Back to Journals » Infection and Drug Resistance » Volume 18
Effectiveness of Susceptibility-Guided Therapy for Helicobacter pylori Infection: A Retrospective Analysis by Propensity Score Matching
Authors Zhou W, Cheng H, Li M, Zhang R, Li Z, Sun G, Zhang D, Liu X, Pei Y
Received 26 September 2024
Accepted for publication 19 February 2025
Published 25 February 2025 Volume 2025:18 Pages 1149—1159
DOI https://doi.org/10.2147/IDR.S498052
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wenyue Zhou,1 Haoxuan Cheng,1 Miaomiao Li,1 Ruian Zhang,1 Zhiren Li,1 Guangyong Sun,2 Dong Zhang,2 Xinjuan Liu,1 Yanxiang Pei1
1Department of Gastroenterology, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Medical Research Center, Beijing Chao-Yang Hospital, Capital Medical University, Chaoyang District, Beijing, 100024, People’s Republic of China
Correspondence: Xinjuan Liu; Yanxiang Pei, Department of Gastroenterology, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, People’s Republic of China, Email [email protected]; [email protected]
Purpose: This study evaluates and compares the eradication rates of Helicobacter pylori (H. pylori) achieved through susceptibility-guided therapy (SGT) based on resistance genotyping and empirical therapy (ET).
Patients and Methods: A retrospective study was conducted at Beijing Chaoyang Hospital (2021– 2023) on patients with H. pylori infection receiving initial eradication therapy. Resistance genotypes for clarithromycin and levofloxacin were identified using fluorescent PCR of gastric biopsy samples. Patients underwent a 14-day bismuth-containing quadruple therapy (BQT) and were evaluated via the C13 urea breath test (UBT). Based on genotyping or clinical judgment, 550 patients were assigned to SGT (n = 125) or ET (n = 425). The SGT group received personalized treatment based on genotype testing results, avoiding the use of antibiotics to which the bacteria were resistant. The ET group received the standard bismuth-containing quadruple therapy (BQT). Additionally, 29 ET patients underwent follow-up genotypic testing and eradication rates were analyzed retrospectively.
Results: SGT achieved higher eradication rates than ET (ITT: 94.4% vs 86.1%, P = 0.012; PP: 95.2% vs 87.6%, P = 0.016). In levofloxacin-resistant strains, SGT showed significantly higher eradication rates in the PP analysis (95.7% vs 50.0%, P = 0.049).
Conclusion: SGT exhibited remarkably superior eradication rates, notably in levofloxacin-resistant strains, proposing a compelling alternative for the treatment of H. pylori, particularly in instances of antimicrobial resistance.
Keywords: Helicobacter pylori infection, susceptibility-guided therapy, bismuth-containing quadruple regimen, propensity score matching
Introduction
Helicobacter pylori (H. pylori) infection represents a profound global health challenge. As of 2022, the worldwide prevalence of H. pylori infection stood at 43.1%, with an infection rate of 43.9% among adults.1,2 H. pylori is a principal etiological agent in chronic gastritis, peptic ulcer disease, gastric cancer, and gastric mucosa-associated lymphoid tissue lymphoma.3–5 A large prospective cohort study has identified H. pylori infection as a pivotal risk factor for both cardia and non-cardia gastric cancer.6 Furthermore, compelling evidence confirms that the eradication of H. pylori markedly reduces the incidence and mortality of gastric cancer.7
In China, national guidelines recommend bismuth-containing quadruple therapy (BQT) for first-line eradication of H. pylori, comprising a proton pump inhibitor (PPI), bismuth, and two antibiotics, such as amoxicillin, clarithromycin, levofloxacin, metronidazole, tetracycline, and furazolidone.8
However, antibiotic resistance remains a formidable obstacle to the effective eradication of H. pylori.9 As of 2022, primary resistance rates in the Asia-Pacific region were reported as follows: clarithromycin (30%), metronidazole (61%), levofloxacin (35%), tetracycline (4%), and amoxicillin (6%).10 Clarithromycin resistance was found to be strongly correlated with treatment failure in regimens containing clarithromycin, with an odds ratio of 6.97 (95% CI [5.23–9.28]; P < 0.001).9 The Maastricht V/Florence Consensus Report (2021) advocates for susceptibility testing in regions with high clarithromycin resistance prior to prescribing antibiotics, thereby guiding treatment decisions.3 After the failure of the initial eradication therapy, Helicobacter pylori acquires nearly total resistance to clarithromycin and levofloxacin, rendering repeated use of these antibiotics in rescue therapy inadvisable. This resistance markedly diminishes the effectiveness of eradication regimens that incorporate both antibiotics.11
Bacterial culture and antimicrobial susceptibility testing remain the gold standard for assessing phenotypic resistance in Helicobacter pylori.12 In recent years, rapid, straightforward, and cost-effective molecular methods—most notably real-time PCR, whole-genome sequencing, and digital PCR—have emerged as invaluable tools for detecting antibiotic resistance in H. pylori.3,13,14 Recent investigations have revealed that the resistance genes for clarithromycin and levofloxacin are in concordance with phenotypic resistance, demonstrating sensitivity values of 81.2% and 69.7%, and specificity values of 88.9% and 93.7%, respectively.14 Clarithromycin exerts its bactericidal effect by binding to the 23S rRNA of H. pylori, thereby disrupting bacterial protein synthesis; however, point mutations in the binding region confer resistance.12,15,16 Levofloxacin functions by inhibiting type II topoisomerases (DNA gyrase and topoisomerase IV) in H. pylori, with mutations in the gyrA gene conferring resistance to this antibiotic.12,16,17
Nevertheless, the capacity to conduct antibiotic resistance testing varies widely across healthcare settings, with many institutions lacking the requisite resources to perform these assessments.8 Moreover, both H. pylori culture and molecular testing methods can occasionally fail,18 with resistance genotype reports typically requiring 3–7 days, while phenotypic susceptibility testing based on bacterial culture takes even longer.19 In contrast, conventional empirical therapy presents notable advantages. Given the rarity of resistance to amoxicillin and tetracycline, prescriptions may be confidently guided by clinical judgment. Furthermore, in rescue therapy, eradication regimens meticulously tailored to the patient’s medication history can attain an almost complete eradication rate.11 Both the Toronto Consensus and the revised Maastricht V Consensus strongly advocate the selection of empirically guided antibiotics, informed by the outcomes of prior eradication regimens.3,20
Existing research suggests that susceptibility-guided therapy (SGT) generally achieves superior eradication rates compared to empirical therapy (ET).21–24 However, outcomes vary depending on the eradication regimen and the methods employed for resistance testing. A 2022 randomized controlled trial conducted in South Korea found no significant difference in eradication rates between ET and SGT when using the DPO-PCR method (ITT: P=0.262; PP: P=0.198).25 A recent meta-analysis indicated that ET had superior eradication rates compared to SGT under BQT (ITT: p = 0.001, RR: 0.93; 95% CI: 0.89–0.97; PP: p = 0.009, RR: 0.95; 95% CI: 0.92–0.99).26 However, the majority of studies in this analysis relied on culture-based methods for antimicrobial susceptibility testing, with only a single RCT incorporating molecular detection techniques. Therefore, further studies are imperative to validate these findings and evaluate the prospective clinical significance of genotyping. Additionally, studies frequently employ non-bismuth triple regimens when treating patients identified as antibiotic-sensitive via SGT. Consequently, robust clinical evidence remains insufficient to definitively establish the superiority of SGT over ET when using PCR-based antibiotic resistance detection with BQT.
This study utilized propensity score matching (PSM) and PCR-based detection methods to compare eradication rates between ET and SGT in the first-line eradication of H. pylori using BQT.
Materials and Methods
Study Design
A retrospective, single-center analysis was meticulously conducted at Beijing Chaoyang Hospital on patients diagnosed with H. pylori infection who underwent initial eradication therapy between 2021 and June 2023. The patients were stratified into two cohorts: one receiving susceptibility-guided therapy (SGT), the other empirical therapy (ET). The study was sanctioned by the Ethics Committee of Beijing Chaoyang Hospital, affiliated with Capital Medical University (No.2024-7-17-3). This study was a retrospective, single-center analysis that used pre-existing clinical data and routine procedures. Patient information and medical records were anonymized and exclusively used for research purposes. The Ethics Committee determined that the study posed no additional risks or interventions, waiving the informed consent requirement. Furthermore, the data collection process did not alter patient treatment or affect their health status, which made informed consent unnecessary.
Patients
Inclusion criteria were as follows: (1) age ≥ 18 years; (2) a confirmed diagnosis of H. pylori infection, verified within 4 weeks through at least one of the following diagnostic modalities: 13C/14C urea breath test (UBT), stool antigen test, rapid urease test, or histological analysis; (3) naïve to H. pylori treatment.
Patients were excluded if any one of the following criteria was present: (1) hypersensitivity to any drug employed in this study; (2) pregnancy or lactation; (3) incomplete endoscopic examination; (4) absence of a post-treatment urea breath test to evaluate eradication; (5) presence of significant systemic diseases, including but not limited to severe cardiac, pulmonary, or hepatic dysfunction.
Procedures
All enrolled patients underwent esophagogastroduodenoscopy (EGD), during which biopsy samples were collected for resistance genotyping where indicated. In the SGT group, treatment was meticulously tailored based on susceptibility testing, thereby circumventing the use of antibiotics with confirmed resistance. Conversely, the ET group comprised patients who either did not undergo resistance genotype testing and received ET or had resistance phenotyping completed, but ET was initiated before the report became available. All patients were administered bismuth-containing quadruple regimens (BQT), with the following antibiotics: PBAC (amoxicillin and clarithromycin), PBAF (amoxicillin and furazolidone), PBAM (amoxicillin and metronidazole), PBAL (amoxicillin and levofloxacin), PBAT (amoxicillin and tetracycline), PBFT (furazolidone and tetracycline), and PBMT (metronidazole and tetracycline). Following the initial eradication therapies, participants underwent UBT to evaluate the efficacy of the treatment.
Determination of Genotypic Resistance
Resistance genotyping was conducted using fluorescent PCR on DNA extracted from formalin-fixed, paraffin-embedded gastric mucosal biopsy specimens obtained during endoscopy. The analysis centered on resistance phenotypes for clarithromycin and levofloxacin antibiotics. Single-nucleotide variations at positions 2142, 2143, and 2142 within the 23S rRNA gene of Helicobacter pylori (A2142G, A2143G, A2142C) serve as reliable markers for clarithromycin resistance. Mutations at loci A260T, T261G, T261A, G271A, G271T, and A272G within the gyrA gene correlate with levofloxacin resistance. In accordance with the reference sequence of HP U27270, the following primers were employed: the HP-23S forward primer (5′-ATGAATGGCGTAACGAGATG-3′) and HP-23S reverse primer (5′-ACACTCAACTTGCGATTTCC-3′) for the detection of mutations within the 23S rRNA gene at loci 2142, 2143, and 2142; as well as the HP-gyrA forward primer (5′-GATCATAGGGCGCGCTTTACC-3′) and HP-gyrA reverse primer (5′-AAGTCGCCATCCCTACAGCGA-3′) for detecting mutations within the gyrA gene at loci A260T, T261G, T261A, G271A, G271T, and A272G. The PCR assay was performed in 25-µL reaction mixtures, comprising 12.5 µL of 2× GS Taq PCR Master Mix, 2 µL of template DNA, 1 µL of forward primer, 1 µL of reverse primer, and 8.5 µL of double-distilled water (ddH2O). The thermal cycling conditions were as follows: an initial denaturation step at 94°C for 3 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes.
Propensity Score Matching
To mitigate potential confounding and selection bias, propensity score matching (PSM) was employed to compare patients undergoing SGT with those receiving ET. Variables such as age, sex, and comorbidities, which could influence the eradication rate of H. pylori, were incorporated into the PSM. Binary logistic regression, with a caliper of 0.02, was applied for matching, resulting in a 1:4 ratio between the SGT and ET groups. This analysis was executed using R version 4.2.1.
Statistical Analyses
We endeavored to establish the noninferiority of bismuth-containing quadruple therapy (BQT) as a first-line regimen for H. pylori eradication. A previous study documented eradication success rates of 86% for SGT and 78% for ET.24 We presumed a 1:4 ratio in sample sizes between the SGT and ET groups. The calculated sample size necessitated a minimum of 97 participants in the SGT group and 388 in the ET group to detect a 12% difference, ensuring 80% statistical power at a 5% significance level in a two-sided test. The primary endpoint was the eradication rate derived from intention-to-treat (ITT) analysis. All PSM-matched subjects were incorporated into the ITT analysis. In the ITT framework, patients with incomplete treatment courses or those who completed UBT within 28 days post-treatment were designated as treatment failures. The per-protocol (PP) analysis included only those who completed the entire treatment regimen and underwent UBT after 28 days. The χ²-test or Fisher’s exact test was employed for the analysis of categorical data, while the Student’s t-test was utilized for continuous data. Normally distributed data were expressed as mean ± standard deviation (Mean ± SD), whereas skewed data were represented as median (IQR). Multiple logistic regression analysis was conducted to assess factors influencing eradication rates. Statistical analyses were performed using SPSS software (version 21, IBM Corp, Armonk, NY). A p-value of less than 0.05 was considered indicative of statistical significance.
Results
Baseline Data
We conducted a retrospective, single-center analysis at Beijing Chaoyang Hospital involving patients diagnosed with H. pylori infection who underwent initial eradication therapy between 2021 and June 2023. A total of 802 patients were included in the study. Among them, 661 received ET, with 48 undergoing genotypic testing during follow-up endoscopy after the completion of eradication therapy, while the remaining 613 proceeded with ET directly. The remaining 141 patients were managed with SGT. Statistically significant distinctions were discerned between the two groups with respect to age and the prevalence of chronic atrophic gastritis and peptic ulcer disease (Table 1). Following PSM at a 1:4 ratio between the SGT and ET groups, 550 patients were included in the final analysis. Of these, 125 received SGT and 425 received ET (Figure 1). The clinical characteristics, encompassing the distribution of age, sex, and comorbid conditions such as chronic gastritis, peptic ulcer disease, early gastric cancer, and MALT lymphoma, were comparable between the SGT and ET cohorts (Table 2).
 |
Table 1 Baseline Demographics and Clinical Characteristics Before Propensity Score Matching (PSM) |
 |
Table 2 Baseline Demographics and Clinical Characteristics After Propensity Score Matching (PSM) |
 |
Figure 1 Flowchart of the process of patient enrollment. |
Notably, after PSM, 29 patients in the ET group underwent H. pylori genotypic testing during follow-up endoscopy post-eradication. Drawing from these findings, we conducted a comprehensive analysis encompassing all patients who underwent genotypic testing. A total of 154 patients were included in the final analysis, with 125 receiving SGT and 29 receiving ET. The clinical profiles of the SGT and ET groups were comparable (Table 3).
 |
Table 3 Baseline Demographics and Clinical Characteristics Among Antibiotic Resistance Phenotypes |
Eradication Regimens
All enrolled patients were administered a 14-day course of BQT (Supplementary Figure 1). In the SGT group (n=125), treatment was meticulously tailored based on resistance genotyping. For CLR-resistant patients (n=32), clarithromycin was excluded, and they were treated with regimens such as PBAL, PBAM, PBAT, or PBAF. LVX-resistant patients (n=24) received regimens omitting levofloxacin, including PBAC, PBAM, PBAT, or PBAF. In cases of dual resistance (n=20), both antibiotics were omitted, with treatment options comprising PBAM, PBAT, or PBAF. For patients with no detected resistance (n=49), a variety of regimens were employed, including PBAC, PBAL, PBAM, PBAT, PBAF, or PBFT. In the ET group (n=425), patients were administered a diverse array of therapeutic regimens, including PBAC, PBAL, PBAM, PBAF, PBAT, and PBFT.
Antibiotic Resistance Rates
The overall resistance rates to clarithromycin, levofloxacin, and dual antibiotics were recorded at 26.6% (41/154), 18.2% (28/154), and 15.6% (24/154) (Supplementary Figure 2), respectively. In the SGT cohort, the prevalence of resistance to clarithromycin, levofloxacin, and dual antibiotics was 25.6% (32/125), 19.2% (24/125), and 16.0% (20/125), respectively. In contrast, the ET group exhibited resistance rates of 31.0% (9/29), 13.8% (4/29), and 13.8% (4/29), respectively. No statistically significant differences in resistance rates were observed between the two treatment strategies (P=0.909) (Supplementary Table 1).
Eradication Rates of H. Pylori
The eradication rates for SGT and ET were 94.4% (118/125) and 86.1% (366/425) in the ITT analysis (p=0.012). Similarly, in the PP analysis, the rates were 95.2% (118/124) and 87.6% (366/418) (p=0.016) (Table 4). Noteworthy differences were observed in the eradication rates between the two therapeutic strategies for the primary eradication of H. pylori in both the ITT analysis (odds ratio [OR], 2.717; 95% confidence interval [CI], 1.208–6.112; p=0.016) and the PP analysis (odds ratio [OR], 2.794; 95% confidence interval [CI], 1.170–6.670; p=0.021).
 |
Table 4 Eradication Rates Comparing SGT With ET |
Among the patients who underwent genotypic testing, the eradication rates for SGT and ET in the ITT analysis were 94.4% (118/125) and 75.9% (22/29), respectively (p=0.006). In the PP analysis, the corresponding rates were 95.2% (118/124) and 75.9% (22/29), with statistical significance observed (p=0.003) (Table 5). Noteworthy differences were observed in the eradication rates between the two therapeutic strategies of H. pylori in both the ITT analysis (odds ratio [OR], 5.364; 95% confidence interval [CI], 1.712–16.809; p=0.004) and the PP analysis (odds ratio [OR], 6.258; 95% confidence interval [CI], 1.920–20.397; p=0.002).
 |
Table 5 Eradication Rates Comparing SGT With ET Among Antibiotic Resistance Phenotypes |
Eradication Efficacy Among Antibiotic Resistance Phenotypes
Among different antibiotic-resistant phenotypes, eradication rates were consistently superior in the SGT group compared to the ET group (Table 6). For clarithromycin-resistant phenotypes, the eradication rates in both ITT and PP analyses were 96.9% (31/32) in the SGT group and 77.8% (7/9) in the ET group, with no statistically significant difference between the two groups (P=0.116). For levofloxacin-resistant phenotypes, eradication rates were 91.7% (22/24) in the SGT group and 50.0% (2/4) in the ET group (P=0.086). In the PP analysis, the eradication rates were 95.7% (22/23) in the SGT group and 50.0% (2/4) in the ET group (P=0.049). The efficacy of susceptibility-guided therapy (SGT) was markedly superior in levofloxacin-resistant phenotypes compared to empirical therapy (ET) (odds ratio [OR], 22; 95% confidence interval [CI], 1.334–362.916; p=0.031). For patients exhibiting dual resistance to clarithromycin and levofloxacin, the eradication rates in both ITT and PP analyses were 90.0% (18/20) in the SGT group and 75.0% (3/4) in the ET group, with no statistically significant difference observed (P=0.437). For phenotypes susceptible to both clarithromycin and levofloxacin, the eradication rates in the ITT and PP analyses were 95.9% (47/49) in the SGT group and 83.3% (10/12) in the ET group, with no statistically significant difference observed (P=0.170).
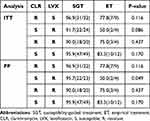 |
Table 6 Eradication Rates Comparing SGT With ET in Different Antibiotic Resistance Phenotypes |
Treatment Regimens for Eradication Failure
Among the 550 participants enrolled in the study, 66 experienced treatment failure. Of these, merely 7 patients underwent SGT, while the remaining 59 individuals failed to achieve eradication with ET. The therapeutic regimens administered to the 66 patients who experienced treatment failure, ranked from most to least, were as follows: PBAF (N=20), PBAC (N=14), PBAL (N=13), PBAM (N=10), PBAT (N=8), and PBFT (N=1). Among the 59 patients who failed ET, 7 underwent post-treatment resistance genotype analysis. Remarkably, a single patient exhibiting clarithromycin resistance was administered a regimen that included clarithromycin.
To gain deeper insight into the influence of antibiotic resistance on eradication success, we scrutinized the regimens that failed across various resistance phenotypes (Figure 2). Among patients sensitive to both clarithromycin and levofloxacin, four individuals failed eradication using the PBAL regimen. In individuals exhibiting resistance to clarithromycin alone, the regimens that resulted in failure included: PBAC (N=1), PBAL (N=1), and PBAF (N=1). In individuals resistant to levofloxacin alone, the regimens that failed included: PBAC (N=1), PBAF (N=1), PBAM (N=1), and PBAT (N=1). Among individuals with dual resistance to both clarithromycin and levofloxacin, three patients failed eradication with the PBAF regimen, while one failed with the PBAT regimen.
Discussion
The heightened rate of antibiotic resistance is a pivotal factor contributing to the suboptimal eradication of Helicobacter pylori.27 According to 2021 data from the Chinese Center for H. pylori Molecular Medicine (CCHpMM), the primary resistance rates to clarithromycin and levofloxacin in China were reported at 37% and 34.21%, respectively.28 A prominent prospective cohort study conducted in Beijing (2013–2014) revealed primary resistance rates of 52.6% for clarithromycin and 18.2% for levofloxacin.29 In our study, resistance rates to clarithromycin and levofloxacin were 26.6% and 18.2%, respectively, with a dual resistance rate of 15.6%. In alignment with the 2022 Maastricht guidelines, which designate China as a region of high clarithromycin resistance, antibiotic susceptibility testing should be undertaken prior to prescribing treatment to guide therapy.3
In accordance with the 2022 Chinese guidelines for H. pylori management, the recommended antibiotic combinations within the bismuth-containing quadruple (BQT) regimen for first-line eradication include amoxicillin paired with clarithromycin, levofloxacin, metronidazole, or tetracycline; tetracycline in combination with metronidazole; and, in cases where eradication is anticipated to be challenging, bismuth-containing quadruple regimens incorporating furazolidone, such as furazolidone combined with amoxicillin or tetracycline.8 In this study, the bismuth-containing quadruple regimens were meticulously tailored to the prescribing practices of the treating physicians and the clinical context, while adhering rigorously to the established guidelines.
Most comparative analyses between SGT and ET have predominantly centered on the detection of clarithromycin resistance. For instance, recent studies from South Korea observed no statistically significant difference in eradication rates between SGT and ET when evaluating clarithromycin-resistant genotypes.30–32 However, a randomized controlled trial (RCT) led by Meng-Shu Hieh, employing PCR-RFLP to detect resistance to both clarithromycin and levofloxacin, demonstrated that SGT achieved markedly higher eradication rates compared to ET (ITT: 89% vs 75.8%; PP: 91% vs 79.3%), with statistically significant differences (ITT: p<0.031; PP: p<0.034).23 Notably, this study employed a triple therapy regimen, whereas our research implemented a bismuth-containing quadruple regimen. In contrast to these findings, our study not only evaluated resistance to both clarithromycin and levofloxacin, but also utilized a bismuth-containing quadruple regimen, which is recognized for its superior efficacy compared to triple therapy.
Our results show that SGT achieved significantly higher eradication rates than ET. This aligns with the conclusions of most existing research and further supports the idea that antibiotic resistance severely reduces the efficacy of clarithromycin and levofloxacin.21–24 Notably, 29 patients in our cohort who initially received ET underwent resistance genotype testing after completing their treatment. A retrospective analysis was therefore conducted to compare the eradication rates of ET and SGT among patients who had completed H. pylori resistance genotype testing. However, significant discrepancies were observed only in cases of levofloxacin resistance across the various genetic phenotypes (PP: p=0.031, OR: 22, 95% CI: 1.334–362.916). This may be due to the relatively small number of patients in the empirical therapy cohort who underwent resistance genotype testing. To address this issue, it is essential to increase the number of patients in the ET group undergoing resistance genotype testing. Furthermore, improving patient recruitment and testing protocols is crucial to ensuring a higher completion rate for the testing process.
We further investigated the possible factors contributing to eradication failure. Among the 66 patients who failed eradication, the treatment regimens were: PBAF (N=20), PBAC (N=14), PBAQ (N=13), PBAM (N=10), PBAT (N=8), and PBFT (N=1). The regimen containing furazolidone was the most common among those who failed eradication. We propose that this may be attributed to the following factors: First, although H. pylori resistance to furazolidone is generally low, it is increasing in certain regions. Reports suggest that the resistance rate in Asia has increased to 62.5%.33 H. pylori resistance substantially diminishes the efficacy of furazolidone.34 Second, furazolidone may cause a range of adverse effects, including gastrointestinal symptoms (eg, nausea, vomiting, diarrhea), neurotoxicity (eg, headache, dizziness), and hepatotoxicity.35 These side effects may reduce patient adherence, thereby lowering eradication rates. Importantly, this study does not include data on patient adherence and adverse reactions. Other antibiotics, such as clarithromycin and levofloxacin, found in the failure regimens, were also used in the ET group, indirectly supporting the benefits of SGT. Interestingly, one patient who received clarithromycin-based therapy was later found to have a clarithromycin-resistant phenotype during follow-up. This is primarily due to the fact that only a limited number of patients underwent resistance genotype testing after ET. Additionally, reports suggest that, besides resistance factors, poor adherence, smoking, alcohol consumption, and cytochrome P450 (CYP2C19) gene polymorphisms may also affect H. pylori eradication rates.36–38
This study has several limitations that require attention. First, this study is a single-center retrospective analysis. The patient cohort may have regional and demographic limitations, which could hinder its ability to fully represent therapeutic outcomes across different geographical areas or populations. Future large-scale, multi-center, prospective studies will be necessary to confirm these findings and enhance the broader applicability of the conclusions. Additionally, this study did not include data on patient adherence or adverse events. It is well known that patients with low adherence and frequent adverse events may overestimate the therapeutic efficacy of certain regimens. Therefore, future studies should carefully document the nature and frequency of adverse events and incorporate these data into the analysis to assess their impact on treatment outcomes. Specifically, for patients experiencing frequent adverse events, alternative regimens with fewer side effects should be considered to optimize both therapeutic efficacy and patient compliance. Finally, this study did not include a cost-effectiveness analysis. While SGT shows promise in improving H. pylori eradication rates, the additional costs associated with testing and personalized treatment may increase overall treatment expenses. Therefore, a cost-effectiveness analysis is essential to assess the feasibility of SGT in different economic contexts, especially in areas with limited healthcare resources. Future studies should integrate such analyses to better assess the overall value and long-term sustainability of SGT within a comprehensive health economics framework.
This study suggests that susceptibility-guided therapy (SGT) based on resistance genotyping should be widely adopted in clinical practice, particularly for patients with high resistance rates or unsatisfactory responses to standard treatments. To effectively implement SGT, it is essential to expand resistance gene testing and provide additional training for physicians, ensuring that test results can rapidly and precisely guide treatment decisions. Clinicians should select appropriate antibiotic combinations based on test results to optimize treatment efficacy and reduce unnecessary antibiotic use. In addition, patient adherence and potential side effects must be closely monitored. As genotyping technologies progress, the availability and timeliness of resistance testing will improve, leading to higher eradication rates and a reduction in resistance transmission.
Conclusion
SGT exhibited markedly superior eradication rates compared to ET, particularly in strains resistant to levofloxacin, thereby underscoring its considerable promise in the treatment of H. pylori infection. Nonetheless, the study’s modest sample size and its narrow temporal scope diminish the broader applicability of its findings. More expansive and longitudinal studies are imperative to validate the long-term efficacy and wider clinical applicability of SGT.
Ethics Approval and Informed Consent
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Beijing Chaoyang Hospital, Capital Medical University (No. 2024-7-17-3). Written informed consent was waived due to the retrospective nature of the study and the absence of personal identifiers in the analysis and reporting. All patient data were kept confidential.
Acknowledgments
We extend our gratitude to Dr. Jianyu Hao (Department of Gastroenterology, Beijing Chao-Yang Hospital, Capital Medical University) for his invaluable guidance and support throughout the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by Beijing Key Clinical Specialty Project.
Disclosure
The authors report no conflicts of interest in this research.
References
1. Li Y, Choi H, Leung K, Jiang F, Graham DY, Leung WK. Global prevalence of Helicobacter pylori infection between 1980 and 2022: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2023;8(6):553–564. doi:10.1016/s2468-1253(23)00070-5
2. Chen YC, Malfertheiner P, Yu HT, et al. Global Prevalence of Helicobacter pylori Infection and Incidence of Gastric Cancer Between 1980 and 2022. Gastroenterology. 2024;166(4):605–619. doi:10.1053/j.gastro.2023.12.022
3. Malfertheiner P, Megraud F, Rokkas T, et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. 2022;71:1724–1764. doi:10.1136/gutjnl-2022-327745
4. Malfertheiner P, Camargo MC, El-Omar E, et al. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9(1):19. doi:10.1038/s41572-023-00431-8
5. Watari J, Chen N, Amenta PS, et al. Helicobacter pylori associated chronic gastritis, clinical syndromes, precancerous lesions, and pathogenesis of gastric cancer development. World J Gastroenterol. 2014;20(18):5461–5473. doi:10.3748/wjg.v20.i18.5461
6. Yang L, Kartsonaki C, Yao P, et al. The relative and attributable risks of cardia and non-cardia gastric cancer associated with Helicobacter pylori infection in China: a case-cohort study. Lancet Public Health. 2021;6(12):e888–e896. doi:10.1016/s2468-2667(21)00164-x
7. Li WQ, Zhang JY, Ma JL, et al. Effects of Helicobacter pylori treatment and vitamin and garlic supplementation on gastric cancer incidence and mortality: follow-up of a randomized intervention trial. BMJ. 2019;366:l5016. doi:10.1136/bmj.l5016
8. Zhou L, Lu H, Song Z, et al. Chinese national clinical practice guideline on Helicobacter pylori eradication treatment. Chin Med J. 2022;135(24):2899–2910. doi:10.1097/cm9.0000000000002546
9. Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori. A Systematic Review and Meta-Analysis in World Health Organization Regions Gastroenterology. 2018;155(5):1372–1382.e17. doi:10.1053/j.gastro.2018.07.007
10. Hong TC, El-Omar EM, Kuo YT, et al. Primary antibiotic resistance of Helicobacter pylori in the Asia-Pacific region between 1990 and 2022: an updated systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2024;9(1):56–67. doi:10.1016/s2468-1253(23)00281-9
11. Gisbert JP. Empirical or susceptibility-guided treatment for Helicobacter pylori infection? A comprehensive review. Therap Adv Gastroenterol. 2020;13:1756284820968736. doi:10.1177/1756284820968736
12. Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6(11):699–709. doi:10.1016/s1473-3099(06)70627-2
13. Pohl D, Keller PM, Bordier V, Wagner K. Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J Gastroenterol. 2019;25(32):4629–4660. doi:10.3748/wjg.v25.i32.4629
14. Cui R, Song Z, Suo B, et al. Correlation Analysis Among Genotype Resistance, Phenotype Resistance and Eradication Effect of Helicobacter pylori. Infect Drug Resist. 2021;14:1747–1756. doi:10.2147/idr.S305996
15. Hu Y, Zhang M, Lu B, Dai J. Helicobacter pylori and Antibiotic Resistance, A Continuing and Intractable Problem. Helicobacter. 2016;21(5):349–363. doi:10.1111/hel.12299
16. Tshibangu-Kabamba E, Yamaoka Y. Helicobacter pylori infection and antibiotic resistance - from biology to clinical implications. Nat Rev Gastroenterol Hepatol. 2021;18(9):613–629. doi:10.1038/s41575-021-00449-x
17. Egli K, Wagner K, Keller PM, Risch L, Risch M, Bodmer T. Comparison of the Diagnostic Performance of qPCR, Sanger Sequencing, and Whole-Genome Sequencing in Determining Clarithromycin and Levofloxacin Resistance in Helicobacter pylori. Front Cell Infect Microbiol. 2020;10:596371. doi:10.3389/fcimb.2020.596371
18. Schuetz AN, Theel ES, Cole NC, Rothstein TE, Gordy GG, Patel R. Testing for Helicobacter pylori in an era of antimicrobial resistance. J Clin Microbiol. 2024;62(2):e0073223. doi:10.1128/jcm.00732-23
19. Wang YH, Wang FF, Gong XL, et al. Genotype profiles of Helicobacter pylori from gastric biopsies and strains with antimicrobial-induced resistance. Therap Adv Gastroenterol. 2020;13:1756284820952596. doi:10.1177/1756284820952596
20. Mahachai V, Vilaichone RK, Pittayanon R, et al. Helicobacter pylori management in ASEAN: the Bangkok consensus report. J Gastroenterol Hepatol. 2018;33(1):37–56. doi:10.1111/jgh.13911
21. Cosme A, Lizasoan J, Montes M, et al. Antimicrobial Susceptibility-Guided Therapy Versus Empirical Concomitant Therapy for Eradication of Helicobacter pylori in a Region with High Rate of Clarithromycin Resistance. Helicobacter. 2016;21(1):29–34. doi:10.1111/hel.12231
22. Lin K, Huang L, Wang Y, et al. Efficacy of genotypic susceptibility-guided tailored therapy for Helicobacter pylori infection: a systematic review and single arm meta-analysis. Helicobacter. 2023;28(6):e13015. doi:10.1111/hel.13015
23. Hsieh MS, Kuo FC, Wu MC, et al. Tailored susceptibility-guided therapy via gastric juice PCR for the first-line H. pylori eradication, a randomized controlled trial. J Formos Med Assoc. 2022;121(8):1450–1457. doi:10.1016/j.jfma.2021.10.011
24. Ouyang Y, Zhang W, He C, Zhu Y, Lu N, Hu Y. Susceptibility-Guided Therapy vs. Bismuth-Containing Quadruple Therapy as the First-Line Treatment for Helicobacter pylori Infection: a Systematic Review and Meta-Analysis. Front Med Lausanne. 2022;9:844915. doi:10.3389/fmed.2022.844915
25. Cho JH, Jin SY, Park S. Comparison of tailored Helicobacter pylori eradication versus modified bismuth quadruple therapy in Korea: a randomized controlled trial. Expert Rev Anti Infect Ther. 2022;20(6):923–929. doi:10.1080/14787210.2022.2017280
26. Li M, Wang X, Meng W, Dai Y, Wang W. Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis. Therap Adv Gastroenterol. 2023;16:17562848231196357. doi:10.1177/17562848231196357
27. Zhong Z, Zhan B, Xu B, Gao H. Emphasizing the importance of successful eradication of Helicobacter pylori on initial treatment. Am J Cancer Res. 2022;12(3):1215–1221.
28. Zhong Z, Zhang Z, Wang J, et al. A retrospective study of the antibiotic-resistant phenotypes and genotypes of Helicobacter pylori strains in China. Am J Cancer Res. 2021;11(10):5027–5037.
29. Zhang YX, Zhou LY, Song ZQ, Zhang JZ, He LH, Ding Y. Primary antibiotic resistance of Helicobacter pylori strains isolated from patients with dyspeptic symptoms in Beijing: a prospective serial study. World J Gastroenterol. 2015;21(9):2786–2792. doi:10.3748/wjg.v21.i9.2786
30. Choi YI, Chung JW, Kim KO, et al. Tailored eradication strategy vs concomitant therapy for Helicobacter pylori eradication treatment in Korean patients. World J Gastroenterol. 2021;27(31):5247–5258. doi:10.3748/wjg.v27.i31.5247
31. Cha B, Bang BW, Shin JB, et al. Bismuth containing quadruple therapy versus tailored therapy as first-line treatments for Helicobacter pylori infection in a high clarithromycin resistance area. Scand J Gastroenterol. 2021;56(9):1017–1022. doi:10.1080/00365521.2021.1948606
32. Kim SJ, Jee SR, Park MI, et al. A randomized controlled trial to compare Helicobacter pylori eradication rates between the empirical concomitant therapy and tailored therapy based on 23S rRNA point mutations. Medicine. 2022;101(33):e30069. doi:10.1097/md.0000000000030069
33. Zamani M, Rahbar A, Shokri-Shirvani J. Resistance of Helicobacter pylori to furazolidone and levofloxacin: a viewpoint. World J Gastroenterol. 2017;23(37):6920–6922. doi:10.3748/wjg.v23.i37.6920
34. Biskamp J, Bartos M, Sauer JF. Organization of prefrontal network activity by respiration-related oscillations. Sci Rep. 2017;7:45508. doi:10.1038/srep45508
35. Zhuge L, Wang Y, Wu S, Zhao R-L, Li Z, Xie Y. Furazolidone treatment for Helicobacter Pylori infection: a systematic review and meta-analysis. Helicobacter. 2018;23(2):e12468. doi:10.1111/hel.12468
36. Suzuki T, Matsuo K, Ito H, et al. Smoking increases the treatment failure for Helicobacter pylori eradication. Am J Med. 2006;119(3):217–224. doi:10.1016/j.amjmed.2005.10.003
37. Yu J, Lv Y, Yang P, Jiang Y, Qin X, Wang X. Alcohol increases treatment failure for Helicobacter pylori eradication in Asian populations. BMC Gastroenterol. 2023;23(1):365. doi:10.1186/s12876-023-03002-z
38. Kuo CH, Lu CY, Shih HY, et al. CYP2C19 polymorphism influences Helicobacter pylori eradication. World J Gastroenterol. 2014;20(43):16029–16036. doi:10.3748/wjg.v20.i43.16029
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


