Back to Journals » International Journal of Nanomedicine » Volume 19
FoxO1-Overexpressed Small Extracellular Vesicles Derived from hPDLSCs Promote Periodontal Tissue Regeneration by Reducing Mitochondrial Dysfunction to Regulate Osteogenesis and Inflammation
Authors Niu Q , Lin C, Yang S , Rong S , Wei J, Zhao T, Peng Y , Cheng Z, Xie Y , Wang Y
Received 24 March 2024
Accepted for publication 18 August 2024
Published 27 August 2024 Volume 2024:19 Pages 8751—8768
DOI https://doi.org/10.2147/IJN.S470419
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Krishna Nune
Qingru Niu,1– 3,* Chuanmiao Lin,1– 3,* Shuqing Yang,1– 3 Shuxuan Rong,1– 3 Junbin Wei,1– 3 Tingting Zhao,1– 3 Yingying Peng,1– 3 Zhilan Cheng,1– 3 Yunyi Xie,1– 3 Yan Wang1– 3
1Hospital of Stomatology, Guanghua School of Stomatology, Guangzhou, People’s Republic of China; 2Sun Yat-Sen University, Guangzhou, People’s Republic of China; 3Guangdong Provincial Key Laboratory of Stomatology, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yunyi Xie; Yan Wang, Email [email protected]; [email protected]
Purpose: Periodontitis is a chronic infectious disease characterized by progressive inflammation and alveolar bone loss. Forkhead box O1 (FoxO1), an important regulator, plays a crucial role in maintaining bone homeostasis and regulating macrophage energy metabolism and osteogenic differentiation of mesenchymal stem cells (MSCs). In this study, FoxO1 was overexpressed into small extracellular vesicles (sEV) using engineering technology, and effects of FoxO1-overexpressed sEV on periodontal tissue regeneration as well as the underlying mechanisms were investigated.
Methods: Human periodontal ligament stem cell (hPDLSCs)-derived sEV (hPDLSCs-sEV) were isolated using ultracentrifugation. They were then characterized using transmission electron microscopy, Nanosight, and Western blotting analyses. hPDLSCs were treated with hPDLSCs-sEV in vitro after stimulation with lipopolysaccharide, and osteogenesis was evaluated. The effect of hPDLSCs-sEV on the polarization phenotype of THP-1 macrophages was also evaluated. In addition, we measured the reactive oxygen species (ROS) levels, adenosine triphosphate (ATP) production, mitochondrial characteristics, and metabolism of hPDLSCs and THP-1 cells. Experimental periodontitis was established in vivo in mice. HPDLSCs-sEV or phosphate-buffered saline (PBS) were injected into periodontal tissues for four weeks, and the maxillae were collected and assessed by micro-computed tomography, histological staining, and small animal in vivo imaging.
Results: In vitro, FoxO1-overexpressed sEV promoted osteogenic differentiation of hPDLSCs in the inflammatory environment and polarized THP-1 cells from the M1 phenotype to the M2 phenotype. Furthermore, FoxO1-overexpressed sEV regulated the ROS level, ATP production, mitochondrial characteristics, and metabolism of hPDLSCs and THP-1 cells in the inflammatory environment. In the in vivo analyses, FoxO1-overexpressed sEV effectively promoted bone formation and inhibited inflammation.
Conclusion: FoxO1-overexpressed sEV can regulate osteogenesis and immunomodulation. The ability of FoxO1-overexpressed sEV to regulate inflammation and osteogenesis can pave the way for the establishment of a therapeutic approach for periodontitis.
Keywords: periodontal ligament stem cells, small extracellular vesicles, periodontitis, bone homeostasis, mitochondrial function
Graphical Abstract:

Introduction
Periodontitis is a chronic infectious disease characterized by pathological loss of alveolar bone and periodontal ligament, resulting in the formation of “periodontal pockets”.1 The development of this disease involves ecological dysbiosis of the oral microflora, host immune defense, and other relevant risk factors.2 The incidence and prevalence of periodontitis have recently shown an increasing trend in Asian countries such as China, India, and Japan.3 Novel nanoparticle delivery systems, such as cell membrane-mimetic nanoparticles and small extracellular vesicles (sEV), hold significant promise for localized antimicrobial and immunomodulatory treatments and tissue regeneration in periodontitis.4,5
The application of dental mesenchymal stem cells (MSCs) for tissue regeneration and homeostasis has been widely studied.6 Human periodontal ligament stem cells (hPDLSCs) are the ideal seed cells for periodontal tissue regeneration. Extracellular vesicles are a heterogeneous group of membrane-bound vesicles released by nearly all kinds of cells.7 sEV typically refers to extracellular vesicles with a diameter smaller than 200 nm, which can transfer bioactive components for intercellular communication.8 Moreover, the supernatant of hPDLSCs has been shown to have a similar ability to inhibit periodontitis-related inflammation and promote periodontal tissue regeneration.9 sEV isolated from MSCs supernatants are promising therapeutic products because of their effective delivery potential and low immunogenicity.10 More importantly, compared to sEV from other cell sources, sEV derived from MSCs can play a similar crucial role as MSCs in immunomodulation and regeneration.11 Considering the characteristics of sEV, the development of engineering methods to modify them internally and externally is necessary to improve their efficiency and therapeutic potential.12,13
Genetic engineering technology can be used to edit sEV-producing cells as well as provide additional possibilities for the generation of sEV.14,15 In general, the method involves transfecting sEV-source cells using viral or non-viral vectors. Through genetic modification, the target molecules are first transferred to the parent cells and then modified onto the sEV membrane or contents through natural packaging processes to enhance the functionality of sEV.16 For example, transfecting cells with a single-chain variable fragment antibody specific to the HIV-1 envelope protein and a vector containing the fusion protein PDGFR transmembrane domain, the collected sEV can selectively target infected cells and tissues.17 In another research, sEV derived from WNT3A-overexpressing cells are heavily enriched in WNT3A. In comparison with recombinant WNT3A, WNT3A-loaded sEV could more effectively improve the repair of cartilage defects in mice.18 Therefore, the use of genetic engineering technology to optimize the properties of sEV for bone regeneration shows great potential for effective delivery and targeted therapy.19,20
Increasing evidence suggests that mitochondrial dysfunction and the accumulation of reactive oxygen species (ROS) play an important role in the pathogenesis of periodontitis.21 A recent study found that chronic inflammation leads to the continuous accumulation of damaged mitochondria in hPDLSCs, thereby impairing the differentiation function of cells.22 Macrophages are key participants in the tissue destruction and repair of periodontitis, and mitochondrial metabolism is crucial in regulating macrophage pro-inflammatory or anti-inflammatory states.23,24 Therefore, restoring or clearing the dysfunctional mitochondria in key cells of periodontal tissues may promote the regeneration and repair of periodontal tissues, offering a promising new approach for the treatment of periodontitis.25,26
As one of the important transcription factors in the FoxO family,27 Forkhead box O1 (FoxO1) can promote early progenitor cell and osteoblast osteogenesis, reverse inflammation-induced bone resorption, and regulate the expression of related genes.28,29 Recent studies have shown that targeting FoxO1 to regulate osteogenesis may be beneficial in the treatment of bone regeneration-related diseases. The existing studies on FoxO1 have focused on its role in bacterial–epithelial interactions, immune dysregulation, bone remodeling, and wound healing in periodontitis. FoxO1 has great potential for the treatment of periodontitis.30,31 However, effective targeting of FoxO1 to promote periodontal tissue regeneration in the treatment of periodontitis requires further study.32,33 A previous study focused on the effects of FoxO1 overexpression in hPDLSCs in response to an inflammatory environment, eg, its ability to enhance anti-inflammatory effects, improve antioxidant capacity, and positively regulate osteogenesis in a simulated inflammatory environment.30
Therefore, we hypothesized that FoxO1-overexpressed sEV would be more effective than normal hPDLSCs sEV in regulating osteogenesis and immunity. To confirm whether FoxO1-overexpressed sEV show a greater probability of successful periodontal regeneration, this study explored the functional effects of FoxO1-overexpressed sEV on therapeutic efficacy in experimental periodontitis in mice and further explored the underlying mechanisms in vitro.
Materials and Methods
Cell Culture and Identification
This study was approved by the Ethics Committee of the Affiliated Stomatology Hospital of Sun Yat-sen University (ERC-2016-46) and informed consent was obtained from the patients. The study complied with the Declaration of Helsinki. HPDLSCs were isolated as described previously.30 They were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco, USA) and 100 units/mL streptomycin/penicillin (Gibco, USA) at 37°C in 5% CO2. Cells from passages two to six were used in the subsequent studies. The cells were seeded into 10-cm dishes and cultured for 14 d, after which they were stained with 0.1% crystal violet solution (Sigma-Aldrich, USA) to detect colony-formation ability. For tissue origin identification, cells were stained with vimentin (1:100, Abcam, UK) or cytokeratin 18 (1:800, Cell Signaling Technology, USA) and sealed with mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Abcam, UK) for the immunofluorescence test. The samples were imaged using confocal laser-scanning microscopy (Olympus, JAPAN). In addition, hPDLSCs were cultured in a lipogenesis induction solution (Cyagen Biosciences, USA) and osteogenic induction medium to confirm their multipotent differentiation capacity. After induction, the cells were stained with Oil Red O and Alizarin Red and observed under a color-inverted microscope (Zeiss, Germany). The surface markers CD105, CD90, CD45, CD34, CD73, and HLA-DR in hPDLSCs were characterized by flow cytometry.
The insert DNA (FoxO1DNA fragment) was amplified using the following primers: (5′-GCCGGAATTAGATCTCTCGAGAGTTAAGTTCTGGGCTCGC-3′ and 5′-CTCCCCTACCCGGTAGAATTCGTAACCTGCTCACTAACCCT-3′). The pMSCV-puro vector was digested with XhoI and EcoRI (TaKaRa, Japan). Both the PCR product and the linearized vector were purified by a Gel Extraction Kit (Omega, China), and the PCR product was inserted into the vector with a ClonExpress II One Step Cloning Kit (Vazyme, China) to create the pMSCV-FoxO1 plasmid. HEK-293T cells were transfected with 5 μg of the pMSCVFoxO1 expression vector and 5 μg of the PIK packaging vector using the calcium phosphate method. An empty vector without the FoxO1 transgene was transfected into 293T cells as a negative control. At 48 h post transfection, the supernatant containing the viral particles was harvested and centrifuged at 8000 rpm for 10 minutes at 4°C to clear cellular debris. The passage 3 hPDLSCs were infected with a viral particle solution containing 8 μg/mL polybrene and centrifuged at 2500 rpm for 1 h. The same passage 3 hPDLSCs without infection were considered as normal cells under the regular culture. Then, the virus‐containing medium was replaced with fresh medium at 6 h viral infection. At 48 h post infection, cells were selected with puromycin‐containing medium. Finally, we obtained three types of hPDLSCs cells of the passage 5. Normal hPDLSCs were named hPDLSCs-Nor; hPDLSCs transfected with the FoxO1 plasmid were named hPDLSCs-Ov-FoxO1 (FoxO1-overexpressed hPDLSCs), and those transfected with the empty vector were named hPDLSCs-NC (negative control hPDLSCs). Lipopolysaccharide (LPS; Sigma-Aldrich, USA) was used at a concentration of 1 μg/mL to simulate the inflammatory microenvironment in vitro.
THP-1 cells were provided by PricellaLife Science & Technology Company (Pricella, China) and cultured in RPMI 1640 (Gibco, USA) containing 10% FBS, 100 units/mL streptomycin/penicillin, and 0.1% β-mercaptoethanol and maintained in 5% CO2 at 37 °C. To observe THP-1 cell polarization, the cells were differentiated into macrophage-like cells with 100 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, USA) for 48 h and induced with 1 µg/mL LPS for 24 h. The cells were then treated with different sEV as described above for 48 h.
Isolation and Characterization of sEV
sEV were isolated separately from above cells under the same culture conditions and at the same passage (passages 6 to 8), and identified using the previously described methods.34 The supernatant was collected and centrifuged at 300 × g for 10 min, 2000 × g for 30 min, and 10,000 × g for 30 min at 4°C to remove cellular debris. Then, the supernatant was filtered through a 0.22-μm filter and ultracentrifuged at 100000 × g for 70 min at 4°C to collect the precipitate. The precipitate was resuspended with phosphate-buffered saline (PBS) and collected by ultracentrifugation at 100000 × g for 70 min at 4°C (Beckman, Germany). The protein concentration of the sEV was quantified using a BCA protein assay kit (Life Technologies, USA). sEV from normal hPDLSCs were named Nor-sEV, while sEV from hPDLSCs carrying empty plasmids were designated NC-sEV. sEV from hPDLSCs overexpressed FoxO1 were named FoxO1-sEV. All the sEV were used at a concentration of 50 μg/mL and within 6 h to treat THP-1 cells and hPDLSCs.
sEV morphology was visualized using a transmission electron microscope (Hitachi, Japan). Nanosight 3000 (Malvern, UK) was used to measure the nanoparticle size. The expression of surface markers, including FoxO1 (1:1000, Cell Signaling Technology, USA), CD63 (1:1000, Abcam, UK), CD81 (1:400, Santa Cruz, USA), α-tubulin (1:5000, Abcam, UK), and Alix (1:1000, Abcam, UK) was detected using Western blotting.
Intracellular Uptake of sEV
PKH26 (Sigma-Aldrich) fluorescent staining was performed to label sEV from the three groups (Nor-sEV, NC-sEV, and FoxO1-sEV). THP-1 cells and hPDLSCs were cultured and treated as described previously. The cells were then co-cultured with different sEV for 24 h. The cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and then incubated with TRITC-labeled cyclic peptide solution in darkness to stain the cytoskeleton. The slides were sealed with mounting medium containing DAPI (Abcam, Cambridge, UK), and nail polish was applied around the coverslips. The samples were imaged using a confocal laser-scanning microscope (Olympus, Tokyo, JAPAN).
Western Blot Analysis
Protein was extracted from the samples using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (CWBIO, China). The protein concentrations of the cells were quantified using a Pierce BCA Protein Assay Kit (Thermo Scientific, USA). Then, electrophoresis of protein was performed using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Biosharp, China), and the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were then blocked using 5% skimmed milk for 1 h and then incubated with the primary antibodies at 4°C overnight. Finally, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Emar, China) at room temperature for 1 h. Antibodies against FoxO1 (1:1000, Cell Signaling Technology, USA), CD63 (1:1000, Abcam, UK), CD81 (1:400, Santa Cruz, USA), α-tubulin (1:5000, Abcam, UK), and Alix (1:1000, Abcam, UK) were used in this study. Protein signals were detected using an ECL chemiluminescent substrate (Biosharp, China), and the membranes were visualized using a ChemiDoc Imaging System (Bio-Rad).
Osteogenic Differentiation
hPDLSCs were inoculated in 24-well plates and cultured in osteogenic induction medium (DMEM containing 10% FBS and 100 units/mL streptomycin/penicillin, 10 nM dexamethasone, 10 mM β-glycerophosphate, and 0.2 mM ascorbic acid). LPS (1 μg/mL) was used to simulate the inflammatory microenvironment. After 21 days of osteogenic induction with or without LPS and sEV, the cells were fixed in 4% paraformaldehyde and stained with 1% Alizarin Red. After Alizarin Red staining, the liquid was aspirated, and 350 μL of 10% cetyl pyridinium chloride solution (Sigma-Aldrich, USA) was used to dissolve it. When the Alizarin Red was completely dissolved, 100 μL/well of the mixture was transferred to a 96-well plate, and the plate was then analyzed by an enzyme meter (Biotek, Germany) to measure the absorbance values (optical density; OD) of each group at a wavelength of 562 nm.
Flow Cytometry
HPDLSCs were collected, washed twice with PBS, and incubated with anti-CD105, anti-CD90, anti-CD45, anti-CD34, anti-HLA-DR, and anti-CD73 (1:100, BioLegend, USA) for 30 min in the dark. The cells were washed and centrifuged twice, suspended, and analyzed using a flow cytometer (BD LSRFortessa Flow Cytometry, USA), and the data were analyzed using FlowJo 10.8.1 software.
THP-1 cells were seeded into 12-well plates at a density of 5×105 cells/well and treated as described above. The cells were washed twice with PBS, harvested using a scraper, collected by centrifugation, and maintained on ice. Subsequently, the cells were blocked with an Fc Receptor Blocking Solution (BioLegend, San Jose, CA, USA) for 15 min on ice. The cells were then stained with a mixture of PE-conjugated anti-CD86 (1:100, BioLegend, USA) and APC-conjugated anti-CD163 (1:100, BioLegend, USA) for 30 min in the dark, and then analyzed using a flow cytometer. Data were analyzed using FlowJo version 10.8.1.
Immunofluorescence Staining
To identify the tissue origin of hPDLSCs, cells were seeded on a confocal Petri dish, and then subjected to immunofluorescence staining when the cell density reached 50%. The cells were fixed, permeabilized, and blocked for staining with rabbit anti-human vimentin (1:100, Abcam, UK) or mouse anti-human cytokeratin 18 (1:800, Cell Signaling Technology, USA) antibodies, incubated with Alexa Fluor-555- and Alexa Fluor-488-labeled secondary antibodies (1:200–1000, Abcam, UK) for 1 h, and sealed using Mounting Medium with DAPI (Abcam, UK). The samples were imaged using a confocal laser-scanning microscope (Olympus, JAPAN).
To detect changes in macrophage polarization phenotypes, the cells were incubated with the primary antibodies against iNOS (1:500, Cell Signaling Technology, USA) and arginase1 (Arg1) (1:500; ProteinTech Group, USA) overnight at 4°C. The cells were then incubated with anti-rabbit Alexa Fluor-488 and anti-mouse Alexa Fluor-555 conjugated secondary antibodies (EMAR, CHINA) and sealed with mounting medium containing DAPI (Abcam, UK). Stained cells were imaged using a confocal laser-scanning microscope (Olympus, JAPAN).
Quantitative RT-PCR Analysis
To analyze the expression of osteogenesis-related genes in hPDLSCs and inflammation-related genes in THP-1 cells, an ultrapure RNA kit (CWBIO, China) was used to isolate total RNA samples from hPDLSCs and THP-1 cells. Reverse transcription was performed using StarScript III RTase (Genstar, China) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) primer sequences are listed in Table 1. PCR was performed with the SYBR Green Kit (GenStar, China) on the LightCycler® 480 Real-Time PCR System (Roche, Basel, Switzerland). The amplification procedure was set as follows: 95°C for 5 min, 45 cycles of denaturation at 95°C for 10s, annealing at 58°C for 20s, and final extension at 72°C for 20s. The gene expression levels were calculated and analyzed using the 2−ΔΔCt method Table 1.
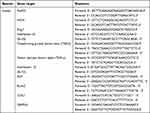 |
Table 1 Primer Sequences Used in Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction |
Transmission Electron Microscopy of Mitochondria
THP-1 cells and hPDLSCs were treated as described previously. The cell culture medium was replaced with 2.5% glutaraldehyde solution at room temperature in darkness. The cells were gently scraped off with a cell scraper, centrifuged at 3000 rpm for 2 min at 25°C to collect the cell clumps, and fixed in fresh 2.5% glutaraldehyde solution, followed by cell dehydration and resin embedding. After re-staining, the sections were imaged under a transmission electron microscope (Hitachi, Tokyo, Japan) to record the mitochondrial morphology.
Evaluation of Intracellular Reactive Oxygen Species Production
The cellular levels of reactive oxygen species (ROS) were measured using a DCFH-DA probe (Sigma-Aldrich). The fluorescence intensity was imaged using a confocal laser-scanning microscope and measured using a flow cytometer.
Mitochondrial Membrane Potential Analysis
The mitochondrial membrane potential was detected using a mitochondrial membrane potential assay kit with JC-1 (Beyotime, CHINA) in accordance with the manufacturer’s protocol. THP-1 cells and hPDLSCs were resuspended and incubated in a freshly prepared JC-1 working solution at 37°C for 20 min. The cells were then analyzed by confocal laser-scanning microscopy.
Analysis of the Oxygen Consumption Rate
Analysis of the mitochondrial oxygen consumption rate (OCR) was performed on a Seahorse XF-96 flux analyzer (Seahorse Biosciences, USA) using a mitochondrial stress test (MST) assay kit (Agilent, USA). THP-1 cells were seeded at a density of 3×104 cells/well in XFe 96-well cell plates, whereas hPDLSCs were seeded at a density of 2×103 cells/well. The cells were treated according to the experimental groups described above. The probe plate was hydrated at 37°C without CO2 overnight the day before experiment. Next day, the sterile water in the probe plate was replaced with hydration solution, and the plate was incubated at 37°C without CO2 for 1 h. Preheated assay solution was used to wash the cells twice gently, and 180 μL of the assay solution was added to the cells, which were incubated at 37°C without CO2 for 1 h. Oligomycin, carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), and rotenone/antimycin were diluted according to the kit instructions and added into wells A, B, and C of the probe plate respectively before testing on the machine. Subsequently, the OCR values (pmol/min) for each well were recorded and averaged for data analysis. The cells were prepared for BCA quantification after detection. The Wave software was used to analyze the data.
Induction of Experimental Periodontitis in Mice
Animals used in this study were maintained in accordance with the Guideline for ethical review of animal welfare of laboratory animals published by the China National Standardization Management Committee (publication No. GB/T 35892–2018). Animal experiments were approved by the Animal Ethical and Welfare Committee of Sun Yat-Sen University (SYSU-IACUC-2022-001592). 40 C57BL/6 mice (female, aged 6–8 weeks) were procured from the Guangdong Medical Laboratory Animal Center (Guangzhou, China), maintained in a standard environment, and supplied with a standard diet and water. 8 mice were selected randomly as the control group, with the remaining 32 used for ligature-induced periodontitis model. To induce periodontal bone loss in mice, a silk ligature was tied around both maxillary second molars under general anesthesia, and the mice were observed every two days to ensure that the silk threads were dislodged and evaluate the periodontal tissue status. 2 weeks later, the ligated mice were randomly divided into the following four groups (n = 8 per group): (1) ligated and PBS treatment (Induced), (2) ligated and Nor-sEV treatment (Induced + Nor-sEV), (3) ligated and NC-sEV treatment (Induced + NC-sEV), and (4) ligated and FoxO1-sEV treatment (Induced + FoxO1-sEV). And 2 mice from each group were randomly selected for CT scanning to confirm the success of the modeling. For sEV treatment, 10 μL (2 μg/μL) of sEV or 10 μL of PBS were injected with a microinjector (Fujian, CHINA) under the proximal and distal mucosa of the palatal side of the second molar in mice three times a week for 4 weeks. Finally, 3 mice from each group were randomly selected for in vivo bioluminescence imaging and CT scanning, while another 3 mice were randomly selected for histological staining.
Micro-Computed Tomography Examination
The fixed maxilla samples were scanned by a micro-computed tomography (CT) scanner (µCT50; SCANCO, Switzerland). Three-dimensional images were obtained and analyzed using the Mimics software (Mimics 17.0; Materialize, Leuven, Belgium). In the reconstructed mouse maxilla, the distances from the proximal-medial, intermediate-medial, and distal-medial cementoenamel junctions to the pinnacle of the alveolar bone (CEJ-ABC) of the second molar were measured, with a total of six measurement sites on the buccal and palatal sides, and the average values were calculated for statistical analyses.
Histological and Immunofluorescence Staining
The maxilla samples were decalcified in 10% ethylenediaminetetraacetic acid (EDTA; GHTECH, China), dehydrated, and subsequently embedded in paraffin at 58°C for 30–60 min. Consecutive sections at a thickness of 4.5 μm were obtained from the defect area and stained with hematoxylin and eosin (H&E, Servicebio, China) or Masson’s trichrome (Servicebio, China) for histological analysis. Sections from each group were subjected to tartrate-resistant acid phosphatase (TRAP) staining using a TRAP Staining Kit (Servicebio, China). Multinucleated TRAP-positive cells were counted as active osteoclasts. Immunohistochemical staining was performed using antibodies against bone sialoprotein (BSP). All samples were observed and images were captured using a digital slice scanner (Leica, Wetzlar, Germany).
For immunofluorescence staining, the sections were incubated with the specific primary antibodies rabbit anti-iNOS (1:500, Cell Signaling Technology, USA) or mouse anti-Arg-1 (1:500, Proteintech, USA) overnight in a wet box at 4°C. The sections were then incubated with anti-rabbit Alexa Fluor-488 or anti-mouse Alexa Fluor-555 conjugated secondary antibodies (EMAR, CHINA) and sealed with mounting medium containing DAPI (Abcam, UK). Stained cells were imaged using a confocal laser-scanning microscope (Olympus, JAPAN).
In vivo Bioluminescence Imaging
To evaluate the ROS levels in vivo, a DCFH-DA probe (Sigma-Aldrich, USA) was injected into the periodontal tissues of mice. The mice were then observed using an IVIS Spectrum in vivo imaging system (PerkinElmer, USA).
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism 8 software package (GraphPad Software, Inc). One-way analysis of variance was conducted, and the post-hoc Bonferroni test was performed for two-group comparisons. All the experimental data were expressed as mean ± standard deviation (SD). Statistical significance was set at p < 0.05.
Results
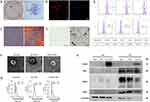
Characterization of hPDLSCs and sEV
The hPDLSCs were characterized as MSCs using a clonogenic proliferative capacity assay (Figure 1A), cell origin identification (Figure 1B), osteogenic lipogenesis induction (Figure 1C and D), and surface-marker identification (Figure 1E). The three groups of sEV (Nor-sEV, NC-sEV, and FoxO1-sEV) were isolated from the media of different hPDLSCs (normal hPDLSCs, negative control hPDLSCs, FoxO1-overexpressed hPDLSCs) by ultracentrifugation. Transmission electron microscopy images showed that all sEV exhibited bilayer membranes and cup-plate-shaped structures with diameters of approximately 100 nm (Figure 1F). Nanosight analysis showed that the average particle sizes were 182.1 (Nor-sEV), 173 (NC-sEV), and 165.1 (FoxO1-sEV) nm (Figure 1G). Western blotting demonstrated that the sEV from FoxO1-overexpressed hPDLSCs were highly overexpressed with the FoxO1 protein, and all sEV showed positive results for the characteristic markers, such as Alix, CD63, and CD81 (Figure 1H). In summary, these results show that hPDLSCs with the three groups of sEV were successfully isolated.
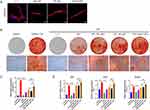
FoxO1-sEV Enhanced Osteogenic Differentiation of hPDLSCs in an Inflammatory Microenvironment
To confirm the uptake of sEV by hPDLSCs, an uptake assay was conducted using different types of sEV. The cytoskeleton was labeled with red fluorescence; the sEV were labeled with green fluorescence; and the nuclei were stained with DAPI. hPDLSCs were able to spontaneously take up the sEV (Figure 2A). To evaluate the effect of FoxO1-sEV on the osteogenic differentiation of hPDLSCs in an inflammatory microenvironment, hPDLSCs were induced in an osteogenesis medium with or without LPS and treated with the same concentration of the three different sEV (50 μg/mL). After 21 days of induction, the cells were assessed using Alizarin Red staining and qRT-PCR. In comparison with the control group, osteogenic induction resulted in a large number of mineralized nodules, indicating the outstanding osteogenic potential of hPDLSCs. In the inflammatory microenvironment induced by LPS treatment, the osteogenic differentiation of hPDLSCs evidently decreased. However, when different sEV were added, osteogenic differentiation of hPDLSCs was strengthened even under inflammatory conditions (Figure 2B). Moreover, FoxO1-sEV had the most significant effect on increasing mineralized nodules and enriching the expression of osteogenesis-related genes among all groups (Figure 2C and D). Altogether, these results suggest that FoxO1-sEV promotes the osteogenesis of hPDLSCs in an inflammatory environment.
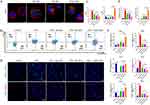
FoxO1-sEV Promoted the Polarization of THP-1 Cells from the M1 Phenotype to the M2 Phenotype
THP-1 cells were able to take up sEV (Figure 3A). To study the effects of FoxO1-sEV on the local inflammatory environment, we treated macrophages with different sEV at the same concentration (50 μg/mL) and evaluated their phenotypic changes by flow cytometry, immunofluorescence analyses, and qRT-PCR. In the absence of induction, few M1 macrophages (CD86) were detected, and M2 macrophages (CD163) accounted for 7.31% of all cells. After induction with LPS, in contrast to the dramatic increase in the number of M1 macrophages, the number of M2 macrophages significantly decreased. Nevertheless, M1 counts significantly decreased with the addition of different sEV, especially with the addition of FoxO1-sEV, whereas M2 counts increased significantly in comparison with those in the control and LPS groups (Figure 3B and C). Next, we evaluated the phenotypic changes in macrophages using immunofluorescence. After LPS induction, the expression of M1 macrophages (iNOS) significantly increased. sEV of distinct origins inhibited M1 formation and enhanced the polarization toward the M2 phenotype (Arg-1). Notably, FoxO1-sEV had the most evident effect on macrophage polarization in comparison with the other sEV (Figure 3D and E). Then, we used qRT-PCR to observe the expression of genes associated with M1 polarization. After LPS treatment, the expression levels of proinflammatory genes such as iNOS, IL-1β and TNF-α, were significantly upregulated, while the expression levels of proinflammatory genes such as IL-10 and TGF-β were downregulated. After treatment with sEV, proinflammatory genes (iNOS, IL-1β and TNF-α) were evidently downregulated, and the expression of the anti-inflammatory gene (Arg-1, IL-10 and TGF-β) increased. Notably, the expression of iNOS, IL-1β, and TNF-α significantly decreased, while the expression of Arg-1, IL-10 and TGF-β dramatically increased in FoxO1-sEV group, in comparison with the groups treated with the other sEV (Figure 3F). These results indicate that FoxO1-sEV efficiently alleviated inflammation by modulating macrophage polarization from M1 to M2.
FoxO1-sEV Stabilized the Mitochondria, Modulated the Antioxidant and Respiratory Function in hPDLSCs
To study the effect of FoxO1-sEV on mitochondria, we observed the structure and dynamics of mitochondria by transmission electron microscopy and found swollen mitochondria with a vacuolar structure and incomplete mitochondrial cristae in hPDLSCs. After treatment with Nor-sEV, NC-sEV, or FoxO1-sEV, the number of swollen and damaged mitochondria decreased by varying degrees (Figure 4A and B). Statistical analyses suggested that the proportion of normal mitochondria significantly increased with the addition of sEV. This effect was particularly evident following FoxO1-sEV treatment.
To detect cellular oxidative stress, we evaluated ATP (Figure 4C) and ROS (Figure 4D and E) production using flow cytometry and confocal microscopy. Results showed that the ROS level stimulated with H2O2 was significantly increased (Additional file 1: Figure S1A). LPS induction increased ROS levels, whereas different kinds of sEV treatments lowered it, particularly treatment with FoxO1-sEV.
To evaluate mitochondrial impairment, we observed the mitochondrial membrane potential using confocal microscopy. After Carbonyl Cyanide 3-ChloroPhenylhydrazone (CCCP) stimulation, the mitochondrial membrane was depolarized, producing a JC-1 monomer that emitted green fluorescence (Additional file 1: Figure S1B). In comparison with the control group, the group treated with LPS showed a decline in mitochondrial membrane potential. However, this downregulation was reversed to different degrees after sEV treatment, especially after FoxO1-sEV treatment (Figure 4F). Thus, FoxO1-sEV can stabilize the mitochondrial membrane potential in hPDLSCs. We also measured the OCR, a key parameter that reflects mitochondrial function. Our results showed that inflammation resulted in decreased basal respiration, maximal respiration, ATP production, and spare-respiratory capacity, indicating mitochondrial dysfunction. Nevertheless, with sEV treatment, particularly treatment with FoxO1-sEV, mitochondrial respiratory function recovered (Figure 4G and H).
These results suggest that FoxO1-sEV effectively stabilized the mitochondria, modulated the antioxidant and protects mitochondrial respiratory function in hPDLSCs.
FoxO1-sEV Stabilized the Mitochondria, Modulated the Antioxidant and Respiratory Function in THP-1 Cells
In our research, we explored the impact of FoxO1-sEV on mitochondria, assessed cellular oxidative stress, and examined mitochondrial respiratory function in THP-1 cells. Notably, we observed a similar phenomenon in THP-1 cells as we did in hPDLSCs. LPS induction disrupted mitochondrial structure (Figure 5A and B), reduced ATP levels (Figure 5C), increased ROS levels (Figure 5D and E), and impaired mitochondrial respiratory function (Figure 5F–H). After treatment with different sEVs, our experimental results revealed that all three groups of sEVs were able to reduce mitochondrial disruption (Figure 5A and B), restore ATP levels (Figure 5C) and decrease ROS levels (Figure 5D and E). In addition, we also observed that the three groups of sEV could restore the mitochondrial membrane potential of THP-1 cells under inflammatory environment (Figure 5F). Moreover, OCR assay showed that sEV weakened the inhibition of LPS on cellular mitochondrial respiratory function (Figure 5G and H). The above results suggested that sEV, particularly FoxO1-sEV, efficiently stabilize the mitochondrial structure, rescue impaired mitochondria and ultimately enhance the antioxidant abilities of THP-1 cells.
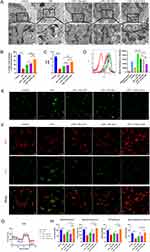
FoxO1-sEV Inhibited Bone Resorption Effectively in Experimental Periodontitis in Mice
To determine whether FoxO1-sEV is effective in treating periodontitis, we established ligature-induced experimental periodontitis in mice. Bone resorption was induced on both the buccal and palatine sides of the oral cavity. Local injections of Nor-sEV, NC-sEV, and FoxO1-sEV inhibited bone resorption to some extent, with the most significant effect observed in the FoxO1-sEV group (Figure 6A and B). Next, we examined the periodontal tissue in each group by using H&E and Masson’s staining. Our results showed that in mice with periodontitis, bone resorption occurred in the alveolar ridge crest, and was characterized by a decrease in height and increased infiltration of local inflammatory cells. Masson’s trichrome staining indicated an evident reduction in the blue staining of collagen fibers. However, after sEV treatment, bone resorption was inhibited, and the number of collagen fibers increased, indicating less severe periodontitis in the sEV groups, particularly in the FoxO1-sEV group (Figure 6C). TRAP staining and immunohistochemical staining with BSP revealed fewer osteoclasts and higher BSP expression in the FoxO1-sEV group than in the other groups (Figure 6D and E). Furthermore, we evaluated the number of iNOS- and Arg-1-positive cells to determine the damage to the periodontal tissue. In mice with periodontitis, the number of iNOS-positive cells increased while the number of Arg-1-positive cells decreased significantly. Nevertheless, after Nor-sEV, NC-sEV, and FoxO1-sEV injections, the number of iNOS-positive cells decreased, whereas the number of Arg-1-positive cells increased, with FoxO1-sEV showing the most evident effect among the sEV (Figure 6F and G). We also measured ROS levels in vivo, and the results showed that the FoxO1-sEV group had lower ROS production (Figure 6H). Taken together, these results show that FoxO1-sEV effectively inhibited bone loss in experimental periodontitis in mice.
Discussion
FoxO1 travels between the cytoplasm and nucleus35 and is involved in the regulation of a variety of physiological processes, such as energy metabolism, apoptosis, self-regulation, and oxidative stress. Therefore, special drugs have been developed for the treatment of related diseases by targeting FoxO1 directly or by modulating its upstream regulators.37,38, Targeted activation of FoxO1 to regulate osteogenesis has been shown to enhance the treatment of bone regeneration-related diseases and promote osteogenesis both in vitro and in vivo.41,42, A related study showed that the FoxO1 protein can help promote periodontal stem cell osteogenic function and periodontal tissue regeneration.30,31 However, since the FoxO family members show similar structural domains, the ability of such targeted drugs to activate other member and induce other effects is unclear.37
MSCs are widely used as therapeutic agents for systemic inflammatory diseases and tissue regeneration.6 Dental MSCs originate from the cranial neural crest-derived mesenchyme, have powerful self-renewal and multilineage differentiation abilities, are easily accessible, and have great clinical potential for periodontal tissue regeneration.45,46 Among dental MSCs, hPDLSCs are considered the most suitable for periodontal tissue engineering.47 Meanwhile, growing evidence from preclinical studies suggests that cell-free therapies such as sEV therapy can be used for treating periodontitis.48,49 A recent study showed that sEV derived from healthy hPDLSCs accelerated bone formation during experimental periodontitis in rats.40 Our study also showed that sEV from normal or negative control hPDLSCs promoted periodontal tissue regeneration in mice with experimental periodontitis.
In previous studies, proteomics analysis and mRNA sequencing of vesicles revealed that they contained FoxO1 proteins and mRNA.39,44, This study is the first to successfully obtain FoxO1-overexpressed sEV by genetic engineering technology. A recent study indicated that IL4-loaded small extracellular vesicles from genetically engineered parent cells showed more potent therapeutic effects.36 The potential of sEV as delivery carriers has been extensively explored and recognized in many studies.50 Therefore, sEV obtained using genetically engineered sEV-producing cells are promising candidates as ideal delivery carriers.43 In this study, we obtained FoxO1-overexpressed sEV using genetic engineering technology and achieved targeted regulation of FoxO1 protein levels. Our study also showed that FoxO1 loading may enhance the therapeutic effects of sEV. In addition, FoxO1 loading had no effect on the morphology, size, or surface markers of the sEV.
In periodontitis, macrophages and MSCs are jointly involved in periodontal tissue bone remodeling.51 Studies have shown that, in the unique immune microenvironment of the alveolar bone, monocytes/macrophages have the most active interactions with MSCs in comparison with other immune cells.52 Cellular endocytosis is the basis of the biological functions of sEV.53 In this study, we demonstrated that labeled PKH26 sEV can be taken up and internalized by hPDLSCs and THP-1 macrophages. Thus, we engineered the sEV to load FoxO1 without altering the physicochemical properties of the sEV. Additionally, our results suggest that in comparison with sEV derived from normal or negative control hPDLSCs, FoxO1-overexpressed sEV showed superior osteogenesis-promoting ability in inflammation-impaired hPDLSCs in vitro. In previous studies, we found that inflammatory conditions downregulate the expression of FoxO1 in cells while inhibiting cellular osteogenic functions in vitro.30 FoxO1 is an important regulator of balanced redox reactions in osteoblasts,54,55 and targeting FoxO1 to regulate the osteogenic process has been demonstrated to be conducive to the enhancement of osteogenic and antioxidant capacities of MSCs.39 Our results suggest that FoxO1-overexpressed sEV can be internalized by target cells and may restore osteogenic function in inflammation-impaired hPDLSCs by affecting intracellular FoxO1 levels. Macrophages, as highly plastic immune cells, play an important role in bone metabolism balance.56 Classical M1 macrophages in an inflammatory environment primarily exhibit pro-inflammatory and antimicrobial functions. M1 macrophages secrete a large number of pro-inflammatory factors, such as TNF-α, which can inhibit bone formation.57 Conversely, alternative M2 macrophages mainly have anti-inflammatory and reparative functions. M2 macrophages not only secrete anti-inflammatory factors to regulate the inflammatory microenvironment but also participate in promoting bone defect repair through the secretion of BMP-2.58 Simultaneously, our experiments showed that FoxO1-overexpressed sEV were more effective in regulating the macrophage phenotype and inhibiting inflammation. In agreement with previous studies, hPDLSCs not only promoted macrophage M2 polarization via the paracrine pathway, but also regulated the periodontal tissue immune microenvironment to promote periodontal tissue regeneration.59
A previous study reported that FoxO1 expression was generally low in clinical samples from patients with periodontitis.60 FoxO1 deficiency affects mitochondrial homeostasis and cellular function.61 Recent studies have also shown that mitochondrial dysfunction is closely associated with the development of periodontitis.21 In this context, the present study is the first to propose and investigate the construction of FoxO1-overexpressed sEV for the targeted treatment of periodontitis. In our study, LPS-stimulated cells showed oxidative stress, mitochondrial structural disruption, and impaired function, consistent with the results of past studies.62 Mitochondrial damage is a key cause of cellular dysfunction, which manifests as oxidative stress, decreased metabolism, disruption of mitochondrial structure, inhibition of oxidative phosphorylation, and impaired performance.63,64 In this study, we confirmed that sEV treatment alleviates cellular inflammatory damage. Moreover, FoxO1-overexpressed sEV were more effective in maintaining mitochondrial homeostasis in an inflammatory microenvironment. Previous studies have concluded that FoxO1 is localized in the mitochondria and regulates various pathways, such as the mitochondrial apoptotic pathway, and mitochondrial ROS, affecting mitochondrial respiration and dynamics and maintaining mitochondrial calcium homeostasis through the mitochondrial apoptotic pathway and mitochondrial function.65 Furthermore, FoxO1 can promote cellular antioxidant defense by upregulating the expression of antioxidant protein genes such as SOD2 and SEPP1.66 Resveratrol has been shown to enhance cellular resistance to oxidative stress by activating the SIRT1/FoxO1 signaling pathway.67 In our study, we found that FoxO1-overexpressed sEV were effective in reducing oxidative stress damage caused by inflammatory stimuli and in maintaining cellular mitochondrial morphology and respiratory function. These results suggested that FoxO1-overexpressed sEV are involved in regulating mitochondrial homeostasis.
Although the effects of other molecules besides FoxO1 in the sEV on the target cells are not yet clear, the present study demonstrated the effect of the direct delivery of FoxO1. However, the specific regulatory mechanism of FoxO1-overexpressed sEV after entering target cells requires further verification. Consistent with the in vitro experiments, we found that periodontal tissues were destroyed and ROS levels were increased in mice with periodontitis. Conversely, periodontitis progression was inhibited and ROS levels were decreased in mice treated with different sEV, and this effect was more pronounced with FoxO1-overexpressed sEV. In summary, these findings suggest that FoxO1-overexpressed sEV may provide new information for the development of treatments for periodontitis associated with the regulation of mitochondrial homeostasis.
Conclusion
In this study, we successfully obtained FoxO1-overexpressed sEV using an engineering technology. We utilized the favorable carrier function and biocompatibility of sEV to deliver the FoxO1 protein to target cells. Our results suggested that FoxO1-overexpressed sEV yield a significantly better in inhibiting bone loss in periodontitis than sEV derived from normal and negative control hPDLSCs. We also found that FoxO1-overexpressed sEV may maintain bone homeostasis and promote bone repair in periodontitis by restoring the mitochondrial function of hPDLSCs and THP-1 cells in inflamed tissues. However, owing to the limitations of this study, the detailed mechanism requires further exploration. In conclusion, FoxO1-overexpressed sEV are expected to be a new approach for the targeted treatment of periodontitis.
Acknowledgments
Graphical abstract were created with BioRender.com. This work was supported by the National Natural Science Foundation of China (Nos. 82071120, 82270978) and Open Project of Guangdong Key Laboratory of Stomatology (No. KF2022120101).
Disclosure
All the authors report no conflicts of interest in this work.
References
1. Slots J. Periodontitis: facts, fallacies and the future. Periodontol 2000. 2017;75(1):7–23. doi:10.1111/prd.12221
2. Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nature Reviews Disease Primers. 2017;3(1):17038. doi:10.1038/nrdp.2017.38
3. Luo LS, Luan HH, Wu L, et al. Secular trends in severe periodontitis incidence, prevalence and disability-adjusted life years in five Asian countries: a comparative study from 1990 to 2017. J Clin Periodontol. 2021;48(5):627–637. doi:10.1111/jcpe.13447
4. Li J, Wang Y, Tang M, et al. New insights into nanotherapeutics for periodontitis: a triple concerto of antimicrobial activity, immunomodulation and periodontium regeneration. J Nanobiotechnol. 2024;22(1):19. doi:10.1186/s12951-023-02261-y
5. Lai H, Li J, Kou X, Mao X, Zhao W, Ma L. Extracellular vesicles for dental pulp and periodontal regeneration. Pharmaceutics. 2023;15(1):282. doi:10.3390/pharmaceutics15010282
6. Li P, Ou Q, Shi S, Shao C. Immunomodulatory properties of mesenchymal stem cells/dental stem cells and their therapeutic applications. Cell. Mol. Immunol. 2023;20(6):558–569. doi:10.1038/s41423-023-00998-y
7. van Niel G, Carter DRF, Clayton A, Lambert DW, Raposo G, Vader P. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat Rev Mol Cell Biol. 2022;23(5):369–382. doi:10.1038/s41580-022-00460-3
8. Jia Y, Yu L, Ma T, et al. Small extracellular vesicles isolation and separation: current techniques, pending questions and clinical applications. Theranostics. 2022;12(15):6548–6575. doi:10.7150/thno.74305
9. Qiu J, Wang X, Zhou H, et al. Enhancement of periodontal tissue regeneration by conditioned media from gingiva-derived or periodontal ligament-derived mesenchymal stem cells: a comparative study in rats. Stem Cell Res Ther. 2020;11(1):42. doi:10.1186/s13287-019-1546-9
10. Lotfy A, AboQuella NM, Wang H. Mesenchymal stromal/stem cell (MSC)-derived exosomes in clinical trials. Stem Cell Res Ther. 2023;14(1):66. doi:10.1186/s13287-023-03287-7
11. Kou M, Huang L, Yang J, et al. Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: a next generation therapeutic tool? Cell Death Dis. 2022;13(7):580. doi:10.1038/s41419-022-05034-x
12. Kim HY, Kwon S, Um W, et al. Functional extracellular vesicles for regenerative medicine. Small. 2022;18(36):e2106569. doi:10.1002/smll.202106569
13. Wang J, Chen D, Ho EA. Challenges in the development and establishment of exosome-based drug delivery systems. J Control Release. 2021;329:894–906. doi:10.1016/j.jconrel.2020.10.020
14. Iaquinta MR, Lanzillotti C, Mazziotta C, et al. The role of microRNAs in the osteogenic and chondrogenic differentiation of mesenchymal stem cells and bone pathologies. Theranostics. 2021;11(13):6573–6591. doi:10.7150/thno.55664
15. Bei HP, Hung PM, Yeung HL, Wang S, Zhao X. Bone-a-Petite: engineering exosomes towards bone, osteochondral, and cartilage repair. Small. 2021;17(50):e2101741. doi:10.1002/smll.202101741
16. Pan Z, Sun W, Chen Y, et al. Extracellular vesicles in tissue engineering: biology and engineered strategy. Adv. Healthcare Mater. 2022;11(21):e2201384. doi:10.1002/adhm.202201384
17. Zou X, Yuan M, Zhang T, et al. Extracellular vesicles expressing a single-chain variable fragment of an HIV-1 specific antibody selectively target Env+ tissues. Theranostics. 2019;9(19):5657–5671. doi:10.7150/thno.33925
18. Thomas BL, Eldridge SE, Nosrati B, et al. WNT3A‐loaded exosomes enable cartilage repair. J Extracell Vesicles. 2021;10(7):e12088. doi:10.1002/jev2.12088
19. Wang W, Liang X, Zheng K, et al. Horizon of exosome-mediated bone tissue regeneration: the all-rounder role in biomaterial engineering. Mater Today Bio. 2022;16:100355. doi:10.1016/j.mtbio.2022.100355
20. Song H, Chen X, Hao Y, Wang J, Xie Q, Wang X. Nanoengineering facilitating the target mission: targeted extracellular vesicles delivery systems design. J Nanobiotechnol. 2022;20(1):431. doi:10.1186/s12951-022-01638-9
21. Jiang W, Wang Y, Cao Z, et al. The role of mitochondrial dysfunction in periodontitis: from mechanisms to therapeutic strategy. J Periodontal Res. 2023;58(5):853–863. doi:10.1111/jre.13152
22. Li X, Tian BM, Deng DK, et al. LncRNA GACAT2 binds with protein PKM1/2 to regulate cell mitochondrial function and cementogenesis in an inflammatory environment. Bone Res. 2022;10(1):29. doi:10.1038/s41413-022-00197-x
23. Yin L, Li X, Hou J. Macrophages in periodontitis: a dynamic shift between tissue destruction and repair. Japanese Dent Sci Rev. 2022;58:336–347. doi:10.1016/j.jdsr.2022.10.002
24. Wculek SK, Dunphy G, Heras-Murillo I, Mastrangelo A, Sancho D. Metabolism of tissue macrophages in homeostasis and pathology. Cell. Mol. Immunol. 2022;19(3):384–408.
25. Zhai Q, Chen X, Fei D, et al. 2022 Nanorepairers Rescue Inflammation-Induced Mitochondrial Dysfunction in Mesenchymal Stem Cells. Adv Sci94 e2103839
26. Peng S, Fu H, Li R, et al. A new direction in periodontitis treatment: biomaterial-mediated macrophage immunotherapy. J Nanobiotechnol. 2024;22(1):359.
27. Santos BF, Grenho I, Martel PJ, Ferreira BI, Link W. FOXO family isoforms. Cell Death Dis. 2023;14(10):702.
28. Chen D, Gong Y, Xu L, Zhou M, Li J, Song J. Bidirectional regulation of osteogenic differentiation by the FOXO subfamily of Forkhead transcription factors in mammalian MSCs. Cell Proliferation. 2019;52(2):e12540. doi:10.1111/cpr.12540
29. Siqueira MF, Flowers S, Bhattacharya R, et al. FOXO1 modulates osteoblast differentiation. Bone. 2011;48(5):1043–1051. doi:10.1016/j.bone.2011.01.019
30. Huang X, Chen H, Xie Y, Cao Z, Lin X, Wang Y. FoxO1 Overexpression Ameliorates TNF-α-induced oxidative damage and promotes osteogenesis of human periodontal ligament stem cells via antioxidant defense activation. Stem Cells Int. 2019;2019:2120453.
31. Huang X, Su X, Ma Q, et al. FoxO1 agonists promote bone regeneration in periodontitis by protecting the osteogenesis of periodontal ligament stem cells. Stem Cells Dev. 2023;32(15–16):491–503. doi:10.1089/scd.2023.0013
32. Ren L, Yang J, Wang J, Zhou X, Liu C. The roles of FOXO1 in periodontal homeostasis and disease. J Immunol Res. 2021;2021:5557095. doi:10.1155/2021/5557095
33. Wang C, Li X, Cheng T, Wang L, Jin L. RNA sequencing reveals the upregulation of FOXO signaling pathway in porphyromonas gingivalis persister-treated human gingival epithelial cells. Int J Mol Sci. 2022;23(10):5728. doi:10.3390/ijms23105728
34. Welsh JA, Goberdhan DCI, O’Driscoll L, et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles. 2024;13(2):e12404. doi:10.1002/jev2.12404
35. Zhang X, Jiang L, Liu H. Forkhead box protein O1: functional diversity and post-translational modification, a new therapeutic target? Drug Des Devel Ther. 2021;15: 1851–1860. doi:10.2147/DDDT.S305016
36. Takenaka M, Yabuta A, Takahashi Y, and Takakura Y. Interleukin-4- carrying small extracellular vesicles with a high potential as anti-inflammatory therapeutics based on modulation of macrophage function. Biomaterials. 2021;278:121160. doi:10.1016/j.biomaterials.2021.121160
37. Orea-Soufi A, Paik J, Bragança J, Donlon TA, Willcox BJ, Link W. FOXO transcription factors as therapeutic targets in human diseases. Trends Pharmacol Sci. 2022;43(12):1070–1084. doi:10.1016/j.tips.2022.09.010
38. Calissi G, Lam EW, Link W. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov. 2021;20(1):21–38. doi:10.1038/s41573-020-0088-2
39. Hong BS, Cho JH, Kim H, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10(1):556. doi:10.1186/1471-2164-10-556
40. Lei F, Li M, Lin T, Zhou H, Wang F, Su X. Treatment of inflammatory bone loss in periodontitis by stem cell-derived exosomes. Acta Biomater. 2022;141:333–343. doi:10.1016/j.actbio.2021.12.035
41. Huang J, Li R, Yang J, et al. Bioadaptation of implants to In vitro and In vivo oxidative stress pathological conditions via nanotopography-induced FoxO1 signaling pathways to enhance Osteoimmunal regeneration. Bioact. Mater. 2021;6(10):3164–3176. doi:10.1016/j.bioactmat.2021.02.023
42. Yang J, Liang J, Zhu Y, et al. Fullerol-hydrogel microfluidic spheres for in situ redox regulation of stem cell fate and refractory bone healing. Bioact. Mater. 2021;6(12):4801–4815. doi:10.1016/j.bioactmat.2021.05.024
43. Yim N, Ryu SW, Choi K, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat Commun. 2016;7(1):12277. doi:10.1038/ncomms12277
44. Skog J, Würdinger T, and van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi:10.1038/ncb1800
45. Cabaña-Muñoz ME, Pelaz Fernández MJ, Parmigiani-Cabaña JM, Parmigiani-Izquierdo JM, Merino JJ. Adult mesenchymal stem cells from oral cavity and surrounding areas: types and biomedical applications. Pharmaceutics. 2023;15(8):2109. doi:10.3390/pharmaceutics15082109
46. Ouchi T, Nakagawa T. Mesenchymal stem cell-based tissue regeneration therapies for periodontitis. Regener Ther. 2020;14:72–78. doi:10.1016/j.reth.2019.12.011
47. Roato I, Chinigò G, Genova T, Munaron L, Mussano F. Oral cavity as a source of mesenchymal stem cells useful for regenerative medicine in dentistry. Biomedicines. 2021;9(9):1085. doi:10.3390/biomedicines9091085
48. Lin H, Chen H, Zhao X, et al. Advances of exosomes in periodontitis treatment. J Transl Med. 2022;20(1):279. doi:10.1186/s12967-022-03487-4
49. Lin H, Chen H, Zhao X, et al. Advances in mesenchymal stem cell conditioned medium-mediated periodontal tissue regeneration. J Transl Med. 2021;19(1):456. doi:10.1186/s12967-021-03125-5
50. Zou Z, Li H, Xu G, Hu Y, Zhang W, Tian K. Current knowledge and future perspectives of exosomes as nanocarriers in diagnosis and treatment of diseases. Int j Nanomed. 2023;18:4751–4778. doi:10.2147/IJN.S417422
51. Hathaway-Schrader JD, Novince CM. Maintaining homeostatic control of periodontal bone tissue. Periodontol 2000. 2021;86(1):157–187. doi:10.1111/prd.12368
52. Hienz SA, Paliwal S, Ivanovski S. Mechanisms of bone resorption in periodontitis. J Immunol Res. 2015;2015:615486. doi:10.1155/2015/615486
53. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. doi:10.1038/s41556-018-0250-9
54. Rached MT, Kode A, Xu L, et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010;11(2):147–160. doi:10.1016/j.cmet.2010.01.001
55. Ambrogini E, Almeida M, Martin-Millan M, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11(2):136–146. doi:10.1016/j.cmet.2009.12.009
56. Hu Y, Huang J, Chen C, et al. Strategies of macrophages to maintain bone homeostasis and promote bone repair: a narrative review. J Funct Biomater. 2022;14(1):18. doi:10.3390/jfb14010018
57. Hu K, Shang Z, Yang X, Zhang Y, Cao L. Macrophage polarization and the regulation of bone immunity in bone homeostasis. J Inflamm Res. 2023;16:3563–3580. doi:10.2147/JIR.S423819
58. Chen S, Ni S, Liu C, et al. Neglected immunoregulation: M2 polarization of macrophages triggered by low-dose irradiation plays an important role in bone regeneration. J Cell & Mol Med. 2023;27(8):1095–1109. doi:10.1111/jcmm.17721
59. Liu J, Wang H, Zhang L, et al. Periodontal ligament stem cells promote polarization of M2 macrophages. J Leukoc Biol. 2022;111(6):1185–1197. doi:10.1002/JLB.1MA1220-853RR
60. Wang Q, Shi W, Lin S, Wang H. FOXO1 regulates osteogenic differentiation of periodontal ligament stem cells through the METTL3 signaling pathway. J Orthopaedic Surg Res. 2023;18(1):637. doi:10.1186/s13018-023-04120-w
61. Cheng Z. FoxO transcription factors in mitochondrial homeostasis. Biochem J. 2022;479(4):525–536. doi:10.1042/BCJ20210777
62. Fock EM, Parnova RG. Protective effect of mitochondria-targeted antioxidants against inflammatory response to lipopolysaccharide challenge: a review. Pharmaceutics. 2021;13(2):144. doi:10.3390/pharmaceutics13020144
63. Zheng CX, Sui BD, Qiu XY, Hu CH, Jin Y. Mitochondrial Regulation of Stem Cells in Bone Homeostasis. Trends Mol Med. 2020;26(1):89–104. doi:10.1016/j.molmed.2019.04.008
64. Liu Y, Xu R, Gu H, et al. Metabolic reprogramming in macrophage responses. Biomarker Res. 2021;9(1):1. doi:10.1186/s40364-020-00251-y
65. Jerome MS, Kuthethur R, Kabekkodu SP, Chakrabarty S. Regulation of mitochondrial function by forkhead transcription factors. Biochimie. 2022;198:96–108. doi:10.1016/j.biochi.2022.03.013
66. Ragu S, Droin N, Matos-Rodrigues G, et al. A noncanonical response to replication stress protects genome stability through ROS production, in an adaptive manner. Cell Death Differ. 2023;30(5):1349–1365. doi:10.1038/s41418-023-01141-0
67. Jiang Y, Luo W, Wang B, Wang X, Gong P, Xiong Y. Resveratrol promotes osteogenesis via activating SIRT1/FoxO1 pathway in osteoporosis mice. Life Sci. 2020;246(117422):117422. doi:10.1016/j.lfs.2020.117422
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.