Back to Journals » International Journal of Nanomedicine » Volume 19
Disulfiram-Loaded Nanoparticles Inhibit Long-Term Proliferation on Preadipocytes
Authors Lorenzo-Anota HY , Gómez-Cantú JM, Vázquez-Garza E, Bernal-Ramirez J, Chapoy-Villanueva H, Mayolo-Deloisa K, Benavides J, Rito-Palomares M, Lozano O
Received 9 March 2024
Accepted for publication 3 November 2024
Published 10 December 2024 Volume 2024:19 Pages 13301—13318
DOI https://doi.org/10.2147/IJN.S467909
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Jie Huang
Helen Yarimet Lorenzo-Anota,1,2 José María Gómez-Cantú,2 Eduardo Vázquez-Garza,1 Judith Bernal-Ramirez,1 Héctor Chapoy-Villanueva,1,2 Karla Mayolo-Deloisa,1,3 Jorge Benavides,1,3 Marco Rito-Palomares,1,2 Omar Lozano1,2
1Tecnologico de Monterrey, Institute for Obesity Research, Monterrey, México; 2Tecnologico de Monterrey, Escuela de Medicina y Ciencias de la Salud, Monterrey, México; 3Tecnologico de Monterrey, Escuela de Ingeniería y Ciencias, Centro de Biotecnología-FEMSA, Monterrey, México
Correspondence: Omar Lozano, Email [email protected]
Introduction: Disulfiram (DSF) reduces insulin resistance and weight gain in obese mice. However, the effect on adipose tissue is unexplored due to their high instability under physiological conditions, limiting clinical applications. Thus, it is meaningful to develop a DSF carrier for sustained release to adipose tissue. We optimized the synthesis of poly-ϵ-caprolactone (PCL) nanoparticles (NPs) loaded with DSF and analyzed their effect on adipose tissue cells in vitro.
Methods: The NPs were synthesized by nanoprecipitation method, varying its solvent, either acetone or acetone/dichloromethane (60:40) (v/v), and ratio PCL:DSF (w/w) 1:2, 1:1, 2:1 and, 1:0; finding the best condition was obtained with acetone/dichloromethane solvent mixture and 2:1 PCL:DSF. Then, NPs toxicity was analyzed on adipose cells (preadipocytes, white-like adipocytes, and macrophages) assessing association and internalization, cell viability, and cell death mechanism.
Results: NPs were spherical with a particle size distribution of 203.2 ± 29.33 nm, a ζ-potential of − 20.7 ± 4.58 mV, a PDI of 0.296 ± 0.084, and a physical drug loading of 18.6 ± 5.80%. Sustained release was observed from 0.5 h (10.94 ± 2.38%) up to 96 h (91.20 ± 6.03%) under physiological conditions. NPs internalize into macrophages, white-like adipocytes and preadipocytes without modifying cell viability on white-like adipocytes and macrophages. Preadipocytes reduce cell viability, inducing mitochondrial damage, increased mitochondrial reactive oxygen species production and loss of mitochondrial membrane potential, leading to effector caspases 3/7 cleaved, resulting in apoptosis. Finally, long-term proliferation inhibition was observed, highlighting the bioequivalent effect of PCL-DSF NPs compared to free DSF.
Conclusion: Our data demonstrated the biological interaction of PCL NPs with adipose cells in vitro. The selective cytotoxicity of DSF towards preadipocytes resulted in milder effects when it was delivered nanoencapsulated compared to the free drug. These results suggest promising pharmacological alternatives for DSF long-term delivery on adipose tissue.
Keywords: polymeric nanoparticles, obesity, adipose tissue, adipocyte, drug loading, cytotoxicity
Introduction
Obesity is a chronic and multifactorial disease characterized by adipose tissue due to a positive imbalance between food intake and energy expenditure.1,2 It is also a notable risk factor for other non-communicable diseases such as heart disease, diabetes, cancer, and other.3 Currently, around 2000 million people are diagnosed, even the World Obesity Federation estimates four billion people globally by 2035.3 Furthermore, complexity and multicausality are directly associated with intrinsic and extrinsic factors, such as genetic, endocrine, neuronal and environmental components that promote an excessive accumulation of fat and hypertrophy of adipose tissue. Adipose tissue is highly metabolically active, allowing it the major function as an energy storage and neuroendocrine organ.4 The specialized lipid handling cells are adipocytes which function as primary energy reservoirs, representing around 80–89% of adipose tissue, and interacting with other cell types such as preadipocytes and immune cells.5 In disturbances on adipose tissue distribution, such as in obesity, the adipocytes become dysfunctional, promoting a pro-inflammatory, hyperlipidemic, and insulin-resistant environment, a determinant factor in the development of metabolic complications.6,7 The conventional obesity treatments include drugs to promote weight loss, better eating habits, and bariatric surgery; unfortunately, they are limited due to the modest effect on weight loss, several gastrointestinal adverse effects and low long-term effectiveness.
Due to multicausality and heterogeneity of obesity, novel alternatives have been proposed to treat it from diverse perspectives. For example, disulfiram (DSF) is an aldehyde dehydrogenase (ALDH) enzyme inhibitor approved by the Food and Drug Administration (FDA) for alcoholism control and exhibits anti-inflammatory and anti-tumoral properties. There is evidence, in vivo and ex vivo, that DSF may have a potential application in the obesity context. For example, in a study DSF at 50 mg/kg/day demonstrated to induce body weight loss in rats fed with a high-fat diet (HFD).8 Particularly, DSF on HFD mice reduces insulin resistance, visceral adiposity and suppresses body weight gain.9 Also, in ALDH2 knockout mice fed with HFD, DSF reduced hepatosteatosis, pancreatic islet hyperplasia, body weight and normalized insulin responsiveness through autophagy.9,10 In addition, in an in vitro fibroblast model derived from Graves orbitopathy patients, DSF showed antiadipogenic and anti-inflammatory effects.11 However, their cytotoxicity on adipose cells is not reported, and its clinical application is limited due to its high instability.12,13 Thus, as a first step towards DSF translation into clinical settings, it is essential to assess whether it can exert cytotoxicity on adipose cells.
In this regard, several biodegradable and biocompatible materials are considered for pharmaceuticals applications. Particularly as drug delivery systems, polymeric nanoparticles (NPs) in whose a drug can be released at a sustained rate for long time intervals, decreasing the drug cyclic dosage.14–16 In this context, poly-ε-caprolactone (PCL), an FDA-approved biopolymer for intended humans, is widely used for multiple biomedical applications due to their high biocompatibility, easy conjugation with a wide range of polymers or molecules, simple processability for fabrication, low cost for fabrication and low biodegradability.17,18 In addition, PCL can be easily functionalized with multiple functional groups, providing modulation of properties such as adhesiveness and hydrophobicity/hydrophilicity. PCL undergoes hydrolysis, similar to other polymers such as PLGA. Due to the slower degradation rate of PCL, compared to other polymers, it has been employed in drug delivery systems with a life span of over 1 year even, and in suture materials.19,20 Additionally, it has been proposed as an alternative administration route for sustained drug release; however, for clinical applications it is paramount to guarantee the safety of novel nanoencapsulations, the base NPs such as PCL and a molecule of interest such as a drug, where in vitro evaluations are needed as a first proof of concept.
In this work, we optimized the synthesis of polymeric PCL NPs loaded with DSF through physicochemical characterization and assessed their in vitro effect on adipose tissue cells including preadipocytes, white-like adipocytes and macrophages.
Materials and Methods
Nanoparticle Synthesis
Poly-ε-caprolactone (PCL) nanoparticles (NPs) were synthesized through an oil-in-water protocol by nanoprecipitation method. Briefly, 20 mg of PCL (PCL, #B6003-1P, Lactel, Birmingham, USA) were dissolved in 2 mL of acetone (CAS 67–64-1, J.T. Baker, USA) or acetone/dichloromethane 60:40 (CAS 75–09-2, DS1432-001, Tedia, Fairfield, USA). Then, the solution was added dropwise into a flask with 8 mL of poly (vinyl alcohol) (PVA, #341584, CAS 9002–89-5) 1% (w/v) under probe sonication (Qsonica Q700, 1/8″ tip) at 20% amplitude for 2 min in an ice bath. Then, the NPs dispersion was added to 30 mL of PVA 0.1% (w/v) under magnetic stirring. After three hours, the NPs were recovered through a centrifugation, washing procedure and lyophilizing procedure, as previously described.21 PCL-DSF NPs were obtained with a similar protocol to PCL NPs, where PCL was mixed with DFS (DSF, #99–77-8, Sigma-Aldrich, St. Louis, MO, USA) at three different ratios and was dissolved with different solvents, see Table 1, including dichloromethane (CAS #75–09-2, DS1432-001, Tedia, Fairfield, USA). Fluorescent PCL NPs were synthetized similarly, mixing PCL with fluorescein isothiocyanate free carboxyl (#F3651, CAS #27072–45-3, Sigma-Aldrich, St. Louis, MO, US) in a ratio 1:10 (w/w) with acetone/dichloromethane 60:40.
Characterization of Nanoparticles
Particle Size Distribution (PSD)
Dynamic light scattering (DLS) was used to quantify the PSD of NPs in ultrapure water. Their PSD was obtained from the NPs hydrodynamic diameter ensemble, which was determined by fitting the intensity autocorrelation function using the Contin method. Measurements were done in a Malvern Zetasizer Nano ZS90 (Malvern Instruments, Malvern, UK).
ζ-Potential
Electrophoretic light scattering (ELS) was used to measure the ζ-potential of the NPs. Measurements were done in a Malvern Zetasizer Nano ZS90 (Malvern Instruments, Malvern, UK), where NPs were loaded in ultrapure water. Its analysis used the Smoluchowski approximation for ζ-potential calculation.
Scanning Electron Microscopy (SEM)
PCL-DSF NPs were examined by scanning electron microscopy (SEM) to determine surface morphology and PSD (Nova NanoSEM 200, LEI, Hillsboro, OR). A water droplet suspension of PCL-DSF NPs was placed on a gold-plated stainless-steel sample holder, and then it was let to dry. Afterwards, the sample was coated by a gold-palladium mixture prior to SEM measurement.
Fourier Transform-Infrared (FTIR) Spectrometry
The FTIR spectra of PCL-DSF NPs was obtained using a Perkin Elmer Spectrum Series 400, along with DSF and PCL-NPs as controls. The spectra were acquired with an attenuated total reflectance (ATR) accessory at a wavelength of 4000–380 cm−1, with resolution of 4 cm−1, and an average of 16 scans per sample. Ten mg of samples were loaded per measurement. Resonance peaks were studied to assess the functional groups present in PCL-DSF NPs.
Nuclear Magnetic Resonance (NMR)
1H (400 MHz) NMR spectra of PCL-DSF NPs were obtained at room temperature with a 400 MHz Bruker Avance III HD 400N spectrometer with a 5 mm multinuclear BBI-decoupling probe with Z grad. Controls included PCL-NPs and DSF. All were studied with dimethyl sulfoxide (DMSO) as a solvent.
DSF Quantification
Absorbance measurements were done to quantify DSF encapsulated in PCL NPs. The NPs were dissolved with DMSO, and the supernatant was thoroughly vortexed before measurement. Absorbance was assessed from 250 to 310 nm, and DSF quantification was done with respect to a calibration curve. Measurements were done in a microplate fluorescence spectrophotometer Synergy HT (BioTek Instruments, Winooski, VT, USA).
Drug Release
PCL-DSF NPs were suspended in phosphate buffer saline (PBS) at pH 7.4 at a concentration 0.5 mg/mL of DSF. The suspension was fractionated in microtubes with 0.5 mL and shaken on a Thermomixer comfort (Eppendorf AG, Hamburg, Germany) at 700 rpm and 37 °C. Each microtube was retrieved at specific times, recovering the NPs by centrifugation (13000 RPM, 4 °C, 20 min), discarding the supernatant, dissolving the NP pellet with DMSO, and quantifying the encapsulated DSF. DSF release was determined compared to the 0 h measurement. Measurements were done in a microplate spectrophotometer Synergy HT (BioTek Instruments, Winooski, VT, USA) in the range of 250–310 nm with respect to a calibration curve.
Cell Culture
Murine embryo fibroblast 3T3-L1 (ATCC® CL-173™), murine macrophages raw 264.7 (ATCC® TIB-71™) and non-small-cell lung cancer cells A549 (ATCC® CCL-185™) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained under suggested conditions. In general, all cells were cultured in plastic sterile flasks (Life Technologies, Grand Island, NY, USA) at 37°C under 5% CO2 atmosphere, using DMEM F-12 medium (Life Technologies, Grand Island, NY, USA) and supplemented with 1 µg/mL penicillin and 2.5×10−3 µg/mL streptomycin, and 10% fetal bovine serum (FBS, Life Technologies, Grand Island, NY).
White-like adipocyte 3T3-L1 differentiation was performed 3T3-L1 cells reached 80% of confluence, where DMEM was replaced by differentiation media (DMI) that contains of 10 mg/mL of insulin (Humulin R; Eli Lilly), 0.5 μM dexamethasone (Allin, Chinoin, China), and 0.5 mM 3-isobutyl-1-methylxanthine (IBMX, I7018, Sigma Aldrich, St. Louis, MO, USA) in supplemented DMEM for 3 days. After, cells were switched to maintenance media (supplemented DMEM with 10 mg/mL of insulin) for 3 days after. Then cells were replaced with supplemented DMEM until day 10.
Nanoparticle Association and Internalization
The association cell-NPs was analyzed by flow cytometry (CytoFLEX, Beckman Coulter). In brief, cells were treated with 300 µg/mL of the fluorescent PCL NPs for 24 h. At the end, cells were trypsinized and resuspended in PBS. Cells were gated on single events and viable cells. Gating and fluorescence analysis was done using the FlowJo software (Tree Star Inc).
To evaluate NPs internalization cells were seeded in glass coverslips. After 24 h of incubation with fluorescent NPs, cells were washed in Tyrode solution (in mM: 128 NaCl, 0.4 NaH2PO4, 6 glucose, 5.4 KCl, 0.5 MgCl-6H2O, 5 creatinine, 5 taurine, and 25 hEPES, pH 7.4), loaded with calcein red (Life Technologies, Grand Island, NY, 2 µM, 30 min, 37°C), and mounted in a superfusion chamber. The nuclei were stained with DRAQ5 (Thermo Fisher, 5 min, 37°C). A Z-stack was acquired using a confocal microscope (TCS SP5, Leica Microsystems, Wetzlar, Germany) recording a xy image with 1 µm step between each image. To collect the fluorescence signal, the following excitation/emission parameters were established to each fluorophore: NPs-fluorescein 488 nm/500-630 nm; calcein-red 543 nm/650-750 nm; and DRAQ5 633 nm/555-630 nm.
Cell Viability Assessment
Relative cell viability was determined by Alamar blue viability test (Life Technologies, Carlsbad, CA); this assay consisted of measuring resazurin reduction by metabolic activity of the cells to resorufin by fluorescence. Cells were seeded at 5×103 cells per well in 96-well plates. Cells were administered during 24 h with PCL-DSF or free DSF (0, 0.1, 1, 5, 10 and 50 μM). Blank PCL NPs were added to cells as a control group based on calculations for PCL-DSF NPs. At the end of the incubation period, relative cell viability was determined. For the next experiments, we employed 10 µM of DSF either encapsulated on PCL NPs or free.
Oil Red Satin
Lipid droplets were analyzed by oil red stain. At the end of the stimuli, cells were washed twice with PBS, then fixed with formaldehyde (Sigma Aldrich, Burlington, Massachusetts, USA) 10% for 15 min at room temperature. Then, cells were stained with oil red (Sigma Aldrich, Burlington, Massachusetts, USA) 0.3% (w/v) at room temperature for 15 min in a shake. Finally, cells were washed with distilled water. Representative images were obtained using Zeiss-AXIO microscopy (Zeiss, Oberkochen, Germany) and the camera Zeiss-Axiocam 208 color (Zeiss, Oberkochen, Germany).
Mitochondrial ROS Production Analysis
Mitochondrial ROS generation was measured using MitoSox red (Invitrogen, St Louis, MO, USA). In brief, 10×105 cells were incubated in 12-well dishes (Corning, NY, USA) with 10 µM of PCL-DSF NPs for 24 h. Cells were then recovered, washed with PBS, stained with 5 µM of MitoSox red, incubated at 37 °C for 20 min, measured using a flow cytometer (CytoFLEX, Beckman Coulter), and analyzed using the FlowJo software (Tree Star Inc). H2O2 (50 μM) was used as a positive control.
Mitochondrial Membrane Potential Analysis
Mitochondrial membrane potential (∆Ψm) was measured using JC-1 staining (Thermo Scientific, Waltham, MA, USA). In brief, 1×104 3T3-L1 cells were seeded in 24-well plates (Corning Inc. Costar®, Corning, NY, USA) and treated with 10 µM of PCL-DSF NPs for 24 h. Afterwards, cells were collected and incubated with 1 μM of JC-1 for 20 min at 37 °C in darkness. Then, cells were washed with PBS and stained with 1 μM of Ghost dye stain (eBioscience, Waltham, MA, USA) for 15 min at 37 °C in darkness. Finally, cells were washed with PBS and analyzed using flow cytometry (CytoFLEX, Beckman Coulter, California, USA), as previously described, and analyzed using Kaluza software (Beckman-Coulter, California, USA).. The levels of ∆Ψm are represented in percentage of red fluorescence intensity and green fluorescence intensity on cell negative for Ghost dye stain, considered as viable cells. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP; 1 µM) was used as a positive control.
Caspases 3/7 Cleaved Quantification
The effector caspases 3/7 cleaved were quantified by luminescent kit (caspase-glo 3/7 assay; Promega, Madison, Wisconsin, USA) and using the microplate reader (SYNERGY HT, Biotek, Winooski, Vermont, USA), following manufacturer’s instructions. Etoposide (3.5 µM) was used as a positive control.
Cell Death Analysis
Cell death was determined by staining cells with Annexin V-allophycocyanin (APC) (AnnV; BD Biosciences Pharmingen, San Jose, CA, USA). In brief, 10×105 cells were seeded in 12-well plates (Corning, NY, USA) with 10 μM of PCL-DSF NPs (concentration that reduced around 50% of cell viability). After cell treatments, cells were detached and washed twice with PBS and then resuspended in 200 μL of binding buffer (10 mM HEPES/NaOH pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) containing AnnV (0.1 μg/mL). Cells were then assessed with a flow cytometer (CytoFLEX, Beckman Coulter) and analyzed using the FlowJo software (Tree Star Inc). Cell heat (56 °C) for 10 min before analysis was used as a positive control.
Clonogenic Assay
A clonogenic assay was used to assess cell proliferation and clonogenicity in 3T3-L1 and A549 cells. In brief, 500 cells were plated in 6-well plates after attached, cells were treated for 24 h with PCL-DSF NPs or free DSF with 10 μM. Afterwards, the medium was changed, and cells were allowed to grow until colony formation (10 days). Then, the colonies were fixed with methanol (100%) and glacial acetic acid (3:1), stained with 0.5% gentian violet. Colonies with >50 cells were counted manually. Finally, cell survival was determined by normalizing the number of colonies in treated wells to the quantification of the control group.
Statistical Analysis
The data were analyzed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). The results given in this study represent the mean ± standard error of the mean (SEM) of at least three independent experiments (n ≥ 3) done in triplicate. Statistical analyses were done with Student´s t-test or ANOVA + posthoc tests. The statistical significance was defined as * p<0.05, ** p<0.01, and *** p<0.001.
Results
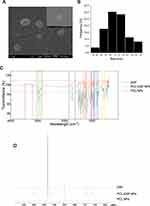
Acetone/Dichloromethane Mixture Improve Physicochemical Properties of PCL-DSF NPs
The optimal PCL-DSF NPs were firstly sought for, as shown in the experimental plan, Figure 1. They were assessed by DLS to calculate PSD and PDI, by ELS to compute the ζ-potential, and by UV-Vis spectroscopy to evaluate its drug loading, see Table 1. The NPs were synthesized under different conditions by varying the solvent employed, either acetone or acetone/dichloromethane 60:40 (v/v), and ratios of PCL:DSF (w/w) at 1:2, 1:1, 2:1 and, 1:0. Table 1 shows that variations in the solvent for ratios 1:2 and 1:1 did not modify the PSD. Interestingly, for ratio 2:1, the acetone/dichloromethane mixture decreased the PSD, 202.1 ± 5.65 nm, compared to acetone, 648.8 ± 56.78 nm. Similarly, PCL NPs show that acetone/dichloromethane mixture diminished PSD, 281.7 ± 42.90 nm, with respect to acetone, 329.9 ± 75.74 nm. This demonstrated that the acetone/dichloromethane mixture and ratio 2:1 (PCL:DSF) favored a reduction of NPs PSD, see Figure 2A for a representative DLS result. Thus, the homogeneity of NPs was evaluated. Table 1 shows that NPs synthesized with acetone at PCL:DSF ratios of 1:2, 1:1, 2:1 and 1:0, presented PDIs of 0.604 ± 0.072, 0.847 ± 0.91, 0.552 ± 0.028 and 0.288 ± 0.02, respectively. Also, in the synthesis with acetone/dichloromethane mixture, the NPs at ratios 1:2, 1:1, 2:1 and 1:0, showed PDIs of 0.625 ± 0.207, 0.525 ± 0.170, 0.296 ± 0.084 and 0.254 ± 0.07, respectively. It resulted, within the conditions here studied, that low PSD NPs have also low PDI. The ζ-potential analysis revealed that all NPs showed less than −12 mV. The NPs synthesized by acetone showed ζ-potential of −15.8 ± 11.05, −23.2 ± 2.4, −20.0 ± 3.19 and −12.0 ± 3.10 mV, for ratios of 1:2, 1:1, 2:1 and 1:0, respectively. Also, NPs synthesized using the acetone/dichloromethane mixture showed ζ-potential of −20.2 ± 5.22, −20.3 ± 6.17, −20.7 ± 4.58 and −14.6 ± 4.04 mV, for ratios of 1:2, 1:1, 2:1 and 1:0, respectively. See Figure 2B for representative results by ELS of NPs synthesized by acetone/dichloromethane mixture and ratios 2:1 (PCL:DSF). Our data demonstrated that solvents employed during synthesis and ratio PCL:DSF impact on physical characteristics. The results suggest that the optimal NPs synthesis for biological application are using the acetone/dichloromethane mixture and 2:1 ratio.
Additionally, the NPs drug loading was evaluated by UV-Vis spectroscopy at 250–310 nm, the maximum absorbance range of DSF, see Figure 2C for a representative result. The NPs synthesized with acetone showed higher drug loading compared to NPs synthesized using acetone/dichloromethane (Table 1). For NPs synthesized with the acetone/dichloromethane mixture, the drug loading was 12.4 ± 2.19, 14.0 ± 6.90 and 18.6 ± 5.80% for 1:2, 1:1, and 2:1 PCL:DSF ratios. Taken together, the optimal synthesis was sought at producing NPs with the smallest PSD and PDI, neutral ζ-potential, and highest drug loading. Therefore, the NPs synthesis with a 60:40 (v/v) acetone/dichloromethane mixture solvent with 2:1 PCL:DSF ratio was selected as the most optimal synthesis, and all further results are presented with it.
Drug release of PCL-DSF NPs was determined in simulated biological conditions. We evaluated the release of DSF from PCL-DSF NPs in physiological media at pH 7.4, see Figure 2D. We observed a sustained release of DSF from 0.5 h (10.94 ± 2.38%), up to 96 h (91.20 ± 6.03%), showing a double response pattern. This result confirmed that PCL-DSF NPs can deliver DSF for extended periods of time under physiological conditions, suggesting potential biomedical applications.
Spherical Poly-ε-Caprolactone-Disulfiram Nanoparticles Encapsulate Physically DSF
The PCL-DSF NPs were probed by SEM to determine its shape and PSD. Figure 3A shows that the NPs have a spheroid shape, with a PSD of 113.4 ± 16.29 nm, see Figure 3B. The functional groups of PCL-DSF NPs were evaluated by FTIR spectroscopy, see Figure 3C. In the case of PCL-DSF NPs, both characteristic peaks present in individual PCL and DSF spectra are shown. The FTIR spectrum of DSF is consistent with previously reported literature.22,23 A notable peak ~3000 cm−1 is attributed to C-H (CH3) stretching, while another characteristic peak at 1496 cm−1 corresponds to C-H symmetrical deformation vibrations. Additionally, absorption peaks spanning the range 720–1465 cm−1 are related to CH2 groups, with a prominent peak observed at 1273 cm−1. Peaks at 1250 cm−1 signify the stretching of C=S bonds and the skeletal vibration of C–C bonds, respectively. Further, FTIR peaks in the range of 967–1062 cm−1 and 818–914 cm−1 are associated with C-N stretching and C-S stretching, respectively. The vibrational peaks in the range of 554–666 cm−1 represent S-S dihedral bending. Similarly, the FTIR spectrum of PCL was examined, revealing characteristic stretching frequencies. A distinct peak at 1730 cm−1 corresponds to C=O stretching, the resonance at 1045 cm−1 indicates C-O stretching, both of them indicative of the PCL block.22,23
The structure of PCL-DSF NPs was assessed by 1H NMR spectroscopy. In Figure 3D, the NMR spectrum of DSF unveils essential features, including sharp peaks associated with CH3 groups at ~1.5 ppm and a distinctive signal arising from the CH2 group bonded to nitrogen at around 4 ppm. Contrastingly, the 1H NMR spectrum of PCL presents one distinct region: chemical shifts between 2 and 2.5 ppm correspond to H atoms attached to C atoms adjacent to C with double-bonded O within the PCL structure;24 while the region centered around 3.2 and 3.5 ppm reveals the presence solvent used, DMSO.
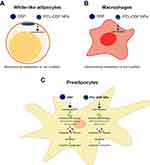
PCL NPs Associate and Internalize into White-Like Adipocytes, Macrophages and Preadipocytes
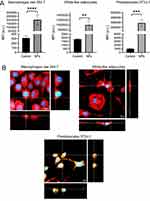
The adipose tissue comprises 80–90% of adipocytes, while the rest includes immune-derived cells and preadipocytes. Thus, the association and internalization of PCL NPs were evaluated by flow cytometry and confocal microscopy, respectively on raw 264.7 (macrophage cell line), white-like adipocytes, and 3T3-L1 preadipocytes. White-like adipocytes were differentiated up to day 10 were obtained as described in the materials and methods section. In Supplementary Figure 1, preadipocytes show fibroblast-like morphology without presence of lipid droplets, contrary white-like adipocytes, which are characterized by less of elongated morphology, increasing rounded body and intracellular lipid droplets. Thus, we synthetized fluorescent PCL NPs with fluorescein, exhibits similar physicochemical properties than non-fluorescent PCL NPs, see Supplementary Table 1. Figure 4A shows that macrophages, white-like adipocyte and preadipocytes treated 24h with the fluorescent PCL NPs exhibit statistically significant higher fluorescence compared to untreated cells, demonstrating NPs association into these cells. The internalization of NPs was confirmed by confocal microscopy. After 24 h of incubation, PCL NPs encapsulating fluorescein were observed in cells, see Figure 4B for a representative orthogonal projection. These findings demonstrated that PCL NPs associated and internalize into cytoplasm of the adipose cells. A549 cells were included as a positive control of cytotoxicity, due to their overexpression of ALDH, a target of DSF. They showed similar association and internalization of PCL NPs, see Supplementary Figure 2.
PCL-DSF NPs Decrease Relative Cell Viability on Preadipocytes but Not in White-Like Adipocytes and Macrophages
Once demonstrated the capability of PCL NPs to associate and internalize into adipose cells, the cytotoxicity of PCL-DSF NPs was evaluated analyzing the cell viability on white-like adipocytes, raw 264.7 and 3T3-L1 preadipocytes. Figure 5A and B shows that DSF and PCL-DSF NPs did not induce a notable cell viability reduction on white-like adipocytes and macrophages, respectively, at 0.1, 1, 5, 10 and 50 μM of DSF compared to the control after 24 h of administration. DSF at low concentrations did not affect cell viability on preadipocytes, but at 5 μM around 20% of cell viability was reduced, continuing the trend at 10 and 50 μM of DSF, see Figure 5C. Similar results were observed in A549 cell lines for 24 up to 72 h of NPs exposure, see Supplementary Figure 3. Interestingly, PCL NPs without DSF did not modify cell viability of adipose cells. Supplementary Figure 4 shows lipid droplets content on white-like adipocytes treated or not with PCL NPs, PCL-DSF NPs (10 µM) or DSF (10 µM) for 24 h. No apparent changes on lipid droplet content were observed on treated cells compared to untreated cells. Taken together, these results demonstrate that PCL-DSF NPs do not induce cytotoxicity on white-like adipocytes and macrophages, but on preadipocytes induce dose-dependent cytotoxicity, similar to lung cancer A549 cells.
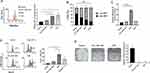
PCL-DSF NPs Demonstrated Long-Term Bioequivalent Cytotoxicity Similar to Free DSF on 3T3-L1
In order to describe the mechanism of cytotoxicity provoked by DSF encapsulated into PCL, mitochondrial damage, effector caspases cleaved, apoptosis and long-term proliferation on preadipocytes 3T3-L1 and A549 cells was analyzed. The mitochondria are the principal cellular reservoir of pro-apoptotic proteins and the major ROS producer. Thus, we analyzed mitochondrial damage induced by assessing mROS production and the ∆Ψm. Figure 6A shows that PCL-DSF NPs did not modify mROS production on 3T3-L1 cells (1.1 ± 0.10) compared to control (1.0 ± 0.0) in contrast to free DSF that increased 3.4 ± 0.28, similar to the positive control H2O2 (3.0 ± 0.52). A similar effect was observed on A549 cells, see Supplementary Figure 5, where PCL-DSF NPs did not promote mROS production (1.1 ± 0.07) compared to untreated cells (1.0 ± 0.0), yet free DSF showed an increase (2.7 ± 0.42) higher than the positive control (1.5 ± 0.07). Likeiwse, Figure 6B shows that PCL-DSF NPs induce 42.81 ± 18.7% of loss of ∆Ψm, DSF 66.28 ± 12.2% and positive control CCCP 90.56 ± 15.5% on preadipocytes 3T3-L1. Similar pattern was observed on A549 cells, see Supplementary Figure 5 where PCL-DSF NPs induce 50.04 ± 14.9%, DSF 68.84 ± 7.78% and CCCP 80.30 ± 12.5 5 of loss of ∆Ψm. Supplementary Figure 6 shows cell viability quantification employed in this analysis using Ghost dye stain. These results demonstrate that the encapsulation of DSF diminished mitochondrial alterations compared to free DSF. Caspases are the principal effector proteases on apoptosis and phosphatidylserine (PS) is an “eat me” signal correlated to regulated cell death.25 Thus, to correlate mitochondrial damage with cytotoxic pathways, effector caspases 3/7 cleaved and PS exposure were evaluated. Figure 6C exhibits that PCL-DSF NPs do not increase caspases 3/7 cleaved compared to free DSF and etoposide (positive control) on preadipocytes 3T3-L1. Figure 6D shows that on 3T3-L1 cells PCL-DSF NPs induced 15.0 ± 3.08% of PS exposure compared to control (4.0 ± 0.87%), while free DSF showed 52.6 ± 6.03% and heat (56 °C, positive control to promote cell death) being 27.5 ± 5.19%. Therefore, DSF encapsulation decreased cytotoxicity. Finally, focused on deciphering the bioequivalence of PCL-DSF NPs compared to free DSF on 3T3-L1 preadipocytes, their recovery post-24 h of stimuli was assessed. The cells were treated for 24 h with the 10 μM (concentration less than 15% of cell death) of PCL-DSF NPs or free DSF, then treatment was removed, and cells were cultured during 10 days, assessing then their capability for colony formation. Figure 6E shows that PCL-DSF NPs have antiproliferative activity, as they reach complete inhibition colony formation similar to free DSF, on 3T3-L1 preadipocytes. Similar results were obtained for A549 cells, see Supplementary Figure 7. This result indicates that PCL-DSF NPs at 10 μM induce mitochondrial damage, leading to low apoptosis that was able to inhibit long-term cell proliferation.
Discussion
Novel anti-obesity solutions are required, even of current pharmacological and non-pharmacological strategies. In this sense, the purpose of this study was to evaluate the cytotoxic effect in vitro of encapsulated DSF into PCL NPs upon adipose tissue cells. First, we optimized the synthesis of NPs based on physicochemical properties. Positive results have been reported in NPs around 200 nm for drug delivery,26 which has been associated with organic solvent employed for synthesis.27,28 Our results showed that a solvent combination of acetone/dichloromethane favored a diminution of PSD and PDI compared to using only acetone (Table 1). Similar results were obtained for other authors where PSD diminution was associated with reduction of acetone to dichloromethane ratio in oil-phase mixture at 40:60 (acetone/dichloromethane) ratio.29 In all conditions NPs showed negative ζ-potential, consistent with polymeric NPs, it may be a result of the carbonyl group of PCL present at the surface of NPs.30,31 NPs synthesized with acetone as a solvent revealed higher drug loading than with the solvent mixture of acetone/dichloromethane. However, such increased drug loading was accompanied by an increased PSD and PDI. In order to elucidate the effects of nanoencapsulated DSF delivery into adipose tissue cells, the ideal synthesis would be that which offers the narrowest PSD to avoid variability due to cellular uptake and DSF release, which directly ties to that of the lowest PDI, and coupled with the highest drug loading to minimize vehicle administration to cells.32 Given this, the synthesis of 2:1 PCL:DSF with the solvent mixture of acetone/dichloromethane was selected, being the smallest PSD of all synthesis, of monodisperse nature, and spheroid morphology, consistent with other PCL NPs studies.33 While the drug loading was not the highest in this synthesis, preference was given to a lower variability of the nanosystem.
Our results showed sustained drug release under physiological conditions, similar to other nanoencapsulation systems.34 The drug release was composed of two phases, a first one of 50% DSF at 24 h, which could be considered as a short-term release, and the other 50% released on a 3-fold longer span, up to 96 h. Such sustained release could result in beneficial in vivo effects, as delivering encapsulated DSF can potentially increase on-site drug presence ~13-fold longer times, as DSF half-life is just 7 h.35 Structural and functional group analyses of PCL-DSF NPs revealed a concordance with studies of its individual components, PCL and DSF.24,36–38 The PCL-DSF NPs did not show evidence of a new functional group, and on the structural analysis the CH2 peak of DSF, conventionally found at around 4 ppm in DSF, underwent a leftward shift in the spectrum. Therefore, both findings suggest that DSF was encapsulated physically into the PCL NPs matrix.
Despite that the biology of obesity is complex, the most commonly therapeutic strategies are lifestyle modification that includes calorie restriction combined with increased physical activity, and bariatric surgery; however, these options normally are frequently disappointing.39,40 Therefore, the panorama forces us to find a novel, efficient, and safe anti-obesity strategy. In this sense, in vitro and in vivo studies showed potential effects of DSF to reduce weight loss, however their effect has not been elucidated on adipose cells, may be due to its instability on gastric juice and blood limits clinical applications.8–10 Therefore, this work focused on DSF encapsulation to analyze the cytotoxic effect in vitro using adipose cells. The intracellular localization of nanosystems is essential for safe and efficient therapeutic applications that guarantee biological effects. First, we demonstrated association and internalization of PCL NPs with cells at 24 h of treatment (Figure 4). These results are consistent with other works where PLGA NPs of 177 ± 6 nm internalize in vitro into preadipocytes derived from stromal vascular fraction (SVF) and in vivo on mature adipocytes through the endocytic pathway.41 The drug delivery based on polymeric NPs offers advantages in enhancing the therapeutic efficacy at time that leveraging cellular internalization and intracellular drug release. Remarkably, we suggest a sustained release of DSF and efficient cellular uptake into adipose cells.
Obesity leads to chronic low-grade inflammation, that increases the risk of cancer development promoting ROS production, DNA damage, and promoting tumor growth and immune evasion, that is directly associated with excessive adipocyte progenitors (preadipocytes) in adipose tissue upregulated by macrophages.42,43 Then, by modulating inflammation, there is potential to reduce the burden of obesity related to metabolic disorders and cancer.44 Here, we did not observe cellular viability affections on macrophages, contrary on preadipocytes where long-term proliferation was inhibited. This could lead into reduced chronic inflammation, impacting positively metabolic disorders and risk of cancer development.
Then we evaluated the cytotoxic effect of PCL-DSF NPs upon adipose cells: preadipocytes, white-like adipocytes and macrophages, also in lung cancer cell line (A549) that overexpress ALDH.45,46 Our results observed that neither free DSF nor PCL-DSF NPs did affect cell viability on white-like adipocytes and macrophages; however, on preadipocytes, 3T3-L1 cell viability decreased inversely proportional to the DSF concentration, similar to A549 cells (Figure 5). Interestingly, PCL-DSF NPs elicited higher cytotoxicity than free DSF on preadipocytes 3T3-L1, while in A549 cells both groups stimulated a similar dose response. We did not observe changes in lipid droplets content on white-like adipocytes stimulated with PCL-DSF Ns or free DSF. There are no previous reports of DSF cytotoxicity free or nanoencapsulated on adipose tissue cells. As an anti-cancer therapy it has been explored,47,48 even when encapsulated in other polymers such as PLGA or PEG.49–51
These cytotoxic differences could be associated with differences in expression and tissue-specific regulation of ALDH. In lung cancer cells, ALDH is involved in proliferation, differentiation, and survival,52 in adipose tissue it regulates adipogenesis, abdominal fat formation, glucose tolerance, and suppression of thermogenesis53 and, in immune cells play a protective role by inducing oncogene suppressors.54 Indeed, ALDH1A1 expression is increased in 3T3-L1 preadipocytes, compared to other enzyme isoforms after adipogenesis.46 Recently, their role on adiposity was explored in ALDHA1-deficient mice that resist to obesity due to increased lipolysis and thermogenesis on visceral adipose tissue.55 Notably, it was suggested that DSF could induce thermogenesis on white-like adipocytes and could induce browning on adipocytes that could efficiently cause adiposity reduction, and most efficiently could improve with the DSF encapsulation. However, our study did not show evidence for thermogenesis, such as reduction of lipid droplets on white-like adipocytes. In addition, the potential of novel nanosystems may be clearly observed up to in vivo studies, where multiple variables play a role on the successful site drug delivery. One recent example was shown recently in a study with a PEG nanosystem for chemotherapy delivery, which showed low in vitro cytotoxicity compared to free drug; however, in vivo assays clearly showed its potential to reduce metastasis and chemotherapy-associated toxicities.56
The elucidated cell death mechanism of PCL-DSF NPs on preadipocytes 3T3-L1 is presented in Figure 7. We observed that free DSF increases mROS production, loss of ∆Ψm and effector caspases 3/7 cleaved while PCL-DSF NPs decrease such effects on preadipocytes 3T3-L1, similar to A549 cells (Figure 6). DSF has been shown to have a similar effect in other cancer cells leading to regulated cell death,57–61 however, on preadipocytes it has not been described yet. Interestingly, for PCL-DSF NPs (10 μM), which reduced ~50% of cell viability, did not induce apoptosis on preadipocytes 3T3-L1 and A549 cells (Figure 6). This reduction of cell viability, which was not associated with cell death, could be due to cell cycle arrest or senescence induction.62 Polymeric NPs could induce cell cycle arrest in G2/M phase blocking G1 and S phase since 12 h, and cell death at 24. 63–65 Finally, PCL-DSF NPs inhibited long-term proliferation similar to free DSF. Successful formulation of PCL NPs or co-polymer NPs provides bioequivalence compared to drug alone to enhance biocompatibility and biological applications in cancer treatments.66,67 These findings suggest a bioequivalence of PCL-DSF NPs compared to DSF in the long-term and their potential biomedical application in obesity context.39 Even, in obese patients, inducing apoptosis is a viable method of removing adipocytes, which can be achieved through suppressing adipogenesis, obstructing fat accumulation, and deleting adipocytes.68 Similar to DSF, other ALDH inhibitors69–71 or copper ionophores72 have reports of anti-obesity effects.
Despite limitations of this study, including limited set of drug release scenarios and cell stimuli, the results encourage further research, such as in vivo settings to validate whether only preadipocytes are affected in adipose tissue, as well as to test whether DSF long-term release from NPs could yield significant difference in weight loss versus free DSF in obesity models.
Conclusion
In summary, this work optimized the synthesis of PCL NPs to encapsulate DSF and characterized the physicochemical properties for biological applications on obesity context. It demonstrated biological interaction, association and internalization, of PCL NPs with macrophages, white-like adipocyte and preadipocytes in vitro. PCL-DSF NPs did not modify the cell viability of white-like adipocytes and macrophages raw 264.7. However, on 3T3-L1 cells decrease cell viability in a dose-dependent manner. The cell death mechanism involved mitochondrial damage and cleavage of effector caspases 3/7, leading to apoptosis. Interestingly, on the control cells for ALDH overexpression A549, there was a decrease of cell viability yet very small increases were observed for apoptosis. Interestingly, in both cases encapsulated DSF inhibited long-term proliferation, similar to free DSF. These results open the door for the design of new pharmacological strategies for DSF long-term delivery to obesity treatments, being preclinical obesity models a natural next step.
Abbreviations
∆Ψm, Mitochondrial membrane potential; ALDH, aldehyde dehydrogenase; Ann-V, Annexin V-allophycocyanin; ATR, Attenuated total reflectance; CCCP, Carbonyl cyanide m-chlorophenyl hydrazone; DSF, Disulfiram; ELS, Electrophoretic light scattering; FBS, Fetal bovine serum; FDA, Food and drug administration; FTIR, Fourier transform infrared spectroscopy; HFD, High fat diet; mROS, Mitochondrial reactive oxygen species; NMR, Nuclear magnetic resonance; NPs, Nanoparticles; PCL, Poly-ε-caprolactone; PDI, Polydispersity index; PS, Phosphatidylserine; PSD, Particle size distribution; SEM, Scanning electron microscopy; SVF, Stromal vascular fraction.
Acknowledgments
The authors thank Jorge Eduardo Galván-Marroquín, José Antonio Sansen-Moreno and Fátima Miroslava Alvarado Monroy for technical help, and J.M.G.C., thanks CONACyT for scholarship funding. Also, we thank Hospital Zambrano Hellion and Centro de Biotecnología FEMSA for the facilities provided to perform this work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Tecnologico de Monterrey through the Challenge-Based Research Funding Program 2022 (Project ID IJXT070-22EG57001).
Disclosure
The authors declare no conflict of interest.
References
1. Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15(5):288–298. doi:10.1038/s41574-019-0176-8
2. Lister NB, Baur LA, Felix JF, et al. Child and adolescent obesity. Nat Rev Dis Primers. 2023;9(1). doi:10.1038/s41572-023-00435-4
3. Barata Cavalcanti O, Barquera S, Baur L, et al. World Obesity 2022; 2022. Available from: www.worldobesity.org#worldobesityatlas.
4. Choe SS, Huh JY, Hwang IJ, Kim JI, Kim JB. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front Endocrinol. 2016;7(APR). doi:10.3389/fendo.2016.00030
5. Sakers A, De Siqueira MK, Seale P, Villanueva CJ. Adipose-tissue plasticity in health and disease. Cell. 2022;185(3):419–446. doi:10.1016/j.cell.2021.12.016
6. Zatterale F, Longo M, Naderi J, et al. Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front Physiol. 2020;10. doi:10.3389/fphys.2019.01607
7. Rohm TV, Meier DT, Olefsky JM, Donath MY. Inflammation in obesity, diabetes, and related disorders. Immunity. 2022;55(1):31–55. doi:10.1016/j.immuni.2021.12.013
8. Omran Z, Sheikh R, Baothman OA, Zamzami MA, Alarjah M. Repurposing disulfiram as an anti-obesity drug: treating and preventing obesity in high-fat-fed rats. Diabetes Metab Syndr Obes. 2020;13:1473–1480. doi:10.2147/DMSO.S254267
9. Bernier M, Harney D, Koay YC, et al. Elucidating the mechanisms by which disulfiram protects against obesity and metabolic syndrome. NPJ Aging Mech Dis. 2020;6(1). doi:10.1038/s41514-020-0046-6
10. Bernier M, Mitchell SJ, Wahl D, et al. Disulfiram treatment normalizes body weight in obese mice. Cell Metab. 2020;32(2):203–214.e4. doi:10.1016/j.cmet.2020.04.019
11. Wang X, Yang S, Ye H, et al. Disulfiram exerts antiadipogenic, anti-inflammatory, and antifibrotic therapeutic effects in an in vitro model of graves’ orbitopathy. Thyroid. 2022;32(3):294–305. doi:10.1089/thy.2021.0246
12. Johansson B. A review of the pharmacokinetics and pharmacodynamics of disulfiram and its metabolites. Acta Psychiatr Scand. 1992;86(S369):15–26. doi:10.1111/j.1600-0447.1992.tb03310.x
13. Cvek B. The promiscuity of disulfiram in medicinal research. ACS Med Chem Lett. 2023;14:1610–1614. doi:10.1021/acsmedchemlett.3c00450
14. Gagliardi A, Giuliano E, Venkateswararao E, et al. Biodegradable polymeric nanoparticles for drug delivery to solid tumors. Front Pharmacol. 2021:12. doi:10.3389/fphar.2021.601626
15. De R, Mahata MK, Kim KT. Structure-based varieties of polymeric nanocarriers and influences of their physicochemical properties on drug delivery profiles. Adv Sci. 2022;9(10). doi:10.1002/advs.202105373
16. Xiao X, Teng F, Shi C, et al. Polymeric nanoparticles—promising carriers for cancer therapy. Front Bioeng Biotechnol. 2022;10. doi:10.3389/fbioe.2022.1024143
17. Kaluzynski K, Pretula J, Lewinski P, Kaźmierski S, Penczek S. Synthesis and properties of functionalized poly(ϵ-caprolactone); chain polymerization followed by polycondensation in one pot with initiator and catalyst in one molecule. synthesis and molecular structures. Macromolecules. 2022;55(6):2210–2221. doi:10.1021/acs.macromol.1c02325
18. El Yousfi R, Brahmi M, Dalli M, et al. Recent advances in nanoparticle development for drug delivery: a comprehensive review of polycaprolactone-based multi-arm architectures. Polymers. 2023;15(8):1835. doi:10.3390/polym15081835
19. Labet M, Thielemans W. Synthesis of polycaprolactone: a review. Chem Soc Rev. 2009;38(12):3484–3504. doi:10.1039/b820162p
20. Homaeigohar S, Boccaccini AR. Nature-derived and synthetic additives to poly(ɛ-Caprolactone) nanofibrous systems for biomedicine; an updated overview. Front Chem. 2022;9. doi:10.3389/fchem.2021.809676
21. Lozano O, Lázaro-Alfaro A, Silva-Platas C, et al. Nanoencapsulated quercetin improves cardioprotection during hypoxia-reoxygenation injury through preservation of mitochondrial function. Oxid Med Cell Longev. 2019;2019:1–14. doi:10.1155/2019/7683051
22. Benkaddour A, Jradi K, Robert S, Daneault C. Grafting of polycaprolactone on oxidized nanocelluloses by click chemistry. Nanomaterials. 2013;3(1):141–157. doi:10.3390/nano3010141
23. Marciniec B, Dettlaff K, Naskrent M, Pietralik Z, Kozak M. DSC and spectroscopic studies of disulfiram radiostability in the solid state. J Therm Anal Calorim. 2012;108(1):33–40. doi:10.1007/s10973-011-1810-4
24. Żółtowska K, Sobczak M, Ole¸dzka E. Novel zinc-catalytic systems for ring-opening polymerization of ε-caprolactone. Molecules. 2015;20(2):2816–2827. doi:10.3390/molecules20022816
25. Shlomovitz I, Speir M, Gerlic M. Flipping the dogma - Phosphatidylserine in non-apoptotic cell death. Cell Commun Signal. 2019;17(1). doi:10.1186/s12964-019-0437-0
26. Thomas OS, Weber W. Overcoming physiological barriers to nanoparticle delivery—are we there yet? Front Bioeng Biotechnol. 2019;7. doi:10.3389/fbioe.2019.00415
27. Dong H, Chen L, Zhang Q, Gao J, Vasanthan T. Optimization of processing parameters to produce nanoparticles prepared by rapid nanoprecipitation of pea starch. Food Hydrocoll. 2021;121. doi:10.1016/j.foodhyd.2021.106929
28. Jameel MS, Aziz AA, Dheyab MA. Impacts of various solvents in ultrasonic irradiation and green synthesis of platinum nanoparticle. Inorg Chem Commun. 2021;128. doi:10.1016/j.inoche.2021.108565
29. Manoochehri S, Darvishi B, Kamalinia G, et al. Surface modification of PLGA nanoparticles via human serum albumin conjugation for controlled delivery of docetaxel. DARU J Pharm Sci. 2013;21(1). doi:10.1186/2008-2231-21-58
30. Rybak E, Kowalczyk P, Czarnocka-śniadała S, Wojasiński M, Trzciński J, Ciach T. Microfluidic-assisted formulation of ε-polycaprolactone nanoparticles and evaluation of their properties and in vitro cell uptake. Polymers. 2023;15(22):4375. doi:10.3390/polym15224375
31. Badran MM, Alanazi AE, Ibrahim MA, Alshora DH, Taha E,H, Alomrani A. Optimization of bromocriptine-mesylate-loaded polycaprolactone nanoparticles coated with chitosan for nose-to-brain delivery: in vitro and in vivo Studies. Polymers. 2023;15(19). doi:10.3390/polym15193890
32. Yang G, Liu Y, Jin S, et al. Phase separation-induced nanoprecipitation for making polymer nanoparticles with high drug loading: special collection: distinguished Australian researchers. Aggregate. 2023;4(2). doi:10.1002/agt2.314
33. Kollarigowda RH, Mathews AS, Abraham S, Montemagno CD. A facile approach of light driven nanoassembly for the controlled accommodation of doxorubicin. Biomed Phys Eng Express. 2017;3(4):045006. doi:10.1088/2057-1976/aa79b6
34. Javaid S, Ahmad NM, Mahmood A, et al. Cefotaxime loaded polycaprolactone based polymeric nanoparticles with antifouling properties for in-vitro drug release applications. Polymers. 2021;13(13):2180. doi:10.3390/polym13132180
35. Stokes M, Patel P, Abdijadid S. Disulfiram. StatPearls Publishing; 2024.
36. Jana S. Spectroscopic characterization of disulfiram and nicotinic acid after biofield treatment. J Anal Bioanal Tech. 2015;6(5). doi:10.4172/2155-9872.1000265
37. Massoumi B, Ramezani M, Jaymand M, Ahmadinejad M. Multi-walled carbon nanotubes-g-[poly(ethylene glycol)-b-poly(ε-caprolactone)]: synthesis, characterization, and properties. J Polym Res. 2015;22(11). doi:10.1007/s10965-015-0863-7
38. Álvarez-Ortega O, Ruiz-Ramírez LR, Garibay-Alvarado JA, et al. Preliminary biocompatibility tests of poly-ε-caprolactone/silver nanofibers in Wistar rats. Polymers. 2021;13(7):1135. doi:10.3390/polym13071135
39. Jakab J, Miškić B, Mikšić Š, et al. Adipogenesis as a potential anti-obesity target: a review of pharmacological treatment and natural products. Diabetes Metab Syndr Obes. 2021;14:67–83. doi:10.2147/DMSO.S281186
40. Angelidi AM, Belanger MJ, Kokkinos A, Koliaki CC, Mantzoros CS. Novel Noninvasive Approaches to the Treatment of Obesity: from Pharmacotherapy to Gene Therapy. Endocr Rev. 2022;43(3):507–557. doi:10.1210/endrev/bnab034
41. Jiang C, Cano-Vega MA, Yue F, et al. Dibenzazepine-loaded nanoparticles induce local browning of white adipose tissue to counteract obesity. Mol Ther. 2017;25(7):1718–1729. doi:10.1016/j.ymthe.2017.05.020
42. Rocha G, Villalobos E, Fuentes C, et al. Preadipocyte proliferation is elevated by calcium sensing receptor activation. Mol Cell Endocrinol. 2015;412:251–256. doi:10.1016/j.mce.2015.05.011
43. Renovato-Martins M, Moreira-Nunes C, Atella GC, Barja-Fidalgo C, de Moraes JA. Obese adipose tissue secretion induces inflammation in preadipocytes: role of toll-like receptor-4. Nutrients. 2020;12(9):1–16. doi:10.3390/nu12092828
44. Kolb R, Sutterwala FS, Zhang W. Obesity and cancer: inflammation bridges the two. Curr Opin Pharmacol. 2016;29:77–89. doi:10.1016/j.coph.2016.07.005
45. Moreb JS, Zucali JR, Ostmark B, Benson NA. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by aldefluor flow cytometry-based assay. Cytometry B Clin Cytom. 2007;72(4):281–289. doi:10.1002/cyto.b.20161
46. Reichert B, Yasmeen R, Jeyakumar SM, et al. Concerted action of aldehyde dehydrogenases influences depot-specific fat formation. Mol Endocrinol. 2011;25(5):799–809. doi:10.1210/me.2010-0465
47. Hwang S, Shin DM, Hong JH. Drug repurposing as an antitumor agent: disulfiram-mediated carbonic anhydrase 12 and anion exchanger 2 modulation to inhibit cancer cell migration. Molecules. 2019;24(18):3409. doi:10.3390/molecules24183409
48. Butcher K, Kannappan V, Kilari RS, et al. Investigation of the key chemical structures involved in the anticancer activity of disulfiram in A549 non-small cell lung cancer cell line. BMC Cancer. 2018;18(1). doi:10.1186/s12885-018-4617-x
49. Fasehee H, Ghavamzadeh A, Alimoghaddam K, Ghaffari SH, Faghihi S. A comparative cytotoxic evaluation of disulfiram encapsulated PLGA nanoparticles on MCF-7 cells. Int J Hematol Oncol Stem Cell Res. 2017;5.
50. Najlah M, Ahmed Z, Iqbal M, et al. Development and characterisation of disulfiram-loaded PLGA nanoparticles for the treatment of non-small cell lung cancer. Eur J Pharm Biopharm. 2017;112:224–233. doi:10.1016/j.ejpb.2016.11.032
51. Rao Madala H, Punganuru SR, Ali-Osman F, Zhang R, Srivenugopal KS. Brain-and brain tumor-penetrating disulfiram nanoparticles: sequence of cytotoxic events and efficacy in human glioma cell lines and intracranial xenografts. Oncotarget. 2018;9:3459.
52. Kang JH, Lee SH, Hong D, et al. Aldehyde dehydrogenase is used by cancer cells for energy metabolism. Exp Mol Med. 2016;48(11):e272–e272. doi:10.1038/EMM.2016.103
53. Petrosino JM, Disilvestro D, Ziouzenkova O. Aldehyde dehydrogenase 1A1: friend or foe to female metabolism? Nutrients. 2014;6(3):950–973. doi:10.3390/nu6030950
54. Bazewicz CG, Dinavahi SS, Schell TD, Robertson GP. Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer. Immunology. 2019;156(1):47–55. doi:10.1111/imm.13016
55. Yasmeen R, Reichert B, Deiuliis J, et al. Autocrine function of aldehyde dehydrogenase 1 as a determinant of diet- and sex-specific differences in visceral adiposity. Diabetes. 2013;62(1):124–136. doi:10.2337/db11-1779
56. Lim M, Fletcher NL, Saunus JM, et al. Targeted hyperbranched nanoparticles for delivery of doxorubicin in breast cancer brain metastasis. Mol Pharm. 2023;20(12):6169–6183. doi:10.1021/acs.molpharmaceut.3c00558
57. Yang Z, Guo F, Albers AE, Sehouli J, Kaufmann AM. Disulfiram modulates ROS accumulation and overcomes synergistically cisplatin resistance in breast cancer cell lines. Biomed Pharmacother. 2019;113. doi:10.1016/j.biopha.2019.108727
58. Yip NC, Fombon IS, Liu P, et al. Disulfiram modulated ROS-MAPK and NFB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br J Cancer. 2011;104(10):1564–1574. doi:10.1038/bjc.2011.126
59. Zha J, Chen F, Dong H, et al. Disulfiram targeting lymphoid malignant cell lines via ROS-JNK activation as well as Nrf2 and NF-kB pathway inhibition. J Transl Med. 2014;12(1). doi:10.1186/1479-5876-12-163
60. O’Brien PS, Xi Y, Miller JR, et al. Disulfiram (Antabuse) activates ROS-dependent ER stress and apoptosis in oral cavity squamous cell carcinoma. J Clin Med. 2019;8(5). doi:10.3390/jcm8050611
61. Qiu C, Zhang X, Huang B, et al. Disulfiram, a ferroptosis inducer, triggers lysosomal membrane permeabilization by up-regulating ros in glioblastoma. Onco Targets Ther. 2020;13:10631–10640. doi:10.2147/OTT.S272312
62. Wang Z. Regulation of cell cycle progression by growth factor-induced cell signaling. Cells. 2021;10(12). doi:10.3390/cells10123327
63. Mayol L, Serri C, Menale C, et al. Curcumin loaded PLGA-poloxamer blend nanoparticles induce cell cycle arrest in mesothelioma cells. Eur J Pharm Biopharm. 2015;93:37–45. doi:10.1016/j.ejpb.2015.03.005
64. Salim EI, Mahfouz ME, Eltonouby EA, Hanafy NAN, Hafez EH. Based polymer nanoparticles from bee pollen attenuate non-small lung cancer through enhancement of apoptosis and cell cycle arrest in vivo. Cancer Nanotechnol. 2023;14(1). doi:10.1186/s12645-023-00229-z
65. Das S, Saha M, Mahata LC, China A, Chatterjee N, Das Saha K. Quercetin and 5-Fu loaded chitosan nanoparticles trigger cell-cycle arrest and induce apoptosis in HCT116 cells via modulation of the p53/p21 axis. ACS Omega. 2023;8(40):36893–36905. doi:10.1021/acsomega.3c03933
66. Peng W, Jiang XY, Zhu Y, et al. Oral delivery of capsaicin using MPEG-PCL nanoparticles. Acta Pharmacol Sin. 2015;36(1):139–148. doi:10.1038/aps.2014.113
67. Danafar H. Study of the composition of polycaprolactone/poly (ethylene glycol)/polycaprolactone copolymer and drug-to-polymer ratio on drug loading efficiency of curcumin to nanoparticles. Jundishapur J Nat Pharm Prod. 2017;12(1). doi:10.5812/jjnpp.34179
68. Shaik Mohamed Sayed UF, Moshawih S, Goh HP, et al. Natural products as novel anti-obesity agents: insights into mechanisms of action and potential for therapeutic management. Front Pharmacol. 2023;14. doi:10.3389/fphar.2023.1182937
69. Kiefer FW, Vernochet C, O’Brien P, et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012;18(6):918–925. doi:10.1038/nm.2757
70. Haenisch M, Nguyen T, Fihn CA, et al. Investigation of an ALDH1A1-specific inhibitor for suppression of weight gain in a diet-induced mouse model of obesity. Int J Obes. 2021;45(7):1542–1552. doi:10.1038/s41366-021-00818-1
71. Stagos D, Chen Y, Brocker C, et al. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38(10):1679–1687. doi:10.1124/dmd.110.034678
72. Meggyesy PM, Masaldan S, Clatworthy SAS, et al. Copper ionophores as novel antiobesity therapeutics. Molecules. 2020;25(21):4957. doi:10.3390/molecules25214957
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.