al. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol. 2020;16(2):91–103. doi:10.1038/s41574-019-0282-7
8. Gao H, Nepovimova E, Heger Z, et al. Role of hypoxia in cellular senescence. Pharmacol Res. 2023;194:106841. doi:10.1016/j.phrs.2023.106841
9. Zhang J, Hu K, Di L, et al. Traditional herbal medicine and nanomedicine: converging disciplines to improve therapeutic efficacy and human health. Adv Drug Deliv Rev. 2021;178:113964. doi:10.1016/j.addr.2021.113964
10. Liu Y, Feng N. Nanocarriers for the delivery of active ingredients and fractions extracted from natural products used in traditional Chinese medicine (TCM). Adv Colloid Interface Sci. 2015;221:60–76. doi:10.1016/j.cis.2015.04.006
11. Hou R, Liu X, Yang H, et al. Chemically synthesized cinobufagin suppresses nasopharyngeal carcinoma metastasis by inducing ENKUR to stabilize p53 expression. Cancer Lett. 2022;531:57–70. doi:10.1016/j.canlet.2022.01.025
12. Apryani E, Ali U, Wang ZY, et al. The spinal microglial IL-10/beta-endorphin pathway accounts for cinobufagin-induced mechanical antiallodynia in bone cancer pain following activation of alpha7-nicotinic acetylcholine receptors. J Neuroinflam. 2020;17(1):75. doi:10.1186/s12974-019-1616-z
13. Terpos E, NTANASIS-STATHOPOULOS I, Gavriatopoulou M, et al. Pathogenesis of bone disease in multiple myeloma: from bench to bedside. Blood Cancer J. 2018;8(1):7. doi:10.1038/s41408-017-0037-4
14. Baek SH, Kim C, Lee JH, et al. Cinobufagin exerts anti-proliferative and pro-apoptotic effects through the modulation ROS-mediated MAPKs signaling pathway. Immunopharmacol Immunotoxicol. 2015;37(3):265–273. doi:10.3109/08923973.2015.1027916
15. Kalluri R, Lebleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478). doi:10.1126/science.aau6977
16. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88(1):487–514. doi:10.1146/annurev-biochem-013118-111902
17. Mashouri L, Yousefi H, Aref AR, et al. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18(1):75. doi:10.1186/s12943-019-0991-5
18. Li W, Li C, Zhou T, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16(1):145. doi:10.1186/s12943-017-0706-8
19. Sun Z, Yang S, Zhou Q, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. doi:10.1186/s12943-018-0831-z
20. Yu L, Sui B, Fan W, et al. Exosomes derived from osteogenic tumor activate osteoclast differentiation and concurrently inhibit osteogenesis by transferring COL1A1-targeting miRNA-92a-1-5p. J Extracell Vesicles. 2021;10(3):e12056. doi:10.1002/jev2.12056
21. Wu K, Feng J, Lyu F, et al. Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nat Commun. 2021;12(1):5196. doi:10.1038/s41467-021-25473-y
22. Jiang LB, Tian L, Zhang ZG. Bone marrow stem cells-derived exosomes extracted from osteoporosis patients inhibit osteogenesis via microRNA-21/SMAD7. Eur Rev Med Pharmacol Sci. 2018;22(19):6221–6229.
23. Wei Y, Ma H, Zhou H, et al. miR-424-5p shuttled by bone marrow stem cells-derived exosomes attenuates osteogenesis via regulating WIF1-mediated Wnt/beta-catenin axis. Aging. 2021;13(13):17190–17201.
24. Liang M, Yin X, Zhang S, et al. Osteoclast-derived small extracellular vesicles induce osteogenic differentiation via inhibiting ARHGAP1. Mol Ther Nucleic Acids. 2021;23:1191–1203. doi:10.1016/j.omtn.2021.01.031
25. Zhao YZ, WANG Y YL, Yu Y. Immunoenhancement effect of cinobufagin on macrophages and the cyclophosphamide-induced immunosuppression mouse model. Int Immunopharmacol. 2024;131:111885. doi:10.1016/j.intimp.2024.111885
26. Yu G, Jung H, Kang YY, et al. Comparative evaluation of cell- and serum-derived exosomes to deliver immune stimulators to lymph nodes. Biomaterials. 2018;162:71–81. doi:10.1016/j.biomaterials.2018.02.003
27. Zheng X, Sun K, Liu Y, et al. Resveratrol-loaded macrophage exosomes alleviate multiple sclerosis through targeting microglia. J Control Release. 2023;353:675–684. doi:10.1016/j.jconrel.2022.12.026
28. Wang Y, Lin Q, Zhang H, et al. M2 macrophage-derived exosomes promote diabetic fracture healing by acting as an immunomodulator. Bioact Mater. 2023;28:273–283. doi:10.1016/j.bioactmat.2023.05.018
29. Liu T, Jing F, Huang P, et al. Thymopentin alleviates premature ovarian failure in mice by activating YY2/Lin28A and inhibiting the expression of let-7 family microRNAs. Cell Prolif. 2021;54(8):e13089. doi:10.1111/cpr.13089
30. Geng Z, Guo H, Li Y, et al. Stem cell-derived extracellular vesicles: a novel and potential remedy for primary ovarian insufficiency. Front Cell Dev Biol. 2023;11:1090997. doi:10.3389/fcell.2023.1090997
31. DELLA TORRE S, Benedusi V, Pepe G, et al. Dietary essential amino acids restore liver metabolism in ovariectomized mice via hepatic estrogen receptor alpha. Nat Commun. 2021;12(1):6883. doi:10.1038/s41467-021-27272-x
32. Lorenzo J. From the gut to bone: connecting the gut microbiota with Th17 T lymphocytes and postmenopausal osteoporosis. J Clin Invest. 2021;131(5). doi:10.1172/JCI146619
33. Zhan X, Wu H, Wu H, et al. Metabolites from Bufo gargarizans (Cantor, 1842): a review of traditional uses, pharmacological activity, toxicity and quality control. J Ethnopharmacol. 2020;246:112178. doi:10.1016/j.jep.2019.112178
34. Long Y, Fan J, Zhou N, et al. Biomimetic Prussian blue nanocomplexes for chemo-photothermal treatment of triple-negative breast cancer by enhancing ICD [J]. Biomaterials. 2023;303:122369. doi:10.1016/j.biomaterials.2023.122369
35. Yin JQ, Wen L, Wu LC, et al. The glycogen synthase kinase-3beta/nuclear factor-kappa B pathway is involved in cinobufagin-induced apoptosis in cultured osteosarcoma cells. Toxicol Lett. 2013;218(2):129–136. doi:10.1016/j.toxlet.2012.11.006
36. Wang J, Zhang Y, Cao J, et al. The role of autophagy in bone metabolism and clinical significance. Autophagy. 2023;19(9):2409–2427. doi:10.1080/15548627.2023.2186112
37. Thummuri D, Jeengar MK, Shrivastava S, et al. Thymoquinone prevents RANKL-induced osteoclastogenesis activation and osteolysis in an in vivo model of inflammation by suppressing NF-KB and MAPK Signalling. Pharmacol Res. 2015;99:63–73. doi:10.1016/j.phrs.2015.05.006
38. Xu R, Shen X, Si Y, et al. MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment. Aging Cell. 2018;17(4):e12794. doi:10.1111/acel.12794
39. Fulzele S, Mendhe B, Khayrullin A, et al. Muscle-derived miR-34a increases with age in circulating extracellular vesicles and induces senescence of bone marrow stem cells. Aging. 2019;11(6):1791–1803. doi:10.18632/aging.101874
40. Zhang L, Wang Q, Su H, et al. Exosomes from Adipose Tissues Derived Mesenchymal Stem Cells Overexpressing MicroRNA-146a Alleviate Diabetic Osteoporosis in Rats. Cell Mol Bioeng. 2022;15(1):87–97. doi:10.1007/s12195-021-00699-4
41. Yin P, Jiang Y, Fang X, et al. Cell-Based Therapies for Degenerative Musculoskeletal Diseases. Adv Sci. 2023;10(21):e2207050. doi:10.1002/advs.202207050
42. Song H, Li X, Zhao Z, et al. Reversal of Osteoporotic Activity by Endothelial Cell-Secreted Bone Targeting and Biocompatible Exosomes. Nano Lett. 2019;19(5):3040–3048. doi:10.1021/acs.nanolett.9b00287
43. Murphy C, Withrow J, Hunter M, et al. Emerging role of extracellular vesicles in musculoskeletal diseases. Mol Aspects Med. 2018;60:123–128. doi:10.1016/j.mam.2017.09.006
44. Xu R, Shen X, Xie H, et al. Identification of the canonical and noncanonical role of miR-143/145 in estrogen-deficient bone loss. Theranostics. 2021;11(11):5491–5510. doi:10.7150/thno.55041
45. Koide T, Mandai S, Kitaoka R, et al. Circulating Extracellular Vesicle-Propagated microRNA Signature as a Vascular Calcification Factor in Chronic Kidney Disease. Circ Res. 2023;132(4):415–431. doi:10.1161/CIRCRESAHA.122.321939
46. Jin S, Gao J, Yang R, et al. A baicalin-loaded coaxial nanofiber scaffold regulated inflammation and osteoclast differentiation for vascularized bone regeneration. Bioact Mater. 2022;8:559–572. doi:10.1016/j.bioactmat.2021.06.028
47. Giannandrea D, Parolini M, Citro V, et al. Nanoplastic impact on bone microenvironment: a snapshot from murine bone cells. J Hazard Mater. 2024;462:132717. doi:10.1016/j.jhazmat.2023.132717
48. Xie X, Cheng P, Hu L, et al. Bone-targeting engineered small extracellular vesicles carrying anti-miR-6359-CGGGAGC prevent valproic acid-induced bone loss. Signal Transduct Target Ther. 2024;9(1):24. doi:10.1038/s41392-023-01726-8
49. Zhang C, Li H, Li J, et al. Oxidative stress: a common pathological state in a high-risk population for osteoporosis. Biomed Pharmacother. 2023;163:114834. doi:10.1016/j.biopha.2023.114834
50. Manolagass SC. From estrogen-centric to aging and oxidative stress: a revised perspective of the pathogenesis of osteoporosis. Endocr Rev. 2010;31(3):266–300. doi:10.1210/er.2009-0024
51. Walters JLH, De iuliis GN, Nixon B, et al. Oxidative Stress in the Male Germline: a Review of Novel Strategies to Reduce 4-Hydroxynonenal Production. Antioxidants. 2018;7(10). doi:10.3390/antiox7100132.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.