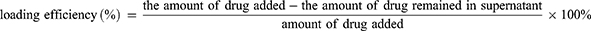Back to Journals » International Journal of Nanomedicine » Volume 20
Developing the Strategy to Use Silk Spheres for Efficient, Targeted Delivery of Oligonucleotide Therapeutics to Cancer Cells
Authors Molenda S, Deptuch T , Sikorska A, Lorenc P, Smialek MJ, Florczak-Substyk A , Pawlak P, Dams-Kozlowska H
Received 28 January 2025
Accepted for publication 16 May 2025
Published 23 June 2025 Volume 2025:20 Pages 8023—8039
DOI https://doi.org/10.2147/IJN.S519906
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Lijie Grace Zhang
Sara Molenda,1– 3 Tomasz Deptuch,1,2 Agata Sikorska,1,2 Patryk Lorenc,1– 3 Maciej Jerzy Smialek,1,2,* Anna Florczak-Substyk,1,2 Piotr Pawlak,4 Hanna Dams-Kozlowska1,2
1Department of Cancer Immunology, Poznan University of Medical Sciences, Poznan, Poland; 2Department of Diagnostics and Cancer Immunology, Greater Poland Cancer Centre, Poznan, Poland; 3Doctoral School, Poznan University of Medical Sciences, Poznan, Poland; 4Department of Genetics and Animal Breeding, Poznan University of Life Sciences, Poznan, Poland
*Current address: Biozentrum, University of Basel, Basel, Switzerland, Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland
Correspondence: Hanna Dams-Kozlowska, Department of Cancer Immunology, Poznan University of Medical Sciences, 15 Garbary St, Poznan, 61-866, Poland, Email [email protected]
Introduction: Oligonucleotide-based drugs, such as siRNA, hold great promise for disease treatment, including cancer. However, their clinical application has challenges related to cell-specific delivery and susceptibility to degradation. The use of drug delivery systems (DDS) may address these problems. Nanoparticles of bioengineered spider silk demonstrate significant potential as DDS due to their biocompatibility and biodegradability. Another advantage of this material is the possibility of functionalization, which allows the control of its property. The main objective of this study was to develop a strategy for targeted delivery of oligonucleotide-based therapeutics into cancer cells using bioengineered silk technology.
Materials and Methods: Two spider silk spheres that bind oligonucleotides and target cancer cells that overexpress HER2 (HER2+) were constructed. One type of sphere was made of a newly designed silk, H2.1MS1KN, which contained two functional peptides: H2.1 for binding HER2 and KN for binding oligonucleotide. The second type of sphere was formed of a blend of two previously described proteins, H2.1MS1 and MS2KN; these proteins differed not only in the functional domain (H2.1 vs KN) but also in the sequence of silk (MS1 vs MS2). The ability of proteins to bind oligonucleotides was analyzed via gel electrophoresis. The biophysicochemical properties of particles were analyzed using an SEM, NanoSight, ZetaSizer, flow cytometry, and scanning confocal microscopy. The silk particle potential was analyzed using siRNA for silencing STAT3 expression in the HER2+ breast cancer model.
Results: Both H2.1MS1KN and H2.1MS1:MS2KN proteins efficiently bound nucleic acid. H2.1MS1:MS2KN formed smaller spheres than H2.1MS1KN. Although both H2.1MS1KN and blended H2.1MS1:MS2KN spheres were effectively loaded with oligonucleotides, only H2.1MS1:MS2KN spheres delivered siRNA to HER2+ cancer cells that successfully silenced STAT3 expression.
Conclusion: Not only the selection of functional peptides but also their quantity and type of silk is crucial when developing an effective silk-based DDS for delivering active siRNA.
Keywords: bioengineered spider silk, spheres, siRNA-delivery, targeted delivery, cancer therapy, STAT3
Introduction
Cancers are currently one of the most significant challenges in modern medicine. It is treated using various methods, including surgery with chemotherapy and/or radiation therapy, immunotherapy, targeted therapy, or hormone therapy.1 Recently, one of the rapidly advancing methods is gene therapy, which involves controlling the expression of genes that play a crucial role in the development and progression of cancer.2 Targeting STAT3 using oligonucleotides such as siRNA, shRNA, or ASO is an example of this type of therapy.3–8 The molecule siRNA STAT3 (siSTAT3) silenced STAT3 expression leading to the induction of apoptosis and the inhibition of the proliferation and migration of breast cancer cells9,10 indicating its potential as therapeutics.
Nevertheless, the gene therapy faces considerable challenges. Due to the therapeutic mechanism, most nucleic acid-based therapeutics must be delivered inside the cells. The large size and negative charge of oligonucleotide therapeutics hinder their ability to penetrate cell membranes effectively.11 Another limitation is that RNAses and DNAses rapidly degrade nucleic acid-based therapeutics when applied in vivo.12 A further challenge is that nucleic acid-based therapeutics are target-specific (regarding mRNA sequence) but not selective for the type of cells they should affect.13
Some limitations associated with applying oligonucleotide-based drugs can be overcome through chemical modifications such as adding phosphorothioate (PS), locked and unlocked nucleotides, and substituting the ribose 2’-OH group.4,14–16 Another approach is using a Drug Delivery System (DDS); however, a highly effective DDS should meet several essential criteria. It should be capable of specifically targeting and interacting with the intended cell type, even within a complex tissue environment. Moreover, DDS should be internalized by cells via endocytosis, allowing the drug to escape from the endosome, and its transport functions must be carried out without causing an adverse toxic and immunological response. Desirable is also that the carrier is biodegradable. It should also exhibit appropriate particle size and stability.11,17
DDS has already shown numerous advantages for drug application, including reducing the systemic side effects of medication, enhancing the efficiency of drug delivery, enabling controlled release, and improving the drug’s specificity.18 DDS based on different kinds of materials were tested in the delivery of various types of therapeutics, such as small molecule drugs, chemotherapeutics, gene therapy agents, antibodies, or enzymes.19–21 Despite the numerous advantages associated with the use of drug delivery systems, they also face certain limitations and challenges. Most DDS utilize the Enhanced Permeability and Retention (EPR) effect to deliver drugs to tumors. However, the limited specificity of this approach often leads to drug toxicity in healthy cells. As a result, there is a growing focus on developing DDS that can selectively target cancer cells or their specific components/characteristics while reducing the exposure of healthy cells to the drugs.22,23 Further challenges associated with the use of drug delivery systems include low bioavailability, limited drug loading capacity, and stability issues.24 Another limitation is that many drug delivery systems require complex and costly manufacturing processes.25 DDS may require expensive components or complex methods for their production or modification.
Drug delivery systems (DDS) are increasingly being applied in clinical settings.26 Currently, many therapeutics are administered using dedicated carriers. Nowadays, highly toxic drugs used in cancer treatment, such as doxorubicin (Doxil) and paclitaxel (Abraxane or Pazenir), may be delivered to tumors through drug delivery systems.27,28 This paclitaxel delivery method significantly reduces its drawbacks, increasing its stability, blood circulation time, and tumor uptake.29,30 However, nanomedicines face significant limitations, including high production costs, lengthy commercialization timelines, and complex manufacturing challenges, particularly for biologically based formulations. Clinical translation is also hindered by regulatory delays and inconsistent toxicological data, limiting their broader adoption.29
DDS for oligonucleotide-based therapeutics can be based on viral and non-viral carriers.31 Currently, non-viral systems are considered more promising.32 Polymeric, lipid-based, or inorganic nanostructures are most commonly utilized.21,33,34 These nanostructures can be functionalized with aptamers, peptides or antibodies to provide specificity toward target cells.35,36
Spider silk is regarded as a material with significant potential for biomedical applications, including cancer treatment.37 Silk can be obtained from nature, but the procedure is inefficient.38 Spider breeding is impossible due to their territorial and cannibalistic behavior. Moreover, silk harvested from nature can be a mixture of several types of spider silk proteins, which is inappropriate for some application (including medical) due to the problem of lack of reproducibility. Therefore, bioengineered silk was proposed as a more applicable material due to control over silk composition, purity, and reproducibility. Recombinant spider silk proteins were used to produce various biomaterials, including drug carriers, films, foams, and scaffolds.39–41 The process of obtaining recombinant spider silk protein begins with constructing a synthetic gene encoding the desired silk. The frequency of codon sequences occurring in the new gene is consistent with the codon frequency occurring in the future host, which can improve protein production yield. The prepared expression vector containing a gene encoding bioengineered silk is then inserted into the chosen expression system. The bacterial expression system is the most frequently employed for producing recombinant silk proteins due to its relatively simple production, rapid bacterial generation time, cost-effectiveness, and scalability of the process, which are key advantages of this method.42,43
An additional advantage of constructing synthetic genes of bioengineered proteins is the ability to introduce sequences that encode functional peptides.42 Such DNA modification results in each protein molecule containing a functional peptide. The functional peptides can modify the properties of the carrier and influence tumor targeting, cellular binding, or inorganic molecules binding.42,44 Various studies have shown that modified spider silk spheres can be efficient drug delivery carriers.40,45–49
In addition to the wide range of possibilities for producing different variants of bioengineered silk, the silk blending strategy broadens its potential applications. To generate more advanced materials with multiple functions, two types of silk designed for different purposes can be combined.37 In such strategy, silk functionalized to enhance its specificity to target cell can be blended with silk functionalized with a peptide that control drug-loading.50,51 Moreover, blending two types of silk can modify the physicochemical property of spheres.52
Our team previously demonstrated that spheres made of functionalized spider silk can effectively deliver doxorubicin to the HER2-overexpressing cancer cells.53 The human epidermal growth factor receptor 2 (HER2) belongs to the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases, which are situated on the cell membrane.54 It is not only a target for therapy itself (important clinically55–57) but is often used as a ligand for functionalized particles for intracellular drug delivery.58–62 In our system, the spider silk spheres modified with the H2.1 peptide (H2.1MS1) were effectively internalized and delivered the drug to HER2-overexpressing cancer cells.63 Once inside the cells, the silk spheres accumulate in lysosomes and undergo degradation; the lysosomal function is essential for silk-based carrier elimination.63 The therapeutic efficacy of the drug delivered by H2.1MS1 spheres was indicated in both in vitro and in vivo studies.53,64 The spheres did not cause toxic and immune adverse side effects, confirming its biocompatibility.65–67 We also demonstrated in another study that incorporating the poly-lysine peptide (KN) into different type of spider silk protein MS2 (MS2KN) made it an effective carrier for oligonucleotide-based drugs.68 Adding the KN peptide to silk significantly enhanced its ability to bind nucleic acids. Furthermore, encapsulating the oligonucleotide therapeutic within the spheres protected it from degradation by nucleases and extended its target gene silencing property.68 Although the oligonucleotide-loaded spheres were effectively internalized by macrophages, it should be pointed out that such system delivers oligonucleotide therapeutics to cells in a non-specific manner.
In the presented study, we generated a DDS that binds to HER2-overexpressing cancer cells through the H2.1 peptide but also effectively binds nucleic acids by KN peptide. Such DDS should specifically deliver oligonucleotide-based therapeutics to HER2+ cancer cells. We compared two systems designed to fulfill this objective. In the first system, we constructed an H2.1MS1KN silk containing both peptides to produce nanocarriers. The second strategy involved using a blend of two proteins, H2.1MS1 and MS2KN, each containing one of the desired peptides. We compared these two strategies regarding nanoparticle properties including their efficiency in binding nucleic acids and HER2-overexpressing cancer cells. Finally, we evaluated the effectiveness of STAT3 silencing as a result of siSTAT3 delivery by both types of spheres to the HER2+ cancer cells. We indicated that not only the selection of functional peptides, but also their quantity and type of silk is crucial when developing an effective silk-based DDS for delivering active siRNA.
Materials and Methods
Construction of Expression Plasmids pETNX-H2.1MS1KN
MS1, H2.1MS1, and MS2KN constructs have been previously designed.53,68,69 The new construct H2.1MS1KN was generated by cloning an oligonucleotide sequence encoding poly-lysine to bind nucleic (KN) into the SpeI restriction site of the H2.1MS1 (13mer) construct for its 3′-terminal functionalization. The sequences of the added oligonucleotides were: KN.F:5′-CTAGCAAAAAGAAAAAAAAGAAAAAGAAAAAAAAGAAAAAGAAAAAGAAAA,KN.R:3′-GTTTTTCTTTTTTTTCTTTTTCTTTTTTTTCTTTTTCTTTTTCTTTTGACT. The KN.F and KN.R oligonucleotides were annealed to form double-stranded DNA fragments with cohesive ends complementary to those generated by NheI and SpeI. The sequence of the resulting plasmid was confirmed by sequencing at the Adam Mickiewicz University Core Facility in Poznan. The restriction enzymes and alkali phosphatase were supplied by Fermentas (Thermo Fisher Scientific Inc., Waltham, MA), while ligase by New England Biolabs, Inc. (Ipswich, MA).
Production and Purification of Bioengineered Spider Silks
Lab-scale production of MS1, H2.1MS1, H2.1MS1KN, and MS2KN proteins was performed in a Bioflo 415 fermentor (New Brunswick Scientific, Edison, NJ). MS1 and H2.1MS1 silk proteins were purified by using the thermal method 80:20, as previously described.69 In brief, the bacterial pellet was resuspended in lysis buffer containing 100 mm NaCl, 20 mm HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.5, and Pierce™ Protease Inhibitor Tablets, EDTA-free (ThermoFisher, Waltham, MA). Lysozyme (Sigma, St. Louis, MO), 0.2 mg/mL, was added to the solution to lyse the bacteria; then the mixture was incubated at 4°C with agitation for 30 min. Additionally, the bacteria and their structures were damaged by sonication. DNA was degraded by incubation with DNaseI (Sigma, St. Louis, MO; 0.1 mg/mL) supplemented with 3 mm MgCl2 (Sigma, St. Louis, MO). Then, bacterial proteins were denatured at 80°C for 10 min and removed by sedimentation at 21,000 × g for 30 min at 4°C. The thermal treatment was repeated at 80°C for 20 min, followed by centrifugation as described above. Next, the spider silk protein was precipitated overnight at 4°C with 20% ammonium sulfate (VWR, West Chester, US). The next day, the protein was sedimented (30 min, 7000 × g) and washed with 20% ammonium sulfate. The precipitated silk was dissolved in 6 M guanidine thiocyanate and dialyzed using Cellulose Dialysis Tubing cutoff 14,000 Da (Sigma, St. Louis, MO) against 10 mm TRIS-HCl buffer pH 7,5- or 50 mm Sodium Borate pH 8,5 depending on further application.
To purify proteins with a nucleic acid-binding peptide (KN), the propionic acid-based method was employed as described before with a modification.69 Five milligrams of bacterial pellets were thoroughly disrupted in 5 mL of 13.43 N propionic acid. The resulting mixture was then diluted with 19 mL of sterile water and incubated at 4°C for one hour with agitation. The precipitated proteins and cells debris were centrifuged at 21,000 × g for 30 min, and the supernatant was collected. Next, NaCl was added to the supernatant at a final concentration of 2 M, incubated at 4°C for 1 h, and then centrifuged at 21,000 × g, for 30 min. After collection of supernatants, silk proteins were precipitated using 20% ammonium sulfate, and further steps of protein purification were processed according to the 80:20 method. The predicted molecular weight of the H2.1MS1KN protein is 38.56 kDa, and the extinction coefficient is 40340 M-1 cm-1.
All proteins underwent a rigorous analysis using the standard scientific method of SDS-PAGE gel electrophoresis. A 12.5% gel was used, and the proteins were stained with colloidal blue (Roti-Blue; Carl Roth, Karlsruhe, Germany). This method provided us with a comprehensive understanding of the proteins’ characteristics and purity.
Nucleic Acid Binding Assay
To test the protein’s ability of nucleic acids binding, 1 µL of 50 µM siRNA Luc (siLuc) (Table 1) was mixed with H2.1MS1, H2.1MS1KN, MS2KN, or H2.1MS1:MS2KN blend (80:20) at a ratio of 1:0, 1:1, 1:10, 1:100, and 1:200, respectively. The binding was performed in 10 mm Tris-HCl pH 7.5. After 5 min of incubation at RT, complexes were analyzed in 2% agarose gel stained with 1,5% Ethidium Bromide (Sigma, St. Louis, MO). The gel was imaged using a G-box transilluminator (Syngene, Frederick, MD) and analyzed using GeneTools software (Syngene, Frederick, MD).
 |
Table 1 List of siRNA Sequences Used for Treatment |
Protein Labeling
MS1, H2.1MS1, or H2.1MSKN protein (1 mg) in 50 mm Sodium Borate, pH 8,5, was mixed with Cy®5 Mono NHS Ester (50 µg) (Cytiva, Danaher Corporation, Washington, DC) and incubated in the dark for 1 hour, according to manufacturer’s protocol. After incubation, the excess dye was removed using PD10 desalting columns (Cytiva, Danaher Corporation, Washington, DC) following the producer’s instructions.
Silk Spheres Production
The spheres were formed using the micropipette method or by a micromixing device. For the blended spheres, H2.1MS1 protein [0,5 mg/mL] was mixed with MS2KN silk [0,5 mg/mL] at a ratio of 80:20. In the micropipette method, 100 µL silk solution at 0.5 mg/mL concentration was mixed by pipetting with 1000 µL of 2 M Potassium Phosphate, pH 8 (Sigma, St. Louis, MO). In the micromixing method, 1 mL of protein at a concentration of 0.5 mg/mL was mixed with 10 mL of 2 M Potassium Phosphate, pH 8, using a Nemesys ultra high-pressure syringe pump (CETONI GmbH, Korbußen, Germany). The silk and 2 M Potassium Phosphate (pH 8) solutions were delivered through tubing with a diameter of 250 nm at flow rates of 10 µL/s and 100 µL/s, respectively, and subsequently mixed at a 1:10 ratio in T-element with a diameter of 500 nm. The obtained particles were incubated in 2 M Potassium Phosphate, pH 8 overnight at room temperature. After dialyzing against ultrapure water, the spheres were centrifuged at 21,000 × g for one hour and redispersed in PBS or deionized water for further analysis.
Loading of siRNA into Silk Spheres
Spheres loaded with siRNA were formed by mixing the silk and nucleic acid complexes with a 2 M Potassium Phosphate, pH 8. First, 50 μL of H2.1MS1KN 1 mg/mL [26µM] soluble silk protein and 50 μL of 20 μM iFam-si Scrambled (iFAM-siScr), siLuc, or siSTAT3 (Table 1) were mixed and incubated at room temperature for 5 min. The following production strategy was employed for blended spheres: first, 10 μL of MS2KN 1 mg/mL [20µM] soluble silk protein and 50 μL of 20 μM iFam-siScr, siLuc, or siSTAT3 were mixed and incubated at room temperature for 5 min. Then, 40 µL of H2.1MS 1 mg/mL [23µM] soluble silk protein was added. The complexes silk:siRNA were then used to form sphere as described at point 2.5.
For loading efficiency determination, the iFam- siScr loaded spheres were used. After loading and spheres sedimentation, the supernatant was measured for fluorescence of unloaded iFam- siScr using Victor X2 Multilabel Microplate Reader (PerkinElmer, Waltham, MA). A standard calibration curve for the iFam- siScr was used for drug quantification. To calculate the efficiency of iFam- siScr loading into spheres, the following formula was used:
Characterization of Silk Spheres by Scanning Electron Microscopy (SEM)
The suspension of silk spheres in water was applied onto coverslips (Thermo Scientific/Nunc, Langenselbold, Germany) and allowed to air-dry. Samples were sputtered with gold under a vacuum in a Quorum Sputter Coater Q150T ES (Quorum Technologies, Ringmer, UK) and analyzed using SEM (JEOL JSM-7001F, JEOL Ltd., Peabody, USA) at 10 kV of accelerating voltage.
Analysis of the Size of Silk Spheres
The sphere size was measured using two distinct methods. The first method involved analyzing images acquired through Scanning Electron Microscopy (SEM), with particle sizes determined using ImageJ 1.46r software. The size analysis was conducted based on three SEM images taken from three independent silk sphere preparation. Thirty measurements were taken from each photo. The second method utilized the NanoSight instrument (Malvern Panalytical, Malvern, GB). To measure the particle size, the spheres were suspended in 3 mL of distilled water at a concentration of 50 µg/mL and sonicated for 5 minutes in an ultrasonic water bath. The experiment was performed three times.
Zeta Potential (ZP) Analysis of Silk Spheres
To measure the zeta potential of particles, 5 μL of spheres (1 mg/mL) were suspended in 10 mL of distilled water and sonicated for 5 min in a sonic water bath. The spheres were then analyzed using a ZetaSizer (Malvern Panalytical, Malvern, GB). The experiment was conducted three times in triplicate.
Cell Cultures
Human HER2 overexpressing ovarian cancer cells SKOV3 (ATCC, Manassas, VA) and HER2 negative breast cancer cells MDA-MB-231 (ATCC, Manassas, VA) were used in the study. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM; Coring, New York, NY) supplemented with 10% FBS (Biowest, Nuaillé, France) and 80 μg/mL gentamicin (KRKA, Novo Mesto, Slovenia).
Cytotoxicity of Silk Spheres
A total of 2×104 SKOV3 cells per well were seeded into a 96-well plate and incubated overnight. The following day, varying H2.1MS1KN or H2.1MS1:MS2KN silk spheres concentrations were introduced into the cell cultures. Untreated cells served as a negative control. After 3 hours, the unbound particles were removed from the medium, and the cells were cultured for 48 hours. Mitochondrial activity was then measured using the MTT assay. A solution of 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) at a concentration of 1 mg/mL (VWR, Radnor,r, PA) was added to the cells, followed by incubation at 37°C for 4 hours. After incubation, the medium was removed, and the resulting formazan crystals were dissolved in Dimethyl Sulfoxide (DMSO) (Chempur, Piekary Slaskie, Poland). The relative cell viability (%) was calculated as (test sample/negative control) × 100%. This experiment was performed in triplicate and repeated three times.
Cell Binding Assay of Silk Spheres
For flow cytometry analysis, SKOV3 cells were washed with PBS and detached using a non-enzymatic cell dissociation solution (Sigma, St. Louis, MO). Next, 10 µg/mL spheres labeled with Cy5 were mixed with 1×105 cells suspended in PBS with BSA [1%] and incubated for one hour at 4°C in the dark. After three times washing with PBS, cells were investigated with a Navios flow cytometer (Beckman Coulter, Brea, CA) and analyzed using FlowJo Software (Becton Dickinson, Franklin Lakes, NJ).
Cellular Uptake of Silk Spheres
The SKOV3 and MDA-MB-231 cancer cell lines were seeded at a quantity of 3×104 on 8-well Lab-Tek® II chambered coverglass (Nunc, Rochester, NY) 24 hours before the experiment. Next, cells were incubated for 4 hours with 5 µg/mL of Cy5-labeled spheres at 37°C, washed twice with PBS, and finally fixed with 4% paraformaldehyde (Agar Scientific, Rotherham, UK) for 10 min. After fixation, the cell membrane was stained with 50 μg/mL FITC-labeled Concanavalin A (Sigma, St. Louis, MO) for 30 min and then coated with a FluoroShield containing DAPI (Sigma, St. Louis, MO). The cells were imaged using a Confocal Laser Scanning Microscope Zeiss LSM 880 Airyscan system (Jena, Germany). Filter sets used included a 488 nm filter with band pass 491–553 nm (laser Argon2), 410–480 nm DAPI (laser Diode 405), and 543 nm with band pass 584–632 nm (laser HeNe1). Plan-Apochromat 63×/1.4 Oli DIC M27 objective was used.
Real-Time Quantitative PCR
The SKOV3 cells were seeded at 1×105 per well of a 12-well plate and cultured for 24 h. Subsequently, cells were treated with siSTAT3 delivered by spheres at a dosage of 250 nM. The spheres loaded with siLuc were used as a control. After 48 hours, RNA was extracted using Fenozol (A&ABiotechnology, Gdansk, Poland) according to the manufacturer’s protocol. RNA was quantified using NanoDrop (Thermo Scientific, Wilmington, DE), and 1 μg of total RNA was used for cDNA synthesis with iScript Reverse Transcription Supermix (Bio-Rad, Irvine, CA). QPCR primers specific for human STAT3 and GAPDH were designed using Primer3Plus software and are listed in Table 2. Sequence-specific amplification was performed with Maxima SYBR Green/ROX qPCR Master Mix (ThermoFisher Scientific, Waltham, MA) and LightCycler 480 (Roche, Basel, Switzerland). Data was normalized to GAPDH expression, and the relative expression levels compared to siLuc were calculated using the 2−ΔΔCt method. The experiment was repeated six times.
 |
Table 2 List of Primer Sequences Used for qPCR |
Statistics
To analyze the significant differences between H2.1MS1KN and H2.1MS1:MS2KN groups, for samples of normal distribution, the t-test was used. In cases of non-normal distribution, the Mann–Whitney test was employed to establish the level of significance between the groups. The differences were considered significant when p < 0.0001 (****), p < 0.001 (***), p < 0.01 (**), p < 0.05 (*). The data presented are expressed as mean ± standard error of the mean. The statistical differences were calculated with GraphPad Prism 8 software.
Results and Discussion
Production and Purification of Bioengineered Spider Silk Variants
The aim of this study was the construction of an efficient, stable, and cell-specific silk-based delivery system for therapeutic siRNA molecules to HER2-overexpressing cancer cells. Our team previously designed, produced, and purified two bioengineered silk variants, MS1 and MS2.69,70 The bioengineered silk MS1 was based on the MaSp1 protein and the MS2 silk on MaSp2 from N. clavipes spider. Both MS1 and MS2 proteins can be modified by the addition of short peptides, which control their properties.45,50,52,68 Previous studies have shown that spheres made of the MS1 protein functionalized with H2.1 peptide (H2.1MS1, MYWGDSHWLQYWYE) can effectively bind and internalize into HER2+ cancer cells,53 while the modification of MS2 silk with poly-lysine KN (K15) peptide enables it to bind nucleic acids.68 In this study, we examined the possibility for construction of DDS based on properties of H2.1MS1 and MS2KN for delivery oligonucleotide therapeutics into HER2+ cells. The first strategy involved adding a KN peptide into H2.1MS1 spider silk protein sequence. The second strategy involved mixing both spider silk proteins to obtain blend of protein with desired properties.
The schematic representation of the spider silk protein constructs used in this study is illustrated in Figure 1A. As mentioned above, the proteins MS1, H2.1MS1 and MS2KN have been previously characterized.53,68,69 A novel bioengineered spider silk H2.1MS1KN was constructed by adding oligonucleotide sequence encoding KN peptide to H2.1MS1 (13mer) silk. The average yield of purified MS1 silk protein was approximately 1.5 mg, H2.1MS1 0.8 mg, MS2KN 0.6 mg, and H2.1MS1KN 0.2 mg per gram of bacterial pellets. The SDS-PAGE analysis indicated that except for the H2.1MS1 variant, silks did not show a sense of degradability, and all proteins exhibited high purity (Figure 1B). Moreover, the proteins MS1, H2.1MS1, and H2.1MS1KN migrate in accordance with expected molecular weight, however, MS2KN displayed higher molecular weight than expected (Figure 1B). Previously, we indicated that various types of MS2 silk (9-mer or 15-mer), although migrated in the SDS-PAGE gel not in accordance with expected molecular weights, the MALDI-TOF results aligned with the predicted values.70,71 One of the possible explanations for the impaired migration on the SDS-PAGE gel of the MS2-type silk is a relatively high content of proline residues (15.7%) in contrast to the MS1-type protein. It is considered that high proline content affects the conformation of the protein/SDS complex, causing low mobility on SDS-PAGE.72 The cyclic structure of proline imparts rigidity to the polypeptide chain, hindering the complete unfolding of the protein under SDS-PAGE conditions and altering its electrophoretic migration. Overestimation of molecular mass based on SDS-PAGE analysis has been reported for proline-rich proteins such as gliadin, hordein, secalin, calf thymus histone I, and collagen fragments.72
To obtain all silk variants, we applied two purification protocols. The standard method utilizes high temperature (80°C) to denature the bacterial proteins, while the soluble silk in the supernatant is then precipitated with 20% ammonium sulfate (method named 80/20). Although initially, we purified all silk variants using a thermal method, the procedure had to be changed due to the high endogenous nucleic acid contamination of H2.1MS1KN and MS2KN silks (data not shown, manuscript in preparation). The KN peptide could bind the bacterial nucleic acids, and during the standard purification protocol, the nucleic acids did not separate from silk, ultimately causing contamination. Modifying the purification protocol using an acid environment69 proved to be a superior method for the purification of the proteins with the KN peptide (manuscript in preparation). Shortly, the application of a high concentration of NaCl in acid condition led to the dissociation of the nucleic acids from the protein, which then allowed silk to be salted out using ammonium sulfate. The silk-purification protocol was modified based on the method used for the extraction of proteins, which are known to interact with DNA in cells, like histones.73–75 High salt decreases a non-specific interaction between the protein and impurities (like DNA) due to the interruption of the electrostatic interactions between them. Such an approach improved the H2.1MS1KN quality since the silk purity, expressed by the ratio of absorbance measured at 260 and 280 nm (260/280), was less than 0.6.
Additionally, the H2.1MS1KN silk demonstrated a considerably lower (four times) final yield than the H2.1MS1 protein. It could result from the double functionalization of MS1 silk, especially adding a poly-lysine peptide, which could be toxic to bacterial hosts.76–78
Analysis of siRNA Binding by Soluble Silk Variants
The study aimed to construct carriers for nucleic acid-based drug delivery. Thus, we compared the silks in terms of their nucleic acid binding capabilities. The ability of MS1, H2.1MS1KN, H2.1MS1, and H2.1MS1:MS2KN soluble proteins to bind oligonucleotides was tested in agarose gel assay. As indicated in Figure 2, the MS1 silk variant cannot form complexes with siLuc. Although adding H2.1 peptide to MS1 silk constructs slightly increased the nucleic acid binding property, the KN peptide was the one that considerably increased the oligonucleotide binding to both tested silk variants H2.1MS1KN and the blend of H2.1MS1:MS2KN (Figure 2).
Interestingly, the H2.1 peptide increased the nucleic acid binding potential. Based on the H2.1 peptide composition, the additional negatively charged amino acids were introduced to the construct (Asp and Glu), which should decrease the oligonucleotide binding due to electrostatic repulsion. On the other hand, additional histidine (His) was also introduced, which could increase oligonucleotide attachment. However, the reaction was performed at 10 mm Tris pH 7.5, and H should be relatively neutral at such pH. Nevertheless, the most pronounced effect induced poly-lysine peptide. At the pH neutral, the positively charged Lysine (pK 10.5) effectively interacts with nucleic acid. It should be pointed out that despite only 20% content of the MS2KN protein in the H2.1MS:MS2KN blend, the oligonucleotide binding property was similar to one with silk with both functionalizations, in which each molecule possesses one KN peptide.
The Properties of H2.1MS1KN and H2.1MS1:MS2KN Silk Spheres
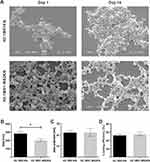
Both H2.1MS1KN and a blend of H2.1MS1:MS2KN proteins formed spheres, indicating that neither the KN peptide nor the addition of MS2KN protein impeded sphere formation. The morphology of the H2.1MS1KN and blend of H2.1MS1:MS2KN spheres was quite different (Figure 3A). Although both H2.1MS1KN and blended spheres indicated the spherical shape, the spherical form of the blended H2.1MS1:MS2KN spheres was more clearly distinguished. These spheres were more defined and separated. Additionally, based on the analysis of SEM images, it can be concluded that the H2.1MS1KN spheres exhibited lower stability. After two weeks of storage, they exhibited a less spherical shape and reduced structural distinction. Moreover, the mean size of spheres indicated by analysis of the SEM images was approximately 336.2 ± 20 nm (mean ± standard error of the mean) for H2.1MS1:MS2KN and 393.1 ± 29 nm for H2.1MS1KN spheres, while by using NanoSight analyzer 148 ± 12 nm and 190 ± 8 nm, respectively. The discrepancies in the obtained mean size of particles may result from difference in sample preparation and the experimenter’s subjective selection of larger, well-defined spheres during SEM image analysis. Since the mean ± standard error of the mean value did not show the particle size distribution, we presented the D90 obtained from the NanoSight analyzer in Figure 3B. Analysis indicated that 90% of the H2.1MS:MS2KN particles were of size 205 ± 30 nm or smaller, while H2.1MS1KN spheres 286 ± 50 nm (Figure 3B). Regardless of the measurement method, H2.1MS1:MS2KN particles were consistently smaller than H2.1MS1KN spheres, and analysis of D90 values indicated a significant difference in particle size.
Our previous results indicated that MS1- and MS2-based spheres differed in shape and size.52,71 The SEM picture analysis indicated that the MS2-based spheres were smaller (426 (±109) nm vs 246 (±66) nm for MS1 vs MS2), and the spherical morphology was more distinct compared with MS1-type particles.52 The observed differences were probably due to their higher beta-sheet content (crystallinity) and the presumed presence of beta-spirals in MS2 spheres, which could lead to the tight packing of the silk molecules into particles.71 Additionally, we performed the study for blended silk particles (MS1:MS2), indicating that 20% MS2 content in the MS1:MS2 spheres resulted in the smaller and more spherical in shape spheres formation comparing with MS1, however the content of beta-sheet in blended spheres was similar to the plain MS1 particles.52 The data indicated in this study are in agreement with our previous result in terms of the size and morphology of spheres; the addition of the MS2 component to MS1 silk influenced the characteristics of silk particles. On the other hand, the analyzed spheres also contained KN peptides, which could be critical for their physicochemical characterization. Indeed, the addition of poly-lysine molecules decreased the size of MS2 spheres from 246 (±66) nm to 182.7 nm (±6.4 nm).52,68 Moreover, Numata et al indicated that the size of plain MS1-type particles changed from 570 nm to 210 nm after adding poly-lysine peptide, and their size depended on the number of Lysine repeats.79 Thus, the size and morphology of both spheres presented in this study were probably modified by poly-lysine peptide and MS2 silk, resulting in more favorable characteristics for blended H2.1MS1:MS2KN particles.
The zeta potential of the H2.1MS1KN and H2.1MS1:MS2KN particles was similar. The average of ZP for H2.1MS1:MS2KN was 23 (± 4) mV and for H2.1MS1KN 23 (± 6) mV (Figure 3C). It was interesting that blended spheres indicated similar ZP to H2.1MS1KN spheres. The H2.1MS1KN particles are built of silk in which each molecule contains poly-lysine peptide (KN), while in the blended H2.1MS1:MS2KN spheres, only 20% of silk is functionalized with KN. It indicates that the number of positively charged residues in the silk particles is not a dominant factor for its overall ZP value, but rather their distribution and manner of packing the silk molecules into spheres. A similar ZP in both types of spheres is also associated with a comparable potential for loading nucleic acids. The loading efficiency of oligonucleotides was similar, approximately 53% for H2.1MS1KN and 56% for the H2.1MS1:MS2KN spheres (Figure 3D). There was an increase in a negatively charged oligonucleotide embedding into blended particles compared with plain H2.1MS1KN, but the difference was insignificant.
The Cytotoxicity Analysis H2.1MS1KN and H2.1MS1:MS2KN Spheres
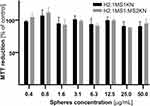
Spheres made of H2.1MS1KN and a blend of H2.1MS1 and MS2KN2 silks were not cytotoxic when examined at a wide range of concentrations (Figure 4). There was no significant difference between H2.1MS1KN and H2.1MS1:MS2KN spheres in terms of cytotoxicity. As demonstrated in our previous study, plain MS2KN spheres unloaded with siRNA exhibited toxic properties when tested at a high concentration in contrast to MS2 particles, indicating potential cytotoxicity of KN peptide.68 Using only 20% of MS2KN protein in the H2.1MS1:MS2KN blend did not induce a similar effect. Interestingly, H2.1MS1KN particles were also not cytotoxic despite containing a high KN content. It may suggest that MS1-type protein may mask the cytotoxic effects of the KN peptide.
The Binding Property of H2.1MS1KN and H2.1MS1:MS2KN Silk Spheres to HER2+ Cancer Cells
To assess the binding property of functionalized spheres to HER2+ cancer cells, we incubated HER2-overexpressing SKOV3 ovarian cancer cells with fluorescently labeled (Cy5) spheres for 1 hour without transfection reagents. Flow cytometric analysis confirmed that the H2.1 peptide increased the binding of silk spheres to HER2-overexpressing cancer cells (Figure 5) Additionally, adding positively charged KN peptide increased the cell-binding potential of H2.1MS1KN and H2.1MS1:MS2KN particles and the percentage of cells that bound fluorescently labeled spheres were higher than controls (Figure 5). However, the MFI analysis indicated the considerable difference between cells that interact with the H2.1MS1KN and H2.1MS1:MS2KN spheres, the H2.1MS1:MS2KN being one significantly more accepted by cells (number of bound-spheres per cell).
The Internalization of H2.1MS1KN and H2.1MS1:MS2KN Silk Spheres into HER2+ Cancer Cells
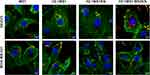
The flow cytometry analysis indicated unequal distribution of cells that bound of H2.1MS1KN vs H2.1MS1:MS2KN spheres suggesting different efficiency of their internalization. Thus, the cellular internalization of silk spheres into HER2- (MDA-MB-231) and HER2+ (SKOV3) cancer cells was analyzed using confocal microscopy (Figure 6). The singular MS1 particles (control) were visible inside the HER2- and HER2+ cancer cells. Some H2.1MS1KN spheres were internalized into HER2+ cancer cells and were present on the cell surface of HER2- cells; however, their fluorescent signal (red) was co-localized with FITC-labelled membrane structures. The H2.1MS1:MS2KN spheres were present in the cytosol of the HER2+ cancer cells at the highest quantity compared with other particles. Although H2.1MS1:MS2KN spheres indicated some binding to the HER2- MDA-MB-231 breast cancer cells, most of the particles remained on the cell surface and did not internalize (Figure 6).
We indicated previously that functionalization with the H2.1 peptide significantly increased the cellular internalization of silk spheres.53,63 The flow cytometry analysis suggested that further functionalization with KN peptide increased the cell-binding potential of silk spheres (Figure 5). The positively charged peptides, like poly-lysine or poly arginine, can penetrate the cell membrane, and they are often used for the functionalization of different types of nanoparticles to enable their cellular internalization.80–83 In our previous study, we indicated that MS2KN silk spheres (without a specific targeting peptide) were internalized by macrophages.68 However, the main macrophage’s function is phagocytosis of various molecules, including bacteria, viruses, cells, and others; thus, the internalization of the MS2KN particles could be related to such macrophage’s properties. In this study, we indicated that adding poly-lysine peptide caused distinct effects depending on sphere type. Indeed, there was more abounded number of the blended H2.1MS1:MS2KN particles in the cytoplasm of SKOV3 cancer cells than H2.1MS1 one, suggesting the synergistic effect of KN and H2.1 peptides. However, the H2.1MS1KN internalized at a much lower level into SKOV3 cells. Moreover, based on the CLSM pictures, it can be suggested that the silk particles stuck into the cell membrane (the co-localization of the fluorescent signals of spheres and cell membrane). Although the MTT assay did not indicate the influence of silk particles on cellular metabolism, the analysis of microscopic pictures may suggest the influence of H2.1MS1KN particles on cellular morphology. It was reported previously that poly-lysine, as a cationic polymer, could be toxic to cells when applied at a high concentration.80,83 Summarizing, the obtained data suggests that a high number of poly-lysine residues (again, each H2.1MS1KN silk molecule contains KN peptide) is not beneficial for cell interaction. Interestingly, some parts of the blended H2.1MS1:MS2KN particles also co-localized with the cell membrane, contrary to H2.1MS1 spheres (Figure 6). Probably, it was an intermediate state since a much larger number of them occurred in the cytoplasm.
Analysis of the Gene Silencing Effect Exerted of siRNA Delivered Using H2.1MS1KN and H2.1MS1:MS2KN Spheres
The siSTAT3 (250 nM) and control siLuc (250 nM) were encapsulated into H2.1MS1KN and H2.1MS1:MS2KN silk spheres, applied on the SKOV3 HER2+ cancer cells, and the STAT3 silencing effect was determined by quantitative real-time PCR analysis. The six time repeated experiments indicated that H2.1MS1:MS2KN spheres delivered siSTAT3 more effectively than H2.1MS1KN particles, which caused significantly higher downregulation of STAT3 expression into SKOV3 cells (Figure 7A). The same preparation of silk spheres was used three times with a one-week interval, and the silencing effect of STAT3 was repetitively achieved for siRNA delivered by H2.1MS1:MS2KN spheres (Figure 7B; I–III). The efficacy of H2.1MS1KN particles for delivering functional siSTAT3 decreased in time (Figure 7B; I–III). The experimental cycle was repeated, and the results were similar (data not shown). The reason for the loosing time-related potential of H2.1MS1KN spheres for delivery active siRNA was not examined in detail. However, it may be related to the unstable morphology of the spheres, releasing of siRNA, exposure more cytotoxic poly-lysine residues or combination of these. As the MTT study indicated that none silk particles modified the cellular metabolism, the cytotoxicity was not the direct reason for the lower performance of H2.1MS1KN particles. However, SEM image analysis indicated that the spheres’ instability may be responsible for the reduction in siRNA delivery efficiency of H2.1MS1KN over time. The unstable morphology of the spheres may contribute to the release of siRNA and inadequate interaction/recognition of the cells, decreasing the overall performance of H2.1MS1KN particles in siRNA delivery. It needs further detailed explanation; however, it was not within the scope of this study.
Conclusion
We constructed an efficient, stable, and cell-specific delivery system for therapeutic siRNA molecules to HER2+ cancer cells using bioengineered silk spheres vehicles. The blended H2.1MS1:MS2KN spheres were effectively loaded with oligonucleotides and delivered siSTAT3 to HER2+ cancer cells that successfully silenced STAT3 expression. The blended H2.1MS1:MS2KN spheres are better choice than H2.1MS1KN particles due to i) higher efficiency of protein yields, ii) smaller size, iii) more defined spherical morphology, iv) higher HER2+ cell-binding potential, v) higher specific internalization into HER-overexpressing cancer cells, and finally vi) more efficient and stable delivery of functional siRNA. The superior property of the H2.1MS1:MS2KN spheres may be due to lower number of poly-lysine residues in the particles and stabilizing influence of MS2-type silk. H2.1MS1:MS2KN particles have emerged as a promising siRNA targeted delivery system with great potential for use in the treatment of HER2-overexpressing cancers. The reasonable next step is in vivo evaluation of the safety and efficiency of the developed DDS. In the future, the silk-based DDS that precisely delivers oligonucleotide therapeutics can be tested alongside other strategies to treat cancer, including chemo-, photodynamic, thermal, immune-, or hormone therapy. Nowadays, various DDSs, including silk-based ones, are being tested to fulfill the requirements of such strategies. Undoubtedly, targeted delivery systems are key to optimizing safe, side-effect-minimized treatment and will continue to evolve. Silk is very promising for these applications as a biocompatible and degradable material.
Acknowledgments
The project was supported by the grants from the National Science Centre, Poland (NCN 2020/37/B/NZ6/03435).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kaur R, Bhardwaj A, Gupta S. Cancer treatment therapies: traditional to modern approaches to combat cancers. Mol Biol Rep. 2023;50(11):9663–9676. doi:10.1007/s11033-023-08809-3
2. Cesur‐Ergün B, Demir‐Dora D. Gene therapy in cancer. J Gene Med. 2023;25(11):e3550. doi:10.1002/jgm.3550
3. Chung SS, Giehl N, Wu Y, Vadgama JV. STAT3 activation in HER2-overexpressing breast cancer promotes epithelial-mesenchymal transition and cancer stem cell traits. Int J Oncol. 2014;44(2):403–411. doi:10.3892/ijo.2013.2195
4. Molenda S, Sikorska A, Florczak A, Lorenc P, Dams-Kozlowska H. Oligonucleotide-based therapeutics for STAT3 targeting in cancer—drug carriers matter. Cancers. 2023;15(23):5647. doi:10.3390/cancers15235647
5. Hossain DMS, Dos Santos C, Zhang Q, et al. Leukemia cell–targeted STAT3 silencing and TLR9 triggering generate systemic antitumor immunity. Blood. 2014;123(1):15–25. doi:10.1182/blood-2013-07-517987
6. Kunigal S, Lakka SS, Sodadasu PK, Estes N, Rao JS. Stat3-siRNA induces Fas-mediated apoptosis in vitro and in vivo in breast cancer. Int J Oncol. 2009;34(5):1209–1220.
7. Rahmati M, Johari B, Kadivar M, Rismani E, Mortazavi Y. Suppressing the metastatic properties of the breast cancer cells using STAT3 decoy oligodeoxynucleotides: a promising approach for eradication of cancer cells by differentiation therapy. J Cell Physiol. 2020;235(6):5429–5444. doi:10.1002/jcp.29431
8. Moreira D, Adamus T, Zhao X, et al. STAT3 inhibition combined with CpG immunostimulation activates antitumor immunity to eradicate genetically distinct castration-resistant prostate cancers. Clin Cancer Res. 2018;24(23):5948–5962. doi:10.1158/1078-0432.CCR-18-1277
9. Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33(13):1670–1679. doi:10.1038/onc.2013.115
10. McDaniel JM, Varley KE, Gertz J, et al. Genomic regulation of invasion by STAT3 in triple negative breast cancer. Oncotarget. 2017;8(5):8226–8238. doi:10.18632/oncotarget.14153
11. Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23(5):265–280. doi:10.1038/s41576-021-00439-4
12. Haupenthal J, Baehr C, Kiermayer S, Zeuzem S, Piiper A. Inhibition of RNAse A family enzymes prevents degradation and loss of silencing activity of siRNAs in serum. Biochem Pharmacol. 2006;71(5):702–710. doi:10.1016/j.bcp.2005.11.015
13. Mittal M, Kumari A, Paul B, et al. Challenges and opportunities of gene therapy in cancer. OBM Gene. 2024;08(01):1–501. doi:10.21926/obm.genet.2401219
14. Shaw BR, Dobrikov M, Wang X, et al. Reading, writing, and modulating genetic information with boranophosphate mimics of nucleotides, DNA, and RNA. Ann N.Y Acad Sci. 2003;1002(1):12–29. doi:10.1196/annals.1281.004
15. Kurreck J. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30(9):1911–1918. doi:10.1093/nar/30.9.1911
16. Bramsen JB, Pakula MM, Hansen TB, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38(17):5761–5773. doi:10.1093/nar/gkq341
17. Altuntaş E, Özkan B, Güngör S, Özsoy Y. Biopolymer-based nanogel approach in drug delivery: basic concept and current developments. Pharmaceutics. 2023;15(6):1644. doi:10.3390/pharmaceutics15061644
18. Zhang Y, Liu X, Li X. Development and Advantages Of Drug Delivery Systems. In: Teng L, Yang Z, Li C editors. Drug Delivery to Tumors. Springer Nature Singapore; 2025:35–60. doi:10.1007/978-981-19-8930-8_2.
19. Nie Y, Li D, Peng Y, et al. Metal organic framework coated MnO2 nanosheets delivering doxorubicin and self-activated DNAzyme for chemo-gene combinatorial treatment of cancer. Int J Pharm. 2020;585:119513. doi:10.1016/j.ijpharm.2020.119513
20. Castellino SM, Pei Q, Parsons SK, et al. Brentuximab vedotin with chemotherapy in pediatric high-risk Hodgkin’s lymphoma. N Engl J Med. 2022;387(18):1649–1660. doi:10.1056/NEJMoa2206660
21. Yu X, Liu S, Cheng Q, et al. Lipid‐modified aminoglycosides for mRNA delivery to the liver. Adv Healthcare Mater. 2020;9(7):1901487. doi:10.1002/adhm.201901487
22. Ezike TC, Okpala US, Onoja UL, et al. Advances in drug delivery systems, challenges and future directions. Heliyon. 2023;9(6):e17488. doi:10.1016/j.heliyon.2023.e17488
23. Yang H, Li Q, Chen X, et al. Targeting SOX13 inhibits assembly of respiratory chain supercomplexes to overcome ferroptosis resistance in gastric cancer. Nat Commun. 2024;15(1):4296. doi:10.1038/s41467-024-48307-z
24. Loke YH, Jayakrishnan A, Mod Razif MRF, et al. A comprehensive review of challenges in oral drug delivery systems and recent advancements in innovative design strategies. CPD. 2025;31(5):360–376. doi:10.2174/0113816128338560240923073357
25. Milewska S, Niemirowicz-Laskowska K, Siemiaszko G, Nowicki P, Wilczewska AZ, Car H. Current trends and challenges in pharmacoeconomic aspects of nanocarriers as drug delivery systems for cancer treatment. IJN. 2021;16:6593–6644. doi:10.2147/IJN.S323831
26. Patra JK, Das G, Fraceto LF, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnol. 2018;16(1):71. doi:10.1186/s12951-018-0392-8
27. Moskowitz AJ, Shah G, Schöder H, et al. Phase II trial of pembrolizumab plus gemcitabine, vinorelbine, and liposomal doxorubicin as second-line therapy for relapsed or refractory classical Hodgkin lymphoma. JCO. 2021;39(28):3109–3117. doi:10.1200/JCO.21.01056
28. Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–2121. doi:10.1056/NEJMoa1809615
29. Rodríguez F, Caruana P, De La Fuente N, et al. Nano-based approved pharmaceuticals for cancer treatment: present and future challenges. Biomolecules. 2022;12(6):784. doi:10.3390/biom12060784
30. Linxweiler H, Thiesen J, Krämer I. Physicochemical stability of nab-paclitaxel (Pazenir) infusion dispersions in original glass vials and EVA infusion bags. Pharmaceutics. 2024;16(11):1372. doi:10.3390/pharmaceutics16111372
31. Torres-Vanegas JD, Cruz JC, Reyes LH. Delivery systems for nucleic acids and proteins: barriers, cell capture pathways and nanocarriers. Pharmaceutics. 2021;13(3):428. doi:10.3390/pharmaceutics13030428
32. Dume B, Licarete E, Banciu M. Advancing cancer treatments: the role of oligonucleotide-based therapies in driving progress. Mol Ther Nucleic Acids. 2024;35(3):102256. doi:10.1016/j.omtn.2024.102256
33. Pan J, Ruan W, Qin M, et al. Intradermal delivery of STAT3 siRNA to treat melanoma via dissolving microneedles. Sci Rep. 2018;8(1):1117. doi:10.1038/s41598-018-19463-2
34. Morgan E, Wupperfeld D, Morales D, Reich N. Shape matters: gold nanoparticle shape impacts the biological activity of siRNA delivery. Bioconjugate Chem. 2019;30(3):853–860. doi:10.1021/acs.bioconjchem.9b00004
35. Seleci DA, Seleci M, Jochums A, Walter JG, Stahl F, Scheper T. Aptamer mediated niosomal drug delivery. RSC Adv. 2016;6(91):87910–87918. doi:10.1039/C6RA19525C
36. Jiang S, Wang X, Zhang Z, et al. CD20 monoclonal antibody targeted nanoscale drug delivery system for doxorubicin chemotherapy: an in vitro study of cell lysis of CD20-positive Raji cells. Int J Nanomed. 2016;11:5505–5518. doi:10.2147/IJN.S115428
37. Florczak A, Deptuch T, Kucharczyk K, Dams-Kozlowska H. Systemic and local silk-based drug delivery systems for cancer therapy. Cancers. 2021;13(21):5389. doi:10.3390/cancers13215389
38. Ramezaniaghdam M, Nahdi ND, Reski R. Recombinant spider silk: promises and bottlenecks. Front Bioeng Biotechnol. 2022;10:835637. doi:10.3389/fbioe.2022.835637
39. Agostini E, Winter G, Engert J. Water-based preparation of spider silk films as drug delivery matrices. J Control Release. 2015;213:134–141. doi:10.1016/j.jconrel.2015.06.025
40. Herold HM, Döbl A, Wohlrab S, Humenik M, Scheibel T. Designed spider silk-based drug carrier for redox- or pH-triggered drug release. Biomacromolecules. 2020;21(12):4904–4912. doi:10.1021/acs.biomac.0c01138
41. Schacht K, Vogt J, Scheibel T. Foams made of engineered recombinant spider silk proteins as 3D scaffolds for cell growth. ACS Biomater Sci Eng. 2016;2(4):517–525. doi:10.1021/acsbiomaterials.5b00483
42. Deptuch T, Dams-Kozlowska H. Silk materials functionalized via genetic engineering for biomedical applications. Materials. 2017;10(12):1417. doi:10.3390/ma10121417
43. Tokareva O, Michalczechen‐Lacerda VA, Rech EL, Kaplan DL. Recombinant
44. Trossmann VT, Scheibel T. Design of recombinant spider silk proteins for cell type specific binding. Adv Healthcare Mater. 2023;12(9):2202660. doi:10.1002/adhm.202202660
45. Kucharczyk K, Florczak A, Kaminska A, et al. MMPs-responsive silk spheres for controlled drug release within tumor microenvironment. Int J Biol Macromol. 2024;269:132016. doi:10.1016/j.ijbiomac.2024.132016
46. Huang T, Kumari S, Herold H, et al. Enhanced antibacterial activity of se nanoparticles upon coating with recombinant spider silk protein eADF4(κ16). Int J Nanomed. 2020;15:4275–4288. doi:10.2147/IJN.S255833
47. Lammel A, Schwab M, Hofer M, Winter G, Scheibel T. Recombinant spider silk particles as drug delivery vehicles. Biomaterials. 2011;32(8):2233–2240. doi:10.1016/j.biomaterials.2010.11.060
48. Mulinti P, Shreffler J, Hasan R, Dea M, Brooks AE. Infection responsive smart delivery of antibiotics using recombinant spider silk nanospheres. Pharmaceutics. 2021;13(9):1358. doi:10.3390/pharmaceutics13091358
49. Numata K, Reagan MR, Goldstein RH, Rosenblatt M, Kaplan DL. Spider silk-based gene carriers for tumor cell-specific delivery. Bioconjugate Chem. 2011;22(8):1605–1610. doi:10.1021/bc200170u
50. Kucharczyk K, Kaczmarek K, Jozefczak A, Slachcinski M, Mackiewicz A, Dams-Kozlowska H. Hyperthermia treatment of cancer cells by the application of targeted silk/iron oxide composite spheres. Mater Sci Eng C. 2021;120:111654. doi:10.1016/j.msec.2020.111654
51. Kucharczyk K, Florczak A, Deptuch T, et al. Drug affinity and targeted delivery: double functionalization of silk spheres for controlled doxorubicin delivery into Her2-positive cancer cells. J Nanobiotechnol. 2020;18(1):56. doi:10.1186/s12951-020-00609-2
52. Florczak A, Jastrzebska K, Mackiewicz A, Dams-Kozlowska H. Blending two bioengineered spider silks to develop cancer targeting spheres. J Mat Chem B. 2017;5(16):3000–3011. doi:10.1039/C7TB00233E
53. Florczak A, Mackiewicz A, Dams-Kozlowska H. Functionalized spider silk spheres as drug carriers for targeted cancer THERAPY. Biomacromolecules. 2014;15(8):2971–2981. doi:10.1021/bm500591p
54. Van Der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Ann Rev Cell Biol. 1994;10(1):251–337. doi:10.1146/annurev.cb.10.110194.001343
55. Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov. 2023;22(2):101–126. doi:10.1038/s41573-022-00579-0
56. Baselga J. Clinical trials of Herceptin® (trastuzumab). Eur J Cancer. 2001;37:18–24. doi:10.1016/S0959-8049(00)00404-4
57. Ryan Q, Ibrahim A, Cohen MH, et al. FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. oncologist. 2008;13(10):1114–1119. doi:10.1634/theoncologist.2008-0816
58. Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci. 2013;110(20):8170–8175. doi:10.1073/pnas.1302594110
59. Kim B, Shin J, Wu J, et al. Engineering peptide-targeted liposomal nanoparticles optimized for improved selectivity for HER2-positive breast cancer cells to achieve enhanced in vivo efficacy. J Control Release. 2020;322:530–541. doi:10.1016/j.jconrel.2020.04.010
60. Quinn Z, Mao W, Xia Y, John R, Wan Y. Conferring receptors on recipient cells with extracellular vesicles for targeted drug delivery. Bioact Mater. 2021;6(3):749–756. doi:10.1016/j.bioactmat.2020.09.016
61. Hashemi-Moghaddam H, Zavareh S, Karimpour S, Madanchi H. Evaluation of molecularly imprinted polymer based on HER2 epitope for targeted drug delivery in ovarian cancer mouse model. React Funct Polym. 2017;121:82–90. doi:10.1016/j.reactfunctpolym.2017.10.025
62. Satpathy M, Wang L, Zielinski RJ, et al. Targeted drug delivery and image-guided therapy of heterogeneous ovarian cancer using HER2-targeted theranostic nanoparticles. Theranostics. 2019;9(3):778–795. doi:10.7150/thno.29964
63. Florczak A, Mackiewicz A, Dams-Kozlowska H. Cellular uptake, intracellular distribution and degradation of Her2-targeting silk nanospheres. Int J Nanomed. 2019;14:6855–6865. doi:10.2147/IJN.S217854
64. Florczak A, Deptuch T, Lewandowska A, et al. Functionalized silk spheres selectively and effectively deliver a cytotoxic drug to targeted cancer cells in vivo. J Nanobiotechnol. 2020;18(1):177. doi:10.1186/s12951-020-00734-y
65. Deptuch T, Florczak A, Lewandowska A, et al. Ms1-type bioengineered spider silk nanoparticles do not exhibit toxicity in an in vivo mouse model. Nanomedicine. 2021;16(18):1553–1565. doi:10.2217/nnm-2021-0029
66. Deptuch T, Penderecka K, Kaczmarek M, Molenda S, Dams-Kozlowska H. In vivo study of the immune response to bioengineered spider silk spheres. Sci Rep. 2022;12(1):13480. doi:10.1038/s41598-022-17637-7
67. Deptuch T, Kucharczyk K, Florczak A, Dams‐Kozlowska H. Endotoxin reduction from biotech silk material inhibits the production of anti‐silk antibodies in mice. J Biomed Mater Res Part A. 2024;112(3):463–472. doi:10.1002/jbm.a.37644
68. Kozlowska AK, Florczak A, Smialek M, et al. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomater. 2017;59:221–233. doi:10.1016/j.actbio.2017.07.014
69. Dams‐Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag‐free bioengineered spider silk proteins. J Biomed Mater Res Part A. 2013;101A(2):456–464. doi:10.1002/jbm.a.34353
70. Jastrzebska K, Felcyn E, Kozak M, et al. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci Rep. 2016;6:28106. doi:10.1038/srep28106
71. Jastrzebska K, Florczak A, Kucharczyk K, et al. Delivery of chemotherapeutics using spheres made of bioengineered spider silks derived from MaSp1 and MaSp2 proteins. Nanomedicine. 2018;13(4):439–454. doi:10.2217/nnm-2017-0276
72. Bunce NAC, White RP, Shewry PR. Variation in estimates of molecular weights of cereal prolamins by SDS-PAGE. J Cereal Sci. 1985;3(2):131–142. doi:10.1016/S0733-5210(85)80023-0
73. Smart JE, Bonner J. Selective dissociation of histones from chromatin by sodium deoxycholate. J Mol Biol. 1971;58(3):651–659. doi:10.1016/0022-2836(71)90030-1
74. Rodriguez-Collazo P, Leuba SH, Zlatanova J. Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Res. 2009;37(11):e81–e81. doi:10.1093/nar/gkp273
75. Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nature Protocols. 2007;2(6):1445–1457. doi:10.1038/nprot.2007.202
76. Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56(3):395–411. doi:10.1128/mr.56.3.395-411.1992
77. Tan Z, Shi Y, Xing B, Hou Y, Cui J, Jia S. The antimicrobial effects and mechanism of ε-poly-lysine against Staphylococcus aureus. Bioresources Bioprocess. 2019;6(1):11. doi:10.1186/s40643-019-0246-8
78. Švedienė J, Novickij V, Žalnėravičius R, et al. Antimicrobial activity of L-Lysine and Poly-L-Lysine with pulsed electric fields. Appl Sci. 2021;11(6):2708. doi:10.3390/app11062708
79. Numata K, Subramanian B, Currie HA, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2009;30(29):5775–5784. doi:10.1016/j.biomaterials.2009.06.028
80. Granier F, Marie S, Al Amir Dache Z, et al. Assessment of dendrigrafts of poly-
81. Riveros AL, Eggeling C, Riquelme S, et al. Improving cell penetration of gold nanorods by using an amphipathic arginine rich peptide. Int J Nanomed. 2020;15:1837–1851. doi:10.2147/IJN.S237820
82. Zhang H, Men K, Pan C, et al. Treatment of colon cancer by degradable rrPPC nano-conjugates delivered STAT3 siRNA. Int J Nanomed. 2020;15:9875–9890. doi:10.2147/IJN.S277845
83. Mohamed A, Korzhikov-Vlakh V, Zhang N, et al. Effect of Poly(L-lysine) and heparin coatings on the surface of polyester-based particles on prednisolone release and biocompatibility. Pharmaceutics. 2021;13(6):801. doi:10.3390/pharmaceutics13060801
 © 2025 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a
Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the
original author and source are credited.
© 2025 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a
Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the
original author and source are credited.