Back to Journals » International Journal of Nanomedicine » Volume 19
Enhancing Transfection Efficacy in Glioma Cells: A Comparison of Microfluidic versus Manual Polypropylenimine Dendriplex Formation
Authors Ali-Jerman H , Al-Quraishi Z, Muglikar A, Perrie Y, Tate RJ , Mullin M, McNeill G, Mackenzie G, Dufès C
Received 10 August 2024
Accepted for publication 6 November 2024
Published 21 November 2024 Volume 2024:19 Pages 12189—12203
DOI https://doi.org/10.2147/IJN.S490936
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Eng San Thian
Hawraa Ali-Jerman,1 Zainab Al-Quraishi,1 Ashish Muglikar,1 Yvonne Perrie,1 Rothwelle J Tate,1 Margaret Mullin,2 Gayle McNeill,1 Graeme Mackenzie,1 Christine Dufès1
1Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, Glasgow, G4 0RE, UK; 2Cell Analysis Facility, Medical and Veterinary & Life Sciences Shared Research Facilities, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, G12 8QQ, UK
Correspondence: Christine Dufès, Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, 161 Cathedral Street, Glasgow, G4 0RE, UK, Tel +44 1415483796, Fax +44 1415522562, Email [email protected]
Background: Gene therapy is a promising therapeutic approach for treating various disorders by introducing modified nucleic acids to correct cellular dysfunctions or introduce new functions. Despite significant advancements in the field, the effective delivery of nucleic acids remains a challenge, due to biological barriers and the immune system’s ability to target and destroy these molecules. Due to their branched structure and ability to condense negatively charged nucleic acids, cationic dendrimers have shown potential in overcoming these challenges. Despite this, standardized scalable production methods are still lacking. This study investigates the use of microfluidics to formulate generation 3-diaminobutyric polypropylenimine (DAB) dendriplexes and compares their characteristics and in vitro gene delivery efficacy to those prepared using conventional manual mixing.
Methods: DAB dendriplexes were produced by both microfluidic and manual approaches and characterized. Their cellular uptake and gene expression were evaluated on C6 glioma cancer cells in vitro.
Results: Dendriplexes formed using microfluidics at the optimal flow rate and ratio demonstrated enhanced DNA condensation over time, achieving up to 97% condensation at 24 hours. Both preparation methods produced positively charged dendriplexes, indicating stable formulations. However, dendriplexes prepared through hand mixing resulted in smaller particle sizes, significantly higher cellular uptake and gene expression efficacy compared to those prepared by microfluidics. Nonetheless, microfluidic preparation offers the advantage of standardized and scalable production, which is essential for future applications.
Conclusion: This study highlights the potential of microfluidic technology to improve precision and scalability in gene delivery, paving the way for future advancements in gene therapy. Our findings suggest that, with further optimization, microfluidic systems could provide superior control over dendriplex formation, expanding their potential use in gene therapy applications.
Keywords: polypropylenimine dendrimer, dendriplex, glioma, microfluidics, cellular uptake, transfection
Graphical Abstract:

Introduction
Gene therapy is an innovative therapeutic approach aimed at treating various disorders through the introduction of modified exogenous nucleic acids to correct cellular dysfunctions or introduce new cellular functions.1 Over recent years, significant advancements have been made in the field of gene therapy for the treatment and prevention of a wide range of diseases, including cancer, beta-thalassemia, spinal muscular atrophy, and inherited retinal disorders.2 A recent example of gene therapy for infection prevention is the COVID-19 vaccine, which uses mRNA payloads encoding the SARS-Cov-2 spike protein to stimulate antibody production.3,4 However, the delivery of nucleic acids to cells remains a challenge due to biological barriers, the size and charge of nucleic acids, and their susceptibility to destruction by the immune system.5,6 To overcome these challenges, a delivery system is required to protect and facilitate the transfer of therapeutic genes to their target site.7
Both viral and non-viral delivery vectors have been developed for nucleic acid delivery. While viral vectors exhibit high levels of gene expression, non-viral vectors, such as liposomes, nanoparticles, and dendrimers, offer lower toxicity and immunogenicity and allow for significant modifications to their structures.8 Among these non-viral delivery systems, cationic dendrimers such as polypropylenimine dendrimer have emerged as promising delivery systems for gene therapy due to their highly branched structure, which enables them to effectively condense negatively charged nucleic acids.9–13 Typically, therapeutic nucleic acids are complexed to dendrimers to form dendriplexes, a process traditionally carried out by adding one solution to another followed by vortexing, or through manually hand-mixing of dendrimer and nucleic acid solutions while pipetting.
Although dendriplexes prepared through manual methods have shown success in various applications, scaling up their preparation remains a significant challenge.14 The manual preparation process is not only labor-intensive but also prone to batch-to-batch variability, leading to inconsistencies in dendriplex behavior and, consequently, affecting their transfection efficiency and therapeutic outcomes.15 Moreover, the intricate balance between complex stability and the effective release of DNA within target cells requires meticulous control, which can be challenging to achieve with manual methods. These limitations underscore the need for a preparation tool that enables standardization and scalability in the production of dendriplexes.16
By contrast, microfluidics, defined as the manipulation of nanoliter volumes of fluids through micro-channels, has gained considerable attention in the field of nanomedicine for the precise formulation of nanoparticles.17–22 Microfluidic systems offer a continuous, standardized, and controllable environment for the formulation of particles at the nanoscale, allowing for the fine-tuning of experimental parameters such as flow rate and flow ratio of fluids in microfluidic channels, and leading to more uniform and stable complexes.23,24 Furthermore, microfluidics systems are highly scalable, allowing for the rapid production of large quantities of gene delivery complexes, which is a significant advantage for both research and clinical applications. The scalability, combined with the high level of control microfluidics provides over particle size, composition, and condensation efficiency, makes it particularly advantageous for gene therapy to overcome many of the limitations associated with traditional methods of nucleic acid complex preparation, including batch variability and scalability issues. This technology has been extensively researched for the formation of lipid nanoparticles and liposomes for drug and gene therapy, demonstrating significant advantages over traditional methods in terms of reproducibility and scalability. Despite the growing body of research on microfluidic technologies, there is a limited number of studies focusing on the synthesis and the efficacy of dendriplexes formed using microfluidics for gene delivery.25–27
The aim of this study is therefore to formulate generation 3-diaminobutyric polypropylenimine (DAB) dendriplex using microfluidics and to evaluate their physicochemical characteristics and in vitro gene delivery efficacy in glioma cancer cells, in comparison with dendriplexes formed through conventional manual mixing.
Materials and Methods
Cell Lines and Reagents
Generation-3 diaminobutyric polypropylenimine (DAB) dendrimer was purchased from SyMO-Chem (Eindhoven, Netherlands). Alexa Fluor® 647 probe and the expression plasmid encoding β-galactosidase (pCMVsport β-galactosidase) were obtained from Invitrogen (Paisley, UK), with the plasmid DNA purified using an Endotoxin-free Giga Prep® Plasmid Kit (Qiagen, Hilden, Germany). Passive lysis buffer was sourced from Promega (Southampton, UK). Quant-IT® PicoGreen® dsDNA reagent was procured from Invitrogen (Paisley, UK). The Label IT® Fluorescein Nucleic Acid Labelling kit was obtained from Cambridge Biosciences (Cambridge, UK). Vectashield® mounting medium containing 4′,6-diamidino-2- phenylindole (DAPI) was purchased from Vector Laboratories (Newark, US). The C6 rat glioma cell line was acquired from the European Collection of Cell Cultures (Salisbury, UK), while Dulbecco’s Modified Eagle’s Medium (DMEM) was obtained from ThermoFisher Scientific (Massachusetts, US). All other chemicals not specifically mentioned were purchased from Sigma Aldrich (Poole, UK).
Preparation of Dendriplexes
Dendriplexes were prepared using both microfluidics and hand mixing. Briefly, stock solutions of DAB dendrimer at a concentration equivalent to a 5:1 dendrimer: DNA weight ratio and expression plasmid encoding β-galactosidase at concentrations of 2.5, 10, 12.5, or 50 µg/mL were prepared in either glucose 5% solution or Tris-EDTA (TE) buffer, depending on the specific assay protocol. This 5:1 dendrimer: DNA weight ratio was chosen based on previous optimization studies, which showed that this ratio achieved the highest cellular uptake and transfection efficacy in C6 cells and other cell lines when the dendriplexes were prepared by manual mixing.28,29
For the microfluidics preparation, dendriplexes were prepared using a NanoAssemblr® Benchtop system (Precision NanoSystems, Vancouver, Canada). Both the dendrimer solution (organic phase) and the plasmid DNA solution (aqueous phase) were loaded into separate 3-mL syringes. The dendrimer solution was introduced into the right inlet channel, while the DNA solution was introduced into the left inlet channel. The samples were processed at various total flow rates (TFR) of 10, 15 or 20 mL/min, and various flow rate ratios (FRR) of 1:1, 2:1, and 3:1 (aqueous phase to organic phase). The TFR and FRR parameters used in this study were chosen based on previous publications utilizing similar microfluidic techniques for other types of delivery systems.17–22 The resulting dendriplex dispersions were then collected from the NanoAssemblr® system outlet channel.
For the hand-mixing method, 500 µL of the DNA solution at the specified concentrations was added to 500 µL of the dendrimer stock solution by pipetting and gently mixing. The concentration of the dendrimer stock solution was adjusted as needed to achieve a final dendrimer: DNA weight ratio of 5:1 for all experiments.
DNA Condensation
A gel retardation assay was conducted to evaluate the DNA condensation efficacy of dendrimers prepared using both methods. Dendriplex solutions were prepared in 5% glucose solution using either the microfluidic system or manual mixing, with a DNA concentration of 10 µg/mL. The samples were mixed with 2 µL of gel loading buffer, and 15 µL of each sample was then loaded onto a 0.8% (w/v) agarose gel buffered with 1× Tris-Borate-EDTA (TBE) (containing 89 mm Tris base, 89 mm boric acid, and 2 mm Na2-EDTA, pH 8.3) and stained with ethidium bromide (0.4 µg/mL). HyperLadder® 1kb was used as the DNA size marker. The gel was run at 50 V for 1 hour and subsequently photographed under UV light.
The ability of DAB dendrimer to condense DNA was evaluated using a PicoGreen® assay, a highly sensitive fluorescence-based method. PicoGreen® selectively binds to free DNA, forming a highly luminescent complex compared to the free dye in solution. The assay measures the fluorescence intensity of PicoGreen®, which is directly proportional to the amount of free DNA in the solution. One mL of dendriplex solution was prepared in Tris-EDTA buffer (10 mm Tris, 1 mm EDTA, pH 7.5) using either the microfluidic system or manual mixing, with a DNA concentration of 10 µg/mL. Following preparation, 1 mL of PicoGreen® reagent, diluted 200-fold in the same buffer, was added to each dendriplex solution. The fluorescence intensity of the PicoGreen® reagent in the presence of the dendriplexes was then measured at various time points (0, 1, 2, 4, 6, and 24 hours), using a Varian Cary Eclipse® spectrofluorimeter (Palo Alto, CA, USA), with excitation at 480 nm and emission at 520 nm. The results were presented as a percentage of DNA condensation.
Determination of Size and Zeta Potential of Dendriplexes
The size, polydispersity index (PDI), and zeta potential of dendriplexes prepared as described above were measured in quadruplet using photon correlation spectroscopy and laser Doppler electrophoresis. These measurements were conducted with a Malvern Zetasizer Nano-ZS® (Malvern Instruments Ltd, Malvern, UK) at 37°C, immediately after complexation and again after 24 hours. For all samples, the DNA concentration was kept constant at 50 µg/mL.
Morphology of Dendriplexes
The morphology of DAB dendriplexes was observed using transmission electron microscopy (TEM). Dendriplexes (dendrimer: DNA weight ratio of 5:1) were prepared using both hand mixing and microfluidics, with the latter conducted at a flow rate of 20 mL/min and a flow ratio of 1:1 aqueous: organic phase, as the parameters yielding the best condensation. Samples were prepared at a dendrimer concentration of 400 μg/mL in glucose 5% solution and diluted 100-fold in distilled water. Five microliters of the final solutions were drop-cast onto a carbon copper grid (grid size of 400 mesh) and air-dried overnight. The samples were imaged using a Jeol JEM-1400 Flash® transmission electron microscope (Jeol, Peabody, MA, USA), operating at an accelerating voltage of 80 kV. The images were captured using TEM Center® software version 1.7.26.3016 and a Jeol Flash® CCD camera (Jeol, Peabody, MA, USA).
Cellular Uptake
The cellular uptake of fluorescein-labelled DNA complexed with DAB dendrimer, prepared by hand mixing and microfluidics, was assessed qualitatively using confocal microscopy and quantitatively using flow cytometry. C6 rat glioma cells were grown as monolayers in DMEM supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) L-glutamine, and 0.5% (v/v) penicillin–streptomycin. The cell culture flasks were kept at 37 °C in a 5% carbon dioxide humid atmosphere.
Firstly, plasmid DNA was labelled with the fluorescent probe fluorescein using Label IT® Nucleic Acid Labelling kit, as described by the manufacturer. For confocal microscopy, cells were seeded at a density of 100,000 cells/well on coverslips in 6-well plates and incubated for 24 hours before treatment. Dendriplexes were prepared either 24 hours or immediately before treatment using microfluidics and hand mixing, with parameters yielding best size, zeta potential, PDI, and condensation efficiency (total flow rate of 20 mL/min and flow rate ratio of 1:1 aqueous phase to organic phase). Hand-mixed samples were prepared in DMEM, while microfluidics samples were prepared in glucose 5% solution, and then diluted in DMEM to the same final DNA concentration of 2.5 µg/well. Slides were prepared for imaging 24 hours post-treatment. The cells were washed three times with 3 mL of phosphate buffer saline (PBS) and then fixed with 3 mL of 3.7% w/v formaldehyde solution for 15 min at 25°C. Following fixation, the cells were washed three more times with 3 mL PBS and incubated with 3 mL of 0.1% Triton-X solution for 15 min. To reduce non-specific binding, they were incubated with 3 mL of 1% w/v bovine serum albumin in PBS for 30 min at 25°C. Alexa Fluor® 647 dye (diluted in 200 μL of PBS) was then added, and the cells were incubated for 45 min at 25°C. They were then mounted on a glass slide with Vectashield® mounting medium containing DAPI and incubated, while protected from light, at 25°C for 2 hours before imaging with a Leica TCS SP5® confocal microscope (Wetzlar, Germany). DAPI, used to stain the nucleus, was excited with a 405 nm laser line (emission bandwidth: 415–491 nm). Fluorescein, which labelled the DNA, was excited with a 514 nm laser line (emission bandwidth: 550–620 nm), while Alexa Fluor® 647, used to stain the cell membranes, was excited by a 633 nm laser line (emission bandwidth: 650–685 nm).
For quantitative assessment of cellular uptake, cells were seeded at a density of 100,000 cells/well in 6-well plates, incubated for 24 hours before treatment, and treated under the same conditions used for confocal microscopy. Twenty-four hours post-treatment, wells were washed three times using 3 mL of PBS, and single-cell suspensions were prepared by adding 250 µL of trypsin per well and incubating for 5 min at 37°C, followed by 500 µL of flow cytometry buffer (PBS with 0.5% bovine serum albumin (BSA) and 2 mm EDTA). Fluorescence intensity was measured using an Attune NxT® flow cytometer (Thermo Fisher Scientific, Waltham, MA), counting 10,000 cells (gated events) for each sample.
Gene Expression
The gene expression efficacy of DNA complexed with DAB dendrimer, prepared either 24 hours or immediately before treatment using microfluidics and hand mixing, was evaluated on C6 rat glioma cells plasmid DNA encoding β-galactosidase. The preparation parameters, optimized for size, zeta potential, PDI, and condensation efficiency (TFR of 20 mL/min and FFR of 1:1 aqueous phase to organic phase), were maintained throughout the study.
Cells were seeded at a density of 2,000 cells/well in 96-well plates, with experiments conducted in triplicate. After 72 hours of incubation at 37 °C in a 5% CO2 atmosphere, the cells were treated with the various dendriplex formulations, with the DNA concentration kept constant at 1 ng/well. They were then incubated for an additional 72 hours before analysis. To assess β-galactosidase expression, cells were lysed with 50 μL of 1X passive lysis buffer (PLB) for 30 min. Following this, ortho-nitrophenyl-β-D-galactosidase (ONPG) solution (1.33 mg/mL) in an assay buffer (containing 2 mm magnesium chloride, 100 mm mercaptoethanol, and 200 mm sodium phosphate buffer, pH 7.3) was added to the lysates. Fifty microliters of ONPG solution were added to each well, and the plates were incubated under the same conditions for 2 hours. The absorbance of the resulting solutions was then measured at 405 nm, using a Multiskan Ascent® plate reader (MTX Lab Systems, Bradenton, FL, USA).
Statistical Analysis
The results were expressed as means ± standard error of the mean (SEM). Statistical analysis was assessed by one-way analysis of variance (ANOVA) followed by Tukey multiple comparison post-test. Unpaired t-test was performed for paired comparisons (Minitab® software, State College, PE). Differences were considered statistically significant for P values lower than 0.05.
Results
DNA Condensation
The ability of dendrimers to condense DNA using microfluidics and manual mixing at various flow rates and flow ratios was assessed using a gel retardation assay (Figure 1). The results demonstrated a high condensation efficacy for dendriplexes formed using both techniques. No migration of free DNA was observed in the gel for dendriplexes formed using various microfluidics parameters, including NanoAssemblr® flow rates of 10, 15, and 20 mL/min, and flow ratios of 1:1, 2:1, and 3:1 aqueous phase to organic phase, or by hand mixing. In contrast, significant migration is observed for naked DNA, which serves as a control. The consistent lack of DNA migration across different conditions suggests that the dendrimers effectively condense DNA regardless of the specific parameters used in the dendriplex formation, effectively minimizing the amount of free DNA available to migrate through the gel (Figure 1). This outcome indicates the robustness and efficacy of the dendrimer-based system for DNA condensation, making it a potentially reliable method for gene delivery.
The results were further confirmed and quantified using PicoGreen® assay. At least 75% of the negatively charged DNA was complexed with DAB dendrimer at the three tested flow rates of 10, 15, and 20 mL/min. Over a period of 24 hours, the condensation increased, with the highest condensation of 97 ± 3% observed for dendriplexes formed using microfluidics at a flow rate of 20 mL/min after 24 hours. By contrast, DNA condensation following the preparation of dendriplexes by hand mixing was slightly lower within the first 7 h after preparation but reached similar levels thereafter (Figure 2A).
The condensation of dendriplexes formed using microfluidics at the optimal flow rate of 20 mL/min was evaluated at various flow ratios of 1:1, 2:1, and 3:1 (DNA to dendrimer) to determine the optimal ratio. At Time zero, the highest condensation was achieved at a flow ratio of 1:1 DNA to dendrimer, with a condensation rate of 76 ± 0.7%, followed by a condensation efficacy of 56 ± 4% and 48 ± 2% at flow ratios of 2:1 and 3:1, respectively (Figure 2B). The DNA condensation achieved by hand mixing was initially lower than that observed at a 1:1 ratio for the first 7 h following preparation of the dendriplex. It gradually increased over time to reach a maximum of at least 95% after 24 hours for all prepared dendriplexes.
Determination of Size and Zeta Potential of Dendriplexes
The smallest dendriplex sizes were observed for those formed using hand mixing, an average size of 101 ± 6 nm, followed by dendriplexes formed using a NanoAssemblr® at a flow rate of 20 mL/min, with an average size of 212 ± 13 nm (Figure 3A). The largest size (429 ± 22 nm) was observed for dendriplexes formed at a NanoAssemblr® flow rate of 10 mL/min, which was significantly higher than those formed at flow rates of 15 and 20 mL/min (which had sizes of 222 ± 6 nm and 212 ± 13 nm, respectively).
To determine the optimal flow ratio, dendriplexes were formed using a NanoAssemblr® at a flow rate of 20 mL/min, the rate that yielded the best size and zeta potential. Various DNA to dendrimer flow ratios of 1:1, 2:1, and 3:1 were tested while maintaining a final dendrimer to DNA weight ratio of 5:1 (Figure 3C). Dendriplexes formed at DNA to dendrimer flow ratios of 2:1 and 3:1 had the smallest sizes, with average diameters of 93 ± 4 nm and 83 ± 6 nm, respectively. In contrast, dendriplexes produced at a flow ratio of 1:1 had a larger average size of 212 ± 14 nm. A PDI of less than 0.3 was recorded for dendriplexes formed using hand mixing and microfluidics, indicating low heterogeneity in size. All dendriplexes exhibited a positive zeta potential. Those formed using microfluidics showed zeta potential values ranging from 23 ± 2 mV to 26 ± 1 mV, while those formed by hand mixing were slightly more positive, with a zeta potential of 32 ± 2 mV.
Over a 24-hour period, minimal changes in size and zeta potential were observed for all dendriplexes, indicating stability (Figure 3B and D).
Morphology of Dendriplexes
Transmission electron microscopy imaging revealed spherical nanoparticles with sizes smaller than 200 nm for dendriplexes prepared using hand mixing and microfluidics (Figure 4). The images show that the nanoparticles maintain a consistent spherical shape, with no significant size variation between the two preparation methods.
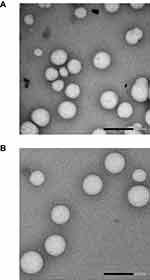 |
Figure 4 TEM images of dendriplexes formed using microfluidics at a flow rate of 20 mL/min and a DNA: dendrimer flow ratio of 1:1 (A), and dendriplexes formed using hand mixing (B). |
Cellular Uptake
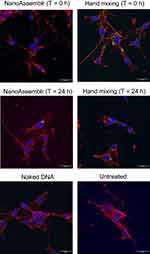
The cellular uptake of DAB dendriplexes complexed to fluorescein-labelled DNA was evaluated using confocal microscopy in the C6 glioma cell line. The intensity of the green fluorescence, indicating the presence of DNA, was higher in cells treated with dendriplexes prepared using hand mixing both immediately and 24 hours prior to treatment, compared to those prepared using microfluidics (Figure 5). This suggests that dendriplexes formed by hand mixing may have higher cellular uptake efficacy or stability over time.
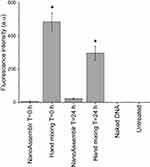
Cellular uptake was further confirmed and quantified using flow cytometry (Figure 6). Dendriplexes prepared using hand mixing exhibited significantly higher cellular uptake compared to those prepared using microfluidics. Specifically, a fluorescence intensity of 486 ± 55 a.u. was recorded for dendriplexes prepared using hand mixing immediately before treatment, compared to only 7 ± 4 a.u. for those prepared using microfluidics. A decrease in cellular uptake was observed for dendriplexes prepared using hand mixing 24 hours prior to treatment, with a recorded intensity of 297 ± 41 a.u. Dendriplexes prepared using microfluidics 24 hours before treatment showed a 3.2-fold increase in cellular uptake (22 ± 4 a.u.) compared to those prepared immediately after preparation (Figure 6).
Gene Expression
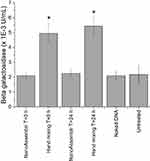
The gene expression efficacy of DAB dendriplex encoding β-galactosidase was evaluated in the C6 glioma cell line (Figure 7). Cells treated with dendriplexes prepared immediately and 24 hours before treatment using microfluidics exhibited gene expression levels of 2.1 ± 0.2 mU/mL and 2.3 ± 0.6 mU/mL, respectively. These levels were not significantly different from the gene expression observed with naked DNA (2.1 ± 0.3 mU/mL). In contrast, treatment with dendriplexes prepared using hand mixing immediately before and 24 hours before treatment resulted in significantly higher gene expression, with levels of 5.4 ± 0.6 mU/mL and 4.9 ± 0.6 mU/mL, respectively. This represents an approximate 2.3-fold and 2.6-fold increase in gene expression compared to dendriplexes prepared using microfluidics and naked DNA.
Discussion
Microfluidics has emerged as a cutting-edge technique for nanoparticle preparation, offering precise control over particle size, shape, and composition, and thereby surpassing the variability often encountered with manual production methods.22 This precision and reproducibility are particularly critical for the scalable synthesis of nanoparticles for drug and gene delivery applications.
The application of microfluidics in the preparation of liposomes and lipid nanoparticles for drug delivery has been extensively studied, showing improved efficacy and reproducibility.30 This study explored the use of microfluidics for the complexation of plasmid DNA with cationic DAB dendrimer, aiming to optimize parameters for gene delivery.
Critical parameters in microfluidic nanoparticle synthesis include flow rate and flow ratio, both of which significantly influence nanoparticle characteristics.31 Condensation efficacy, particle size, and zeta potential of dendriplexes produced using microfluidics at various flow rates and flow ratios were investigated to identify optimal conditions for gene delivery. When using a DNA-to-dendrimer flow ratio of 1:1, dendrimers effectively condensed at least 75% of the DNA across flow rates of 10, 15, and 20 mL/min. However, dendriplexes formed at 10 mL/min were significantly larger than those formed at 15 and 20 mL/min, likely due to differences in shear forces at the aqueous-organic (DNA-dendrimer) interface. Higher flow rates increase the shear forces, leading to the formation of smaller particles.15 Additionally, the residence time, defined as the time a particle spends within a microchannel, is another critical concept in microfluidics affecting the size of particles. Higher flow rates result in shorter residence time limiting the growth phase of a particle and leading to smaller sizes.32 Given these findings, a flow rate of 20 mL/min was selected to further explore the optimal aqueous-to-organic flow ratio.
Dendriplexes formed at flow ratios of 3:1 and 2:1 showed significantly lower DNA condensation (48 ± 2% and 56 ± 4%, respectively) compared to a 1:1 ratio (76 ± 0.2%). Dilution of the DNA solution reduces the availability of the negative charge in the microchannel while mixing, essential for the electrostatic interaction between cationic dendrimers and negatively charged DNA, and therefore decreasing DNA condensation. In contrast, the equal volumes of both phases when using a flow ratio of 1:1 enhances mixing and interaction between the negative charge of the DNA and the positive charge of cationic dendrimers, resulting in higher condensation.33
Using the optimal conditions identified (20 mL/min flow rate and a 1:1 aqueous phase to organic phase flow ratio), we compared the in vitro efficacy of dendriplexes formed using microfluidics with those prepared by hand mixing, by assessing their cellular uptake and gene expression efficacy. The cellular uptake of fluorescein-labelled DNA complexed with dendrimers was assessed in C6 rat glioma cells. Dendriplexes formed using microfluidics immediately before treatment exhibited minimal cellular uptake, which was 60-fold lower than those prepared by hand mixing. The predominant pathways for the internalization of non-targeted dendrimer-based nanocarriers include clathrin-mediated endocytosis, which favors nanoparticles sized between 100 and 200 nm, and caveolae-mediated endocytosis, which favors particles in the 50 to 80 nm range.34,35 The relatively larger size of dendriplexes prepared using microfluidics immediately before treatment (212 ± 13 nm) likely limited their internalization efficiency through these pathways. Conversely, the smaller size of dendriplexes formed by hand mixing (average size 101 ± 6.5 nm) and those prepared by microfluidics 24 hours before treatment (average size 155 ± 2 nm) resulted in higher cellular uptake.
The reduced cellular uptake observed for dendriplexes prepared by microfluidics is likely the main factor limiting gene expression for plasmid DNA encoding β-galactosidase. Additionally, the strong condensation of DNA over 24 hours for dendriplexes formed by microfluidics may restrict DNA dissociation from the carrier within the cell, further contributing to the lower gene expression observed with these dendriplexes.36
Future studies should focus on optimizing key microfluidic parameters to enhance the efficacy of dendriplex formation. Adjustments to flow rates, dendrimer to DNA weight ratios, and residence time within the microfluidic channels could yield more favorable particle sizes and improved DNA condensation, enhancing cellular uptake and gene expression. Investigating additional factors, such as the conjugation of targeting moieties, may also influence the ionic strength of the dendriplex, further enhancing the transfection efficacy of microfluidic-based dendriplexes. As microfluidic systems offer great potential for scalability and reproducibility, these refinements will be essential for future applications in gene delivery.
Conclusion
This study demonstrates that microfluidics facilitates efficient DNA condensation and uniformity in nanoparticle formulation when optimal flow rates and ratios are applied. However, in vitro studies revealed that dendriplexes formed using hand mixing yielded superior cellular uptake and gene expression in C6 rat glioma cells. These findings highlight the need for further optimization of microfluidic parameters to enhance the therapeutic efficacy of dendrimer-based nanocarriers in gene delivery systems. Despite current limitations, microfluidic systems hold significant potential for clinical applications due to their scalability, precision, and reproducibility in nanoparticle formulation. To fully harness this potential, future research should focus on refining microfluidic parameters, such as improving transfection efficacy and enhancing particle stability. With these advancements, microfluidic platforms could emerge as a viable platform for large-scale, dendrimer-based gene delivery in clinical settings.
Abbreviations
ANOVA, analysis of variance; a.u., arbitrary units; BSA, bovine serum albumin; DAB, generation 3-diaminobutyric polypropylenimine dendrimer; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco’s modified Eagle’s medium; DNA, deoxyribonucleic acid; dsDNA, double strand DNA; EDTA, ethylenediaminetetraacetic acid; FRR, flow rate ratio; mRNA, messenger ribonucleic acid; ONPG, ortho-nitrophenyl-β-D-galactosidase; PBS, phosphate buffer saline; PDI, polydispersity index; PLB, passive lysis buffer; SARS-Cov-2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of the mean; TBE, tris-borate-ethylenediaminetetraacetic acid; TE, tris- ethylenediaminetetraacetic acid; TFR, total flow rate; TEM, transmission electron microscopy.
Acknowledgments
This work was financially supported by a PhD studentship from Kuwait University (Kuwait) to Hawraa Ali-Jerman and by a Global Research Scholarship from the University of Strathclyde to Zainab Al-Quraishi.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shchaslyvyi A, Antonenko S, Tesliuk M, et al. Current State of Human Gene Therapy: approved Products and Vectors. Pharmaceuticals. 2023;16:1416–1466.
2. Henderson ML, Zieba JK, Li X, et al. Gene Therapy for Genetic Syndromes: understanding the Current State to Guide Future Care. BioTech. 2024;13(1):1. doi:10.3390/biotech13010001
3. Li Y, Lu SM, Wang JL, et al. Progress in SARS-CoV-2, diagnostic and clinical treatment of COVID-19. Heliyon. 2024;10(12):e33179. doi:10.1016/j.heliyon.2024.e33179
4. Cullis PR, Felgner PL. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat Rev Drug Discov. 2024;23(9):709–722. doi:10.1038/s41573-024-00977-6
5. Dume B, Licarete E, Banciu M. Advancing cancer treatments: the role of oligonucleotide-based therapies in driving progress. Mol Ther Nucleic Acids. 2024;35(3):102256. doi:10.1016/j.omtn.2024.102256
6. Taghdiri M, Mussolino C. Viral and Non-Viral Systems to Deliver Gene Therapeutics to Clinical Targets. Int J Mol Sci. 2024;25(13):7333. doi:10.3390/ijms25137333
7. Santhanes D, Wilkins A, Zhang H, et al. Microfluidic formulation of lipid/polymer hybrid nanoparticles for plasmid DNA (pDNA) delivery. J Nanobiotechnol. 2022;20(3):123–134.
8. Zu H, Gao D. Non-viral Vectors in Gene Therapy: recent Development, Challenges and prospects. AAPS J. 2021;23(4):78. doi:10.1208/s12248-021-00608-7
9. Altwaijry N, Somani S, Dufès C. Targeted nonviral gene therapy in prostate cancer. Int J Nanomed. 2018a;13:5753–5767. doi:10.2147/IJN.S139080
10. Altwaijry N, Somani S, Parkinson JA, et al. Regression of prostate tumors after intravenous administration of lactoferrin-bearing polypropylenimine dendriplexes encoding TNF-α, TRAIL, and interleukin-12. Drug Delivery. 2018b;25(1):679–689. doi:10.1080/10717544.2018.1440666
11. Almowalad J, Laskar P, Somani S, et al. Lactoferrin- and Dendrimer-Bearing Gold Nanocages for Stimulus-Free DNA Delivery to Prostate Cancer Cells. Int J Nanomed. 2022;17:1409–1421. doi:10.2147/IJN.S347574
12. Laskar P, Dufès C. Emergence of cationic polyamine dendrimersomes: design, stimuli sensitivity and potential biomedical applications. Nanoscale Adv. 2021;3(21):6007–6026. doi:10.1039/D1NA00536G
13. Laskar P, Somani S, Mullin M, et al. Octadecyl chain-bearing PEGylated poly(propyleneimine)-based dendrimersomes: physicochemical studies, redox-responsiveness, DNA condensation, cytotoxicity and gene delivery to cancer cells. Biomater Sci. 2021;9(4):1431–1448. doi:10.1039/D0BM01441A
14. Li X, Naeem A, Xiao S, et al. Safety Challenges and Application Strategies for the Use of Dendrimers in Medicine. Pharmaceutics. 2022;14(6):1292–1329. doi:10.3390/pharmaceutics14061292
15. Wu J, Yadavali S, Lee D, et al. Scaling up the throughput of microfluidic droplet-based materials synthesis: a review of recent progress and outlook. Appl Phys Rev. 2021;8(3):31304–31321. doi:10.1063/5.0049897
16. Patel D, Patel N, Patel J. Nanomedicine Scale-Up Technologies: feasibilities and Challenges. In: Patel JK, Pathak YV, editors. Emerging Technologies for Nanoparticle Manufacturing. Cham: Springer; 2021:511–539.
17. Anderluzzi G, Perrie Y. Microfluidic Manufacture of Solid Lipid Nanoparticles: a Case Study on Tristearin-Based Systems. Drug Delivery Lett. 2020;10(3):197–208. doi:10.2174/2210303109666190807104437
18. Roces CB, Lou G, Jain N, et al. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics. 2020;12(11):1095. doi:10.3390/pharmaceutics12111095
19. Schmidt ST, Christensen D, Perrie Y. Applying Microfluidics for the Production of the Cationic Liposome-Based Vaccine Adjuvant CAF09b. Pharmaceutics. 2020;12(12):1237. doi:10.3390/pharmaceutics12121237
20. Shah S, Dhawan V, Holm R, et al. Liposomes: advancements and innovation in the manufacturing process. Adv Drug Delivery Rev. 2020;154–155:102–122. doi:10.1016/j.addr.2020.07.002
21. Webb C, Ip S, Bathula NV, et al. Current Status and Future Perspectives on MRNA Drug Manufacturing. Mol Pharm. 2022;19(4):1047–1058. doi:10.1021/acs.molpharmaceut.2c00010
22. Meewan J, Somani S, Almowalad J, et al. Preparation of Zein-Based Nanoparticles: nanoprecipitation versus Microfluidic-Assisted Manufacture, Effects of PEGylation on Nanoparticle Characteristics and Cellular Uptake by Melanoma Cells. Int J Nanomed. 2022;17:2809–2822. doi:10.2147/IJN.S366138
23. Protopapa G, Bono N, Visone R, et al. A new microfluidic platform for the highly reproducible preparation of non-viral gene delivery complexes. Lab Chip. 2022;23(1):136–145. doi:10.1039/D2LC00744D
24. Gimondi S, Ferreira H, Reis R, et al. Microfluidic Devices: a Tool for Nanoparticle Synthesis and Performance Evaluation. ACS Nano. 2023;17(15):14205–14228. doi:10.1021/acsnano.3c01117
25. Agnoletti M, Bohr A, Thanki K, et al. Inhalable siRNA-loaded nano-embedded microparticles engineered using microfluidics and spray drying. Eur J Pharm Biopharm. 2017;120:9–21. doi:10.1016/j.ejpb.2017.08.001
26. Doosti-Telgerd M, Mahdavi FS, Moradikhah F, et al. Nanofibrous Scaffolds Containing Hydroxyapatite and Microfluidic-Prepared Polyamidoamin/BMP-2 Plasmid Dendriplexes for Bone Tissue Engineering Applications. Int J Nanomed. 2020;15:2633–2646. doi:10.2147/IJN.S244416
27. Agha A, Waheed W, Stiharu I, et al. A review on microfluidic-assisted nanoparticle synthesis, and their applications using multiscale simulation methods. Discov Nano. 2023;18(1):18. doi:10.1186/s11671-023-03792-x
28. Zinselmeyer BH, Mackay SP, Schatzlein AG, et al. The lower-generation polypropylenimine dendrimers are effective gene-transfer agents. Pharm Res. 2002;19(7):960–967. doi:10.1023/A:1016458104359
29. Dufès C, Keith WN, Bilsland A, et al. Synthetic anticancer gene medicine exploits intrinsic antitumor activity of cationic vector to cure established tumors. Cancer Res. 2005;65(18):8079–8084. doi:10.1158/0008-5472.CAN-04-4402
30. Liu Y, Yang G, Hui Y, et al. Microfluidic Nanoparticles for Drug Delivery. Small. 2022;18(36):2106580–2106613. doi:10.1002/smll.202106580
31. Ripoll M, Martin E, Enot M, et al. Optimal self-assembly of lipid nanoparticles (LNP) in a ring micromixer. Sci Rep. 2022;12(1):9483. doi:10.1038/s41598-022-13112-5
32. Shrimal P, Jadeja G, Patel S. Microfluidics nanoprecipitation of telmisartan nanoparticles: effect of process and formulation parameters. Chem Papers. 2020;75(1):205–214. doi:10.1007/s11696-020-01289-w
33. Feng Q, Sun J, Jiang X. Microfluidics-mediated assembly of functional nanoparticles for cancer-related pharmaceutical applications. Nanoscale. 2016;8(25):12430–12443. doi:10.1039/C5NR07964K
34. Torres-Vanegas J, Cruz J, Reyes L. Delivery Systems for Nucleic Acids and Proteins: barriers, Cell Capture Pathways and Nanocarriers. Pharmaceutics. 2013;13(3):428–466. doi:10.3390/pharmaceutics13030428
35. Sztandera K, Rodríguez-García J, Ceña V. In Vivo Applications of Dendrimers: a Step toward the Future of Nanoparticle-Mediated Therapeutics. Pharmaceutics. 2024;16(4):439–461. doi:10.3390/pharmaceutics16040439
36. Ramawat KG, Mérillon JM, editors. Polysaccharides: Bioactivity and Biotechnology. Cham: Springer International Publishing; 2015.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.