Back to Journals » Cancer Management and Research » Volume 16
Comprehensive Analysis of the Significance of Breast Cancer Gene 1 (BRCA-1) in Bladder Cancer
Authors Zhang X, Tao X, Zhou Y, Shi G, Wang T
Received 22 July 2024
Accepted for publication 18 September 2024
Published 30 September 2024 Volume 2024:16 Pages 1305—1319
DOI https://doi.org/10.2147/CMAR.S467817
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Xinyu Zhang,1,* Xiaoxuan Tao,2,* Yuxin Zhou,1,* Guangyue Shi,1 Tianjiao Wang1
1Department of Internal Medicine-Oncology, Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin, People’s Republic of China; 2Department of Radiotherapy, Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guangyue Shi; Tianjiao Wang, Email [email protected]; [email protected]
Background: Bladder carcinoma (BLCA) is characterized by high morbidity, mortality, and treatment costs. Breast cancer gene 1 (BRCA1), a tumor suppressor gene, inhibits the development of malignant tumors. However, research on the significance of BRCA1 in BLCA is limited. This study aims to explore the importance of BRCA1 in BLCA using bioinformatic methods and immunohistochemistry.
Methods: Gene expression, clinical, and survival data were collected from the TCGA databases through the UCSC Xena platform (http://xena.ucsc.edu/). The TPM data from the TCGA and GETEx databases were integrated using the GEPIA database (http://GEPIA.cancer-pku.cn). The study then explored the differential expression, survival prognosis, functional enrichment, and immune cell infiltration analyses of BRCA1 in BLCA. A PPI network of BRCA1 was constructed using the STRING database, and a BRCA1-associated gene-gene interaction network was generated using the GeneMANIA database. Immunohistochemistry (IHC) assays were performed to verify the expression levels of BRCA1 in bladder tumour tissues and adjacent normal tissues.
Results: BRCA1 is associated with BLCA. Differential analysis indicated that BRCA1 acts as a risk factor for BLCA but does not show significant expression differences across genders, stages, tumor stages, lymph node stages, or metastasis stages. Additionally, staging was based on the eighth edition of the American Joint Committee on Cancer (AJCC) for BLCA. Co-expression network and Gene Set Enrichment Analysis (GESA) confirmed that BRCA1 is involved in various BLCA pathways. Furthermore, BRCA1 expression was also linked to immune cell infiltration. However, survival prognosis analysis revealed no significant correlation between the prognosis of BLCA and BRCA1.
Conclusion: We demonstrated that BRCA1 is a prospective predicted and immunological biomarker in BLCA, offering new avenues for potential therapies.
Keywords: bladder cancer, BRCA1, expression, immune cell infiltration, prognosis
Introduction
Bladder carcinoma (BLCA) is a common urological malignancy worldwide1 and represents the second most prevalent neoplasm of the male urinary tract, being a significant cause of death with approximately 550,00 new cases annually in 2020.2 Recent studies have shown an increase in the incidence of BLCA.3 About, 70% of patients with superficial BLCA tumors are treated with transurethral resection of the bladder (TURB), although these tumors are typically not life-threatening, they have high risks of relapse and disease progression.4 In contrast, patients with muscle-invasive BLCA tumors have poor prognoses and higher rates of distant metastasis.5 The emergence of drug resistance continues to severely impact human life,6 highlighting the need for new gene targets to improve BLCA treatment and patient management.
Constitutional BRCA1 methylation is recognized as a cancer risk factor for beast and ovarian cancers.7 BRCA1 is linked to an increased risk of breast cancer, its expression levels are correlated with tumor size, axillary nodal status, histological grade, Ki-67 expression, and molecular subtypes.8 BRCA1, a tumor suppressor gene, operates through a complex mechanism.9 Previous research has shown that BRCA1’s key cancer-suppressive activity lies in its capacity for homologous recombination repair of DNA double-strand breaks.10–12 The intricate role of BRCA1 in cellular functions has been elucidated through its extensive research history.13 Recent studies have highlighted BRCA1’s significant role in various malignant tumors. For example, BRCA1 expression has shown no correlation with survival rates in breast cancer.14 Conversely, positive BRCA1 expression correlates with advanced tumor stages and poorer prognostic outcomes in ovarian serous cancer,15 while its presence in BLCA is rarely reported. Investigating the pathogenesis and potential drug targets for BLCA could lead to novel therapeutic and diagnostic developments. Hence, the exploration of BRCA1’s role is critical, potentially enhancing survival rates and diagnostic accuracy in BLCA.
In our study, we assessed BRCA1’s function in BLCA by analyzing expression levels and differences. Additionally, survival analyses were conducted to determine its prognostic significance. We explored its biological functions through gene-gene interaction and GSEA enrichment analyses. Furthermore, we examined the relationships between BRCA1 and immunological characteristics. Overall, our findings suggest that BRCA1 may be a potential target in BLCA.
Materials and Methods
Data Collection and Clinical Cases
TCGA-BLCA gene expression and clinical information were obtained from the UCSC Xena platform (http://xena.ucsc.edu/).16 TPM data from the TCGA and GTEx databases were integrated using the GEPIA platform. In accordance with the eighth edition of the American Joint Committee on Cancer (AJCC), BLCA classification was standardized. Additional survival data were downloaded from the UCSC Xena platform (http://xena.ucsc.edu/). The STRING database was accessed to analyze the biological functions of BRCA1, and the GeneMANIA database was used to examine gene-gene interaction networks.17
Difference Analysis
The “DESeq2” package (v1.40.2) was employed to identify differentially expressed genes (DEGs) with a log fold change cutoff of 1 and an adjusted p-value below 0.05 among BRCA1 upregulated, downregulated, and non-regulated groups. Visualization of the results was performed using the “ggpubr” (v0.6.0) and “ggthemes” (v4.2.4) packages.18 The “pheatmap” package (v1.0.12) depicted the expression levels of various genes in BLCA, including BRCA1, in a heatmap. The “Rcircos” package (v4.30) categorized BLCA patients and normal individuals into 01A and 11A groups, respectively. “DESeq2” (v1.40.2) was again utilized to screen results (fold change >1, p < 0.05). Clinical data and data from the 01A group were merged. Lastly, XianTao Academic (https://www.xiantaozi.com/literatures) provided an analysis of BRCA1 expression across different groups. It was noted that the database did not account for patients who had received any standard intravesical treatment.
Functional Enrichment Analysis
The dataset of differentially expressed genes was imported into the “Rcircos” package (v4.30) with reference to the Molecular Signature Database gene set (msigdb.v7.0.symbols.gmt). The “ClusterProfiler” package (v4.8.3) was employed to investigate the regulatory functions of BRCA1, such as Gene Set Enrichment Analysis (GSEA).19,20 The “enrichplot” package (v1.20.1) depicted the results of the GSEA, which included four main data points: enrichment score (ES), normalized ES, error detection rate, and p-value.
Survival Prognosis Analysis of BRCA1
Survival data from UCSC was analyzed using the “survival” (v3.5–5) package (v3.5–5). The “DESeq2” package (v1.40.2) calculated differentially expressed genes (DEGs) with a fold change of 1 and p < 0.05 between the BRCA1-high and BRCA1-low groups. Univariate Cox regression analysis identified DEGs with prognostic significance. A risk score model was developed based on these prognostic DEGs. Cox regression analysis was performed using the “survival” package (v3.5–5). Patients were stratified into low or high-risk groups based on the median risk score. Model accuracy was assessed using receiver operating characteristic curves and time-dependent ROC curves with corresponding AUC values, conducted by the “ROCR” (v1.0–11) and “timeROC” (v0.4) packages. The hazard ratio (HR) with a 95% confidence interval (CI) was calculated using Cox proportional hazards regression. The prognostic role of BRCA1 was further evaluated by a forest plot, generated by the “forestplot” package (v3.1.1), to calculate p-values, HR, and 95% CI. Differences in overall survival (OS) between low and high BRCA1 expression groups were determined using the Kaplan-Meier survival analysis method.21
Immune Cell Infiltration Analysis
CIBERSORT analysis was first utilized to assess the proportions of tumor-infiltrating immune cells (TICs) in BLCA cases, considering p-values <0.05 as statistically significant.22 Subsequently, correlations between BLCA and immune cell infiltration were explored using the UCSC database. The composition of immune infiltrating cells included B cells, CD8+ T cells, CD4+ T cells, NK cells, macrophages, neutrophils, plasma cells, and dendritic cells. Additionally, correlations between immune infiltrating cells and several genes, including BRCA1, were estimated. Ultimately, the relevance of BRCA1 expression to immune infiltrating cells in BLCA patients was visualized using “ggsci” packages (v 3.0.0). (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns=no significance).
Construction of PPI, Gene-Gene Interaction Network
The STRING database (https://string-db.org/) was accessed to analyze the biological function of BRCA1, from which a visualization result was obtained. The GeneMANIA database (http://genemania.org/) was used to construct a gene-gene interaction network. This network helped determine the correlation between BRCA1 and other genes for co-expression network construction.
Immunohistochemistry
Human bladder cancer tissues were collected from 13 patients at Harbin Medical University Cancer Hospital. The inclusion criteria were as follows: diagnosis of BLCA at Harbin Medical University Affiliated Cancer Hospital; no other primary malignant tumors; survival time exceeding 30 days; non-metastatic; absence of other cancer cell variants; no prior intravesical treatment; and no perineural invasion.
Immunohistochemistry (IHC) analysis was conducted on both the bladder tumor tissues and adjacent normal tissues.23 Tissue sections, 3μm thick, were prepared and incubated with BRCA1 rabbit polyclonal antibody (1:100, Affinity Biosciences, Polyclonal Rabbit Antibody, AF-6289) overnight at 4°C, followed by a secondary antibody for 30 minutes at 37°C. Visualization was achieved using DAB reagent for 10 minutes, and nuclei were counterstained with DAPI (blue).
Statistical Analysis
The Wilcoxon test was used to compare two groups, while the Kruskal–Wallis test analyzed differences among three groups. The chi-square test assessed the objective response rate. Survival differences were evaluated through Kaplan–Meier analysis, and Pearson’s correlation analysis examined the relationship between two variables.
All statistical analyses were performed using R (v4.3.0), with p<0.05 considered statistically significant.
Results
Expression Levels of BRCA1 in BLCA and Normal Tissues
The GEPIA database was used to analyze the expression of BRCA1 across various cancer types, revealing elevated expression in BLCA (Figure 1A). Further analysis confirmed high BRCA1 expression in most BLCA patients (Figure 1B and C). Comparative analysis between bladder carcinoma tissues and normal tissues showed significant differences in BRCA1 expression (Figure 1D). Subsequent verification of BRCA1’s differential expression in BLCA was conducted using the GEPIA database (Figure 1E). Expression differences of BRCA1 were also analyzed across different genders, stages, tumor stages, lymph node stages, and metastasis stages (Figure 1F). According to the eighth edition of the AJCC, the staging was standardized. A negligible number of patients at T0 stage precluded meaningful expression analysis, and no significant differential expression was observed (Figure 1G). Similarly, the small sample sizes in T0 and T1 stages limited the display of expression data.
Survival Prognosis Analysis of BRCA1
Survival data from the UCSC database were utilized to explore the relationship between BRCA1 expression and BLCA survival, employing a Receiver Operating Characteristic Curve. BRCA1 showed modest predictive value for overall survival in BLCA (AUC=0.52) (Figure 2A). Further analysis indicated that BRCA1 predicted first-year survival moderately (AUC=0.56); however, predictions for third and fifth-year survival were less accurate than random chance (AUC=0.48, AUC=0.46) (Figure 2B). Cox regression analysis was performed on several randomly selected genes, including BRCA1, which was not considered an independent prognostic factor for survival (Cox HR=1.034, P=0.771) (Figure 2C). To assess the significance of BRCA1 expression in BLCA prognosis, TCGA cohort patients were divided into “BRCA1-high” and “BRCA1-low” groups based on median expression levels; no significant difference in overall survival (OS) time was found between these groups (P=0.666) (Figure 2D).
BRCA1 Associated Gene-Gene Interaction and Co-Expression Network
To explore the biological function of BRCA1, a PPI network was established in the STRING database (Figure 3A). A gene-gene interaction network relevant to BRCA1 was also constructed using the GeneMANIA database (Figure 3B). It was noted that genes such as BARD1 and PARP1 were involved in both the PPI and gene-gene interaction networks. The Gene Set Enrichment Analysis (GSEA) of the BRCA1 co-expression network identified the top seven enriched GO terms, including epidermis development, epidermal cell differentiation, epithelium development, skin development, epithelial cell differentiation, muscle system process, and muscle contraction (all p<0.001), which are associated with BLCA.
Figure 3 Continued. Figure 3 Continued. Figure 3 Continued.


Immunological Analysis of BRCA1 in BLCA
The impact of immune cell infiltration on BRCA1 expression in UCSC subsets was investigated (Figure 4A). The relevance of immune cell infiltration in BLCA was further identified (Figure 4B). Analysis revealed that resting NK cells, activated CD4 memory T cells, and follicular helper T cells exhibited high relevance, while activated NK cells, resting CD4 memory T cells, resting mast cells, and Tregs showed a negative correlation (Figure 4C). Additionally, the influence of BRCA1 expression on immune cell infiltration in BLCA patients was examined; it was found that activated CD4 memory T cells, resting NK cells, and M1 macrophages were positively correlated, whereas plasma cells, resting mast cells, resting CD4 memory T cells, and Tregs showed negative relevance in the BRCA1-high group (Figure 4D).
Validation of BRCA1 Expression in BLCA
Bladder carcinoma tissues and adjacent normal tissues were collected from 13 patients who had never received intravesical treatment. Clinical information was compiled in Table 1. Immunohistochemistry (IHC) revealed strong and diffuse cytoplasmic staining in tumor tissues, whereas weaker staining was observed in adjacent normal tissues (Figure 5). IHC confirmed that BRCA1 expression was higher in bladder tumor tissues compared to adjacent normal tissues.
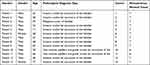 |
Table 1 Patient Information Participating in Immunohistochemistry Experiments |
 |
Figure 5 Typical IHC images illustrating BRCA1 staining in bladder carcinoma and adjacent normal tissues. |
Discussion
BLCA is classified as a high-risk malignant tumor.24 Early-stage diagnosis and prognostic prediction are crucial. Most current prediction models rely on mRNAs, miRNAs, or clinical characteristics.25–27 Moreover, infiltrating immunocytes significantly influence BLCA pathogenesis.28 BLCA responds to immunotherapy, such as Bacillus Calmette-Guérin therapy, and is characterized by a relatively high tumor mutational burden.29 For BLCA patients, a high density of tumor-infiltrating CD8+ T cells has been identified as a beneficial prognostic factor, whereas programmed death-ligand 1 expression and tumor-associated macrophages are linked to poorer outcomes.30–32 Therefore, studying the interaction between BLCA and tumor-infiltrating immunocytes could be beneficial for treatment.
In our analysis, bioinformatics tools such as UCSC, GEPIA, STRING, and GeneMANIA were utilized to investigate BLCA. We noted elevated expression levels of BRCA1 in BLCA tissues. Additionally, our findings indicate that BRCA1 expression does not significantly differ across genders or stages, including various tumor stages and metastatic statuses. Patients undergoing intravesical treatment were excluded from the standard analysis. Cox regression analysis identified the prognostic data, suggesting that BRCA1 has a moderate predictive value for BLCA survival. The hazard ratio (HR) with a 95% confidence interval (CI) was calculated, and patients were categorized into low or high-expression groups based on the median score to compare survival times between the two groups. Ultimately, BRCA1 was not considered an independent risk factor for BLCA survival, indicating the need for more effective models.
GSEA results showed that BRCA1 is involved in pathways such as epidermis development, epidermal cell differentiation, epithelium development, skin development, epithelial cell differentiation, muscle system process, and muscle contraction,33 which are implicated in BLCA. Previous research has highlighted the significant role of these pathways in BLCA, indicating BRCA1’s involvement in BLCA development through these mechanisms.
It has been reported that tumors can evade immune surveillance if the tumor microenvironment loses immune function.34 Xiao et al found that immune-related genes involved in T-cell activation, NK cell activity, and other biological processes were strongly correlated with overall survival (OS).35 In this study, positive correlations were observed between BLCA and activated CD4 memory T cells, resting NK cells, and M1 macrophages, while negative correlations were noted with resting plasma cells, mast cells, resting CD4 memory T cells, and Tregs. Recent research has indicated that understanding the impact of immune-related genes can improve early-stage prognostic assessments of BLCA.36 Thus, our study may provide new insights for evaluating and treating early-stage BLCA. Immunohistochemistry (IHC) analysis showed that BRCA1 expression was more pronounced in bladder tumor tissues than in adjacent normal tissues. Exclusion of patients receiving intravesical treatment was maintained as a standard.
Several biological processes, including DNA damage repair, transcription, ubiquitylation, cell-cycle checkpoints, and centrosome duplication, are associated with the tumor suppressor gene BRCA1’s protein product.37–40 Large rearrangements or whole gene deletions of BRCA1 are linked to an increased risk of hereditary breast and/or ovarian cancer.41–44 However, the relationship between BRCA1 and BLCA has been minimally explored. Our research initially assessed the diagnostic values of BRCA1 in BLCA, revealing significant differences between patients and normals; such difference analysis could enhance BLCA diagnosis in clinical practice. Our survival prognosis analysis indicated no significant difference in OS time between high and low BRCA1-expressing patients, although BRCA1 showed some predictive value for first-year survival in BLCA. Exploring the reasons for these results may provide insights for future clinical therapies. Additionally, we investigated the biological function of BRCA1 and its correlation with the immunological characteristics of BLCA, finding a strong positive correlation with immune cell infiltration and immune checkpoints. These findings could enable more precise therapies for patients in the future, assisting physicians in making effective treatment decisions based on pathways influenced by high BRCA1 expression.
Overall, our research comprehensively highlighted the diagnostic and prognostic significance of BRCA1 in BLCA, as well as its relationship with immunological characteristics, potentially enhancing therapeutic approaches. However, our study has limitations. Using public databases for prognostic and therapeutic analysis may introduce biases. Further exploration of BRCA1’s role in BLCA is needed through cellular function experiments. Lastly, validating our findings with a small, unbalanced cohort (10 of 13 cases being invasive disease) is insufficient. More clinical data and experimental research are required to confirm our conclusions.
Conclusions
Bioinformatic methods and clinical validation were employed to conduct a systematic and comprehensive analysis of BRCA1 in BLCA. Our study suggests that BRCA1 acts as a predictive factor for BLCA, potentially aiding in its diagnosis. Furthermore, due to its high accuracy in immunological analysis, BRCA1 may guide personalized therapies in BLCA. Additionally, our findings indicate that GSEA of BRCA1 could elucidate its role in BLCA, potentially improving treatment accuracy. This research is expected to further our understanding of BRCA1’s function and contribute to more precise therapies for BLCA.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are not publicly available due to patient confidentiality requirements, but are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study adhered to the Declaration of Helsinki and received approval from the Ethics Committee of Harbin Medical University Cancer Hospital (No. KY2024-40). All procedures were performed in accordance with relevant guidelines and regulations, and written informed consent was obtained from all participants.
Author Contributions
All authors made significant contributions to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by the Beijing Medical Award Foundation (YXJL-2022-0080-0014).
Disclosure
Xinyu Zhang, Xiaoxuan Tao, and Yuxin Zhou are co-first authors for this study. Guangyue Shi and Tianjiao Wang are corresponding authors. Among them, Guangyue Shi is the main corresponding author. All authors declare no competing interests in this work.
References
1. Xie R, Xie M, Zhu L, Chiu JWY, Lam W, Yap DYH. The relationship of pyroptosis-related genes, patient outcomes, and tumor-infiltrating cells in bladder urothelial carcinoma (BLCA). Front Pharmacol. 2022;13:930951. doi:10.3389/fphar.2022.930951
2. Yang Y, Yu J, Xiong Y, Xiao J, Dai D, Zhang F. Prognostic analysis of differentially expressed DNA damage repair genes in bladder cancer. Pathol Oncol Res. 2022;28:1610267. doi:10.3389/pore.2022.1610267
3. Li H, Chen S, Mi H. A multiomics profiling based on online database revealed prognostic biomarkers of BLCA. Biomed Res Int. 2022;2022:2449449. doi:10.1155/2022/2449449
4. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi:10.1016/j.eururo.2016.06.010
5. Ding Y, Liu N, Chen M, et al. Overexpressed pseudogene MT1L associated with tumor immune infiltrates and indicates a worse prognosis in BLCA. World J Surg Oncol. 2021;19(1):133. doi:10.1186/s12957-021-02231-4
6. Wu Z, Feng Z, Wei H, Lin C, Chen K. Development and validation of prognostic index based on purine metabolism genes in patients with bladder cancer. Front Med Lausanne. 2023;10:1193133. doi:10.3389/fmed.2023.1193133
7. Al-Moghrabi N, Al-Showimi M, Al-Yousef N; MicroRNA-155-5p AL. Reduced by curcumin-re-expressed hypermethylated brca1, is a molecular biomarker for cancer risk in BRCA1-methylation carriers. Int J Mol Sci. 2023;24(10):9021. doi:10.3390/ijms24109021
8. Shehaj I, Krajnak S, Almstedt K, et al. BRCA1, BRCA2 and PALB2 mRNA expression as prognostic markers in patients with early breast cancer. Biomedicines. 2024;12:1361. doi:10.3390/biomedicines12061361
9. Nelson N, Jigo R, Clark GJ. BRCA1 and NORE1A Form a Her2/ras regulated tumor suppressor complex modulating senescence. Cancers. 2023;15(16):4133. doi:10.3390/cancers15164133
10. Diabate M, Islam MM, Nagy G, et al. DNA repair function scores for 2172 variants in the BRCA1 amino-terminus. bioRxiv. 2023. doi:10.1101/2023.04.10.536331
11. Hou M, Sun L, Xiuzhang Y, et al. The landscape of BRCA1 and BRCA2 alterations in Chinese ovarian cancer patients. J clin oncol. 2023;41:e17565–e17565. doi:10.1200/JCO.2023.41.16_suppl.e17565
12. Wenjing L, Witus S, Wang M, Brzovic P, Klevit R, Zhao W. Abstract 6096: BRCA1-BARD1 ubiquitylates histones for genome maintenance. Cancer Res. 2023;83:1.
13. Zhong AX, Chen Y, Chen PL. BRCA1 the versatile defender: molecular to environmental perspectives. Int J Mol Sci. 2023;24(18):14276. doi:10.3390/ijms241814276
14. Chida K, Oshi M, Roy A, et al. Enhanced cancer cell proliferation and aggressive phenotype counterbalance in breast cancer with high BRCA1 gene expression. Breast Cancer Res Treat. 2024. doi:10.1007/s10549-024-07421-8
15. Amin N, Ahmed B, Abou-Bakr A, et al. The impact of BRCA1 expression on survival status in ovarian serous carcinoma of Egyptian patients. Asian Pac J Cancer Prev. 2023;24:3613–3620. doi:10.31557/APJCP.2023.24.10.3613
16. Deng D, Li X, Qi T, Dai Y, Liu N, Li H. A novel platelet risk score for stratifing the tumor immunophenotypes, treatment responses and prognosis in bladder carcinoma: results from real-world cohorts. Front Pharmacol. 2023;14:1187700. doi:10.3389/fphar.2023.1187700
17. Zhang X, Zhou Y, Hu J, et al. Comprehensive analysis identifies cuproptosis-related gene DLAT as a potential prognostic and immunological biomarker in pancreatic adenocarcinoma. BMC Cancer. 2023;23(1):560. doi:10.1186/s12885-023-11042-7
18. Khan SM, Das T, Chakraborty S, et al. A transcriptome study of p53-pathway related prognostic gene signature set in bladder cancer. Heliyon. 2023;9(10):e21058. doi:10.1016/j.heliyon.2023.e21058
19. Wang X, Bai Y, Zhang F, et al. Prognostic value of COL10A1 and its correlation with tumor-infiltrating immune cells in urothelial bladder cancer: a comprehensive study based on bioinformatics and clinical analysis validation. Front Immunol. 2023;14:955949. doi:10.3389/fimmu.2023.955949
20. Hezhen L, Yue S, Yang Z, Chengyan H, Pan J. Prognostic value of iron metabolism-related genes in bladder urothelial carcinoma. Oncologie. 2023;25:10.1515/oncologie–2023–0145.
21. Zhang MZ, Hu J, Zhang P, et al. Development and validation of cancer-associated fibroblasts-related gene landscape in prognosis and immune microenvironment of bladder cancer. Front Oncol. 2023;13:1174252. doi:10.3389/fonc.2023.1174252
22. Chen S, Ran J, Fan Z, et al. Functional status analysis of RNH1 in bladder cancer for predicting immunotherapy response. Sci Rep. 2023;13(1):12625. doi:10.1038/s41598-023-39827-7
23. Lai Z, Bai Z, Yang S, Zhang R, Xi Y, Xu J. Hub genes in adenocarcinoma of the esophagogastric junction based on weighted gene co-expression network analysis and immunohistochemistry. Transl Oncol. 2023;37:101781. doi:10.1016/j.tranon.2023.101781
24. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388:2796–2810. doi:10.1016/S0140-6736(16)30512-8
25. Jin K, Qiu S, Jin D, et al. Development of prognostic signature based on immune-related genes in muscle-invasive bladder cancer: bioinformatics analysis of TCGA database. Aging. 2021;13:1859–1871. doi:10.18632/aging.103787
26. Hofbauer SL, de Martino M, Lucca I, et al. A urinary microRNA (miR) signa- ture for diagnosis of bladder cancer. Urol Oncol. 2018;36:531.e531–8. doi:10.1016/j.urolonc.2018.09.006
27. Zhang Y, Hong YK, Zhuang DW, He XJ, Lin ME. Bladder cancer survival nomogram: development and validation of a prediction tool, using the SEER and TCGA databases. Medicine. 2019;98:e17725. doi:10.1097/MD.0000000000017725
28. Schneider AK, Chevalier MF, Derré L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol. 2019;16:613–630. doi:10.1038/s41585-019-0226-y
29. Chalasani V, Chin JL, Izawa JI. Histologic variants of urothelial bladder cancer and nonurothelial histology in bladder cancer. Can Urol Assoc J. 2009;3:S193–8.
30. Yu A, Mansure JJ, Solanki S, et al. Presence of lymphocytic infiltrate cytotoxic T lymphocyte CD3+, CD8+, and immunoscore as prognostic marker in patients after radical cystectomy. PLoS One. 2018;13:e0205746.
31. Yang M, Yu Q, Liu J, et al. T‑cell immunoglobulin mucin‑3 expression in bladder urothelial carcinoma: clinicopathologic correlations and association with survival. J Surg Oncol. 2015;112:430–435. doi:10.1002/jso.24012
32. Wang B, Liu H, Dong X, et al. High CD204+ tumor‑infiltrating mac‑rophage density predicts a poor prognosis in patients with urothelial cell carcinoma of the bladder. Oncotarget. 2015;6:20204–20214. doi:10.18632/oncotarget.3887
33. Jiang S, Ma J, Wei S, et al. Identification of hub genes associated with progression and prognosis of bladder cancer by integrated bioinformatics analysis. Arch Esp Urol. 2022;75(9):779–790. doi:10.56434/j.arch.esp.urol.20227509.114
34. Lawson KA, Sousa CM, Zhang X, et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586(7827):120–126. doi:10.1038/s41586-020-2746-2
35. Xiao Y, Dong Y, Yu T, et al. Characterization of the immune related lncRNAs in bladder cancer to aid immunotherapy. Front Immunol. 2022;13:941189. doi:10.3389/fimmu.2022.941189
36. Tang R, Wang H, Liu J, et al. (2023). Identification of hypoxia- and immune- based prognostic signature and validation of TFRC as a potential biomarker and therapeutic target in BLCA. 10.21203/rs.3.rs–3640530/v1.
37. Yoshino Y, Fang Z, Qi H, Kobayashi A, Chiba N. Dysregulation of the centrosome induced by BRCA1 deficiency contributes to tissue-specific carcinogenesis. Can-cer Sci. 2021;112:1679–1687. doi:10.1111/cas.14859
38. Zhao W, Wiese C, Kwon Y, Hromas R, Sung P. The BRCA tumor suppressor network in chromosome damage repair by homologous recombination. Annu Rev Biochem. 2019;88:221–245. doi:10.1146/annurev-biochem-013118-111058
39. Zhang X, Li R. BRCA1-dependent transcriptional regulation: implication in tissue-specific tumor suppression. Cancers. 2018;10(12):513. doi:10.3390/cancers10120513
40. Witus SR, Stewart MD, Klevit RE. The BRCA1/BARD1 ubiquitin ligase and its substrates. Biochem J. 2021;478(18):3467–3483. doi:10.1042/BCJ20200864
41. Fallat ME, Katz AL, Mercurio MR; Committee On Bioethics; Committee On Genetics, And; American College Of Medical Genetics And; Genomics Social; Ethical; Legal Issues Committee. Ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131(3):620–622. PMID: 23428972. doi:10.1542/peds.2012-3680
42. American College of Obstetricians and Gynecologists. Committee Opinion No. 693: counseling about genetic testing and communication of genetic test results. Obstet Gynecol. 2017;129(4):e96–e101. doi:10.1097/AOG.0000000000002020
43. Garcia-Casado Z, Romero I, Fernandez-Serra A, et al. A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer. BMC Med Genet. 2011;12:134. doi:10.1186/1471-2350-12-134
44. Hercher L, Uhlmann WR, Hoffman EP, Gustafson S, Chen KM. Public Policy Committee of NSGC. Prenatal testing for adult-onset conditions: the position of the national society of genetic counselors. J Genet Couns. 2016;25(6):1139–1145. doi:10.1007/s10897-016-9992-3
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





