Back to Journals » International Journal of Nanomedicine » Volume 19
Enhanced Efficacy of Some Antibiotics in the Presence of Silver Nanoparticles Against Clinical Isolate of Pseudomonas aeruginosa Recovered from Cystic Fibrosis Patients
Authors Al-Momani H , Albalawi H, Al Balawi D, Khleifat KM, Aolymat I , Hamed S , Albiss BA, Khasawneh AI, Ebbeni O, Alsheikh A, Zueter AM , Pearson JP, Ward C
Received 29 July 2024
Accepted for publication 1 November 2024
Published 23 November 2024 Volume 2024:19 Pages 12461—12481
DOI https://doi.org/10.2147/IJN.S479937
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Eng San Thian
Hafez Al-Momani,1 Hadeel Albalawi,2 Dua’a Al Balawi,2 Khaled M Khleifat,3 Iman Aolymat,4 Saja Hamed,5 Borhan Aldeen Albiss,6 Ashraf I Khasawneh,1 Ola Ebbeni,7 Ayman Alsheikh,8 AbdelRahman M Zueter,9 Jeffrey Peter Pearson,10 Christopher Ward11
1Department of Microbiology, Pathology and Forensic Medicine, Faculty of Medicine, The Hashemite University, Zarqa, 13133, Jordan; 2Faculty of Applied Medical Sciences, The Hashemite University, Zarqa, 13133, Jordan; 3Biology Department, College of Science, Mutah University, Mutah, Karak, 61710, Jordan; 4Department of Anatomy, Physiology and Biochemistry, Faculty of Medicine, The Hashemite University, Zarqa, 13133, Jordan; 5Department of Pharmaceutics & Pharmaceutical Technology, Faculty of Pharmaceutical Sciences, The Hashemite University, Zarqa, 13133, Jordan; 6Nanotechnology Institute, Jordan University of Science & Technology, Irbid, 22110, Jordan; 7Department of Pharmacology and Public Health, Faculty of Medicine, The Hashemite University, Zarqa, 13133, Jordan; 8Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Zarqa University, Zarqa, 13110, Jordan; 9Department of Medical Laboratory Sciences, Faculty of Applied Medical Sciences, The Hashemite University, Zarqa, 13133, Jordan; 10Biosciences Institute, Newcastle University Medical School, Newcastle Upon Tyne, UK; 11Translational and Clinical Research Institute, Newcastle University Medical School, Newcastle Upon Tyne, NE2 4HH UK
Correspondence: Hafez Al-Momani, Department of Microbiology, Pathology and Forensic Medicine, Faculty of Medicine, The Hashemite University, Zarqa, 13133, Jordan, Email [email protected]
Introduction: Given the increasing frequency of drug-resistant bacteria and the limited progress in developing new antibiotics, it is necessary to explore new methods of combating microbial infections. Nanoparticles, particularly silver nanoparticles (Ag-NPs), have shown exceptional antibacterial characteristics; however, elevated concentrations of Ag-NPs can produce noticeable levels of toxicity in mammalian cells.
Aim: This study examined the potential synergistic effect of combining a low dosage of Ag-NPs and anti-pseudomonas drugs against Pseudomonas aeruginosa (ATCC strain) and eleven clinical isolates from cystic fibrosis patients.
Methods: The Ag-NPs were chemically produced by utilizing a seed extract from Peganum Harmala and characterized via ultraviolet-visible spectroscopy and scanning electron microscopy. The broth microdilution technique was utilized to investigate the minimum inhibitory concentration (MIC) of Ag-NPs and eight antibiotics (Piperacillin, Ciprofloxacin, Levofloxacin, Meropenem, Amikacin, Ceftazidime, Gentamicin, Aztreonam). The fractional inhibitory concentration index (FICI) was determined via the checkerboard method to evaluate the synergistic effects of Ag-NPs and various antibiotics.
Results: The biosynthesized Ag-NPs were uniformly spherical and measured around 15 nm in size. When combined with antibiotics, Ag-NP produced statistically significant reductions in the amount of antibiotics required to completely prevent P. aeruginosa growth for all strains. The findings revealed that the MIC of Ag-NPs was 15 ug/mL for all strains which decreased substantially when administered with antibiotics at a dose of 1.875– 7.5 ug/mL. The majority of Ag-NP and antibiotic combinations exhibited a synergistic or partially synergistic impact. This was particularly noticeable in combinations containing Meropenem, Ciprofloxacin, and Aztreonam (in which the FIC index was less than or equal to 0.5).
Conclusion: The findings revealed that combining Ag-NPs with antibiotics was more effective than using Ag-NPs or antibiotics in isolation and that combinations of Ag-NPs and antimicrobial agents displayed synergistic activity against the majority of strains assessed.
Keywords: cystic fibrosis CF, biofilm, Pseudomonas aeruginosa, nanoparticles, silver nanoparticles, antibiotic resistance, synergy
Introduction
The abundant Gram-negative bacteria Pseudomonas aeruginosa is part of the Pseudomonadaceae family and can survive in many different environments.1 Studies have identified P. aeruginosa as a common opportunistic pathogen associated with nosocomial infections and ventilator-associated pneumonia.2 Healthy people rarely contract this bacteria; however, it is associated with elevated mortality rates in patients with cystic fibrosis (CF) and individuals with compromised immune systems.3
CF is defined as a genetic disorder resulting from mutations in both copies of the gene-encoding cystic fibrosis transmembrane conductance regulator (CFTR) which results in the development of a thick layer of mucus on the surface of the airways in CF patients.4 This process prevents the body from performing mucociliary clearance, reduces bacterial internalization by epithelial cells in the lungs and inhibits the actions of antimicrobial peptides;5–7 therefore (in CF patients), the lungs serve as environments that promote bacterial growth and colonization. The prevalent cause of lung infections among CF patients is the pathogen P. aeruginosa which is resistant to existing antibiotics, reduces pulmonary function, and results in death for patients with the condition.7 Furthermore, P. aeruginosa is responsible for 5% of infectious exacerbations in chronic obstructive pulmonary disease (COPD) patients.8 Globally, COPD is the third highest cause of death (3.23 million deaths in 2019) and research has revealed that P. aeruginosa may increase mortality rates in such individuals.9
Monotherapy and combination therapy form part of the empirical antibiotic therapy for P. aeruginosa and can reduce mortality in patients with severe infections. However, because P. aeruginosa is resistant to many existing antibiotics, treating these infections is becoming increasingly problematic. P. aeruginosa is resistant to quinolones, aminoglycosides, and β-lactam antibiotics and other similar drugs.10 Moreover, P. aeruginosa develops an adaptive resistance via the production of biofilm in the lungs of infected individuals.11 This biofilm acts as a diffusion barrier that prevents antibiotics from reaching the bacterial cells.12 Furthermore, the biofilm may give rise to multidrug-tolerant persister cells that can withstand antibiotic treatment and cause persistent and recurring infections in CF patients.13 Additionally, the overuse of antibiotics during treatment hastens the emergence of drug-resistant strains of P. aeruginosa and makes empirical antibiotic therapy unusable against this bacteria.10,14 Therefore, the development of innovative drugs or other therapeutic approaches is critical and urgently needed to treat infections caused by P. aeruginosa infections.
The current advancements in antimicrobial nanotechnologies possess the potential to eradicate resistant pathogens and inhibit the development of additional antibiotic-resistant strains.15 Nanoparticles (NPs) exhibit physiochemical properties which can be programmed to be bactericidal, antifouling, immunomodulating, and to accurately distribute antibacterial elements to the infection site.15,16 Additionally, NPs can negate antibiotic resistance via the utilization of unique bactericidal pathways which instigate the required antimicrobial activity.17 For example, Ag+ ions form the active component of antimicrobials based on silver nanoparticles (Ag-NPs) and can disrupt electron transport across the bacterial membrane and damage DNA.18 Contemporary research indicates that metal and metal oxide NPs (including zinc oxide, platinum, gold and Ag) exhibit beneficial antibacterial capabilities against numerous infections.19 Recently, the utilization of Ag-NPs has intensified and this can be attributed to two factors: cost effectiveness and their efficacy against numerous Gram-positive and -negative bacterial strains.20
Numerous studies have reported on the antimicrobial properties of Ag-NPs and noted their effectiveness against multidrug-resistant bacteria including Escherichia coli,21 P. aeruginosa22,23 and extended-spectrum β-lactam (ESBL) producing bacteria.24 Additionally, Ag-NPs are a cost-effective and efficient treatment for antibiotic-resistant Methicillin-resistant Staphylococcus aureus (MRSA).25 In concordance with Waseem, Naveed, Rehman, Makhdoom, Aziz, Alharbi, Alsahammari and Alasmari25 study which reported that Ag-NPs inhibit virulent genes of MRSA (such as Spa, LukD, FmhA, and hld), it has been noted that at an Ag-NP concentration of 7.5 µg/mL among P. aeruginosa clinical isolate from CF patients, the expression of the quorum-sensing genes were substantially reduced.20 These findings can be explained by the fact that these NPs are ultra-small and have an elevated surface-to-volume ratio which facilitates direct interaction at the molecular level.26
Previous research has demonstrated that Ag-NPs spontaneously generate reactive oxygen species (ROS), interact directly with biological membranes (causing them to rupture) and that the internalized NPs release silver ions (Ag+) to mediate their antimicrobial activity in a Trojan horse mechanism.27,28 Notably, earlier studies have reported that Ag-NPs can eradicate Multidrug resistance (MDR) bacteria and negate resistance mechanisms such as biofilm formation and modulating microbial influx/efflux pumps formed by bacteria in response to antibiotics.29 A primary concern in the field of nanomedicine is that the benefit of employing NPs to demonstrate toxicity against MDR bacteria may be negated by its harmful effect on mammalian cells.28 To mitigate the cytotoxicity of metal NPs, several studies have noted that pairing metal NPs with traditional antimicrobials can reduce the toxicity of both substances to mammalian cells by lowering the need for high dosage schedules and improving the bactericidal effects.30 Therefore, combining these two treatment approaches could provide a viable strategy, particularly when treating MDR bacterial strains.
A wide range of physical and chemical methodologies have been employed for the creation of metal NPs. However, it is globally recognized that biological, synthetic processes are benign, uncomplicated, and non-toxic (when compared to chemical and physical processes which are costly, toxic, and hazardous).31,32 The challenges presented by chemical and physical production can be mitigated via biological synthesis32 involving bacteria, fungi, or plant extracts.33–35 Several plant-derived components (such as leaves, roots, fruits, stems, and seeds) have been used in the production of NPs35,36 and these extracts can produce NPs of specific size, structure, and configuration. Additionally, these extracts contain numerous phytochemicals (such as flavones, phenolics, proteins, polysaccharides, terpenoids, alkaloids, enzymes, amino acids, and alcohols) which function as natural stabilizers and/or reducing agents for the production of NPs.36,37 The use of plant-derived NPs minimizes harmful side effects in humans (compared to chemically synthesized NPs) and possesses the biological potential to be applied in agriculture, food science and technology, bioengineering, cosmetic or nanomedicine, and human health protection.37,38 These factors have resulted in increasing levels of interest regarding the use of biogenic techniques for the ecological production of NPs.39,40
This research employed Peganum harmala as a reducing agent for the biosynthesis of Ag-NPs. P harmala is a glabrous, perennial which propagates in sandy soil located in semi-arid regions such as the desert regions in Jordan.41 It grows to between 0.2 and 0.9m in length, produces spherical seed capsules (each containing approximately forty seeds), and produces white blooms. In the Middle East, it is primarily employed for its antiseptic and disinfectant qualities (obtained by burning or boiling its seeds).41 Additionally, it has been employed in the treatment of several ailments such as lumbago, asthma, colic, and jaundice, and as an emmenagogue,42,43 and research indicates that it possesses antiviral, antibacterial, and antifungal properties.43,44
Previous research has examined Ag-NP-based combination therapy using different antimicrobial agents to test their effectiveness at treating a wide range of bacteria (ie, MDR bacteria).42 However, no studies have examined the combination of different anti-pseudomonal antibiotics with biosynthesized Ag NPs using different clinical strains of P. aeruginosa taken from the sputum of people with CF. Therefore, this research hypothesizes that Ag-NPs can reinforce (or strengthen) the antimicrobial efficacy of traditional antimicrobial agents when used against clinical P. aeruginosa isolates.
Methods
Ethical Approval
The Ethics Service Committee of Hashemite University and Prince Hamza Hospital has provided ethical permission for this study (reference number 7/10/2019-2020) and the tests were conducted in compliance with all applicable rules and legislation. The P. aeruginosa clinical isolates were collected from sputum samples of CF patients following the acquisition of informed consent. This study complies with the Declaration of Helsinki.
Green Synthesis of the Ag-NPs
Preparing the Aqueous Peganum harmala Seed Extract
The first researcher (H A-M) obtained complete P. harmala plants from their farm which is located in the Abbin region of the Ajloun Governorate in the Kingdom of Jordan. To confirm the identification of the P. harmala seeds, they were compared to a sample located in the Faculty of Agriculture at Jerash University (Jordan). The identification process was corroborated by an assistant professor in agriculture (Dr M. Bani-Hani) and a voucher sample (AM/2024/01/002) was stored at the herbarium in the university’s Plant Protection Department. The P. harmala plants were harvested in line with national and international guidelines.
The seeds were dried at room temperature, washed with double-distilled water (Sigma-Aldrich), and pulverized into a coarse powder. 50 mL of double-distilled water and 5 g of powder were combined and left at 80 °C for 120 minutes following which they were filtered twice through Grade 2 Whatman filter paper. The process of microfiltration with a 0.22 μm syringe membrane filter produced a clear aqueous extract with a pH of 6.8 at 25 °C.
Measuring the Total Phenolic and Total Flavonoid Content of the P. harmala Extract
To determine the presence of phenolic compounds (ie, plant-based secondary metabolites that can scavenge radicals due to their redox potentials)43 in the P. Harmala extracts, this study employed the Folin-Ciocalteu approach.44 This process consists of phenolic moiety oxidation following the addition of Folin-Ciocalteu reagent (comprised of phosphomolybdic and phosphotungstic acids).45
This study employed a modified version of the method published by Singleton, Orthofer and Lamuela-Raventós:44 100 μL Folin-Ciocalteu reagent was added to 20 μL P. harmala extract and after ten minutes, a 300 μL 25% sodium carbonate solution was added to the mixture and a wavelength of 765nm was employed to determine the solution’s absorbance. This experiment was performed in triplicate and the mean absorbance value was computed. Gallic acid solutions varying in strength from 0–500 mg/L were also applied to preduce the calibration curve. Data were presented as gallic acid equivalents (GAE) per 100 gm extract.
The level of flavonoids in the sample was determined by creating a flavonoid-aluminum complex46 with a wavelength of maximum absorption of 430 nm. Quercetin was used to create a calibration curve. P. harmala extract and a 2% aluminum chloride methanolic solution were combined in 1 mL, and the mixture was incubated for ten minutes at room temperature. At the aforementioned wavelength, the absorbance of the solution was measured and Quercetin equivalents (QE) per 100 grams of extract were employed to express the data.
Green Synthesis of Ag-NPs
For Ag-NP synthesis, a 50 mL of a 3 mM silver nitrate (AgNO₃) solution was prepared by computing the necessary quantity for 50 mL at 3 mM, which equals 0.15 mM of AgNO₃, equating to 0.0255 g based on its molar mass of 169.88 g/mol.
hen, a 50 mL solution of 3 mm AgNO3 was placed in a 100 mL foil-lined Erlenmeyer flask and stirred at 1100 rpm at room temperature. During this, 5 mL of the aqueous P. harmala seed extract was added via micropipette at a rate of 40 µL/min with stirring until the colour changed from orange to dark brown. UV spectrophotometry, which measures the absorbance of a reaction medium in the 300–800 nm wavelength range, was used to monitor the reduction of pure Ag ions (which produce Ag-NPs). The synthesized Ag-NPs were centrifuged for 15 minutes at 10,000 rpm which enabled them to be purified. After allowing the particles to settle in the supernatant, centrifugation was conducted repeatedly with a cooled microfuge in a clean and dry container. This process produced a dry, pure product which facilitated the characterization of the Ag-NPs. To get the optimum size of the Ag-NP, the procedure was repeated using varying quantities of P. harmala seed extract and AgNO3 concentrations at room temperature.
Fourier-Transform Infrared Spectroscopy (FT-IR)
An FT-IR analysis of the Ag-NPs and P. harmala seeds extract was conducted in the University of Hashemite’s chemistry department. This process was used to confirm the relationship between the functional groups of the P. harmala extract and the synthesized Ag-NPs existed within a wavenumber range of 3600–400 cm−1.
Characterizing Silver Nanoparticles
Ultraviolet-Visible Spectroscopy
To monitor the bio-reduction of the AgNO3 salt solution and examine the production of Ag-NPs using P. harmala seed aqueous extract, this study employed UV-visible (UV-Vis) spectroscopy (UV-1900, Shimadzu, Japan). Double-distilled water was used to create the control sample, a quartz cuvette containing a 1 cm path length was employed in all of the spectrophotometric analyses, and a wavelength range of 800–200 nm was implemented to measure the surface plasmon resonance (SPR).
Scanning Electron Microscopy
A scanning electron microscope (SEM) was employed to measure the surface size and features of the biosynthesized Ag-NPs. The surfaces of the sample were coated with a 4 nm layer of gold, and a low vacuum of 50 Pa and 3 KV (with a working distance of 8–10 mm) was used to evaluate the samples.
X-Ray Diffraction
To determine the X-ray diffraction (XRD) spectra of the Ag-NPs, this study employed an X-ray diffractometer (XRD-6000, Shimadzu) (containing a copper K-α radiation source) and a 0.154 nm wavelength. The sample holder measured 0.5mm by 2cm.
 |
Figure 1 FT-IR spectra of P. harmala extracts. |
Particle Size Analysis
To measure the particle sizes, this study employed a Malvern Zetasizer Nano ZS90. Ag-NPs were diluted in distilled water and left for five minutes to reach 25 °C and ensure temperature homogeneity. Following this, three independent trials were conducted across a series of duplicate tests. The solution under consideration had a viscosity of 0.8872 cP and a refractive index of 1.59. The testing and investigations were conducted in the Nanotechnology Institute at the Jordan University of Science and Technology.
Identifying Bacterial Isolates
In this work, 11 P. aeruginosa clinical isolates (PA1–PA11) were employed alongside the reference strain identified as PA 27853 (from the American Type Culture Collection (ATCC)).47 The clinical isolates of P. aeruginosa were obtained from adult patients with CF and were provided by Jordan University’s Microbiology Laboratory and the Microbiology Department at Jordan’s Prince Hamza Hospital. Sputum samples were obtained from CF patients and several tests were employed to identify P. aeruginosa bacteria (including the Gram-Stain test, Oxidase test, motility, growth on selective medium-cetrimide agar, formation of green pigments on nutrient agar, growth on MacConkey agar, and the ability to grow at 42°C). The VITEK2 (Bio Merière, Lyon, France) computerized automatic bacterium identification system was used for confirmation.
Culture Conditions
Pseudomonas Cetrimide Agar (Oxoid TM) medium was applied to isolate P. aeruginosa. To enrich the samples, a brain-heart infusion (BHI) broth medium (Oxoid TM) was used and the samples were cultured using Cetrimide Agar and the pour plate and streak plate methods. Pseudomonas Cetrimide Agar plates were cultivated at 37°C using single colonies of each strain as the starting point for subcultures and the samples were incubated aerobically in 5 mL of Luria-Bertani (LB) medium at 37 °C.
Antibacterial Agent
In this research, several pure-powdered antibiotics were employed including Piperacillin, Ciprofloxacin, Levofloxacin, Meropenem, Amikacin, Ceftazidime, Gentamicin, and Aztreonam (Sigma– Aldrich, St. Louis, USA). The powders were weighed and dissolved into corresponding diluents to produce the desired concentrations using the following equation:
Weight (mg) = Volume (mL)*Concentration (μg/mL)/Potency (μg/mg)
This process resulted in the creation of the following stock antibacterial solution: Piperacillin (256 μg/mL), Ciprofloxacin (4 μg/mL), Levofloxacin (8 μg/mL), Meropenem (16 μg/mL), Amikacin (128 μg/mL), Ceftazidime (64 μg/mL), Gentamicin (32 μg/mL), and Aztreonam(64 μg/mL).
Preparing the Ag-NP Suspension
The Ag-NPs/water suspension was freshly prepared before the start of each experiment. Once the NPs were weighed and powdered, 24 milligrams of the powder were added to 100 mL of sterile distilled water (240 μg/mL). The resulting suspension was vortexed vigorously before each use.
Minimum Inhibitory Concentration (MIC)
The MIC of Ag-NPs and antibiotics for each bacterial sample was ascertained via the utilization of the broth microdilution approach (Clinical Laboratory Standards Institute (CLSI 2006, M7-A7)). To initiate broth microdilution, the microtiter plate was prepared by adding 95 μL of MHB to each of the twelve columns (leaving the first column unfilled). Following this, 285 µL of the antibiotic stock solution and the Ag-NPs were applied to the well in the first column on the microtiter plate. A multichannel pipette was used to extract 95 µL from the first column and place it in the second (new tips were used for each dilution step) until the tenth column was reached.
At each stage, the mixing was performed independently via the use of the multichannel pipette. Finally, 10 µL of the bacterial suspension was added to each plate (except for column 12 which served as the negative control and only contained 100 µL MHB to ensure that the media remained sterile at all times). Column 11 served as the positive control to ensure that the growth conditions were optimal for the bacteria. To calculate the MIC, the test bacteria growth in the wells was compared with the positive and negative controls.
To ensure quality control in all of the experimental stages, this study used referential isolated strains PA ATCC 27853. This selection ensured that the reference strains had MIC values that fell within the acceptable range based on CLSI guidelines. Triphenyl tetrazolium chloride (TTC) served as an indicator in the experiments and MIC was established to be the lowest concentration preventing color change.48
Minimal Bactericidal Concentration (MBC) Test for Ag-NPs
The MBC is defined as the minimum bactericidal concentration required to guarantee the eradication of all microbes following contact with an antimicrobial. Following the establishment of the Ag-NP MICs, 50 µL aliquots were inoculated from any well not exhibiting any obvious bacterial growth. Each aliquot was placed on BHI agar plates and incubated at 37°C for 24 hours. The MBC endpoint was established via the eradication of 99.9% of bacteria by the minimal concentration of Ag-NPs.
Synergistic Impacts of Combining Ag-NP with Other Antimicrobial Agents
Using the checkerboard approach, the potential synergistic effects of Ag-NPs and the antimicrobial drugs were assessed against the clinical isolates and ATCC strain. The MIC values were employed to determine the tested concentrations in the checkerboard. Two concentrations above the MIC value were selected to account for the antagonistic effects, and the remaining concentrations accounted for the synergistic and indifference effects. There was 1*105 cfu/mL in the inoculum. In preparation for the tests, antimicrobial solutions were created and freshly diluted and each test was conducted in triplicate.
The methodology of the checkerboard assay is similar to the microdilution approach for calculating MIC values. To facilitate the intersection of concentrations, the antimicrobials and Ag-NPs were diluted in two different directions. The concentration of Ag-NPs and the antibiotics decline in the vertical and horizontal directions, respectively. Subsequently, the identical method was utilized to apply bacterial inoculum to the MIC plate and the untreated media or bacteria in the wells were employed as positive and negative controls.
The plates were incubated at 37°C for 24 hours and the growth of the test strain on the plates was monitored. The combination of the Ag-NPs and antimicrobial agents in which the growth is completely inhibited was considered to be an effective MIC. To confirm the results, bacterial growth was examined after incubation by streaking a loopful from each well in sterile nutrient agar plates.
To interpret the findings of the tests, this research utilized the Fractional Inhibitory Concentration (FIC) index which is a mathematical model that can be applied to assess the effects of drug combinations and identify the existence of any synergistic effects.49
The following equation was used to calculate the ΣFICs (fractional inhibitory concentrations):
∑FIC=FIC A+FIC B
Here, FIC A denotes Drug A’s MIC (antibiotics such as ciprofloxacin) in the combination/MIC of Drug A (antibiotics eg, ciprofloxacin), while FIC B refers to the MIC of Drug B (Ag-NPs) in the combination/MIC of Drug B (Ag-NPs).
To be deemed synergistic, the ΣFIC of the combination must be ≤0.5. Meanwhile, a partial synergistic effect is achieved when the ΣFIC falls between >0.5 to <1. Finally, the effect is considered indifferent if the ΣFIC falls between >1 to <2, and antagonistic if the ΣFIC is ≥2.
Ag-NP Cytotoxicity Assay
The methyl tetrazolium (MTT) test was used to evaluate the cytotoxicity of Ag-NPs at varying concentrations (15–0.225 μg/mL) on the HT29-MTX cell line.50 Although HT29-MTX is derived from colon goblet cells, it secretes the mucin MUC5AC, which is specific to goblet cells in the respiratory system, as opposed to the MUC2 mucin that is specific to goblet cells in the colon. T75 flasks were used to culture the HT29-MTX cells with 10% CO2 at 37°C in 12 mL Dulbecco's Modified Eagles Medium (DMEM, Sigma, UK). Subsequently, 10% FCS was added to the solution (Sigma, UK), as well as 50mg/mL streptomycin (Sigma, UK), 50mg/mL gentamicin (Lonza, USA), 50 µg/mL amphotericin B (Lonza, USA), and 50U/mL penicillin.
The Ag-NPs were added to the solution at concentrations of 7.5–0.225 µg/mL after the HT29 MTX cells had been stored for 24 hours in serum-free DMEM with 50U/mL penicillin, 50mg/mL gentamicin, 50 µg/mL amphotericin B, and 50mg/mL streptomycin.
After removing the medium, the cells were rinsed with PBS, 25 milliliters of MTT solution (5 mg/mL) were added, and the wells were incubated for four hours. Following this, 100 mL of DMSO was added, and the mixture was gently stirred for 15 minutes. The enzyme-linked immunosorbent assay plate reader was utilized to determine the 570 nm absorbance and estimate the viable cell percentage for the Ag-NP-treated and control cells to evaluate the impacts that Ag-NPs have on cell viability. After computing the cell survival rates, the percentage viability was plotted using the relative absorbance equation:
Results
Quantification of Phenolic and Flavonoid Contents
The Folin-Ciocalteu technique quantified the total phenolic content of the samples as 9.31 ± 2.1 mg GAE/100 mg extract. The spectrophotometric examination, consisting of the colorimetric aluminum chloride test, revealed that the samples had a total flavonoid concentration of 0.81 ± 0.43 mg QE / 100 mg extract.
Fourier-Transform Infrared Spectroscopy (FT-IR)
To ascertain the interactions between Ag atoms and bioactive chemicals which affect the stability of Ag-NPs, this study employed FT-IR which revealed the existence of parallel FT-IR spectra peaks and a minor shift in the P. harmala extract (Figure 1) and the biosynthesized Ag-NPs (Figure 2) as shown in Table 1.
 |
Table 1 FTIR Peak Values of P. Harmala Seed Extract and Biosynthesized Ag-NPs with the Functional Groups of Their Active Components, Which are Identified Based on Peak Values |
 |
Figure 2 FT-IR spectra of biosynthesized AgNPs. |
 |
Figure 3 Green synthesis of Ag-NPs, a change in the color of reaction mixture from Orange to dark brown after addition of seedsextract into AgNO3 solution. |
Ag-NPs Characterization
When synthesizing the NPs, the formation of Ag-NP instigates a color change in the reaction mixture from orange to dark brown which implies that there is a reduction in AgNO3 from SPR excitation in the Ag-NPs (Figure 3). A UV-Vis spectrophotometer with a wavelength ranging from 200–800 nm was utilized to measure the SPR of the Ag-NPs and revealed that the Ag-NPs SPR occurs between 450 and 500 nm. Figure 4 illustrates that the UV-Vis spectrum has a significant absorption peak of approximately 460 nm.
 |
Figure 4 Ag-NPs characterization using ultraviolet-visible spectroscopy. |
 |
Figure 5 SEM image (x 500(A1), x300(A2) and x200(A3) magnification) of Ag-NP morphology with histogram (B) of Ag-NPs particle size distribution. |
SEM was used to evaluate the size and surface shape of the Ag-NPs (Figure 5A1-A3) and revealed that they are approximately 15 nm in diameter, uniformly spherical and do not aggregate. The Ag-NP size histograms (Figure 5B) indicate that the primary particle size for the Ag-NPs in the present research was 13.6 ± 2.6 nm.
 |
Figure 6 X-ray diffraction presenting crystalline phases of the synthesized Ag-NPs. |
To examine the crystalline formations of the Ag-NPs, the specimens were placed at an ambient temperature of 298 Kelvin (24.85°C) and copper K-α radiation with a wavelength of 1.5406 Å in the (2θ) range was applied. As illustrated by Figure 6, the Ag-NPs displayed four peaks at the 2θ values of 38.2901, 44.5583, 64.8185, and 77.4383. These values (111, 200, 220, and 311, respectively) were taken to represent the Ag metal and the related Ag plane values for the HKL. The diffractogram (Figure 6) and the JCPDS standard power diffraction card (silver file No. 04–0783) were compared. The face-centered cubic structure of the Ag-NPs is confirmed by the diffraction resultant pattern. When calculated using the Scherrer equation,51 the Ag-NPs’ predicted particle size was found to be 15.9 nm.
After conducting the DLS analysis, the average particle sizes for the Ag-NPs were 21 nm (Figure 7). The electrical potential that develops at the solid-liquid interface as a reaction to the relative movements of the solvent and Ag-NPs is known as the zeta potential. The resultant A-NPs were dissolved in double-distilled water which facilitated an examination of the zeta potential. Ag-NP stability is contingent upon both surface charge and electrical potential. As the zeta potential rises, so does the surface potential of a charged particle. As illustrated by Figure 8, the prepared Ag-NPs have a zeta potential of −25.3 Mv.
 |
Figure 7 Dynamic light scattering (DLS) spectrum presenting the distribution of particle sizes in the Ag-NPs. |
 |
Figure 8 Zeta potential analysis of Ag-NPs. |
MIC of Ag-NP
Following a 24-hour aerobic incubation period at 37°C with Ag-NPs at concentrations varying between 0.93–240 μg/mL, turbidity was observed in the samples containing between 0.93 to 7.5 µg/mL of Ag-NPs. With these Ag-NP concentrations, bacteria were thriving (as evidenced by the turbidity); however, at doses between 15 and 240 µg/mL, no turbidity was visible, implying that bacterial growth had been suppressed.
For 24 hours, BHI agar plates were inoculated with the suspensions (ranging from 15 to 240 µg/mL) and no bacterial growth was observed in any of the concentrations which confirms the bactericidal properties of the Ag-NPs (Figure 9). The results indicated a MIC and MBC of 15 µg/mL Ag-NPs for all P. aeruginosa strains.
MIC of the Selected Antibiotics Against P. aeruginosa Clinical Isolates and ATCC 2785 Strains
In this research, the minimum inhibitory concentrations of eight antibiotics and Ag-NPs were compared with those of 11 clinically isolated P. aeruginosa strains and the P. aeruginosa ATCC 2785 strain (Table 2). For seven isolated P. aeruginosa strains (PA2, PA3, PA4, PA5, PA7, PA9 and PA10) and the ATCC, the MIC values of Piperacillin were identified as highly effective. Conversely, strains PA1, PA6, PA8 and PA11 were found to have intermediate sensitivity and exhibited a MIC value of 32 ug/mL with SD ± 0. Good sensitivity to Levofloxacin was evident for seven strains (ATCC, PA1, PA2, PA7, PA8, PA9 and PA11) and strains PA4 and PA6 had an intermediate sensitivity at a MIC of 2 ug/mL. Additionally, it should be noted that PA3, PA5 and PA10 were found to be resistant to Levofloxacin with a MIC of 4 ug/mL.
The strains PA3, PA4, PA5, PA6, and PA10 were found to be resistant to Ciprofloxacin and to have a MIC of 2 ug/mL. However, all of the strains under investigation (except for PA5 and PA11) exhibited sensitivity to Ceftazidime and Amikacin. The results also revealed that Aztreonam was effective against all strains except PA9 (intermediate) and PA10 (resistance) and that three clinical strains were resistant to Meropenem (PAPA3, PA10 and PA11).
Impact of Antibiotic and Ag-NP Combinations on P. aeruginosa
When combined with Levofloxacin, Ceftazidime, and Piperacillin, Ag-NP produced statistically significant reductions (F (11, 48) = 32.85, P<0.0001) in the amount of antibiotics required to completely prevent P. aeruginosa growth for all strains examined by this research (Figure 10). This significant reduction in antibiotic concentration that completely inhibited the growth of the P. aeruginosa strain was also observed with other antibiotics for the majority of strains examined by this study.
 |
Figure 10 MIC of the eight anti-pseudomonal antibiotic in the presence of Ag-NPs. ****P<0.0001, ***P<0.0001, **P<0.001, *P<0.01, ns: not statistically significant. |
In eight of the studied strains (ATCC, PA1, PA2, PA4, PA6–9), combining Meropenem with Ag-NPs did not cause a statistically significant reduction in the MIC of Meropenem. Additionally, Figure 10 demonstrates that the MIC of Amikacin combined with Ag-NPs was statistically significantly lower for the ATCC strain and clinical strains PA3, PA5-7, and PA10-11.
Figure 11 shows that the reduction in concentration of Ag-NPs required to prevent P. aeruginosa growth when used in combination with antibiotics was statistically significant. Additionally, these findings reveal that the MIC of Ag-NPs was 15 ug/mL for all strains and decreased substantially when administered with antibiotics (at a dose of 1.875–7.5 ug/mL).
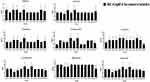 |
Figure 11 MIC of the Ag-NPs against P. aeruginosa strains in the presence of Ag-NPs. |
Fractional Inhibitory Concentration Index (FICI) of Ag NPs When Combined with Antibiotics
In the present study, the impacts of Ag-NPs when combined with selected antibiotics were evaluated via the checkerboard assay and the extent of the interactions between the Ag-NPs was examined to calculate the FIC index.
Table 3 demonstrates that some Ag-NP combinations exhibit either partial synergistic (0.5 < FICI < 1) or full synergistic (FIC ≤ 0.5) effects when employed against the tested bacteria. This was particularly noticeable in the combinations of Meropenem, Ciprofloxacin, and Aztreonam, in which the FIC index was less than or equal to 0.5 (in the majority of strains). Only the combination of Ceftazidime and Ag-NPs demonstrated indifferent effects on three strains of P. aeruginosa. The findings revealed that combining Ag-NPs with antibiotics was more effective than using Ag-NPs or antibiotics alone.
 |
Table 3 Fractional Inhibitory Concentration (FICI) for Combination of Ag-NPs with Antibiotics |
Cytotoxicity of Ag-NPs
To perform the initial screenings of Ag-NP cytotoxicity on the HT29-MTX cancer cell lines, MTT assays were employed. HT29-MTX cell lines were found to be resistant to Ag NPs with 1.8 ug/mL concentrations, and no statistically significant declines in cell viability could be identified (F (6, 14) = 0.2871, P=0.9334). However, as shown in Figure 12, there was a significant decrease in cell viability when the Ag-NP concentration exceeded 7.5 µg/mL.
Discussion
Antibiotic resistance is an acute global health concern that adversely impacts the efficacy of different medications.52 Drug resistance compels the use of high doses of antibiotics which results in intolerable toxicity, the need for new antibiotic research, and time, labor, and financial investments.53,54 If current trends continue, by 2050, it is anticipated that antibiotic failure will cause ten million deaths per year.55
The use of NPs is one of the most promising methods for combating microbial resistance.56 NP resistance requires numerous simultaneous gene alterations in a single microbial cell and this is unlikely to occur.57 This study presents new data regarding the effects of Ag-NP on clinical isolates of P. aeruginosa and how NPs interact with widely administered antimicrobials to inhibit P. aeruginosa.
In this study, P. harmala seed extract was used to biosynthesize Ag-NPs via green synthesis. This process facilitates the cost-effective supply of capping and reducing agents which are employed to stabilize the generated NPs. It also enabled the application of a green synthesis strategy that is ecologically friendly and minimizes hazardous byproducts and chemical waste materials.58,59
Phenolic compounds and flavonoids were found to be present in the P. harmala extract. Phenolic compounds are secondary metabolites that have redox potentials that enable them to scavenge radicals and perform antioxidant actions.43 Consequently, they can be employed as reducing agents, singlet oxygen quenchers and hydrogen donors.60 This study demonstrated that P. harmala contains both phenolic and flavonoid content and previous studies revealed that the P. harmala extract contains flavonoids, saponins, tannins, compound reducers, volatile oils, anthraquinones, triterpenes, sterols, and alkaloids.61 Several of these chemical compounds have been linked to therapeutic effects.62,63 The variety of phytochemical components that plants contain reflects their almost limitless ability to produce aromatic molecules and their by-products. Many traditional healers have employed the P harmala due to its antiseptic and pain-relieving qualities, and its ability to combat bacterial infections.64
P harmala extract was used in this work to green-synthesize Ag-NPs, and it served as both a reducing and stabilizing agent.65 The color change (from bright orange to a reddish-brown) of the silver nitrate solution indicates the development of NPs and this finding aligns with those of previous studies.22,23 Furthermore, UV-visible spectroscopy was performed to confirm the production of NPs.66 The Ag-NP exhibited absorption peaks of approximately 460 which is relevant to the SPR of Ag-NPs. The findings are in line with those of Singh, Kim, Singh, Wang, Hwang, Farh and Yang67 and Pryshchepa, Pomastowski and Buszewski,68 who noted that Ag-NPs exhibited spectral bands in the range of 450 to 500 nm. This research compared its findings with these earlier studies and found that both Ag-NPs were successfully created. There is a clear and robust correlation between the XRD peaks and the crystalline structure of Ag-NPs and the XRD findings in this study emphasize the purity of the Ag-NP production process. The positions of the diffraction peaks formed a pattern identical to that of the JCPDS.
The values of NPs’ zeta potential control their stability.69 These numbers indicate that the NPs’ surface is negatively charged which enables them to scatter in the medium with a high degree of stability. Tran, Pham, Luu, Nguyen and Nguyen70 state that particles with zeta potential values between +35 and −35 mV are categorized as stable suspensions which indicates that there was stability among the NPs in this study. By generating an electric repulsion force between molecules with similar negative charges, the presence of negative charges inhibits the agglomeration of particles.71 The particles in their colloidal solution are stabilized by this force.72 The generation of NPs and the confirmation of their roughly spherical form and consistent dimensions were corroborated by the SEM data.
The MIC values obtained during the broth microdilution indicate that the Ag-NPs created via this green synthesis methodology possess strong anti-bacterial effects. According to the findings revealed in this work, Ag-NPs were found to be biocidal and effective against all strains of P. aeruginosa at 15 ug/mL. Ag-NPs have been shown to exhibit potent antibacterial activity in numerous studies.23,73,74 This is due to the small particle size producing a high surface-to-volume ratio that makes it easier for NPs to pass through membranes and disrupt bacterial metabolism and cell structure.75
It is recognized that Ag-NPs have significant antibacterial properties by penetrating the cell membrane, inducing oxidative stress, and harming proteins, carbohydrates, lipids, and DNA.76 Additionally, lipid peroxidation can alter the cell membrane which can ultimately interfere with essential cellular processes.77 Moreover, the Ag+ ions already present in Ag-NPs have the potential to breach the cell walls of microorganisms, disable the efflux pump, and leak cytoplasm into the intercellular space through the cell wall. As a result of these events, cell death and lysis occur.78
Ag-NPs possess enormous potential for use as antibacterial agents; however, there are uncertainties regarding their safety when administered to human subjects.79 Ag-NPs’ cytotoxic potential has made it difficult for them to become established as effective chemotherapeutic agents.79,80 Using the neutral red absorption assay, Siddique, Aslam, Imran, Ashraf, Nadeem, Hayat, Khurshid, Afzal, Malik and Shahzad81 investigated the hazardous potential of Ag-NPs against HeLa cell lines at different doses. Their findings indicated that these NPs were hazardous at certain concentrations (ie, between 15 and 120 μg/mL). Furthermore, research evaluating the effects of Ag-NPs on human cell culture in vitro indicates that the hazardous concentrations of Ag-NPs vary between 10 and 100 µg/mL.82–84 These findings are in line with this research which revealed that Ag-NP had significant potential to reduce viable cell numbers in the HT29-MTX cell line at a concentration of 7.5–15 μg/mL implying that the Ag-NPs may be safe if applied in low concentrations. It is possible to reduce the concentration of Ag-NPs to generate antimicrobial effects by combining Ag-NP with an antibiotic.
According to recent research, combining antibiotics with NPs not only improves the antibacterial qualities of the agents but minimizes their toxicity to human cells by reducing the concentrations required for effect.85 The results of the current research indicate that the capacity of antibiotics to eliminate drug-resistant bacteria is restored when they are combined with Ag-NPs. It has also been demonstrated that using NPs in conjunction with antibiotics increases the concentration of the antibiotics at the region of the antibiotic-bacterium interaction and assists the antibiotics in binding with bacteria.86
Several of the antibiotics studied in this work demonstrated superior potential when combined with Ag-NPs. The findings of this work indicate that combining antibiotics with Ag-NPs can have either a synergistic or partially synergistic effect. When Ag-NPs are present, antibiotic activity increases due to the inhibition of antibiotic efflux from the cell which results from the impact that Ag-NPs have on the MexAB-OprM efflux pumps in P. aeruginosa.87 This process activates the antibiotic uptake by stimulating the activity of the Omf protein membrane and causing a binding reaction between antibiotic agents and Ag-NPs stabilizing the antibiotic-Ag-NP complex.86,88 The presence of the Carboxyl and Fluore groups in some antibiotics (such as Ciprofloxacin) can facilitate this process and ciprofloxacin proved to be the most effective antibiotic to combine with the Ag-NPs. The Fluore group present in some antibiotics (including ciprofloxacin) may also interact with the chelating Ag atom which stabilizes the antibiotic-NP combination. Combining the Ag-NPs with an antibiotic may increase the production of the reactive oxygen species (ROS) which makes Gram-negative bacteria more permeable.89 Additionally, Ag-NPs can initiate reactions between oxygen and dehydrogenase enzymes which increases the antimicrobial activity of the antibiotics.90
Moreover, active groups of hydroxyl and amino groups are present in some antibiotic molecules (eg, Levofloxacin) and may react well with Ag-NPs to disrupt the processing of peptidoglycan in the cell wall. When combined with positively charged Ag-NPs, cell-wall inhibitors can disrupt or prevent cell-wall synthesis.91 Additionally, researchers have suggested that silver ions may penetrate the cell and intercalate between pyrimidine and purine which causes the DNA molecule to denature.92 Under the theory proposed by Mikhailova,92 NPs disrupt peptidoglycan in the cell wall by bonding with the cell wall inhibitors. They can damage the cell membrane and obstruct transport channels because they are positively charged and attack the negative charges in the transmembrane proteins.93 They can infiltrate the bacterium and interfere with cellular processes such as protein synthesis, transportation, and nucleic acid function.94
Agreles, Cavalcanti and Cavalcanti95 have reported on the synergistic potential of Ag-NP and how their small size may be critical in minimizing the antibiotic burden of patients with chronic conditions. The same study92 indicates that small citrate-capped Ag-NPs can improve the antimicrobial effects of aztreonam when used to combat P. aeruginosa synergistically. Additionally, when used to treat P. aeruginosa, E. coli, S. aureus, Klebsiella pneumoniae, Micrococcus luteus, and Bacillus spp, the synergistic effect of antibiotics (imipenem, ciprofloxacin, gentamicin, and vancomycin) used in combination with biologically synthesized Ag-NPs was described.93 The results of the current research align with those of Birla, Tiwari, Gade, Ingle, Yadav and Rai96 who reported that Ag-NPs combined with antibiotics such as vancomycin, gentamicin, streptomycin, ampicillin, and kanamycin had increased efficacies in fighting E. coli, P. aeruginosa, and S. Aureus. The antibacterial properties of biologically synthesized Ag-NPs combined with ceftriaxone and ofloxacin were enhanced when used against several species of Salmonella.97
To conclude, an alternative methodology is urgently required to treat P. aeruginosa infections that are resistant to traditional antibiotics. The current research project aims to investigate the impacts of combining certain antibiotics and Ag-NPs to treat strains of P. aeruginosa ATCC 27853 and other clinically isolated bacterial strains. To the best of this author’s knowledge, the data regarding clinical isolates is unique and has not been reported previously. Ag-NPs and most anti-Pseudomonas drugs demonstrated a partial or partial synergistic effect which emphasizes the potential for the combined use of antibiotics and Ag-NPs as a substitute for maintaining and restoring bacterial susceptibility to these medications. According to this study, biosynthesized Ag-NPs may have the ability to work collaboratively with antibiotics to boost their antibacterial efficacy against P. aeruginosa. Additionally, these findings suggest that these combinations may be useful in reversing drug resistance in bacteria which is significant for two reasons: firstly, the concentration of Ag-NPs used to treat the infection can be reduced which limits their known cytotoxic effects and secondly, reducing the concentration of antibiotics required to effectively treat biofilm-bound microorganisms will reduce the development of additional antibiotic resistance. This study could generate new insights regarding the in vivo and in vitro antibacterial activities of antibiotics, which, when combined with metallic nanomaterials, can selectively treat MDR pathogens. These findings are extremely promising, particularly for treatment purposes; however, additional research is required to evaluate the specific mechanisms of action which may be critical in predicting undesirable interactions.
Data Sharing Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.
Acknowledgment
H Al-momani received financial support from the Department of scientific research at The Jordanian Hashemite University (Grant number 2023/81/941). B.A.Albiss acknowledged the deanship of research at Jordan University of Science and Technology for their financial support.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Alhazmi A. Pseudomonas aeruginosa–pathogenesis and pathogenic mechanisms. Int J Biol. 2015;7(2):44–67. doi:10.5539/ijb.v7n2p44
2. Pressler T, Bohmova C, Conway S, et al. Chronic Pseudomonas aeruginosa infection definition: euroCareCF working group report. J Cyst Fibros. 2011;10:S75–S78. doi:10.1016/S1569-1993(11)60011-8
3. Talwalkar JS, Murray TS. The approach to Pseudomonas aeruginosa in cystic fibrosis. Clinics Chest Med. 2016;37(1):69–81. doi:10.1016/j.ccm.2015.10.004
4. Almughem FA, Aldossary AM, Tawfik EA, et al. Cystic fibrosis: overview of the current development trends and innovative therapeutic strategies. Pharmaceutics. 2020;12(7):616. doi:10.3390/pharmaceutics12070616
5. Sosnay PR, Raraigh KS, Gibson RL. Molecular genetics of cystic fibrosis transmembrane conductance regulator: genotype and phenotype. Pediatr Clinics. 2016;63(4):585–598. doi:10.1016/j.pcl.2016.04.002
6. Cantin AM, Hartl D, Konstan MW, Chmiel JF. Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J Cyst Fibros. 2015;14(4):419–430. doi:10.1016/j.jcf.2015.03.003
7. Turcios NL. Cystic fibrosis lung disease: an overview. Respiratory Care. 2020;65(2):233–251. doi:10.4187/respcare.06697
8. Moghoofei M, Azimzadeh Jamalkandi S, Moein M, Salimian J, Ahmadi A. Bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a systematic review and meta-analysis. Infection. 2020;48(1):19–35. doi:10.1007/s15010-019-01350-1
9. Rodrigo‐Troyano A, Suarez‐Cuartin G, Peiró M, et al. Pseudomonas aeruginosa resistance patterns and clinical outcomes in hospitalized exacerbations of COPD. Respirology. 2016;21(7):1235–1242. doi:10.1111/resp.12825
10. Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109–119. doi:10.1016/j.gendis.2019.04.001
11. Thi MTT, Wibowo D, Rehm BH. Pseudomonas aeruginosa biofilms. Int J Mol Sci. 2020;21(22):8671. doi:10.3390/ijms21228671
12. Maurice NM, Bedi B, Sadikot RT. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol. 2018;58(4):428–439. doi:10.1165/rcmb.2017-0321TR
13. Mlynarcik P, Kolar M. Starvation-and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2017;161(1).
14. Santi I, Manfredi P, Maffei E, Egli A, Jenal U. Evolution of antibiotic tolerance shapes resistance development in chronic Pseudomonas aeruginosa infections. MBio. 2021;12(1). doi:10.1128/mBio.03482-20
15. Mercan D-A, Niculescu A-G, Grumezescu AM. Nanoparticles for antimicrobial agents delivery—an up-to-date review. Int J Mol Sci. 2022;23(22):13862. doi:10.3390/ijms232213862
16. Sharmin S, Rahaman MM, Sarkar C, Atolani O, Islam MT, Adeyemi OS. Nanoparticles as antimicrobial and antiviral agents: a literature-based perspective study. Heliyon. 2021;7(3):e06456. doi:10.1016/j.heliyon.2021.e06456
17. Imani SM, Ladouceur L, Marshall T, Maclachlan R, Soleymani L, Didar TF. Antimicrobial nanomaterials and coatings: current mechanisms and future perspectives to control the spread of viruses including SARS-CoV-2. ACS nano. 2020;14(10):12341–12369. doi:10.1021/acsnano.0c05937
18. Khina A, Krutyakov YA. Similarities and differences in the mechanism of antibacterial action of silver ions and nanoparticles. Appl Biochem Microbiol. 2021;57(6):683–693. doi:10.1134/S0003683821060053
19. Gold K, Slay B, Knackstedt M, Gaharwar AK. Antimicrobial activity of metal and metal‐oxide based nanoparticles. Adv Ther. 2018;1(3):1700033. doi:10.1002/adtp.201700033
20. Kalwar K, Shan D. Antimicrobial effect of silver nanoparticles (AgNPs) and their mechanism–a mini review. Micro Nano Lett. 2018;13(3):277–280. doi:10.1049/mnl.2017.0648
21. Bates MG, Risselada M, Peña-Hernandez DC, Hendrix K, Moore GE. Antibacterial activity of silver nanoparticles against Escherichia coli and methicillin-resistant staphylococcus pseudintermedius is affected by incorporation into carriers for sustained release. Am J Vet Res. 2024;85(3). doi:10.2460/ajvr.23.10.0229
22. Al-Momani H, Al Balawi D, Hamed S, et al. The impact of biosynthesized ZnO nanoparticles from Olea europaea (Common Olive) on Pseudomonas aeruginosa growth and biofilm formation. Sci Rep. 2023;13(1):5096. doi:10.1038/s41598-023-32366-1
23. Al-Momani H, Almasri M, Al Balawi DA, et al. The efficacy of biosynthesized silver nanoparticles against Pseudomonas aeruginosa isolates from cystic fibrosis patients. Sci Rep. 2023;13(1):8876. doi:10.1038/s41598-023-35919-6
24. Abou-Dobara MI, Kamel MA, El-Sayed AK, El-Zahed MM. Antibacterial activity of extracellular biosynthesized iron oxide nanoparticles against locally isolated β-lactamase-producing Escherichia coli from Egypt. Discover Applied Sciences. 2024;6(3):113. doi:10.1007/s42452-024-05770-z
25. Waseem M, Naveed M, Rehman S, et al. Molecular characterization of spa, hld, fmhA, and l ukD Genes and computational modeling the multidrug resistance of staphylococcus species through Calliandra Harrisii silver nanoparticles. ACS omega. 2023;8(23):20920–20936. doi:10.1021/acsomega.3c01597
26. Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arabian J Chem. 2019;12(7):908–931. doi:10.1016/j.arabjc.2017.05.011
27. Singh R, Shedbalkar UU, Wadhwani SA, Chopade BA. Bacteriagenic silver nanoparticles: synthesis, mechanism, and applications. App. Microbio. Biotechnol. 2015;99(11):4579–4593. doi:10.1007/s00253-015-6622-1
28. Kaweeteerawat C, Na Ubol P, Sangmuang S, Aueviriyavit S, Maniratanachote R. Mechanisms of antibiotic resistance in bacteria mediated by silver nanoparticles. J Toxicol Environ Health Part A. 2017;80(23–24):1276–1289. doi:10.1080/15287394.2017.1376727
29. Mateo EM, Jiménez M. Silver nanoparticle-based therapy: can it be useful to combat multi-drug resistant bacteria? Antibiotics. 2022;11(9):1205. doi:10.3390/antibiotics11091205
30. Panáček A, Smékalová M, Kilianová M, et al. Strong and nonspecific synergistic antibacterial efficiency of antibiotics combined with silver nanoparticles at very low concentrations showing no cytotoxic effect. Molecules. 2015;21(1):26. doi:10.3390/molecules21010026
31. Rónavári A, Igaz N, Adamecz DI, et al. Green silver and gold nanoparticles: biological synthesis approaches and potentials for biomedical applications. Molecules. 2021;26(4):844. doi:10.3390/molecules26040844
32. Huq MA, Akter S. Biosynthesis, characterization and antibacterial application of novel silver nanoparticles against drug resistant pathogenic Klebsiella pneumoniae and Salmonella enteritidis. Molecules. 2021;26(19):5996. doi:10.3390/molecules26195996
33. Tsekhmistrenko S, Bityutskyy V, Tsekhmistrenko O, Horalskyi L, Tymoshok N, Spivak M. Bacterial synthesis of nanoparticles: a green approach. Biosystems Diversity. 2020;28(1):9–17. doi:10.15421/012002
34. Alavi M. Bacteria and fungi as major bio-sources to fabricate silver nanoparticles with antibacterial activities. Exp Rev Anti-Infective Ther. 2022;20(6):897–906. doi:10.1080/14787210.2022.2045194
35. Adeyemi JO, Oriola AO, Onwudiwe DC, Oyedeji AO. Plant extracts mediated metal-based nanoparticles: synthesis and biological applications. Biomolecules. 2022;12(5):627. doi:10.3390/biom12050627
36. Jeevanandam J, Kiew SF, Boakye-Ansah S, et al. Green approaches for the synthesis of metal and metal oxide nanoparticles using microbial and plant extracts. Nanoscale. 2022;14(7):2534–2571. doi:10.1039/D1NR08144F
37. Gebre SH. Bio-inspired synthesis of metal and metal oxide nanoparticles: the key role of phytochemicals. J Clust Sci. 2023;34(2):665–704. doi:10.1007/s10876-022-02276-9
38. Ritu VKK, Das A, Chandra P. Phytochemical-based synthesis of silver nanoparticle: mechanism and potential applications. BioNanoScience. 2023;13(3):1359–1380. doi:10.1007/s12668-023-01125-x
39. Ahmad S, Munir S, Zeb N, et al. Green nanotechnology: a review on green synthesis of silver nanoparticles—an ecofriendly approach. Int j Nanomed. 2019;14:5087. doi:10.2147/IJN.S200254
40. Keat CL, Aziz A, Eid AM, Elmarzugi NA. Biosynthesis of nanoparticles and silver nanoparticles. Bioresources Bioprocess. 2015;2(1):1–11. doi:10.1186/s40643-015-0076-2
41. Atrooz OM, Wietrzyk J, Filip-Psurska B, Al-Rawashdeh I, Soub M, Abukhalil MH. ANTIPROLIFERATIVE, ANTIOXIDANT, AND ANTIBACTERIAL ACTIVITIES OF CRUDE PLANT EXTRACTS OF ASPHODELINE LUTEA L. AND PEGANUM HARMALA L. World J Pharm Res. 2018;7(11):148–167.
42. Sharma P, Goyal D, Chudasama B. Nanostrategies against rising antimicrobial resistance (AMR)-metallic nanoparticles as nanoweapon. In: Emerging Modalities in Mitigation of Antimicrobial Resistance. Springer; 2022:541–561.
43. Vuolo MM, Lima VS, Junior MRM. Phenolic compounds: structure, classification, and antioxidant power. Bioactive Compounds Elsevier. 2019:33–50.
44. Singleton VL, Orthofer R, Lamuela-Raventós RM. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymolo. 1999:152–178.
45. Singleton VL, Rossi JA. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Enol Vitic. 1965;16(3):144–158. doi:10.5344/ajev.1965.16.3.144
46. Pękal A, Pyrzynska K. Evaluation of aluminium complexation reaction for flavonoid content assay. Food Anal Methods. 2014;7(9):1776–1782. doi:10.1007/s12161-014-9814-x
47. Stover C, Pham X, Erwin A, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959. doi:10.1038/35023079
48. Stockert JC, Horobin RW, Colombo LL, Blázquez-Castro A. Tetrazolium salts and formazan products in cell biology: viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018;120(3):159–167. doi:10.1016/j.acthis.2018.02.005
49. Hall M, Middleton R, Westmacott D. The fractional inhibitory concentration (FIC) index as a measure of synergy. J Antimicrob Chemother. 1983;11(5):427–433. doi:10.1093/jac/11.5.427
50. Fotakis G, Timbrell JA. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett. 2006;160(2):171–177.
51. Dorofeev G, Streletskii A, Povstugar I, Protasov A, Elsukov E. Determination of nanoparticle sizes by X-ray diffraction. Colloid J. 2012;74(6):675–685. doi:10.1134/S1061933X12060051
52. Talebi Bezmin Abadi A, Rizvanov AA, Haertlé T, Blatt NL. World Health Organization report: current crisis of antibiotic resistance. BioNanoScience. 2019;9(4):778–788. doi:10.1007/s12668-019-00658-4
53. Banin E, Hughes D, Kuipers OP. Bacterial pathogens, antibiotics and antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):450–452. doi:10.1093/femsre/fux016
54. Chinemerem Nwobodo D, Ugwu MC, Oliseloke Anie C, et al. Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace. J Clin Lab Analysis. 2022;36(9):e24655. doi:10.1002/jcla.24655
55. Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10(10):1310. doi:10.3390/pathogens10101310
56. Munir MU, Ahmed A, Usman M, Salman S. Recent advances in nanotechnology-aided materials in combating microbial resistance and functioning as antibiotics substitutes. Int j Nanomed. 2020;Volume 15:7329–7358. doi:10.2147/IJN.S265934
57. Gómez-Núñez MF, Castillo-López M, Sevilla-Castillo F, et al. Nanoparticle-based devices in the control of antibiotic resistant bacteria. Front Microbiol. 2020;11:563821. doi:10.3389/fmicb.2020.563821
58. Salem SS, Fouda A. Green synthesis of metallic nanoparticles and their prospective biotechnological applications: an overview. Biol Trace Elem Res. 2021;199(1):344–370. doi:10.1007/s12011-020-02138-3
59. Dikshit PK, Kumar J, Das AK, et al. Green synthesis of metallic nanoparticles: applications and limitations. Catalysts. 2021;11(8):902.
60. Olszowy M. What is responsible for antioxidant properties of polyphenolic compounds from plants? Plant Physiol Biochem. 2019;144:135–143. doi:10.1016/j.plaphy.2019.09.039
61. Benbott A, Bahri L, Boubendir A, Yahia A. Study of the chemical components of Peganum harmala and evaluation of acute toxicity of alkaloids extracted in the Wistar albino mice. J Mater Environ Sci. 2013;4:558–565.
62. Moloudizargari M, Mikaili P, Aghajanshakeri S, Asghari MH, Shayegh J. Pharmacological and therapeutic effects of Peganum harmala and its main alkaloids. Pharmacogn revi. 2013;7(14):199. doi:10.4103/0973-7847.120524
63. Miraj S. A review study of therapeutic effects of Peganum harmala. seeds. 2016;4:8.
64. Kutama RM, Abdulkadir S, Kwalli SA, Chiroma G. Phytochemical compositions in some Nigerian medicinal plants and their pharmacological properties: a review. J Anesthesiol. 2018;6(1):15–25. doi:10.11648/j.ja.20180601.14
65. Senhaji S, Lamchouri F, Boulfia M, Lachkar N, Bouabid K, Toufik H. Mineral composition, content of phenolic compounds and in vitro antioxidant and antibacterial activities of aqueous and organic extracts of the seeds of Peganum harmala L. S Afr J Bot. 2022;147:697–712. doi:10.1016/j.sajb.2022.03.005
66. Desai R, Mankad V, Gupta SK, Jha PK. Size distribution of silver nanoparticles: UV-visible spectroscopic assessment. Nanosci Nanotechnol Lett. 2012;4(1):30–34. doi:10.1166/nnl.2012.1278
67. Singh P, Kim YJ, Singh H, et al. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int j Nanomed. 2015;10:2567–2577. doi:10.2147/IJN.S72313
68. Pryshchepa O, Pomastowski P, Buszewski B. Silver nanoparticles: synthesis, investigation techniques, and properties. Adv Colloid Interface Sci. 2020;284:102246. doi:10.1016/j.cis.2020.102246
69. Kumar A, Dixit CK. Methods for characterization of nanoparticles. advances in nanomedicine for the delivery of therapeutic nucleic acids. Elsevier. 2017:43–58.
70. Tran QT, Pham VH, Luu MQ, Nguyen HL, Nguyen HH. Preparation and properties of silver nanoparticles by heat-combined electrochemical method. VNU Journal of Science. 2015;31(2).
71. Shrestha S, Wang B, Dutta P. Nanoparticle processing: understanding and controlling aggregation. Adv Colloid Interface Sci. 2020;279:102162. doi:10.1016/j.cis.2020.102162
72. Bhattacharjee S. DLS and zeta potential–what they are and what they are not? J Control Release. 2016;235:337–351. doi:10.1016/j.jconrel.2016.06.017
73. Tang S, Zheng J. Antibacterial activity of silver nanoparticles: structural effects. Adv Healthcare Mater. 2018;7(13):1701503. doi:10.1002/adhm.201701503
74. Das CA, Kumar VG, Dhas TS, et al. Antibacterial activity of silver nanoparticles (biosynthesis): a short review on recent advances. Biocatal Agri. Biotechnol. 2020;27:101593. doi:10.1016/j.bcab.2020.101593
75. Lv H, Cui S, Yang Q, et al. AgNPs-incorporated nanofiber mats: relationship between AgNPs size/content, silver release, cytotoxicity, and antibacterial activity. Mater Sci Eng C. 2021;118:111331. doi:10.1016/j.msec.2020.111331
76. Sofi MA, Sunitha S, Sofi MA, Pasha SK, Choi D. An overview of antimicrobial and anticancer potential of silver nanoparticles. J King Saud Univ Sci. 2022;34(2):101791. doi:10.1016/j.jksus.2021.101791
77. Xiang -Q-Q, Wang D, Zhang J-L, et al. Effect of silver nanoparticles on gill membranes of common carp: modification of fatty acid profile, lipid peroxidation and membrane fluidity. Environ Pollut. 2020;256:113504. doi:10.1016/j.envpol.2019.113504
78. Kaiser KG, Delattre V, Frost VJ, et al. Nanosilver: an old antibacterial agent with great promise in the fight against antibiotic resistance. Antibiotics. 2023;12(8):1264. doi:10.3390/antibiotics12081264
79. Noga M, Milan J, Frydrych A, Jurowski K. Toxicological aspects, safety assessment, and green toxicology of silver nanoparticles (AgNPs)—critical review: state of the art. Int J Mol Sci. 2023;24(6):5133. doi:10.3390/ijms24065133
80. Alshameri AW, Owais M. Antibacterial and cytotoxic potency of the plant-mediated synthesis of metallic nanoparticles Ag NPs and ZnO NPs: a review. OpenNano. 2022;8:100077. doi:10.1016/j.onano.2022.100077
81. Siddique MH, Aslam B, Imran M, et al. Effect of silver nanoparticles on biofilm formation and EPS production of multidrug-resistant Klebsiella pneumoniae. Biomed Re Int. 2020;2020:1–9. doi:10.1155/2020/6398165
82. Marin S, Mihail Vlasceanu G, Elena Tiplea R, et al. Applications and toxicity of silver nanoparticles: a recent review. Curr Top Med Chem. 2015;15(16):1596–1604. doi:10.2174/1568026615666150414142209
83. Jaswal T, Gupta J A review on the toxicity of silver nanoparticles on human health.
84. Salomoni R, Léo P, Montemor A, Rinaldi B, Rodrigues M. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol Sci Appl. 2017;10:115. doi:10.2147/NSA.S133415
85. Abo-Shama UH, El-Gendy H, Mousa WS, et al. Synergistic and antagonistic effects of metal nanoparticles in combination with antibiotics against some reference strains of pathogenic microorganisms. Infect Drug Resist. 2020;Volume 13:351–362. doi:10.2147/IDR.S234425
86. Ribeiro AI, Dias AM, Zille A. Synergistic effects between metal nanoparticles and commercial antimicrobial agents: a review. ACS Appl Nano Mater. 2022;5(3):3030–3064. doi:10.1021/acsanm.1c03891
87. Shakib P, Saki R, Marzban A, et al. Antibacterial effects of nanocomposites on efflux pump expression and biofilm production in Pseudomonas aeruginosa: a systematic review. Current Pharm Biotechnol. 2024;25(1):77–92. doi:10.2174/1389201024666230428121122
88. Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int J Mol Sci. 2022;23(24):15779. doi:10.3390/ijms232415779
89. Canaparo R, Foglietta F, Limongi T, Serpe L. Biomedical applications of reactive oxygen species generation by metal nanoparticles. Materials. 2020;14(1):53. doi:10.3390/ma14010053
90. Porzani SJ, Lorenzi AS, Eghtedari M, Nowruzi B. Interaction of dehydrogenase enzymes with nanoparticles in industrial and medical applications, and the associated challenges: a mini-review. Mini Reviews in Med Chem. 2021;21(11):1351–1366. doi:10.2174/1570193X17666201119152944
91. Parmar S, Kaur H, Singh J, Matharu AS, Ramakrishna S, Bechelany M. Recent advances in green synthesis of Ag NPs for extenuating antimicrobial resistance. Nanomaterials. 2022;12(7):1115. doi:10.3390/nano12071115
92. Mikhailova EO. Silver nanoparticles: mechanism of action and probable bio-application. J Funct Biomater. 2020;11(4):84. doi:10.3390/jfb11040084
93. Naqvi SZH, Kiran U, Ali MI, et al. Combined efficacy of biologically synthesized silver nanoparticles and different antibiotics against multidrug-resistant bacteria. Int j Nanomed;2013. 3187–3195. doi:10.2147/IJN.S49284
94. Khatoon N, Alam H, Sardar M. Proteomics analysis of Escherichia coli treated with nanosilver: an approach to analyze the bactericidal action. Curr Proteomics. 2020;17(5):404–412. doi:10.2174/1570164617666191204125202
95. Agreles MAA, Cavalcanti IDL, Cavalcanti IMF. Synergism between metallic nanoparticles and antibiotics. Appl Microbiol Biotechnol. 2022;106(11):3973–3984. doi:10.1007/s00253-022-12001-1
96. Birla S, Tiwari V, Gade A, Ingle A, Yadav A, Rai M. Fabrication of silver nanoparticles by Phoma glomerata and its combined effect against Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus. Lett Appl Microbiol. 2009;48(2):173–179. doi:10.1111/j.1472-765X.2008.02510.x
97. Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomed Nanotechnol Biol Med. 2010;6(1):103–109. doi:10.1016/j.nano.2009.04.006
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Detection of Antibiotic Resistance Genes in Pseudomonas aeruginosa by Whole Genome Sequencing
Ahmed OB
Infection and Drug Resistance 2022, 15:6703-6709
Published Date: 18 November 2022
Prevalence of Quorum Sensing and Virulence Factor Genes Among Pseudomonas aeruginosa Isolated from Patients Suffering from Different Infections and Their Association with Antimicrobial Resistance
Ghanem SM, Abd El-Baky RM, Abourehab MAS, Fadl GFM, Gamil NGFM
Infection and Drug Resistance 2023, 16:2371-2385
Published Date: 21 April 2023
Combination of AS101 and Mefloquine Inhibits Carbapenem-Resistant Pseudomonas aeruginosa in vitro and in vivo
Li R, Shen X, Li Z, Shen J, Tang H, Xu H, Shen J, Xu Y
Infection and Drug Resistance 2023, 16:7271-7288
Published Date: 18 November 2023
Multicenter Surveillance of Pseudomonas aeruginosa Isolates From Blood: Clinical Distribution Characteristics and Antibiotic Resistance Trends in Hebei Province, China (2016–2021)
Qiang C, Liu X, Qin P, Wen H, Li Z, Yang J, Niu Y, Wang W, Ouyang Z, Zhao M, Li J, Zhang Y, Zhao J
Infection and Drug Resistance 2025, 18:703-713
Published Date: 5 February 2025





