Back to Journals » International Journal of Nanomedicine » Volume 19
DENV Peptides Delivered as Spherical Nucleic Acid Constructs Enhance Antigen Presentation and Immunogenicity in vitro and in vivo
Authors Zhao J, He J, Ding X, Zhou Y, Liu M, Chen X, Quan W, Hua D, Tong J, Li J
Received 6 May 2024
Accepted for publication 6 September 2024
Published 20 September 2024 Volume 2024:19 Pages 9757—9770
DOI https://doi.org/10.2147/IJN.S467427
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kamakhya Misra
Jing Zhao,* Jiuxiang He,* Xiaoyan Ding, Yuxin Zhou, Minchi Liu, Xiaozhong Chen, Wenxuan Quan, Dong Hua, Jun Tong, Jintao Li
College of Basic Medicine, Army Medical University, Chongqing, 400038, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jintao Li, College of Basic Medicine, Army Medical University, Gaotanyan Street, No. 30, Shapingba District, Chongqing, 400038, People’s Republic of China, Tel +1-86-23-68771389, Fax +1-86-23-68771391, Email [email protected]
Background: The global prevalence of Dengue virus (DENV) infection poses a significant health risk, urging the need for effective vaccinations. Peptide vaccines, known for their capacity to induce comprehensive immunity against multiple virus serotypes, offer promise due to their stability, safety, and design flexibility. Spherical nucleic acid (SNA), particularly those with gold nanoparticle cores, present an attractive avenue for enhancing peptide vaccine efficacy due to their modularity and immunomodulatory properties.
Methods: The spherical nucleic acid-TBB (SNA-TBB), a novel nanovaccine construct, was fabricated through the co-functionalization process of SNA with epitope peptide, targeting all four serotypes of the DENV. This innovative approach aims to enhance immunogenicity and provide broad-spectrum protection against DENV infections. The physicochemical properties of SNA-TBB were characterized using dynamic light scattering, zeta potential measurement, and transmission electron microscopy. In vitro assessments included endocytosis studies, cytotoxicity evaluation, bone marrow-dendritic cells (BMDCs) maturation and activation analysis, cytokine detection, RNA sequencing, and transcript level analysis in BMDCs. In vivo immunization studies in mice involved evaluating IgG antibody titers, serum protection against DENV infection and safety assessment of nanovaccines.
Results: SNA-TBB demonstrated successful synthesis, enhanced endocytosis, and favorable physicochemical properties. In vitro assessments revealed no cytotoxicity and promoted BMDCs maturation. Cytokine analyses exhibited heightened IL-12p70, TNF-α, and IL-1β levels. Transcriptomic analysis highlighted genes linked to BMDCs maturation and immune responses. In vivo studies immunization with SNA-TBB resulted in elevated antigen-specific IgG antibody levels and conferred protection against DENV infection in neonatal mice. Evaluation of in vivo safety showed no signs of adverse effects in vital organs.
Conclusion: The study demonstrates the successful development of SNA-TBB as a promising nanovaccine platform against DENV infection and highlights the potential of SNA-based peptide vaccines as a strategy for developing safe and effective antiviral immunotherapy.
Keywords: nanovaccine, dengue virus, spherical nucleic acid, immunogenicity, peptide antigen
Introduction
Dengue virus (DENV), a member of the flavivirus genus within the Flaviviridae family, is a single-stranded, positive-sense RNA virus primarily transmitted by Aedes aegypti or Aedes albopictus.1 The virus comprises four distinct serotypes (DENV-1 to DENV-4), capable of causing conditions ranging from dengue fever to severe manifestations such as dengue hemorrhagic fever and dengue shock syndrome.1,2 With approximately half of the global population now susceptible to DENV, the risks associated with antibody-dependent enhancement (ADE) heighten the likelihood of severe illness and mortality upon subsequent infection with a heterologous virus.3,4 Despite being partially self-limiting, the need for effective and safe vaccinations is imperative to curb infection and disease dissemination, especially considering the absence of specific therapeutic medications for DENV.5,6
Peptide vaccines emerge as a promising strategy capable of eliciting broad-spectrum immunity across multiple viral serotypes.7 By incorporating immunodominant epitopes highly conserved among different serotypes, peptide vaccines can be designed to target various strains or serological variations effectively.8,9 Noteworthy advantages of peptide-based vaccines include enhanced stability, safety, and cost-effectiveness, as synthetic peptides enable the design of vaccines focusing solely on antigenic regions, mitigating adverse reactions associated with vaccinations.8,10 Additionally, the composition of peptide vaccines can be modified by introducing universal CD4+ T cell epitopes or combining B-cell epitopes, leading to the creation of universal formulations that activate specific human leukocyte antigen-restricted T-cell specificities. In this study, we identified the HLA-DRB epitope (HA308-319 KYVKQNTLKLAT) as the antigenic determinant for T helper cells within a cluster of polypeptides. The HA308-319 epitope, a notable influenza hemagglutinin segment, has been previously reported as a benchmark for positive T-cell epitopes. This choice was substantiated by references to its utilization as a control in related immunological studies.11–13 The B-cell epitope, delineated as VDRGWGNGCGLFGKG, originates from the structural domain II of the DENV E protein. This epitope was pinpointed through computational analyses by Muthusamy et al14 as an immunodominant region that exhibits conservation across all Dengue virus serotypes, thus underscoring its significance due to its high degree of conservation among the four DENV serotypes.15,16 Furthermore, our research employed immunoinformatics to identify a segment representing a shared B-cell epitope (VLGSQEG) across flaviviruses. By employing flexible ligation techniques, we successfully conjugated the T helper cell epitope with these two highly conserved B-cell epitopes, aiming to target all four DENV serotypes effectively. This innovative approach underscores the potential for developing broad-spectrum Dengue virus immunogens through strategic epitope linkage. Compared to conventional total protein immunizations, epitope vaccines demonstrate superior efficacy.17 Despite these advantages, peptide vaccines share common limitations with other vaccine modalities, encompassing low immunogenicity, diminished metabolic stability, and challenges related to pharmacokinetic parameters.18 However, recent research indicates that the immune-boosting efficacy of peptide vaccines can be augmented. This may be achieved by promoting endocytosis and phagocytosis through increased peptide size or modified binding characteristics, directly engaging antigen-presenting cells (APCs), employing adjuvants to activate APCs, or utilizing efficient peptide delivery systems.19,20
Spherical nucleic acid (SNA) represent nanoscale architectures well-suited for immune modulation due to their modularity, tunability, and facile synthesis from chemical building blocks. These structures comprise highly oriented and densely arranged oligodeoxyribonucleotides surrounding a nanoparticle core. In contrast to linear structures, SNAs exhibit enhanced cellular absorption without the need for harmful co-transfection agents.21–23 In this study, gold nanoparticles (AuNPs), chosen for their unique physical and chemical characteristics such as tunable particle size and shape, high specific surface area, and biocompatibility, served as the core of spherical nucleic acid. The advantageous physicochemical features of AuNPs and DNA can be synergistically harnessed through functionalized modification of AuNPs with other biomolecules, broadening their potential applications.24,25 CpG, an effective Toll-like receptor 9 (TLR9) agonist, stimulates dendritic cells (DCs) and facilitates efficient antigen presentation, subsequently activating antigen-specific T lymphocytes. Spherical nucleic acid (SNA) constructed from these sequences demonstrate higher efficacy or an extended therapeutic window compared to linear sequences at equivalent doses.26 Moreover, SNAs enable the creation of diverse structures with subtle nanoscale variations, such as coupling antigens to the SNA surface or embedding them within the SNA, facilitating the exploration of structural characteristics that enhance immune responses against viruses and tumors.27,28
Taking advantage of the distinctive properties of SNAs, we loaded synthetic peptides onto their surface and investigated their effective phagocytosis by dendritic cells (DCs), pivotal specialized antigen-presenting cells (APCs) in vivo.29 DCs, the sole APCs capable of activating primary T-lymphocytes and efficiently initiating immune responses in cytotoxic T-lymphocytes (CTLs), absorb extracellular materials like vaccines and employ Major Histocompatibility Complex (MHC) type II to process protein antigens onto their surfaces.29 Following vaccination with polypeptide vaccines, T cells with T cell receptors (TCRs) detect the MHC/peptide combination, multiply, and develop into effector cells. CD4+ T cells differentiate into numerous subsets of helper T cells (Th) or regulatory T cells (T Reg), providing feedback to enhance and guide B and T cell growth while regulating the immune system’s response to various threats or illnesses.30 Upon attachment to and activation of TCRs, CD8+ T cells undergo differentiation into cytotoxic T lymphocytes (CTLs) capable of eliminating infected cells when their TCRs bind to the MHC I/peptide complex. Vaccines can also directly interact with B cells, APCs with immunoglobulin surface receptors that bind foreign molecules, activating danger signals and leading to increased antigen presentation, proliferation, and antibody secretion.29–32
In this investigation, we leveraged the modularity inherent in the spherical nucleic acid (SNA) architecture, employing AuNPs as the core, to co-functionalize the SNA with a conserved epitope screened against the four serotypes of the Dengue virus, serving as the antigenic source (Figure 1A). Our evaluation centered on scrutinizing the potential of this architectural configuration to activate BMDCs and dissecting the ensuing immune responses in immunized mice. Our findings reveal a discernible augmentation in several antiviral immune parameters associated with the delivered epitope within the SNA structure in comparison to a conventional mixed formulation of free adjuvant and antigen. Specifically, the SNA structure demonstrates heightened efficacy in stimulating BMDCs maturation, elevating the production and secretion of cytokines and chemokines, and enhancing specific antibody expression in mice. This underscores the pertinence of exploring efficient nanocarriers to enhance the in vivo antiviral efficacy of peptide vaccines.
Materials and Methods
Synthesis of SNA-TBB Conjugates
The preparation of SNA-TBB involved a sequential procedure:
- CpG and TBB Conjugation: CpG (Sangong Biotech, Shanghai, China) and TBB (Cy5-KYVKQNTLKLATGGVLGSQEGSMVDRGWGNGCGLFGKG, Zhong peptide Biochemistry, Hangzhou, China) were coupled in a two-step process. Initially, 40 μM CpG underwent reduction using a 4 mm Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Beyotime, China) solution (molar ratio of CpG to TCEP solution at 1:100), followed by a 30-minute incubation at 25°C. Subsequently, a 40 mm 2.2′-Dipyridyl disulfide (DTDP, Beyotime, China) solution was introduced (molar ratio of CpG to DTDP solution at 1:1000), and the mixture underwent a 30-minute incubation at 25°C. The resultant product was purified using nucleic acid purification columns (Cytiva, NAP™, Britain) to eliminate excess TCEP and DTDP. The CpG and TBB conjugate was achieved by combining CpG with a TBB solution in a molar ratio of 1:1, followed by a 30-minute incubation at 25°C. The coupling of CpG and TBB was verified through agarose gel electrophoresis.2. AuNP Surface Modification: AuNPs (BBI SOLUTION, UK) underwent surface modification with CpG-complement (Sangong Biotech, Shanghai, China) (refer to Table S1). Sulfhydrylated CpG-complement (100 μM, 3.3 μL) was mixed with citrate-AuNPs (30 nm, 100 μL), and the resultant mixture was frozen at −20°C for 2 hours, followed by thawing at room temperature. The mixed solution underwent centrifugation twice at 7000 rpm for 20 minutes to eliminate excess binding DNA.33 Surface modification of AuNPs was confirmed using UV-visible absorption spectroscopy.3. Conjugation of CpG-TBB and AuNP-CpG-complement: The products from steps 1 and 2 were combined based on DNA base complementary pairing principles, followed by a 30-minute incubation at 37°C and subsequent storage at room temperature.
Characterization of Nanovaccines
Take AuNP and SNA-TBB 0.5 mL each and add deionized water to 1 mL, mix thoroughly to avoid bubbles and aggregation, and add them into the sample pool respectively. The nanoparticles were characterized by dynamic light scattering technique using Zetasizer Nano ZS (Malvern, UK) at 173° backscattering Angle (25°C) by hydrodynamic particle size and zeta potential. AuNP and SNA-TBB were diluted 5 times with distilled water and ultrasound was performed for 5min. Then put the sealing film on the glass sheet, put the copper mesh clip on the sealing film with tweezers, drop the 10μL sample on the copper mesh, leave it for 10 min, and use a small piece of filter paper to absorb the excess liquid on the copper mesh. Then let it stand at room temperature for 1 h to dry the copper mesh naturally. The dispersion and morphology of the two samples were detected by transmission electron microscopy (TEM, JEOL JEM1200EX, Japan).
Generation of Immature BMDCs
Bone marrow-derived dendritic cells (BMDCs) utilized for in vitro experiments were isolated from the femur of C57BL/6 mice (Tengxin Beer Laboratory Animal Sales Co., Ltd., Chongqing, China). BMDCs were cultured in complete RPMI1640 (Gibco, USA) medium supplemented with 10% fetal bovine serum (Gibco, USA), 10 ng/mL IL-4 (Novoprotein, China), and 20 ng/mL GM-CSF (Novoprotein, China) at 37°C. Immature BMDCs were harvested on day 7 to assess the impact of SNA-TBB on antigen-presenting cells (APCs).
Cellular Uptake
BMDCs were plated in 24-well culture plates with 1×105 cells/well, cultured for 24 hours, and subsequently treated for 2 hours with various concentrations of Cy5-labeled TBB, TBB-CpG, and SNA-TBB formulations. After three washes with PBS, BMDCs were fixed with 4% paraformaldehyde (Beijing Solarbio Science & Technology Co., Beijing, China). Cellular uptake by BMDCs was visualized using a laser confocal microscope (CLSM, Zeiss LSM 800, Germany) and detected by flow cytometry.
Cytotoxicity Assessment
To evaluate the cytotoxicity of SNA-TBB, BMDCs were seeded in a 96-well plate with 5×104 cells/well and co-cultured with PBS, TBB, TBB-CpG, and SNA-TBB at various concentrations for 24 hours. Cell viability was assessed using the Cell Counting Kit-8 (CCK-8, Beyotime Biotechnology, China) assay. Plates were incubated at 37°C for 2 hours, and absorbance at 450 nm was measured with a Varioskan Flash microplate reader (Thermo Scientific). Cell viability (%) was calculated using the formula:Cell viability (%)=(Atreat/Acontrol)×100% where Atreat and Acontrol represent the absorbance of treated cells (TBB/TBB-CpG/SNA-TBB) and untreated cells, respectively.
BMDCs Maturation and Activation
BMDCs were placed in a 24-well culture plate in the form of 1×105 cells/well for 24 h, and then TBB, TBB-CpG and SNA-TBB(The concentration of TBB in each group was 300 nM) were incubated with immature BMDCs at 37°C for 24 h, while lipopolysaccharide (2.5 μg/mL) (Novoprotein, China) was used as a positive control. Subsequent to treatment, cells were collected, and BMDCs were stained with perCP-Cy5.5-labeled anti-CD11c antibody, APC-labeled anti-MHCII antibody, PE-labeled anti-CD80 antibody, and FITC-labeled anti-CD86 antibody (Biolegend, USA). Flow cytometry analysis (BD, FACS-1) was employed to assess the labeling of cells by each antibody. Additionally, the expression levels of cytokines, including interleukin 1β (IL-1β), tumor necrosis factor-α, and interleukin 12p70 (IL-12p70), in the culture supernatant were determined using an ELISA kit from Thermo Fisher Science Inc. This analysis aimed to characterize the activation of SNA-TBB on BMDCs.
RNA Analysis by RT-qPCR
The Tissue/Cell Rapid Extraction Kit (BOER, China) is convenient for the extraction of total RNA from cells, extraction, and an ultra-micro spectrophotometer was utilized to determine the concentration and purity of the isolated cellular RNA. Reverse transcription was conducted using the PrimeScript™ RT reagent Kit with gDNA Eraser (Code No. RR820Q, TAKARA, Japan), with the resulting cDNA diluted five times and used as a template. Fluorescence quantitative PCR, employing TB Green® Premix Ex Taq™ II (Code No. RR820A/BTB, TAKARA, Japan) and the LightCycler® 96 system, was carried out to detect CD80, CD86, IL12b, IL13, IL22, and ccl5. The primer sequences can be found in Table S2. Relative quantifications were determined using the 2-∆∆CT method.
RNA Sequencing and Bioinformatics Analysis
BMDCs were placed in a 10cm petri dish in the form of 1×106 cells/well for 24 hours, and then TBB and SNA-TBB were incubated with immature BMDCs at 37°C for 24 hours to extract RNA samples from BMDCs. Elaborate RNA libraries were prepared using Illumina V2’s VAHTS single-strand mRNA-seq library preparation kit. To identify DEGs (differential expression genes) between two different samples, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM was used to quantify gene abundances. Essentially, differential expression analysis was performed using the DESeq2 or DEGseq. DEGs with |log2FC| ≧ 1 and FDR ≤ 0.05(DESeq2) or FDR ≤ 0.001(DEGseq) were considered to be significantly different expressed genes. In addition, functional-enrichment analysis including GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-value ≤0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analysis were carried out by Goatools and KOBAS, respectively.
Mice Immunization Study
Female Balb/c mice (6 weeks old) were stratified into four groups, each comprising six mice. Subcutaneous vaccination, administered through the loose skin of the neck, was conducted on day 0, with subsequent booster immunizations employing various vaccine candidates (TBB, TBB-CpG, and SNA-TBB) at a dose of 30 µg of immunogen on days 14 and 28. The PBS group served as the control. On the 7th day post the last injection, blood collection involved the removal of eyes, and antisera were prepared and stored at −20°C for subsequent assessments.
Assessment of Antigen-Specific Systemic IgG Antibodies
The quantification of immunoglobulin antibody efficacy employed the double antibody sandwich method. In specific terms, 15 μg/mL TBB (0.05 M phosphate solution, pH 9.6) was meticulously introduced into 96-well plates at 100 μL per well, followed by an overnight incubation at 4°C. Subsequent to a thorough washing sequence (three times with a wash buffer of 0.05% Tween-20 (Solarbio China) in Tween buffer, totaling 300 μL), the plates were hermetically sealed with a 1% bovine serum albumin carbonate buffer (Biofroxx, Germany) (250 μL) and incubated at 37°C for 2 hours. Following a series of supplementary washing procedures, antiserum was meticulously diluted at ratios of 1:100, 1:200, 1:400, 1:600, and 1:800, respectively. Each dilution was then carefully dispensed into individual wells and subjected to incubation at a temperature of 37°C for a duration of one hour. Post this, a washing buffer treatment ensued, succeeded by the addition of horseradish peroxidase-labeled sheep anti-mouse IgG antibody (diluted 1:5000) at 100 μL per well (Proteintech, Chicago, USA). A subsequent 1-hour incubation at 37°C, followed by four meticulous washes, paved the way for the final steps. Specifically, 100 μL of 3.3’,5,5’-Tetramethylbenzidine (TMB, Sigma-Aldrich, USA) was incorporated into each well, fostering a 30-minute incubation at room temperature. The reaction was then halted with the addition of 0.5 M H2SO4 (50 μL), and absorbance measurements were taken at 450 nm.
Vivo Protective Experiments Against Dengue Virus Challenge
On the 35th day post-immunization, serum samples were extracted from the mice’s tail blood and subsequently inactivated for 30 minutes at 56 °C. Following the inactivation process, 10 μL of the inactivated serum was intricately mixed with 60 PFU/10 μL of DENV-2 or 125 PFU/10 μL of DENV-4. This mixture underwent a one-hour incubation at 37 °C. After this incubation period, the serum was further incubated for an additional hour at 37 °C. Utilizing a microsyringe, twenty microliters of the viral serum mixture were injected into three-day-old Balb/c newborn mice. Subsequent to the infection, survival outcomes were meticulously recorded over a fifteen-day period.
Statistical Analysis
The data were presented in the format of mean ± standard deviation (SD). Statistical differences among groups were assessed using Student’s t-test and ANOVA. A significance level of P < 0.05 was deemed indicative of a meaningful difference. Significance levels were categorized as follows: (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
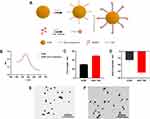
Design, Synthesis, and Characterization of SNAs
In the development of spherical nucleic acid (SNA) structures, we employed gold nanoparticles (AuNPs) to establish Au-S bond interactions with sulfhydrylated CpG complementaries. To bind sulfhydrylated CpG to TBB, which incorporates cysteine, S-S bond interactions were utilized. The construction of a spherical nucleic acid vaccine (SNA-TBB) followed a specific co-incubation period between CpG and CpG complementarities (Figure 1A). The successful binding of the two couplers was confirmed through UV spectrophotometry and agarose gel electrophoresis. Our results demonstrated complete coupling of CpG with TBB, as indicated by the substantially reduced migration rate of TBB-CpG compared to the CpG group, with no observable residual CpG (Figure S1). Furthermore, the conjugation of AuNP and CpG-complement resulted in a significant red-shift in the greatest light absorption peak, indicating the formation of an oligonucleotide sphere adhered to the AuNP surface (Figure 1B). Physicochemical properties of the formulated SNA-TBB vaccine, encompassing hydrodynamic particle size and particle surface charge, were thoroughly characterized. Dynamic light scattering revealed particle sizes of approximately 30 nm for AuNPs and 50 nm for SNA-TBB. The particle sizes of AuNP experienced a significant increase upon binding nucleic acids and antigens (Figure 1C). Zeta potentials of AuNP and SNA-TBB were determined to be around −12 mV and −31.5 mV, respectively (Figure 1D). Modification of AuNP with CpG and TBB rendered the particles negatively charged in pure water, substantially exceeding the charge of AuNPs alone, indicative of CpG’s strong negative charge. This observation suggests improved stability for SNA-TBB in aqueous solutions. Transmission electron micrographs demonstrated the uniform dispersion and spherical nature of SNA-TBB nanoparticles. Compared with AuNP alone, the particle size of SNA-TBB exhibited an increasing trend (Figure 1E and F).
Endocytosis of SNA-TBB
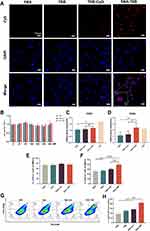
To delve into the in vitro endocytosis of SNA-TBB, we employed laser confocal scanning microscopy and flow cytometry for qualitative and quantitative assessments of TBB. After a two-hour incubation of various groups with bone marrow-derived dendritic cells (BMDCs), we sought to investigate whether SNA-TBB enhances the internalization of the antigen into BMDCs. Maintaining a consistent antigen concentration, confocal microscopy revealed distinct red fluorescent signals in cells of the SNA-TBB group after co-incubation with BMDCs. In contrast, the TBB-CpG group exhibited weaker fluorescent signals, while the TBB group displayed virtually no fluorescent signals at all (Figure 2A). Flow cytometry analysis (Figure S2) showed that the number of cells with TBB uptake by BMDCs was significantly higher than that in the other groups. These findings collectively underscore the potential of SNA-TBB to efficiently transport and present antigens to antigen-presenting cells (APCs), thereby activating an immune response.
In vitro Cell Viability and Maturation of BMDCs Induced by SNA-TBB
To evaluate the potential toxicity of SNA-TBB, bone marrow-derived dendritic cells (BMDCs) underwent a 24-hour incubation with varying concentrations of formulations, utilizing the PBS-treated group as a control. Notably, Figure 2B illustrates that even at a TBB concentration of 400nM in each formulation, no significant cytotoxicity was observed.
To explore whether the peptide vaccine could instigate the maturation of dendritic cells (DCs), a crucial step in initiating antigenic T-cell immunity, BMDCs were co-incubated with distinct peptide vaccine components for 24 hours, alongside lipopolysaccharide (LPS) as a positive control. Subsequent qPCR analysis was employed to discern the molecular expression of co-stimulatory molecules CD80 and CD86 on the surface of BMDCs. As depicted in Figure 2C and D, both CD80 and CD86 expression exhibited an elevation in the SNA-TBB group compared with the TBB group and the TBB-CpG group. Remarkably, CD86 expression in the SNA-TBB group even surpassed that in the LPS group. Flow cytometry was employed to investigate the expression of CD11c, the primary stimulatory molecule MHC-II, and the co-stimulatory molecules CD80 and CD86 on the BMDC surface. The consistently high expression of CD11c, approximately 75%, attested to the high purity of BMDCs (Figure 2E). SNA-TBB notably augmented MHC-II expression, facilitating the presentation of antigens through the MHC-II pathway, a process vital for activating CD4+ T cells and triggering humoral immunity (Figure 2F). Moreover, SNA-TBB significantly enhanced the expression of CD80 and CD86 (Figures 2G, H and Figure S3), affirming the effective promotion of BMDC maturation by the SNA-TBB conjugate.
In vitro Cytokine Analysis
To assess the immunostimulatory impact of the nanovaccine on bone marrow-derived dendritic cells (BMDCs) in vitro, we conducted a comprehensive analysis of various cytokines present in the culture supernatants of BMDCs following incubation with SNA-TBB, utilizing specific enzyme-linked immunosorbent assay (ELISA) assays. As depicted in Figure 3A, the interleukin-12p70 (IL-12p70) levels in cultures treated with SNA-TBB exhibited an 8.6-fold, 9.0-fold, and 4.4-fold increase compared to those treated with PBS, TBB, and TBB-CpG, respectively. Furthermore, the tumor necrosis factor-α (TNF-α) levels in the SNA-TBB-treated group were elevated by 2.0-fold, 1.8-fold, and 1.3-fold compared to the PBS, TBB, and TBB-CpG groups, respectively (Figure 3B). Additionally, the interleukin-1β (IL-1β) levels in the SNA-TBB group surpassed those in the PBS, TBB, and TBB-CpG groups by approximately 1.5 times (Figure 3C). In essence, the results indicate that SNA-TBB induces alterations in antibody expression on the cell surface and the secretion of inflammatory cytokines by BMDCs, ultimately contributing to the enhanced maturation of BMDCs.
Transcriptomic Analysis of TBB and SNA-TBB-Treated BMDCs
In-depth exploration of the mechanistic changes in BMDCs following antigen uptake was conducted by assessing the global transcriptional alterations in TBB- and SNA-TBB-treated BMDCs through RNA-Seq technology. Differential gene expression analysis revealed 347 up-regulated and 597 down-regulated genes, as illustrated in Figure 4A.
Subsequently, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses were employed to unravel potential modifications in cellular processes and signaling pathways (Figure 4B and C). GO analysis unveiled a significant enrichment of differential genes at functional loci associated with immunoreceptor activity, cytokine-mediated signaling pathways, chemokine-mediated signaling pathways, regulation of natural killer cell chemotaxis, regulation of interleukin-12 production, monocyte chemotaxis and migration, among others. These functional loci are intricately tied to processes such as antigen presentation, stimulation of T cell activation, and activation of BMDCs. Further investigation of specific differential genes highlighted elevated expression of IL-12b, Ltb, and CCR7—genes intimately linked to BMDC maturation and activation (Figure 4D).
KEGG analysis unveiled signaling pathways associated with pathogen recognition, such as Toll-like receptor (TLR) signaling pathways, known to facilitate phagocytosis and uptake of vaccination particles. Downstream cytokine and interferon signaling pathways, including JAK-STAT signaling, cytokine-cytokine receptor interactions, and viral protein-cytokine and cytokine receptor interactions, were also overexpressed, potentially inducing the release of chemokines, interferons, and cytokines. Gene enrichment analysis further delved into the molecular mechanism of BMDCs, indicating the up-regulation of IL-22, IL-13, ccl1, ccl5, and ccl4. These genes are primarily associated with inflammatory responses, regulation of cytokine production, and immune responses, aligning with the GO findings (Figure 4E). Collectively, these results suggest profound alterations in the mRNA transcriptome of SNA-TBB-treated BMDCs, with SNA-TBB exerting broader and more potent effects on gene expression related to maturation, activation, and inflammatory responses in BMDCs compared to the TBB group. To validate the transcriptome data, fluorescent quantitative PCR was employed, confirming the consistent expression patterns of IL-12b, IL-13, ccl5, and IL-22 genes (Figure 4F–I), reinforcing the reliability of the transcriptome data.
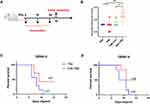
Analysis of IgG Antibody Titers and Serum Protection Against DENV Infection in Neonatal Mice
To assess the immunogenic potential of SNA-TBB following subcutaneous injection in mice, particularly in terms of humoral immunity (Figure 5A), we conducted a comprehensive analysis of serum samples from different vaccinated mouse cohorts to quantify the presence of antigen-specific IgG antibodies (Figure 5B). Upon administering three immunizations, mice immunized with SNA-TBB exhibited significantly higher and statistically elevated antibody levels on day 35 compared to counterparts immunized with PBS, TBB, and TBB-CpG. Subsequently, neutralization experiments were conducted using DENV-2 and DENV-4 with mouse sera collected on the 7th day post the final immunization. Infection studies were carried out on 3-day-old Balb/c neonatal mice using serovar mixtures, and their survival rates were monitored over a 15-day period post-infection. Notably, the SNA-TBB vaccine-immunized group displayed survival rates of 40% and 50%, respectively, within the 15-day observation period. In contrast, the neonatal mice in the TBB group exhibited mortality starting on day 6 following the injection of serum and each virus mixture, ultimately succumbing to infection within 15 days (Figure 5C and D).
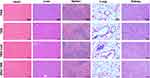
Assessment of in vivo Safety
To scrutinize the potential toxicity of the peptide vaccine in vivo, mice were euthanized, and their hearts, livers, spleens, lungs, and kidneys were examined. As depicted in Figure 6, none of the groups, including TBB, TBB-CpG, and SNA-TBB, displayed abnormal cell death, tissue reactions, or inflammatory infiltration when compared to the PBS group. These observations strongly suggest that neither SNA-TBB nor TBB, TBB-CpG, or TBB alone induced organ damage in vivo, affirming the safety of the peptide vaccination.
Discussion
The absence of specific antiviral medications for dengue fever underscores the need for effective therapeutic approaches. Despite the existence of commercial dengue vaccines, their clinical utilization has been limited due to safety concerns.34,35 Peptide vaccines have gained prominence, capitalizing on the intricate understanding of the immune system and the pivotal role of antigenic epitopes in eliciting robust immune responses. In contrast to conventional vaccines, such as attenuated or inactivated whole-pathogen vaccines, subunit or toxoid vaccines, and carbohydrate-based vaccines, peptide-based vaccines, specifically epitope-pooled vaccines, offer a distinct strategy for developing disease-specific prophylactic and therapeutic solutions. Epitopes, recognized by T and/or B cells governing adaptive immunity, constitute crucial components of antigens. Harnessing multiple epitopes in vaccines holds the advantage of eliciting favorable T and B cell-mediated immune responses.36,37 Notably, epitope vaccines boast a superior safety profile compared to traditional immunizations, significantly reducing the risk of pathogenic or off-target reactions. In this study, a peptide vaccine was meticulously designed to target shared antigenic epitopes across the four dengue serotypes, identified through genetic analysis. However, the effective delivery of this peptide vaccine necessitates a reliable vaccine delivery system. Spherical nucleic acid (SNA) serve as proficient transport carriers, enabling the simultaneous transportation of CpG and peptides. CpG oligonucleotides, known for inducing robust Th1-type immune responses, play a pivotal role.38 The utilization of CpG as an adjuvant in vaccine formulations is an evolving therapeutic strategy, and the integration of CpG into SNAs has progressed to clinical trials.39 In our study, CpG within SNA successfully induced dendritic cell maturation and the production of Th1 cytokines, such as IL-12p70, underscoring the potential of this innovative approach in enhancing vaccine efficacy.40–43
The intricate interplay between dendritic cells (DCs) and peptide-based immunotherapies underscores their collaborative role in augmenting immune responses. DCs, pivotal in enhancing cellular immune responses, play a crucial role in antigen processing. Upon antigen capture, DCs convert these entities into smaller fragments, presenting them on the cell surface for T cell recognition, thereby initiating a robust T cell response.Our well-established spherical nucleic acid (SNA) vector antigen delivery platform has demonstrated remarkable efficacy. The SNA-TBB vaccine, a product of this platform, facilitates enhanced antigen absorption by BMDCs. This heightened absorption, in turn, promotes the expression of co-stimulatory factors CD80 and CD86, stimulating BMDCs to secrete cytokines and chemokines, crucial elements in immune responses.
Furthermore, gene enrichment analysis in SNA-TBB-treated BMDCs reveals a significant up-regulation of downstream cytokine and interferon signaling pathways. Notably, there is a pronounced elevation in IL-22 and IL-13, along with substantial induction of IL-12. IL-22, belonging to the IL-10 cytokine family, is produced by activated DCs and T cells. Its binding to the IL-22 receptor on epithelial and stromal cells promotes the growth, remodeling, and restoration of organs, fortifying innate host defense mechanisms against pathogen invasion.44 IL-13, known for inducing monocyte differentiation, enhancing MHC class II molecule expression, and stimulating B cell proliferation, further contributes to the intricate immune cascade.45 Additionally, mature DCs up-regulate the chemokine receptor CCR7, facilitating their migration to lymph nodes via lymphatic channels.46,47 In the lymph nodes, DCs interact with T cells, delivering primed antigens, thereby initiating a potent immune response. The orchestrated maturation of DCs, coupled with the modulation of antigen internalization and the surface expression of co-stimulatory molecules, critically regulates the amplification of immune response products. This intricate regulatory network underscores the significance of our SNA-TBB vaccine in orchestrating an effective and targeted immune response.
In assessing the immunogenicity of the peptide vaccine, subcutaneous injections were administered to mice in a triple-dose regimen. Evaluation of antigen-specific IgG antibody levels in collected sera revealed a notable enhancement in the production of these antibodies induced by SNA-TBB, surpassing levels observed in both TBB-CpG and TBB groups. Importantly, our findings indicate the safety of all compounds, including SNA-TBB, as they demonstrated no adverse effects on immunized mice and exhibited no toxicity towards normal cells. While the current investigation represents an initial exploration into the immunogenicity of peptide vaccines, future studies can delve deeper into elucidating the cellular immunity elicited in mice. Further investigations may involve refining the immunization strategy, exploring alternative adjuvants, and optimizing the peptide coupling ratio to attain an enhanced immunization effect.
Conclusion
In conclusion, our successful construction of a nanovaccine system based on spherical nucleic acid vectors has demonstrated effective antigen uptake and the stimulation of maturation and activation of BMDCs. This, in turn, has resulted in an augmented antigen-specific immune response with a commendable biosafety profile. The innovative design presented here lays the groundwork for a viable strategy in developing nanovaccines that co-deliver adjuvants and antigens for antiviral immunotherapy. Consequently, it introduces a novel avenue for research in the creation of safe and effective dengue vaccines.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval
All experimental procedures involving mice in this study were approved by the Laboratory Animal Welfare and Ethics Committee of the Army Medical University (AMUWEC20226281) and follow the National Institutes of Health Guidelines for the Care and use of laboratory animals.
Acknowledgments
The project was supported by National Natural Science Foundation of China (Grant No.81570497).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Messina JP, Brady OJ, Scott TW, et al. Global spread of Dengue Virus types: mapping the 70 year history. Trend Microbiol. 2014;22:138–146. doi:10.1016/j.tim.2013.12.011
2. Martina BEE, Koraka P, Osterhaus ADME. Dengue Virus Pathogenesis: an Integrated View. Clin Microbiol Rev. 2009;22:564–581. doi:10.1128/CMR.00035-09
3. Uno N, Ross TM. Dengue Virus and the host innate immune response. Emerging Microbes Infect. 2018;7:1–11. doi:10.1038/s41426-018-0168-0
4. Deng S-Q, Yang X, Wei Y, et al. A Review on Dengue Vaccine Development. Vaccines. 2020;8:63. doi:10.3390/vaccines8010063
5. Yauch LE, Shresta S. Dengue Virus Vaccine Development. In: Advances in Virus Research. Vol. 88. Elsevier; 2014:315–372.
6. Huang C-H, Tsai Y-T, Wang S-F, Wang W-H, Chen Y-H. Dengue vaccine: an update. Exp Rev Anti-Infective Ther. 2021;19:1495–1502. doi:10.1080/14787210.2021.1949983
7. Zottig X, Al-Halifa S, Côté-Cyr M, et al. Self-assembled peptide nanorod vaccine confers protection against influenza A virus. Biomaterials. 2021;269:120672. doi:10.1016/j.biomaterials.2021.120672
8. Chang TZ, Champion JA. Protein and Peptide Nanocluster Vaccines. In: Nanoparticles for Rational Vaccine Design. Cham: Springer International Publishing; 2020:107–130.
9. Tsoras AN, Champion JA. Protein and peptide biomaterials for engineered subunit vaccines and immunotherapeutic applications. Annu Rev Chem Biomol. 2019;10:337–359. doi:10.1146/annurev-chembioeng-060718-030347
10. Malonis RJ, Lai JR, Vergnolle O. Peptide-based vaccines: current progress and future challenges. Chem Rev. 2020;120:3210–3229. doi:10.1021/acs.chemrev.9b00472
11. Braendstrup P, Justesen S, Østerbye T, et al. MHC class II tetramers made from isolated recombinant α and β chains refolded with affinity-tagged peptides. PLoS One. 2013;8:e73648. doi:10.1371/journal.pone.0073648
12. Panigada M, Sturniolo T, Besozzi G, et al. Identification of a promiscuous T-cell epitope in mycobacterium tuberculosis mce proteins. Infect Immun. 2002;70:79–85. doi:10.1128/IAI.70.1.79-85.2002
13. Umamaheswari A, Pradhan D, Hemanthkumar M. Computer aided subunit vaccine design against pathogenic Leptospira serovars. Interdiscip Sci Comput Life Sci. 2012;4:38–45. doi:10.1007/s12539-012-0118-9
14. Muthusamy K, Gopinath K, Nandhini D. Computational prediction of immunodominant antigenic regions & potential protective epitopes for dengue vaccination. Indian J Med Res. 2016;144:587–591. doi:10.4103/0971-5916.200894
15. Falconi-Agapito F, Kerkhof K, Merino X, et al. Dynamics of the magnitude, breadth and depth of the antibody response at epitope level following dengue infection. Front Immunol. 2021;12:686691. doi:10.3389/fimmu.2021.686691
16. Chan Y, Jazayeri SD, Ramanathan B, Poh CL. Enhancement of tetravalent immune responses to highly conserved epitopes of a dengue peptide vaccine conjugated to polystyrene nanoparticles. Vaccines. 2020;8:417. doi:10.3390/vaccines8030417
17. Nelde A, Rammensee H-G, Walz JS. The peptide vaccine of the future. Mol Cell Proteomics. 2021;20:100022. doi:10.1074/mcp.R120.002309
18. Lim HX, Lim J, Jazayeri SD, Poppema S, Poh CL. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomedical Journal. 2021;44:18–30. doi:10.1016/j.bj.2020.09.005
19. Reche P, Flower DR, Fridkis-Hareli M, Hoshino Y. Peptide-Based Immunotherapeutics and Vaccines 2017. J Immunol Res. 2018;2018:1–2. doi:10.1155/2018/4568239
20. Hamley IW. Peptides for Vaccine Development. ACS Appl Bio Mater. 2022;5:905–944. doi:10.1021/acsabm.1c01238
21. Mokhtarzadeh A, Vahidnezhad H, Youssefian L, et al. Applications of spherical nucleic acid nanoparticles as delivery systems. Trends Mol Med. 2019;25:1066–1079. doi:10.1016/j.molmed.2019.08.012
22. Kapadia CH, Melamed JR, Day ES. Spherical nucleic acid nanoparticles: therapeutic potential. BioDrugs. 2018;32:297–309.
23. Zhu S, Xing H, Gordiichuk P, Park J, Mirkin CA. PLGA spherical nucleic acids. Adv Mater. 2018;30:1707113. doi:10.1002/adma.201707113
24. Zhou Q, Zhang Y, Du J, et al. Different-sized gold nanoparticle activator/antigen increases dendritic cells accumulation in liver-draining lymph nodes and CD8+ T cell responses. ACS Nano. 2016;10:2678–2692. doi:10.1021/acsnano.5b07716
25. Wang C, Zhu W, Wang B-Z. Dual-linker gold nanoparticles as adjuvanting carriers for multivalent display of recombinant influenza hemagglutinin trimers and flagellin improve the immunological responses in vivo and in vitro. IJN. 2017;12:4747–4762. doi:10.2147/IJN.S137222
26. Wang S, Qin L, Yamankurt G, et al. Rational vaccinology with spherical nucleic acids. Proc Natl Acad Sci USA. 2019;116:10473–10481. doi:10.1073/pnas.1902805116
27. Qin L, Wang S, Dominguez D, et al. Development of spherical nucleic acids for prostate cancer immunotherapy. Front Immunol. 2020;11:1333. doi:10.3389/fimmu.2020.01333
28. Teplensky MH, Dittmar JW, Qin L, et al. Spherical nucleic acid vaccine structure markedly influences adaptive immune responses of clinically utilized prostate cancer targets. Adv Healthc Mater. 2021;10:2101262. doi:10.1002/adhm.202101262
29. Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012;12:265–277. doi:10.1038/nrc3258
30. Schmid MA, Diamond MS, Harris E. Dendritic cells in dengue virus infection: targets of virus replication and mediators of immunity. Front Immunol. 2014;5. doi:10.3389/fimmu.2014.00647
31. Martins S, Silveira G, Alves L, Dos Santos C, Bordignon J. Dendritic cell apoptosis and the pathogenesis of dengue. Viruses. 2012;4:2736–2753. doi:10.3390/v4112736
32. Liu D, Deng B, Liu Z, et al. Enhanced antitumor immune responses via a self-assembled carrier-free nanovaccine. Nano Lett. 2021;21:3965–3973. doi:10.1021/acs.nanolett.1c00648
33. Liu B, Liu J. Freezing directed construction of bio/nano interfaces: reagentless conjugation, denser spherical nucleic acids, and better nanoflares. Journal of the American Chemical Society. 2017;139(28):9471–9474. doi:10.1021/jacs.7b04885
34. Thisyakorn U, Tantawichien T. Dengue vaccine: a key for prevention. Expert Rev Vaccin. 2020;19:499–506. doi:10.1080/14760584.2020.1775076
35. Wilder-Smith A, Ooi -E-E, Horstick O, Wills B. Dengue. Lancet. 2019;393:350–363. doi:10.1016/S0140-6736(18)32560-1
36. Alharbi N, Skwarczynski M, Toth I. The influence of component structural arrangement on peptide vaccine immunogenicity. Biotechnol Adv. 2022;60:108029. doi:10.1016/j.biotechadv.2022.108029
37. O’Neill CL, Shrimali PC, Clapacs ZE, Files MA, Rudra JS. Peptide-based supramolecular vaccine systems. Acta Biomater. 2021;133:153–167. doi:10.1016/j.actbio.2021.05.003
38. Lai C-Y, Yu G-Y, Luo Y, Xiang R, Chuang T-H. Immunostimulatory Activities of CpG-oligodeoxynucleotides in teleosts: toll-like receptors 9 and 21. Front Immunol. 2019;10:179. doi:10.3389/fimmu.2019.00179
39. Zhang H, Gao X-D. Nanodelivery systems for enhancing the immunostimulatory effect of CpG oligodeoxynucleotides. Mater Sci Eng C. 2017;70:935–946.
40. Kayraklioglu N, Horuluoglu B, Klinman DM. CpG oligonucleotides as vaccine adjuvants. In: Sousa Â, editor. DNA Vaccines. Vol. 2197. New York, NY: Springer US; 2021:51–85.
41. Jin JW, Tang SQ, Rong MZ, Zhang MQ. Synergistic effect of dual targeting vaccine adjuvant with aminated β-glucan and CpG-oligodeoxynucleotides for both humoral and cellular immune responses. Acta Biomater. 2018;78:211–223. doi:10.1016/j.actbio.2018.08.002
42. Leleux JA, Pradhan P, Roy K. Biophysical Attributes of CpG Presentation Control TLR9 signaling to differentially polarize systemic immune responses. Cell Rep. 2017;18:700–710. doi:10.1016/j.celrep.2016.12.073
43. Komastu T, Ireland DDC, Reiss CS. IL-12 and Viral Infections. Cytokine Growth Factor Rev. 1998;9:277–285. doi:10.1016/S1359-6101(98)00017-3
44. Keir ME, Yi T, Lu TT, Ghilardi N. The role of IL-22 in intestinal health and disease. J Exp Med. 2020;217:e20192195. doi:10.1084/jem.20192195
45. Dubin C, Del Duca E, Guttman-Yassky E. The IL-4, IL-13 and IL-31 pathways in atopic dermatitis. Expert Rev Clin Immunol. 2021;17:835–852. doi:10.1080/1744666X.2021.1940962
46. Liu J, Zhang X, Chen K, et al. CCR7 chemokine receptor-Inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50:600–615.e15. doi:10.1016/j.immuni.2019.01.021
47. Hong W, Yang B, He Q, Wang J, Weng Q. New Insights of CCR7 signaling in dendritic cell migration and inflammatory diseases. Front Pharmacol. 2022;13:841687. doi:10.3389/fphar.2022.841687
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.