Back to Journals » International Journal of Nanomedicine » Volume 19
Covalent Organic Framework-Based Theranostic Platforms for Restricting H1N1 Influenza Virus Infection
Authors Ding LG , Ji X, Liu YY, Shi M, Li JD, Liu F, Zhang YY, Yu J, Wu JQ
Received 29 January 2024
Accepted for publication 13 July 2024
Published 22 July 2024 Volume 2024:19 Pages 7399—7414
DOI https://doi.org/10.2147/IJN.S461866
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Luo-Gang Ding,1,* Xiang Ji,2,* Yue-Yue Liu,3,* Min Shi,1 Jian-Da Li,1 Fei Liu,1 Yu-Yu Zhang,1 Jiang Yu,1 Jia-Qiang Wu1
1Shandong Key Laboratory of Disease Control and Breeding, Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Jinan, 250100, People’s Republic of China; 2Key Laboratory of Shandong Microbial Engineering, College of Bioengineering, Qilu University of Technology (Shandong Academy of Sciences), Jinan, 250353, People’s Republic of China; 3Institute of Poultry Science, Shandong Academy of Agricultural Science, Jinan, 250100, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jia-Qiang Wu; Jiang Yu, Shandong Academy of Agricultural Sciences, Jinan, 250100, People’s Republic of China, Email [email protected]; [email protected]
Background: Influenza A (H1N1) virus is a highly contagious respiratory disease that causes severe illness and death. Vaccines and antiviral drugs are limited by viral variation and drug resistance, so developing efficient integrated theranostic options appears significant in anti-influenza virus infection.
Methods: In this study, we designed and fabricated covalent organic framework (COF) based theranostic platforms (T705@DATA-COF-Pro), which was composed of an RNA polymerase inhibitor (favipiravir, T705), the carboxyl-enriched COF (DATA-COF) nano-carrier and Cy3-labeled single DNA (ssDNA) probe.
Results: The multi-porosity COF core provided an excellent micro-environment and smooth delivery for T705. The ssDNA probe coating bound to the nucleic acids of H1N1 selectively, thus controlling drug release and allowing fluorescence imaging. The combination of COF and probe triggered the synergism, promoting drug further therapeutic outcomes. With the aid of T705@DATA-COF-Pro platforms, the H1N1-infected mouse models lightly achieved diagnosis and significantly prolonged survival.
Conclusion: This research underscores the distinctive benefits and immense potential of COF materials in nano-preparations for virus infection, offering novel avenues for the detection and treatment of H1N1 virus infection.
Keywords: crystal porous materials, microenvironment-responsive, controlled release, fluorescence imaging, diagnosis and therapy
Graphical Abstract:

Introduction
The influenza virus outbreak causes severe respiratory system infection and enormous challenges to public health.1–3 History has proved that influenza viruses spread will result in historical pandemics, which may cause hundreds of thousands of infected patients worldwide.4,5 For example, the novel H1N1 in North America caused thousands of deaths in 2009.6 Moreover, asymptomatic cases are hard to detect, aggravating virus transmission, especially in underdeveloped districts. Currently, the primary methods for preventing and controlling the spread of influenza depend on vaccines and antiviral drugs. However, the efficiency of vaccines is severely circumscribed by frequent variations of the influenza virus.7,8 Although the therapeutic drugs of M2 ion channel inhibitor and neuraminidase inhibitor were once effective against influenza virus, such as amantadine, rimantadine, and zanamivir,9–11 the resistance of these drugs was increased with the passage of time.12–15 Favipiravir (T705), as a viral RNA polymerase inhibitor, would play antiviral effects by blocking viral nucleic acid replication, which has received extensive attention since it might be a viable remedy for pandemic influenza virus infections. Nevertheless, its application is also hindered by low tabletability and permeability.16–18 Owing to the advances in nanotechnology and the development of nanomaterials in bio-medicine, intelligent platforms have become an alternative to the therapy of viral infections in recent years.19–22 However, most traditional nanoplatforms are not ideal for loading capacity, release effect, and improvement outcome on chemotherapy (mesoporous silica, nanoparticles, liposomes, etc),23,24 resulting in complicated specific imaging and time-consuming chemotherapy. Therefore, it is crucial to alleviate the therapy method for viral infections by establishing high-performance intelligent diagnostic and therapeutic platforms through a simple and effective strategy.
Covalent organic framework materials (COFs),25–27 a new type of organic porous crystalline materials with a periodic network structure, have attracted much attention given their extraordinary design-ability, permanent porosity, and precise porous structures since 2005. With the size of COFs reducing to the nanoscale, the potential of COFs has been explored for bioanalysis and nanomedicine,28 such as cancer treatment,29,30 drug delivery31,32 and cell imaging,33 which brings new opportunities for the detection and treatment of viral infection.34 In one study, it was reported that quinoline-4-carboxylic acid-linked and porphyrin-enriched COFs exhibited multifunctional antiviral activity via a triple-modal chemo/PDT/PTT synergistic treatment under natural sunlight irradiation, indicating a great potential of COF materials in the treatment of viral infection.35 Compared with effective therapy for influenza virus, COF also play a crucial role in accurately diagnosing influenza virus infection, facilitating the control of infection sources and illness progression.36–38 For instance, Li’s group has fabricated an electrochemical biosensor using magnetic covalent organic frameworks/pillararene hetero-supramolecular nano-composites as the signal probes, which exhibited accuracy and selectivity for detecting human norovirus (HuNoV).39 Although COF materials have exhibited promise in diagnosing and treating viral infection, limited research has focused on COF-based nanoplatforms against pathogenic viruses. To our knowledge, intelligent theranostic platforms with selective diagnosis and restriction of viruses are even rarer.
In this contribution, we reported an intelligent theranostic platform (T705@DATA-COF-Pro) based on a carboxyl-enriched COF (DATA-COF) whose channel was occupied with T705 and the external layer was modified by Cy3-labeled ssDNA.40 When the T705@DATA-COF-Pro encountered the H1N1-infected cell, the superficial Cy3-labeled ssDNA as efficient probes displayed a remarkable fluorescent signal, and the loaded T705 as an RNA polymerase inhibitor exhibited dramatic antiviral activity. Therefore, the obtained T705@DATA-COF-Pro showed specific imaging and efficient treatment for H1N1 virus infection. In vivo experiments, its exceptional theranostic performance has also been thoroughly elucidated via microenvironmental-responsive41–43 and drug delivery.44–48
Material and Methods
Materials
All the chemicals were obtained from commercial sources without further purification. 1,3,5-triformylphloroglucinol (TFP) was synthesized according to a previously reported method, and the characterization data was provided in the ESI (Figures S1 and S2 and Scheme S1). RNA oligonucleotides were synthesized and purified by Sangon Biotechnology Co., Ltd (Shanghai, China) and those oligonucleotide sequences were shown in Table S1.
Synthesis of the DATA-COF
A mixture of 2,5-diaminoterephthalic acid (14.7 mg, 0.075 mmol), 1,3,5-triformylphloroglucinol (10.5 mg, 0.05 mmol) and HOAc (6 M, 0.2 mL) in mesitylene/1,4-dioxane (2.0 mL, 3/1, v/v) was flash frozen in a liquid nitrogen bath and degassed. After capping, the tube was heated at 120°C for 72 h. The obtained precipitate was collected by centrifugation and washed with THF and ethanol. The solid was dried under vacuum at 120°C for 12 h to afford the DATA-COF crystals as black-red solids (20.0 mg, 88.7% yield). IR (KBr): ν = 3456 (m), 3351 (m), 3102 (w), 1710 (m), 1588 (s), 1426 (s), 1360 (s), 1262 (s), 1221 cm−1 (s); Solid-state 13C MAS NMR (δ, ppm): 184.1, 167.1, 145.6, 135.3, 121.6, 118.5, 108.3; Anal. calcd for (C14H8NO6)n: C, 56.05; N, 9.33; H, 2.67; found: C, 58.10; N, 8.79; H, 2.80.
Synthesis of the T705@DATA-COF
The T705 (157 mg, 1 mmol) and DATA-COF (10 mg) were mixed in methanol (25 mL) and stirred for 24 h. The product was collected by centrifugation and washed with methanol thrice for the unloaded. The drug was washed thoroughly and dried in a vacuum (60°C, 12 h) to prepare T705@DATA-COF. The loading capacity of T705 in T705@DATA-COF was given as 9.27 wt% via UV-Vis spectrophotometry.
Synthesis of the T705@DATA-COF-Pro
Based on previous reports,49 after the 3-(3-dimethylaminopropyl)carbodiimide (3 μL, 30 mg/mL) and the N-Hydroxy succinimide (1 μL, 100 mg/mL) were added into T705@DATA-COF (100 μL, 1 mg/mL), the mixture was stirred for 30 min, and collected by centrifugation. The obtained precipitate was redispersed in PBS (496 μL), and 4 μL of the Cy3-labeled ssDNA was added to the above solution. After shaking for 24 h in the dark, T705@DATA-COF-Pro was obtained by centrifugation. The final product was washed with PBS three times, and then the supernatant was collected to calculate the loading density of the Cy3-labeled ssDNA based on the fluorescence standard curve.
Fluorescent Detection of Target ssDNA
According to the references,37 the target ssDNA (50 nM) was added into T705@DATA-COF-Pro solution (100 μg/mL), and the mixture was incubated for 60 min at room temperature to hybridize adequately. The fluorescence was measured every 3 min at room temperature to record the response process (Ex = 525 nm). Establishment of fluorescence dependence curve: The different concentrations of target ssDNA (100–10 nM) were added into T705@DATA-COF-Pro solution for fluorescence detection at 30 min. Selectivity of target ssDNA assay: The fluorescence of T705@DATA-COF-Pro solution was detected at 30 min after adding into target ssDNA (50 nM) and mismatched ssDNA (50 nM).
Drug Controllable Release
To test the controllable release of T705, the T705@DATA-COF-Pro (20 mg) was immersed and shaken in 100 mL PBS solution at 37°C. The 2 mL solution was extracted and replaced by fresh PBS at a prescribed time interval until the concentration of T705 in the extract remained constant. The concentration of T705 in the sample was measured by detecting the UV absorbance at 323 nm.
Cytotoxicity Test
Cytotoxicity of T705@DATA-COF-Pro was determined using a cell counting CCK-8 kit (Boster Biological Technology Co. Ltd., Cat. #AR1160), and six parallel samples were made to eliminate experimental errors in each group. MDCK (Madin-Darby canine kidney cells, ATCC CCL-34), purchase from Shanghai EK-Bioscience Biotechnology Co., Ltd. MDCK cells were treated with sterilized DATA-COF, T705@DATA-COF, and T705@DATA-COF-Pro containing different concentrations (40–240 µg/mL) for 24 h at 37°C, and untreated counterparts served as the control group. The CCK-8 solution (10 µL) was added into each well and further incubated for 30 min. Each group was expressed as the percentage of viable cells relative to the control group.
Acquisition and Culture of H1N1 Virus
Acquisition: SIV (QD-2018 strain) was a H1N1 strain that isolated from the diseased pigs in Shandong province of China, which can be used as one of representative strains for the analysis of variant strains. The virus was continuously passaged in MDCK cells, and its titer was as high as 10–4.48 TCID50/mL. Culture: MDCK cells were maintained with 10% FBS DMEM medium at 37°C under 5% CO2. After seeding MDCK cells into a 24-well plate and culturing them to form a monolayer, the liquid in the plate was removed, and the cells were washed twice with PBS. Then, incubation with the H1N1 virus for 2 h, and the medium was replaced with 2% maintenance medium for 24 h. RNA or protein was collected according to the instructions for subsequent experiments.
Intracellular Prophylactic Test
MDCK cells were cultured in the 24-well plates containing DMEM (10% FBS) at 37°C with 5% CO2. Meantime, the cells were incubated with sterilized T705@DATA-COF-Pro or DMEM at 37°C for 2 h. Next, 25 µL of H1N1 virus (100 TCID50/mL) and 175 µL of DMEM were added to MDCK cells at 37°C, and removed after 1 h of infection. The monolayer cells were cultured separately with 500 µL of T705@DATA-COF-Pro (0, 25, 50, and 75 µg in 2% DMEM, respectively) or T705 (6.9 µg in DMEM, as positive control) at 37°C for 3, 6, 12, 18 and 24 h, respectively. Washed three times with PBS, the cells were collected and repeated freeze-thaw three times to gather the RNA of H1N1 (Cell DNA Isolation Mini Kit). After reverse transcription, the virus content of samples was evaluated via qRT-PCR for intracellular therapeutic effects.
Western Blot
MDCK cells were cultured to 60–80% confluence and exposed to H1N1 virus (100 TCID50/mL) for 1 h in the 6-well plates, and then the cells were incubated with T705@DATA-COF-Pro (0, 50, 100, 150 µg/mL) or 13.9 µg/mL of T705 for 24 h in 2% DMEM, respectively. To intuitively examine the cytopathic effects of virus-infected cells, 250 μL of lysis buffer was added to the cultured cells of each group. After centrifuging at 4°C for 20 min, the supernatant was analyzed by Western blot. Finally, the expression of the H1N1 NP (Abcam, Cat.#ab104870, 1:3000 diluted) protein was determined, and the amount of GADPH (Abcam, Cat.#ab181602, 1:5000 diluted) protein represented a housekeeping protein for control.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from virus-infected cells, and cDNA was carried out by reverse transcription following the kit instructions (Vazyme, Cat.#RC311-01). The antiviral activities of T705@DATA-COF-Pro were analyzed by qRT-PCR (SYBR Green Pro Taq HS, AG). The primers used were as follows: H1N1 forward (5′-ACAGAAGTTATAAGAATGA-3′), H1N1 reverse (5′-ACGGCCAGGTCATCACTATTG-3′); β-actin forward (5′-CCGTATCGGACGCCTGGTTA-3′), β-actin reverse (5′-AGGGGCCGGACTCATCGTA-3′). Fold change in gene expression level was calculated using the 2−ΔΔct method, and all PCR reactions were performed in triplicate.
Hemagglutination Assay
The assay was performed to detect the H1N1 virus by the hemagglutination assay. The process of cells incubated with the H1N1 virus was the same as Western blot. Then, 50 μL of the virus sample supernatant solution was serially diluted (2-fold dilution) and dispensed into each well of a 96-well microtiter plate. The chicken red blood cells (50 μL of 1.0% volume fraction) were added to each well and gently mixed. The maximum agglutination titer was recorded and captured by a camera after standing for 30 min under normal conditions.
Indirect Immunofluorescence Assay
The process of intracellular fluorescence imaging was the same as the intracellular therapy curve. In brief, MDCK cells were cultured to 60–80% confluence and exposed to the H1N1 virus (25 µL, 100 TCID50/mL) for 1 h in a 48-well plate, and then the free viruses were discarded through extensive washing. The mentioned cells were incubated with T705@DATA-COF-Pro (0, 50, 100, 150 µg/mL) or 13.9 µg/mL of T705 for 24 h in 2% DMEM, respectively. Subsequently, the cells were fixed with cold 4% paraformaldehyde, and the permeabilization was performed with 0.1% Triton under the ambient condition. Finally, the cells were incubated with H1N1 NP antibody (Abcam, Cat.#ab104870, 1:3000 diluted) for 1 h at 37°C, and the secondary antibody (HRP-conjugated goat anti-rabbit secondary antibody, Abways, Cat.#ab0141) for 30 min at 37°C. Then, the nucleus was stained with 4’, 6-diamidino-2-phenylindole (DAPI) and washed thrice with PBS. The fluorescence images were measured and analyzed by an Olympus IX73 microscope.
Hemolytic Test
Mice blood cells were collected and exposed to various concentrations of DATA-COF, T705@DATA-COF, and T705@DATA-COF-Pro (ranging from 0 to 800 μg/mL) for 4 h. Negative and positive controls were set using PBS and pure water, respectively. After incubation, the samples were centrifuged at 4000 rpm for 5 min, and the supernatant isolated was measured for absorbance using a Microplate reader.
Animal Infection and Treatment
All the animal experiments were approved by the Shandong Academy of Agricultural Sciences Animal Ethics Committees and conducted strictly in alignment with the “Laboratory Animal – Guidelines for Ethical Review of Animal Welfare” (GB/T 35892–2018) promulgated by the State Ministry of Science and Technology, as well as the ethical standards endorsed by the Ethics Committee of the Shandong Academy of Agricultural Sciences (Ethical Approval Number: IASVM-2023-004). The performance of T705@DATA-COF-Pro on the H1N1 diagnosis and healing process was evaluated by mice models infected with H1N1. Forty-two healthy female BALB/c nude mice (aged 5–6 weeks, Jinan Pengyue Laboratory Animal Breeding CO., Ltd., Shandong, China) were divided into seven groups randomly. The operation was as follows: seven groups were anesthetized via intraperitoneal injection. One of the seven groups was treated with normal saline by nasal dripping as a negative control group, while the other six groups were treated with 50 µL H1N1 virus (100 TCID50/mL) as experimental groups. After 24 h, the experimental groups were anesthetized and treated with 100 µL of T705@DATA-COF-Pro (0, 0.5, 1.0, and 2.0 mg/mL), DATA-COF (2.0 mg/mL), or T705@DATA-COF (2.0 mg/mL) via tail vein injection. The cumulative dose per unit of body mouse weight was calculated as 0, 3.13, 6.36, 13.03, 12.26, and 12.82 mg/kg, respectively. The clinical signs of every group were recorded daily, for instance, survival rate and weight change. All groups were sacrificed by cervical dislocation after chloral hydrate anesthesia on the 3, 7 and 14th days. The samples of lungs were taken, washed with normal saline, and ground to measure the content of H1N1-RNA via qRT-PCR. The remaining lung tissue was fixed with 4% paraformaldehyde for subsequent analysis.
To further assess the impact of the most active COF system on the lung of mice, six healthy female BALB/c nude mice (aged 5–6 weeks) were chosen as the independent control group. Following intraperitoneal anesthetization, the mice received 100 μL of T705@DATA-COF-Pro (2.0 mg/mL, cumulative dose of 12.41 mg/kg per unit body weight) via vein injection. Daily observations were made to document signs such as survival rate and weight changes. On the 3rd, 7th, and 14th day, the mice were euthanized via cervical dislocation post-anesthetization. The lung specimens were washed with normal saline and fixed in 4% paraformaldehyde for subsequent analysis and comparison.
Imaging Assay in vivo
After anesthetizing with 2% isoflurane, the tail vein injected 100 µL of T705@DATA-COF-Pro (2.0 mg/mL). To test the specific imaging of the COF platforms, 100 µL of normal saline, DATA-COF (2.0 mg/mL) and T705@DATA-COF (2.0 mg/mL) groups were added. The mice were randomly set with three mice in each group and observed by a live body imaging system within the specified time.
Histology and Immunohistochemistry
The five samples of the lungs on the 3, 7, and 14th days were fixed in paraformaldehyde, dehydrated, and embedded in paraffin. The tissue sections were cut into 5 µm thick slices for histological and immunohistochemical studies. For hematoxylin and eosin (H&E) staining, histopathological changes of the lungs in each group were observed. The nucleus was stained bright blue, and the cytoplasm exhibits pink to peach red, and the eosinophilic particles in the cytoplasm are strongly bright red. For immunohistochemistry, the tissue sections of the lungs on the 7th day were dewaxed with dimethylbenzene and rehydrated with alcohol. Before incubating with primary antibody (H1N1 NP) overnight at 4°C, the sections were quenched with 3% H2O2 and blocked. The washed sections were incubated with secondary antibody (HRP-labeled Goat Anti-Rabbit lgG) and dehydrated. All microscope images were taken under a Leica digital microscope.
Statistical Analysis
All the experiment results were exhibited as the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) along with Tukey’s multiple comparisons test was applied to evaluate the statistical significance (*P< 0.05, **P< 0.01, ***P< 0.001).
Results and Discussion
Synthesis and Characterization of COF
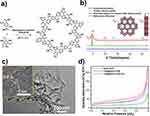
As depicted in Figure 1a, 1,3,5-triformylphloroglucinol (TFP) and 2,5-diaminoterephthalic acid (DATA) were polymerized with the acetic acid (6 M) under solvothermal conditions (mesitylene/1,4-dioxane, 2.0 mL, 3/1, v/v) 120°C for 72 h, the brownish red crystalline solid (named DATA-COF) was synthesized in 88.7% yield. The DATA-COF was collected after washing three times with THF and ethanol and drying under a vacuum at 120°C overnight. The FT-IR spectra showed the characteristic stretching peak at 1646 cm−1 for C=O in TFP disappeared, similar to the stretching mode at 3461 and 3345 cm−1 for N-H in DATA (Figure S3). Concurrently, the stretching vibrations at 1705 cm−1 associated with C=O species in a carboxylic group of DATA-COF were observed. Additionally, the stretching mode at 1596 and 1225 cm−1 were attributed to C=C and N-H species were found, which indicated the existence of β-ketoenamine in DATA-COF. Solid-state 13C CP-MAS NMR (Figure S4) also proved the successful formation of DATA-COF. The existence of resonance peaks at 184.1, 145.6, and 108.3 corresponded to the ketone and enamine carbon species in DATA-COF, respectively.50
The measured powder X-ray diffraction (PXRD) pattern of DATA-COF indicated that a good crystalline material was obtained (Figure 1b). The structural model of the PXRD pattern was simulated by the Material Studio (version 2018), which revealed DATA-COF was a 2D eclipsed AA-type stacking structure preferentially. The information of unit cell suggested DATA-COF was assigned to the P6 space group with the lattice parameters of a = b = 22.83 Å, c = 3.81 Å, α = β = 90.0, γ = 120.0, Rwp=10.14% and Rp = 7.75% (Figure S5 and Table S2). Based on structure analysis, the series of strong peaks in DATA-COF at 4.68, 9.38, 12.11, and 26.28 are associated with (100), (200), (210), and (001) planes, respectively. Moreover, the interlayer distance of DATA-COF was 3.8 Å based on the 2D layered simulated structure, which was proved by high-resolution transmission electron microscope (HRTEM) image (Figure 1c). The transmission electron microscope (TEM) and scanning electron microscopy (SEM) images demonstrated DATA-COF was a nanorod material with the average size of about 460 nm (Figure S6). Furthermore, the N2 adsorption curve of DATA-COF at 77 K was determined to be 230.3 cm3/g, and the Brunauer–Emmett–Teller (BET) specific surface area was 122.2 m2/g (Figure 1d). Based on Barrett–Joyner–Halenda analysis, the pore diameter of DATA-COF was concentrated distribution at 1.40 nm, which was in agreement with the simulated structure at 1.43 nm (Figure S7).
The T705@DATA-COF was prepared by solution impregnation (methanol solution of T705, RT, 24 h), whose loading amount of T705 was 9.27 wt% via the standard curves based on UV–Vis spectroscopy (Figure S8). Subsequently, after washing three times and Cy3-labeled ssDNA modification, T705@DATA-COF-Pro was fabricated successfully as a black-red crystalline solid (Scheme S2). The fluorescence standard curve estimated the DNA coverage as 25.3 nmol/mg (Figure S9). The FT-IR spectra indicated the amide bond formation at 1650 cm−1, which fully verified that Cy3-labeled ssDNA was successfully modified on the T705@DATA-COF (Figure S10). Moreover, in the XPS spectrum, the peak at 135.0 eV corresponding to P2p further evidenced the formation of T705@DATA-COF-Pro (Figure S11). Due to the loading of T705 and the modification of Cy3-labeled ssDNA, the N2 uptake amount and BET-specific surface area of T705@DATA-COF and T705@DATA-COF-Pro were reduced (141.1 cm3/g, 89.2 m2/g and 78.3 cm3/g, 42.7 m2/g), respectively (Figure 1d), and the apertures decreased correspondingly to 1.26 and 1.17 nm according to the Barrett–Joyner–Halenda analysis (Figure S7). As shown in Figure 2a and b, the SEM images and Dynamic light scattering (DLS) data declared that the morphology and size of T705@DATA-COF-Pro remained nanorods. The PXRD patterns of T705@DATA-COF and T705@DATA-COF-Pro were similar to the previous DATA-COF, indicating that the structural crystallinity of DATA-COF was well maintained during subsequent treatment (Figure 2c). Besides, SEM-EDX mapping displayed the homogeneous distribution of C, N, O, and F elements in T705@DATA-COF, and C, N, O, F, and P elements were also uniformly distributed in T705@DATA-COF-Pro, which further confirmed the successful preparation of uniform structure (Figure 2d). In addition, thermal gravimetric analysis (TGA) suggested that they all had good thermal stability, which was intact up to 200°C (Figure S12). Furthermore, the water contact angles of DATA-COF, T705@DATA-COF and T705@DATA-COF-Pro were found to be 0°±2° respectively, implying that they were hydrophilic materials (Figure S13).
Fluorescence Response and Drug Release Properties
The response-ability of T705@DATA-COF-Pro was explored as a fluorescent platform for detecting the target ssDNA (T). When the T (50 nM) was added into the system of T705@DATA-COF-Pro, it interacted with the Cy3-labeled ssDNA on T705@DATA-COF-Pro to form double-stranded DNA by base complementary pairing. Subsequently, the obtained double-stranded DNA is gradually stretched and removed from COF materials.51,52 Consequently, the quenched fluorescence was recovered rapidly and reached equilibrium after 30 min (Figure 3a). As shown in Figure 3b, the Cy3-labeled ssDNA probe exhibited strong fluorescence at 567 nm. Fluorescence emission of T705@DATA-COF-Pro was also maintained significantly in the presence of T (concentration of 50 nM). However, T705@DATA-COF-Pro could not be detected fluorescence basically, which implied its massive fluorescence quenching capability through fluorescence resonance energy transfer.40 Under the optimal conditions, the linear relationship between recovered fluorescence intensity (∆F) and concentration of T (C, nM) was tested from 10 to 100 nM (Figure 3c). The calibration function was calculated as ∆F=6.69 [C]+235.1 with the correlation coefficient (R2) of 0.976, and the detection limit of T was 2 nM, demonstrating intracellular mRNA can be visualized at the nanomolar level.53–55 To further investigate the selectivity of the biosensor, the same concentration of T, partially mismatched ssDNA (T1), and completely mismatched ssDNA (T2 and T3) were introduced into the solution of T705@DATA-COF-Pro separately (Figure 3d). With the increase of the mismatched base pairs, the intensity of the recovered fluorescence decreased sharply, purporting that the fluorescence intensity was closely related to the matching level of target base pair. The above proof indicated that the COF-based sensing platform had high selectivity for target mRNA, demonstrating a potential mRNA assay potential in biological samples.
To further investigate the controllable drug release of T705@DATA-COF-Pro in vitro, it was immersed in PBS solution at 25°C (Figure 3e). In the presence of T, the released rate of T705 in the COF platforms was observed at about 80% after 10 h via UV–Vis spectrum. The release efficiency was consistent with the T705@DATA-COF (10 h, about 86%), demonstrating that the modification of Cy3-labeled ssDNA did not affect drug release. On the contrary, with the disappearance of T, the released efficiency of T705 was less than 10% as wrapped Cy3-labeled ssDNA prevented drug leakage. The results indicated that the COF-based theranostic platforms were a stable drug delivery system with good control behaviors in drug release. In addition, biosafety for MDCK cells was evaluated by the CCK-8 kit. After incubation with different concentrations (40–240 µg/mL) of DATA-COF, T705@DATA-COF, and T705@TATA-COF-Pro for 24 h, the MDCK cells remained highly viable in comparison with the blank control, which indicated the developed COF systems did not exhibit significant cytotoxicity (Figure 3f and Figure S14). These results provided the base property for subsequent diagnosis and restriction of H1N1 virus infection.
Intracellular Antiviral and Locational Effect
With the above results in hand, the antiviral effect of T705@DATA-COF-Pro on the H1N1 virus in vitro was evaluated primarily. MDCK cells were infected with the H1N1 virus and subsequently treated for 24 h with different concentrations of T705@DATA-COF-Pro. To determine the survival rate of the H1N1 virus in MDCK cells, the gene expression levels of H1N1 were conducted and quantified based on RNA extraction and qRT-PCR. As shown in Figure 4a, the antiviral activities of T705@DATA-COF-Pro against H1N1 (25 μL, 100 TCID50/mL) were concentration-dependent, and the survival rates of H1N1 decreased proportionally with increasing dosage (50, 100, 150 μg/mL). The infected MDCK cells were treated with T705@DATA-COF-Pro at 150 μg/mL, and the H1N1 virus was inactivated to ca. 90%, which was basically consistent with the 13.9 μg/mL of the T705 group. Additionally, the antiviral effect of T705@DATA-COF-Pro was evaluated by standard hemagglutination assay (HA). The obtained agglutination patterns decreased with the increase of T705@DATA-COF-Pro content, which quite agreed with the results of gene expression (Figure 4b). Moreover, this argument was also approved by indirect immunofluorescence staining and Western blot. In comparison with positive groups, no green fluorescence was observed in the 150 μg/mL treated group, and it kept pace with the results of the T705 group (Figure 4c). The protein expression of H1N1 declined as well with the increase of T705@DATA-COF-Pro (Figure 4d and e). These results directly displayed that T705@DATA-COF-Pro had sound antiviral properties.
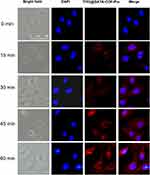
After demonstrating the T705@DATA-COF-Pro as an efficient platform for drug-specific delivery and treatment for H1N1 infection, we applied the theranostic platforms for H1N1 infected-cell imaging. While MDCK cells were incubated with 150 µg/mL of T705@DATA-COF-Pro, the fluorescence signals initially appeared in the cytoplasm, and gradually increased until it reached maximum fluorescence intensity at nearly 60 min (Figure 5). Next, the cellular uptake of T705@DATA-COF-Pro and sequence-specific sensing of target RNA were studied using fluorescence microscopy and flow cytometry. The H1N1 infected MDCK cells were cultured in a 12-well plate and treated with T705@DATA-COF-Pro (150 µg/mL) for 1 h, and observed by fluorescence microscopy and flow cytometry (Figure S15). Cell images showed strong Cy3-corresponding red fluorescence in the perinuclear region of infected cells treated with T705@DATA-COF-Pro. However, almost no Cy3 fluorescence was observed in uninfected cells treated with T705@DATA-COF-Pro, indicating that the ssDNA probe in T705@DATA-COF-Pro could restore fluorescence after hybridization with target H1N1 mRNA. Collectively, the data suggested that T705@DATA-COF-Pro could readily enter cells through the cell membrane without the assistance of transfection reagents, and the recognition of endogenous H1N1 mRNA by ssDNA probe and subsequent fluorescence recovery was sequence-specific.56 In addition, the hemolytic test was carried out to prove the reliability of the DATA-COF, T705@DATA-COF, and T705@DATA-COF-Pro material. Basically, no obvious hemolytic phenomenon was detected even at a high 800 μg/mL concentration, signifying their good biocompatibility (Figure S16).
Fluorescence Imaging and Treatment in vivo
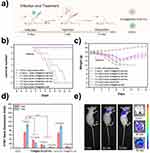
To evaluate the anti-H1N1 virus effect of T705@DATA-COF-Pro in vivo, BALB/c mice were inoculated with the H1N1 virus (50 μL, 100 TCID50/mL) through a nasal drip and treated with physiological saline, T705@DATA-COF-Pro, DATA-COF, or T705@DATA-COF. Mice of each group were observed the clinical symptoms and weight changes in the next 14 days (Figure 6a). In addition to the control group, all the virus-infected mice showed clinical symptoms on the second day, including loss of appetite, inactivity, weight loss and lethargy, and a majority even appeared obvious signs, such as severe dyspnea and skin folds. After showing severe symptoms, the unmedicated (0 mg/mL) and DATA-COF group (2.0 mg/mL) all died at 8 and 9 days after infection (Figure 6b). Compared with the above two groups, the clinical symptoms of T705@DATA-COF-Pro group were significantly improved in the observation period. Due to less dosage, the administration of 0.5 mg/mL group for the H1N1-infected mice only delayed the progression of the disease and failed to save the mice at 13 days. The 1.0 mg/mL group gave similar results, and partial death occurred in the mice for the reason of dosage.
In contrast, the infected mice in the 2.0 mg/mL group displayed milder symptoms except for weight loss at 2–7 days, and all mice survived (Figure 6b and c), which was confirmed with the group of T705@DATA-COF (2.0 mg/mL). The levels of H1N1-mRNA in lungs were conducted based on RNA extraction and qRT-PCR on 3 and 7 days (Figure 6d). In contrast with the unmedicated (0 mg/mL) and DATA-COF groups (2.0 mg/mL), the expression of H1N1 in the T705@DATA-COF-Pro treatment group was significantly less and had concentration-dependent, and the levels of H1N1-mRNA in lungs decreased proportionally with increasing dosage (0.5, 1.0, 2.0 mg/mL). Interestingly, the H1N1-level of lungs in the T705@DATA-COF-Pro group was lower than T705@DATA-COF group, demonstrating that the ssDNA not only performed an imaging function but also had a synergistic therapeutic effect with the T705 via a control behavior in drug release. This point was confirmed by the amount of T705 in infected mice’s blood using HRMS, and the results showed that the T705@DATA-COF-Pro group had a higher level than the T705@DATA-COF group on 3rd day (Figure S17). It might be the reason that the infected mice treated with T705@DATA-COF-Pro showed milder symptoms and survived the lethal challenge of H1N1 virus infection. These results demonstrated the effectiveness of T705@DATA-COF-Pro in combating the virus infection in vivo.
To investigate deeply impact of the COF system on the lung of mice, a distinct control group was established, consisting solely of mice injected with T705@DATA-COF-Pro. The outcomes showed that the body weight of mice in the group increased normally, and all of them survived, which was consistent with the results observed in the negative control group that received only PBS injection. Subsequently, the lung tissue sections were stained with H&E and observed under a fluorescence microscope. No signs of damage or pathological abnormalities were detected. These observations concurred with the results of the hemolysis experiment, confirming the excellent biocompatibility of T705@DATA-COF-Pro and highlighting its potential as a promising platform for viral treatment (Figure S18).
H&E staining and immunohistochemical test were executed to examine the pathological indices of lung tissue sections closely. As revealed in Figure 7, the positive-infected group was observed severe lung injury with diffuse swelling, bronchial edema, alveolar cavity collapse, and infiltration of inflammatory cells at 7 days, indicating that H1N1 infection destroyed the spongy structure and the lungs architecture.57 The DATA-COF treatment group had almost no therapeutic effect compared to the positive-infected group. In contrast, the symptoms of the T705@DATA-COF-Pro and T705@DATA-COF groups were lessened substantially. Critically, T705@DATA-COF-Pro displayed more significantly attenuated histopathological manifestations on alleviating cell infiltration and thrombosis. The results were consistent with the expression level of H1N1 in the lungs, further confirming that the targeted release of COF-based theranostic platforms had a facilitative therapeutic effect (Figure 6d). The pathological features of T705@DATA-COF-Pro group got obviously mitigation with time (Figure S19), suggesting that T705@DATA-COF-Pro had a better protective impact on acute lung injury caused by H1N1 virus infection.
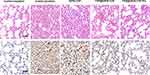 |
Figure 7 Pathological examination of lungs determined by H&E staining and immunohistochemistry (The red arrow refers to H1N1 NP protein, scale bar: 50 μm). |
Moreover, the diagnostic potential of T705@DATA-COF-Pro in vivo was explored with the H1N1-infected nude mice as models. The infected mice of each group were treated with 2.0 mg/mL of T705@DATA-COF-Pro, T705@DATA-COF and DATA-COF via tail vein injection, respectively, and observed by a live body imaging system within the specified time. The results indicated fluorescence signals mainly concentrated in mice’s lungs, trachea, and kidneys (Figure S20). Importantly, in vivo fluorescence imaging analysis of T705@DATA-COF-Pro showed high lung accumulation, purporting the potential of T705@DATA-COF-Pro for H1N1-mRNA imaging in vivo (Figure S21). Besides, no fluorescence signals were found in DATA-COF and T705@DATA-COF groups, indicating that the imaging derived from Cy3-labeled ssDNA modified on the COF. As Figure 6e shows, the fluorescence signal gradually increased with the prolongation of treatment time, which coincided with the fluorescence signal intensity in the lungs. These results revealed that T705@DATA-COF-Pro delivered to lung cells specifically and showed a rapid fluorescence response to H1N1-mRNA in vivo.
It is significant to establish an ideal diagnostic and therapeutic platform against H1N1 pathogens. Based on the above experiments, we presumed the working mechanism of T705@DATA-COF-Pro to explain its potential action in imaging and treatment. Figure S22 shows the preparation procedure of T705@DATA-COF-Pro. The pores of DATA-COF were first filled with T705, and then the surface was modified by Cy3-ssDNA. Due to the COF material could excellently form π-π interaction with single-stranded DNA, the post-modified Cy3-ssDNA was adsorbed on the surface of DATA-COF, and the Cy3-fluorophore exhibited fluorescence quenching behavior through energy transfer. Besides, the tightly wrapped Cy3-ssDNA protects T705 leakage in DATA-COF. According to the literature,58–61 in the presence of the target H1N1-RNA in infected cells, T705@DATA-COF-Pro could selectively bind to the nucleic acid of the virus. Primarily, the Cy3-ssDNA adsorbed on the surface of the DATA-COF would form stable double-stranded with the H1N1-RNA by base complementary pairing. Due to the double-stranded DNA having poor π–π interactions with COF, Cy3-ssDNA begins to stretch, gradually detach from the COF, and the fluorescence of Cy3 recovers.52 Subsequently, the T705 loaded in the pores of T705@DATA-COF-Pro also releases normally.
Conclusion
In summary, we successfully synthesized a carboxyl-enriched COF (DATA-COF) and established smart integrated theranostic platforms (T705@DATA-COF-Pro) via solution impregnation and sequential post-synthetic modification. Cy3-labeled ssDNA, wrapped tightly on the surface of T705@DATA-COF-Pro, could selectively identify the target ssDNA and prevent loaded drug leakage. When target ssDNA was present, T705@DATA-COF-Pro released the drug almost 80% and emitted strong fluorescence following complementary base pairing. Furthermore, T705@DATA-COF-Pro demonstrated dramatic specific imaging and therapeutic effects on H1N1 virus infection, which provided intelligent theranostic platforms for addressing the issue of H1N1 virus infection facilely.
Acknowledgments
We are grateful for financial supports from National Key Research and Development Program (2022YFD1800904, 2022YFD1800905), National Natural Science Foundation of China (22305145), Natural Science Foundation of Shandong Province (ZR2023QB074), Key R&D Program of Shandong Province, China (2022TZXD0041, 2020CXGC010801), The Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2023F10, CXGC2023A21), Shandong Province Pig Industrial Technology System (SDAIT-08-01), Taishan Scholars Programs.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Krammer F, Smith GJD, Fouchier RAM, et al. Influenza. Nat Rev Dis Primers. 2010;15:3. doi:10.1038/s41572-018-0002-y
2. Petrova VN, Russell CA. The evolution of seasonal influenza viruses. Nat Rev Microbiol. 2018;16:47–60. doi:10.1038/nrmicro.2017.118
3. Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7(6):440–451. doi:10.1016/j.chom.2010.05.009
4. Verity R, Okell LC, Dorigatti P, et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect Dis. 2020;20(6):669–677. doi:10.1016/S1473-3099(20)30243-7
5. Lu RJ, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi:10.1016/S0140-6736(20)30251-8
6. Yang Y, Sugimoto JD, Halloran ME, et al. The transmissibility and control of pandemic influenza a (H1N1) virus. Science. 2009;326(5953):729–733. doi:10.1126/science.1177373
7. Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23(4):222–228. doi:10.1016/j.cmi.2017.02.009
8. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev. 2015;14:167–182. doi:10.1038/nrd4529
9. He WJ, Zhang WX, Yan HX, et al. Distribution and evolution of H1N1 influenza a viruses with adamantanes-resistant mutations worldwide from 1918 to 2019. J Med Virol. 2021;93(6):3473–3483. doi:10.1002/jmv.26670
10. Belshe RB, Burk B, Newman F, et al. Resistance of influenza a virus to amantadine and rimantadine: Results of one decade of surveillance. J Infect Dis. 1989;159(3):430–435. doi:10.1093/infdis/159.3.430
11. Zhang WX, Xu HF, Guan SX, et al. Frequency and distribution of H1N1 influenza a viruses with oseltamivir-resistant mutations worldwide before and after the 2009 pandemic. J Med Virol. 2022;94(9):4406–4416. doi:10.1002/jmv.27870
12. Whitley RJ, Monto AS. Resistance of influenza virus to antiviral medications. Clin Infect Dis. 2020;71(4):1092–1094. doi:10.1093/cid/ciz911
13. Holmes EC, Hurt AC, Dobbie Z, et al. Understanding the impact of resistance to influenza antivirals. Clin Microbiol Rev. 2020;34(2):e00224–20. doi:10.1128/cmr.00224-20
14. Poland GA, Jacobson RM, Ovsyannikova IG. Influenza virus resistance to antiviral agents: a plea for rational use. Clin Infect Dis. 2009;48(9):1254–1256. doi:10.1086/598989
15. Li Y, Xiao YS, Chen YC, et al. Nano-based approaches in the development of antiviral agents and vaccines. Life Sci. 2021;265:118761. doi:10.1016/j.lfs.2020.118761
16. Wang XY, Wang L, Yao CL, et al. Novel formulations of the antiviral drug favipiravir: improving permeability and tabletability. Cryst Growth Des. 2021;7(21):3807–3817. doi:10.1021/acs.cgd.1c00150
17. Cain M, Ly H. In vitro and in vivo testing of the antiviral effect of favipiravir (T705) against the neurotropic chandipura virus. J Med Virol. 2023;95(7):e28960. doi:10.1002/jmv.28960
18. Dauby N, Praet SV, Vanhomwegen C, et al. Tolerability of favipiravir therapy in critically ill patients with covid-19: a report of four cases. J Med Virol. 2020;93(2):689–691. doi:10.1002/jmv.26488
19. Jackman JA, Yoon BK, Ouyang L, et al. Biomimetic nanomaterial strategies for virus targeting: antiviral therapies and vaccines. Adv Funct Mater. 2020;31(12):6606–6622. doi:10.1002/adfm.202008352
20. Innocenzi P, Stagi L. Carbon-based antiviral nanomaterials: graphene, C-dots, and fullerenes. a perspective. Chem Sci. 2020;11:6606–6622. doi:10.1039/D0SC02658A
21. Wang WB, Zhai W, Chen Y, et al. Two-dimensional material-based virus detection. Sci China Chem. 2022;65:497–513. doi:10.1007/s11426-021-1150-7
22. Miao XY, Yin YY, Chen YL, et al. Bidirectionally regulating viral and cellular ferroptosis with metastable iron sulfide against influenza virus. Adv Sci. 2023;10(17):e2206869. doi:10.1002/advs.202206869
23. Gao P, Pan W, Li N, et al. Boosting cancer therapy with organelle-targeted nanomaterials. ACS Appl Mater Inter. 2019;30(11):26529–26558. doi:10.1021/acsami.9b01370
24. Xu XC, Zhang J, Liu S, et al. New advances in nanomaterial-based antiviral strategies. Small Struct. 2022;3(7):2200021. doi:10.1002/sstr.202200021
25. Cote AP, Benin AI, Ockwig NW, et al. Porous, crystalline, covalent organic frameworks. Science. 2005;310(5751):1166–1170. doi:10.1126/science.1120411
26. Ding SY, Wang W. Covalent organic frameworks (COFs): from design to applications. Chem Soc Rev. 2013;42:548–568. doi:10.1039/C2CS35072F
27. Liu RY, Tan KT, Gong YF, et al. Covalent organic frameworks: an ideal platform for designing ordered materials and advanced applications. Chem Soc Rev. 2021;50:120–242. doi:10.1039/D0CS00620C
28. Li SM, Zou J, Tan LF, et al. Covalent organic frameworks: from linkages to biomedical applications. Chem Eng J. 2022;446:137418. doi:10.1016/j.cej.2022.137148
29. Ren WX, Kong F, Shao YQ, et al. A covalent organic framework with a self-contained light source for photodynamic therapy. Chem Commun. 2022;58:5245–5248. doi:10.1039/D2CC01397E
30. Gao P, Wang MZ, Chen YY, et al. A COF-based nanoplatform for highly efficient cancer diagnosis, photodynamic therapy and prognosis. Chem Sci. 2020;11:6882–6888. doi:10.1039/D0SC00847H
31. Guo H, Liu YS, Wu N, et al. Covalent organic frameworks (COFs): a necessary choice for drug delivery. ChemistrySelect. 2022;7(34):20220253. doi:10.1002/slct.202202538
32. Ge L, Qiao CY, Tang YK, et al. Light-activated hypoxia-sensitive covalent organic framework for tandem-responsive drug delivery. Nano Lett. 2021;7(21):3218–3224. doi:10.1021/acs.nanolett.1c00488
33. Kang S, Ahn H, Park P, et al. In vivo fluorescence molecular imaging using covalent organic nanosheets without labeling. Adv Sci. 2023;10(16):e2300462. doi:10.1002/advs.202300462
34. Nematollahi MH, Mostafavi E, Iravani S. Covalent organic frameworks and metal-organic frameworks against pathogenic viruses and antibiotic-resistant bacteria: diagnostic and therapeutic applications. J Environ Chem Eng. 2023;11(3):109652. doi:10.1016/j.jece.2023.109652
35. Ding LG, Wang S, Yao BJ, et al. Covalent organic framework based multifunctional self-sanitizing face masks. J Mater Chem A. 2022;10:3346–3358. doi:10.1039/D1TA08743F
36. Pirzada M, Altintas Z. Nanomaterials for virus sensing and tracking. Chem Soc Rev. 2022;51:5805–5841. doi:10.1039/D1CS01150B
37. Tian YY, Lu QP, Guo XX, et al. Au nanoparticles deposited on ultrathin two-dimensional covalent organic framework nanosheets for in vitro and intracellular sensing. Nanoscale. 2020;12:7776–7781. doi:10.1039/C9NR08220D
38. Wang MH, Lin YX, Lu JY, et al. Visual naked-eye detection of SARS-COV-2 RNA based on covalent organic framework capsules. Chem Eng J. 2022;429:132332. doi:10.1016/j.cej.2021.132332
39. Zhao H, Xie W, Zhang RL, et al. Electrochemical sensor for human norovirus based on covalent organic framework/pillararene heterosupramolecular nanocomposites. Talanta. 2022;237:122896. doi:10.1016/j.talanta.2021.122896
40. Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Clin Pharmacol Ther. 2020;209:107512. doi:10.1016/j.pharmthera.2020.107512
41. Xing F, Xu J, Zhou Y, et al. Recent advances in metal-organic frameworks for stimuli-responsive drug delivery. Nanoscale. 2024;16(9):4434–4483. doi:10.1039/d3nr05776c
42. Huang C, Zhou S, Chen C, et al. Biodegradable redox‐responsive AIEgen‐based‐covalent organic framework nanocarriers for long‐term treatment of myocardial ischemia/reperfusion injury. Small. 2022;18(47):2205062. doi:10.1002/smll.202205062
43. Di Cristo F, Valentino A, De Luca I, et al. PLA nanofibers for microenvironmental-responsive quercetin release in local periodontal treatment. Molecules. 2022;27(7):2205. doi:10.3390/molecules27072205
44. Ghosh P, Banerjee P. Drug delivery using biocompatible covalent organic frameworks (COFs) towards a therapeutic approach. Chem Commun. 2023;59(84):12527–12547. doi:10.1039/d3cc01829f
45. Zhang G, Li X, Liao Q, et al. Water-dispersible PEG-curcumin/amine-functionalized covalent organic framework nanocomposites as smart carriers for in vivo drug delivery. Nat Commun. 2018;9(1):2785. doi:10.1038/s41467-018-04910-5
46. Mitra S, Sasmal HS, Kundu T, et al. Targeted drug delivery in covalent organic nanosheets (CONs) via sequential postsynthetic modification. J Am Chem Soc. 2017;139(12):4513–4520. doi:10.1021/jacs.7b00925
47. Zhang W, Xiang S, Long Y, et al. Red-fluorescent covalent organic framework nanospheres for trackable anticancer drug delivery. ACS Appl Mater Inter. 2023;16(1):342–352. doi:10.1021/acsami.3c15521
48. Das S, Sekine T, Mabuchi H, et al. Three-dimensional covalent organic framework with scu-c topology for drug delivery. ACS Appl Mater Inter. 2022;14(42):48045–48051. doi:10.1021/acsami.2c15152
49. Gao P, Tang K, Lou RX, et al. Covalent organic framework-based spherical nucleic acid probe with a bonding defect-amplified modification strategy. Analy Chem. 2021;35(93):12096–12102. doi:10.1021/acs.analchem.1c02602
50. Ding LG, Yao BJ, Li F, et al. Ionic liquid-decorated COF and its covalent composite aerogel for selective CO2 adsorption and catalytic conversion. J Mater Chem A. 2019;7:4689–4698. doi:10.1039/C8TA12046C
51. Gao P, Shen XY, Liu XH, et al. Nucleic acid-gated covalent organic frameworks for cancer-specific imaging and drug release. Analy Chem. 2021;34(93):11751–11757. doi:10.1021/acs.analchem.1c02105
52. Ahmed LR, Gilmanova L, Pan CT, et al. Hollow spherical covalent organic frameworks from nonplanar or planar monomers for the fluorescence detection of telomere DNA: role of the 2-(2-azidoethoxy)ethoxy group. ACS Appl Polym Mater. 2022;12(4):9132–9143. doi:10.1021/acsapm.2c01460
53. Li W, Yang CX, Yan XP. A versatile covalent organic framework-based platform for sensing biomolecules. Chem Commun. 2017;53:11469–11471. doi:10.1039/C7CC06244C
54. Gao P, Shen XY, Liu XH, et al. Covalent organic framework-derived carbonous nanoprobes for cancer cell imaging. ACS Appl Mater Inter. 2021;35(13):41498–41506. doi:10.1021/acsami.1c14998
55. Gao P, Wang MZ, Wan XY, et al. A COF-based anti-interference nanoplatform for intracellular nucleic acid imaging. Chem Commun. 2020;56:14267–14270. doi:10.1039/D0CC05527A
56. Kim S, Ryoo SR, Na HK, et al. Deoxyribozyme-loaded nano-graphene oxide for simultaneous sensing and silencing of the hepatitis C virus gene in liver cells. Chem Commun. 2013;49:8241–8243. doi:10.1039/c3cc43368d
57. Wu Y, Jiang W, Huo SH, et al. Nano-metal-organic-frameworks for treating H2O2-secreting bacteria alleviate pulmonary injury and prevent systemic sepsis. Biomaterials. 2021;279:121237. doi:10.1016/j.biomaterials.2021.121237
58. Samanta D, Ebrahimi SB, Mirkin CA. Nucleic‐acid structures as intracellular probes for live cells. Adv Mater. 2019;32(13):1901743. doi:10.1002/adma.201901743
59. Wu K, Li Q, Yao C, et al. Trojan horse delivery of spherical nucleic acid probes into the cytoplasm for high-fidelity imaging of microRNAs. Analy Chem. 2022;94(31):10942–10948. doi:10.1021/acs.analchem.2c00675
60. Boutorine A, Novopashina D, Krasheninina O, et al. Fluorescent probes for nucleic acid visualization in fixed and live cells. Molecules. 2013;18(12):15357–15397. doi:10.3390/molecules181215357
61. Jayagopal A, Halfpenny KC, Perez JW, et al. Hairpin DNA-functionalized gold colloids for the imaging of mRNA in live cells. J Am Chem Soc. 2010;132(28):9789–9796. doi:10.1021/ja102585v
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.