Back to Journals » Journal of Pain Research » Volume 18
Genetic Evidence for Causal Effects of Immune Cell Subtypes on Postherpetic Neuralgia
Authors Chen T , Tang S, Chen R , Ding W, Chen Y, Jian Z, Wu M, Jia M, Zhang X
Received 30 October 2024
Accepted for publication 21 March 2025
Published 31 March 2025 Volume 2025:18 Pages 1721—1734
DOI https://doi.org/10.2147/JPR.S503748
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Wendy Imlach
Taichang Chen,1,* Songjiang Tang,1,* Rong Chen,1 Wei Ding,2 Ying Chen,1 Zhonglu Jian,1 Min Wu,3 Min Jia,4 Xiuyi Zhang5
1Department of Anesthesiology, First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, People’s Republic of China; 2Department of Urology, First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, People’s Republic of China; 3The First Clinical School of Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, People’s Republic of China; 4Department of Dermatology, First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, People’s Republic of China; 5Anesthesiology Department of Qiandongnan Traditional Chinese Medicine Hospital, Kaili, Guizhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Taichang Chen; Xiuyi Zhang, Email [email protected]; [email protected]
Background: Recent evidence indicates that immune cells are crucial in modulating the pathogenesis of postherpetic neuralgia (PHN), with significant associations identified between immune responses and the development of PHN. However, the specific dynamic immune profile, the underlying molecular mechanisms, and especially the causal relationship between immune cells and PHN have yet to be comprehensively elucidated.
Methods: We implemented a comprehensive analytical framework incorporating two-sample Mendelian randomization (MR), multivariable Mendelian randomization(MVMR), and colocalization analyses to elucidate the causal relationships between immune cell phenotypes and PHN. Utilizing publicly available genetic datasets, we explored potential causal associations between 731 immune cell phenotypes and susceptibility to PHN. Comprehensive sensitivity analyses were performed to assess the robustness of the findings, evaluate heterogeneity, and investigate horizontal pleiotropy.The Steiger directionality test was utilized to address and reduce the likelihood of reverse causation.
Results: After applying the Bonferroni-adjusted, eight immune cell phenotypes exhibited significant causal associations with PHN. Further MVMR analysis revealed a significant positive causal relationship between CD27 on IgD- CD38dim B cell and the risk of PHN, with an odds ratio (OR) of 1.228 (95% confidence interval [CI]: 1.059– 1.566, P = 0.011). Colocalization analysis offered limited evidence supporting a shared genetic architecture.
Conclusion: Our findings present compelling genetic evidence that identifies CD27 on IgD- CD38dim B cell as a potential therapeutic target for the prevention and treatment of PHN. This study reinforces the mechanistic connection between immune cell function and the pathogenesis of PHN, highlighting the necessity for further exploration in this area. These insights provide significant guidance for future clinical research and the development of therapeutic strategies.
Keywords: postherpetic neuralgia, causal relationships, colocalization analysis, MVMR, immune cells, CD27 on IgD- CD38dim B cell
Introduction
Postherpetic neuralgia (PHN), a prevalent and debilitating sequela of herpes zoster (HZ), is clinically delineated by the persistence of pain extending beyond three months subsequent to the initial onset of HZ infection.1,2 This severe form of neuropathic pain is typified by enduring cutaneous discomfort within the affected dermatomes, resulting in a significant decline in patients’ quality of life.3 The condition is characterized by a multifaceted array of symptoms, encompassing sleep disturbances, psychological manifestations such as depression and anxiety, and substantial impairment of physical function. In severe instances, PHN may advance to permanent disability and potentially result in life-threatening complications.4
The pathogenesis of PHN is characterized by a complex and multifactorial process. A critical mechanism in this process is immune activation, during which the immune system produces cytokines and inflammatory mediators that cause neuronal damage, ultimately resulting in chronic pain.5 Additionally, immune dysregulation can significantly modify central pain processing mechanisms, leading to prolonged pain perception even after the initial injury has resolved.6 Immune-mediated inflammatory responses have been identified as critical factors in the pathogenesis and progression of neuropathic pain. A growing body of evidence indicates a substantial role for immune cells in the development of neuropathic pain.7,8 Design-based stereological analyses comparing CD1a(+) Langerhans cell (LC) populations in skin biopsies from pain-affected and non-affected areas in adults with and without PHN demonstrated no significant differences in LC density between herpes zoster-affected sites and control specimens.9 Subsequent study has identified macrophages expressing Agtr2 as the primary immune cell infiltrate at sites of nerve injury, where they play a crucial role in mediating pain sensitization.10 Additionally, patients with PHN demonstrated significantly higher frequencies of PD-1+CD4+ T cells and varicella-zoster virus (VZV)-specific PD-1+CD4+ T cells, along with increased production of tumor necrosis factor-alpha (TNF-α) by VZV-specific T cells, in comparison to individuals without PHN.11 Despite these compelling associations, traditional observational studies exhibit intrinsic limitations, including potential reverse causation and residual confounding variables. As a result, the precise causal relationship between specific immune cell subtypes and the development of PHN has yet to be definitively established. Therefore, a more robust approach is needed to determine which immune cell populations may causally contribute to PHN development.
In order to elucidate the causal relationships between immune cell phenotypes and the development of PHN, we utilized MR methodology. This robust genetic epidemiological approach employs genetic variants as instrumental variables, thereby mitigating the confounding and reverse causation issues commonly associated with traditional observational studies.In the absence of randomized controlled trials (RCTs), MR stands out as a particularly persuasive approach for investigating causal associations between exposures and outcomes.12,13 Through the utilization of genetic variants as instrumental variables (IVs) for exposures, such as the levels of a specific Immune cell, MR analysis can enhance the robustness of causal inference by mitigating unobserved confounding and attenuating reverse causation.14,15 The IVs approach employed in this study closely resembles RCTs by randomly assigning genetic variations at conception. Under specific assumptions, the MR framework also emulates a randomized controlled trial.16 In contrast to traditional epidemiological methods, this approach minimizes the impact of confounding factors such as gender and age, thereby facilitating more reliable causal inference.17 Additionally, the risk of reverse causation is reduced in MR studies due to the formation of genotypes before the onset of disease.
In this study, we employed a comprehensive analytical framework that integrates both univariable Mendelian randomization (UVMR) and Mendelian randomization (MVMR) analyses. By using genetic variants as IVs, we aimed to assess the causal relationships between immune cell phenotypes and PHN. This dual-approach strategy facilitated the evaluation of both direct and pleiotropic effects within the causal pathway.We identified evidence of associations between genetic variants and potential confounding factors influencing immune cell levels, which were subsequently addressed through sensitivity analyses. Assuming that genetic variants affect PHN development exclusively through their impact on exposure (the exclusion restriction assumption), MR analysis offered a robust framework for assessing the causal effect of immune cell characteristics on PHN susceptibility.Additionally, we utilized colocalization analysis to examine the shared genetic architecture and immune-mediated mechanisms underlying the pathogenesis of PHN. This complementary methodology facilitated the identification of potentially causal variants that influence both immune cell phenotypes and PHN risk via common biological pathways.
Methods And Materials
Study Design
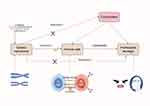
This study utilized comprehensive UVMR and MVMR analyses to investigate potential causal relationships between Immune cells and PHN. The MR analysis and design schematic are depicted in Figure 1.In order to ensure the validity of MR studies, adherence to three key criteria is essential. Firstly, the single nucleotide polymorphisms (SNPs) of the selected IVs must exhibit a significant association with the Immune cells under investigation. Secondly, the IVs should demonstrate independence from other potential confounding factors. Lastly, the IVs must exert their influence on the outcome through the exposure factor rather than through their own effects.18 Given that this study made use of publicly available GWAS data, no additional ethical approval was necessary. The MR analyses conducted in this study were carried out using the Two Sample MR, MRPRESSO, and coloc MR packages within the R software platform (version 4.2.1).
GWAS Data for Immune Cells
Summary statistics from genome-wide association studies (GWAS) pertaining to immune traits were sourced from the GWAS Catalog (ID: GCST0001391-GCST0002121), covering 731 unique immune phenotypes.19 The initial GWAS was executed on a cohort comprising 3757 individuals of European descent, ensuring no population overlap between studies. High-density array genotyping was employed to analyze approximately 22 million SNPs, utilizing a reference panel derived from Sardinian sequences. Association testing was performed with adjustments for pertinent covariates, including sex, age, and age-squared (age²).20 Comprehensive information regarding the aggregated GWAS data can be accessed in the primary publication and the GWAS Catalog. Supplementary Table 1 contains a complete list of the 731 Immune cells, along with their respective names, the GWAS Catalog IDs, and ancestry.
GWAS Data for PHN
Initiated in 2017, the FinnGen study (https://finngen.gitbook.io/documentation/) represents a substantial national cohort endeavor established through a public-private partnership. This initiative integrates genetic data from the Finnish Biobank with digital health records from the Finnish Health Register, thereby providing a distinctive opportunity to examine genetic variations linked to disease trajectories within isolated populations.21 We acquired summary-level estimates of genetic associations with RA from the most recent publicly available R11 data release of the FinnGen study (https://storage.googleapis.com/finngen-public-data-r11/summary_stats/finngen_R11_G6_POSTZOST.gz). PHN diagnoses were classified according to the International Classification of Diseases, Tenth Revision (ICD-10) code B02.2, encompassing 420 cases and 395,832 controls. Genome-wide association analyses for each trait were adjusted for variables including sex, age, genetic ancestry, and genotyping batch.
IVs Selection
In order to test hypothesis (1), we employed a rigorous selection process to identify instrumental variables (IVs) associated with Immune cells from various perspectives. Due to the limited availability of single nucleotide polymorphisms (SNPs) linked to Immune cells, we adjusted the significance threshold to P < 5×10-6 for the selection of relevant SNPs. Subsequently, we pruned the SNPs by excluding those in linkage disequilibrium (LD), defined as R2 > 0.001 within a 10,000 kb range, a criterion commonly utilized in prior research.22,23 To address potential bias stemming from weak IVs, we computed the R2 and F-statistics for each SNP.24 SNPs with F < 10 were defined as weak IVs and were subsequently excluded from the analysis.17 Subsequently, SNPs linked to metabolites were isolated from the findings while SNPs correlated with the outcomes (P < 1×10-5) were excluded. The SNPs were then standardized for both exposures and outcomes by eliminating palindromic SNPs and those with allele discrepancies. In accordance with hypothesis (3), SNPs associated with outcomes (P < 1×10-5) were omitted from the IVs. Ultimately, MR analysis was performed on metabolites with a minimum of two SNPs.25
Statistical Analysis and Secondary Analysis
The principal MR analysis employed the inverse-variance weighted (IVW) method. Specifically, a fixed-effect model IVW was applied in the absence of significant heterogeneity, whereas a random-effect model IVW was utilized when heterogeneity was detected.26 The IVW method synthesizes the Wald estimates of genetic causal associations for individual SNPs to quantify the effect of the exposure on the outcome, contingent upon the assumption that all selected SNPs are valid IVs.27,28 This methodology produces accurate estimates and constitutes the primary statistical technique for evaluating causal relationships.17,29 Therefore, we utilized IVW-based estimates to conduct an initial screening of Immune cells for their potential causal effects on PHN.In order to enhance the robustness of our findings, we employed two supplementary methods, the MR-Egger method and the weighted median (WM) method, to assess metabolites exhibiting significant IVW estimates (P < 0.05). These additional analyses were conducted to improve the reliability of our estimates in less stringent conditions.The WM method permits the inclusion of a maximum of 50% of SNPs that may be invalid, whereas MR-Egger provides capabilities for testing horizontal pleiotropy and detecting heterogeneity in the presence of pleiotropy.30,31 MR-Egger regression is able to yield unbiased estimates under the InSIDE assumption (Instrument Strength Independent of Direct Effect).32,33
The secondary analyses in MR involve assessing heterogeneity, pleiotropy, and sensitivity. Heterogeneity among SNPs associated with exposures was examined through the application of Cochran’s Q test.17 The Q statistic and I² (%) value were utilized to quantify heterogeneity, with I² defined as I² = [Q - (K - 1)] / Q, where K represents the number of SNPs, and Q is the Q statistic. Horizontal pleiotropy was assessed using the MR-Egger intercept method34 and the global test in MR-PRESSO.35 An intercept close to zero suggests a reduced likelihood of horizontal pleiotropy.Following the identification of outliers through MR-PRESSO, a repeat MR analysis was conducted after excluding heterogeneous SNPs. Subsequently, MR-PRESSO was utilized once more to ascertain the presence of heterogeneous SNPs. Sensitivity assessment was carried out through leave-one-out (LOO) analysis,36 systematically removing each SNP to assess its impact on the overall causal estimate. Furthermore, sensitivity analyses involved comparing results from different MR methods to ensure the stability and reliability of the conclusions.37
To strengthen the exclusion restriction assumption in MR analyses, the Steiger test was employed. This test evaluates the direction of causality by comparing the variance explained by genetic variants in the exposure (immune cell subtypes) versus the outcome (PHN), helping confirm that our identified genetic instruments primarily affect PHN through their influence on immune cells rather than the reverse causal pathway.38
In conclusion, we employed a comprehensive multi-step analytical framework to identify immune cell populations with potential causal effects on the development of PHN, adhering to the following stringent criteria: (1) statistical significance in the preliminary analysis, defined as P < 0.05 using the IVW method; (2) consistency in both direction and magnitude across various MR methodologies; (3) absence of heterogeneity or horizontal pleiotropy in MR estimates; and (4) minimal influence of individual SNPs on the overall MR estimates.Subsequent to the initial analysis, positive findings were subjected to multiple testing corrections in accordance with the previously described methods and protocols. Statistical significance was determined using Bonferroni-adjusted P-values, calculated as P < 0.05/N, where N denotes the number of tests conducted.39 The threshold for statistical significance was established at P < 0.0125 (0.05/4), and only results that met this rigorous criterion were deemed statistically significant.
Confounding Analysis and Multivariable MR Analysis
To substantiate our findings, we performed extensive sensitivity analyses to assess horizontal pleiotropy within our MR results and to identify any SNPs that may contravene the assumptions of MR.While recognizing the potential for residual confounding due to a limited number of SNPs, we conducted a systematic investigation of genetic instruments linked to immune cell phenotypes using the GWAS Catalog (https://www.ebi.ac.uk/gwas/). This analysis aimed to identify potential associations with established risk factors for PHN, encompassing demographic variables (such as age and female sex), clinical parameters (including the severity of acute pain and rash), immunological status (such as HIV infection and organ transplantation), timing of treatment, neurological history, psychological factors,40 and disease-specific characteristics. SNPs exhibiting significant associations (P < 1×10⁻5) with any of the confounding variables or the outcome were identified. Subsequently, MR analyses were conducted again, excluding these SNPs, to validate the robustness of our findings and ensure the reliability of our causal inferences.
In order to adhere to the assumptions 2 and 3 of MR, it is imperative to confirm the association of genetic variants with a singular risk factor. Nonetheless, certain genetic variants exhibit associations with multiple risk factors, a concept referred to as pleiotropy. In instances of pleiotropy, MVMR can effectively account for the interactions among genetic variants linked to various exposures that may impact one another.41 MVMR offers a solution to the challenges of independence, dominance, and comparability that single-variable MR is unable to address.Essentially, single-variable MR analyzes the overall impact of exposure on the outcome, whereas MVMR examines the specific impact of each exposure on the outcome, without considering other exposures.In this research, we utilized MVMR to account for the interactions of the Immune cells identified. MVMR was implemented through the IVW,42 MR-PRESSO,35 and LASSO regression43,44 techniques. The IVW method in MVMR entails regressing SNPs for all exposures against the outcome, with weights determined by the inverse variance of the outcome. MR-PRESSO was employed to identify and remove outliers in order to address pleiotropy among IVs. LASSO regression was utilized to eliminate exposures exhibiting collinearity.
Colocalization Analysis
In order to explore the potential relationship between the Immune cells identified in PHN, we conducted a colocalization analysis utilizing the coloc R package.45 This analysis enabled us to identify a shared causal variant locus within a specific genomic region that may drive the association between these factors and the two phenotype. The Coloc method assessed the posterior probabilities (H0, H1, H2, H3, H4) of five hypotheses within a Bayesian framework for each variant locus: (1) no association with either trait; (2) association with trait 1 only; (3) association with trait 2 only; (4) both traits are associated with different causal variants specific to each trait; (5) both traits are correlated and share the same causal variant.46 The colocalization analyses utilized default priors (p1 = 1 × 10–4, p2 = 1 × 10–4, p12 = 1×10-5).pp.H4 > 80% of the results of the colocalization analyses (H4) provide strong evidence in support of the existence of shared causal variants affecting gene expression and the risk of PHN in specific genomic regions.47
Results
Preliminary Analysis
Utilizing a significance criterion of P < 5×10 ^ - 6, this study discovered 10,059 SNPs linked to 731 Immune cells. The F-statistics of these SNPs varied from 10.03 to 6,287,907.51, affirming the statistical soundness and dependability of the chosen phenotype as IVs in the MR analysis. Detailed information regarding the IVs can be found in Table S2. Prior to conducting the formal MR analysis, all outliers were identified and eliminated through the screening of confounders and MR-PRESSO (Table S3). In this extensive MR analysis, which examined 731 immune cell phenotypes in relation to PHN characteristics, we identified significant causal associations utilizing the IVW method. After applying stringent statistical corrections, including the Bonferroni adjustment (P < 0.0125), and excluding one pleiotropic exposure (BAFF-R expression on IgD+ CD38+ B cells), eight immune cell populations exhibited strong causal relationships with the development of PHN (Table S4; Figures 2 and 3). Further details are provided below:CD27 on IgD- CD38dim B cell (OR: 1.240, 95% CI:1.064–1.446, P = 0.006), FSC-A on myeloid Dendritic Cell (OR: 1.134, 95% CI: 1.029–1.249, P = 0.011), BAFF-R on naive-mature B cell (OR: 0.845, 95% CI: 0.747–0.956, P = 0.007), BAFF-R on transitional B cell (OR: 0.845, 95% CI: 0.744–0.961, P = 0.010), CD123 on CD62L+ plasmacytoid Dendritic Cell (OR: 0.780, 95% CI: 0.649–0.936, P = 0.008), CD123 on plasmacytoid Dendritic Cell (OR: 0.779, 95% CI: 0.649–0.935, P = 0.007), CD25 on activated and secreting CD4 regulatory T cell (OR: 0.824, 95% CI: 0.714–0.950, P = 0.008), and CD33+ HLA DR+ CD14- Absolute Count (OR: 0.907, 95% CI: 0.841–0.978, P = 0.011)(Figures 3). The alignment of the MR Egger, Weighted Median, and Weighted Mode methodologies with the IVW approach in the MR analysis underscores the robustness of the findings.
 |
Figure 2 Volcano plot of Immune cells associated with the risk of developing PHN. PHN, postherpetic neuralgia. |
Extensive sensitivity analyses conducted for all positive findings indicated no evidence of SNPs exhibiting horizontal pleiotropy, as assessed through standard pleiotropy evaluations and MR-PRESSO analysis (see Table S5). This supports the reliability of our IVs. Additionally, heterogeneity tests for positive outcomes revealed no significant heterogeneity in MR estimates (refer to Table S6), thereby further validating our results.Furthermore, LOO analyses revealed no significant sources of bias among the positive results (see Supplementary Figure 1). All analyses successfully passed the Steiger directionality tests, indicating the absence of reverse causation in our instrumental variables (refer to Table S7). Additional visual data are provided in Supplementary Figures 2 and 3.
MVMR Analysis
To address potential interactions and confounding effects among immune cell populations, we conducted a MVMR analysis. This approach facilitated a comprehensive examination of the independent contributions of different immune cell populations to the risk of PHN. The MVMR analysis incorporated three robust methodologies: IVW analysis, MR-PRESSO testing, and LASSO regression. These methods were chosen to ensure the reliability and consistency of our findings while effectively mitigating potential bias and confounding variables.
Our analysis identified a single immune cell population (CD27 expression on IgD- CD38dim B cells) that maintained significant association with PHN risk after controlling for other immune cell populations (OR: 1.288, 95% CI: 1.059–1.566, P = 0.011). This finding suggests that genetic predisposition to altered CD27 expression on IgD- CD38dim B cells may independently influence PHN susceptibility. The positive odds ratio suggests that individuals with a genetic predisposition for this immune cell phenotype may have an elevated risk of developing or experiencing more severe PHN. Importantly, these findings were consistent across all three analytical methods used in the MVMR analysis, underscoring the robustness of our results and demonstrating the stability of the observed association across various statistical approaches.(Table S8, Figures 4).
Colocalization Analysis
Colocalization analyses were conducted on all eight immune cell populations that exhibited statistical significance following Bonferroni correction. Robust evidence for a shared genetic architecture between PHN risk and immune cell expression was found in two populations: CD123 expression on plasmacytoid dendritic cells (posterior probability of hypothesis 4 [PP.H4] = 70.65%, with no lead SNP identified) and CD123 expression on CD62L+ plasmacytoid dendritic cells (PP.H4 = 69.50%, with no lead SNP identified)(Table S9, Figure 5).Key loci identified in genome-wide association studies may specifically confer a reduced risk of PHN through the biological regulation of these two immune cell populations. Conversely, other immune cell populations did not exhibit evidence supporting colocalization hypotheses with PHN susceptibility.
Discussion
This study represents several groundbreaking advances in our understanding of PHN pathogenesis. First, while previous observational studies have only demonstrated correlative relationships between immune cells and PHN, our MR analysis provides the first robust evidence of causal relationships between specific immune cell subtypes and PHN development. Second, our comprehensive analysis of multiple immune cell subtypes, combined with colocalization analysis, has revealed a previously unknown hierarchical importance of different immune cells in PHN pathogenesis. This novel finding was not achievable through traditional observational approaches. Third, by identifying causal genetic variants, our study opens new avenues for therapeutic targeting, potentially shifting the treatment paradigm from symptom management to preventive strategies based on immune modulation.
PHN, a prevalent complication of herpes zoster, constitutes its most severe sequela, leading to substantial physical, psychological, functional, and social impairments. Although an increasing body of evidence underscores the essential role of immune cells in the development and progression of PHN, the specific causal relationships and underlying mechanisms involving immune cell populations and PHN remain largely undefined.In this study, we utilized two-sample MR, MVMR, and colocalization analyses to investigate these causal relationships.Our findings revealed that individuals with genetically determined higher levels of CD27 expression on IgD- CD38dim B cells and FSC-A on myeloid dendritic cells showed increased PHN risk, while elevated levels of BAFF-R on naive-mature B cells, BAFF-R on transitional B cells, CD123 on CD62L+ plasmacytoid dendritic cells, CD123 on plasmacytoid dendritic cells, CD25 on activated and secreting CD4 regulatory T cells, and CD33+ HLA DR+ CD14- absolute count were associated with decreased PHN risk.
The MVMR analysis revealed that CD27 expression on IgD- CD38dim B cells may have a detrimental effect on the development of PHN, with each standard deviation increase correlating with a 28.8% heightened risk of PHN. Exploring these mechanistic pathways could offer new insights and potential therapeutic targets for managing PHN. Additionally, colocalization analyses indicated that certain genetic loci identified in GWAS studies might influence PHN susceptibility by regulating CD123 expression on both plasmacytoid dendritic cells and CD62L+ plasmacytoid dendritic cells.This research introduces an innovative methodological framework aimed at examining potential causal relationships between PHN and immune cell populations by employing an integrated approach of MVMR and colocalization analyses.Utilizing comprehensive publicly accessible genetic data, we examined the causal associations between 731 immune cell phenotypes and susceptibility to PHN. To the best of our knowledge, this study constitutes the inaugural MR analysis investigating the causal relationships between multiple immune phenotypes and the development of PHN.
CD27 on IgD- CD38dim B cell constitute a distinct subset of memory B lymphocytes, characterized by the absence of IgD expression (IgD-), intermediate expression of CD38 (CD38^dim), and the presence of CD27, a member of the tumor necrosis factor receptor superfamily. This cellular population represents a subset of antigen-experienced B cells that have undergone immunoglobulin class-switch recombination.CD27 is recognized as a classical marker for memory B cells, which possess the ability to swiftly differentiate into antibody-secreting cells upon subsequent antigenic exposure.48 This process is crucial for immunological memory and the expedited secondary immune response.While no direct evidence has previously established the association between CD27+IgD-CD38dim B cell and PHN, our Mendelian randomization analyses, specifically employing the MVMR approach, have demonstrated a positive causal relationship between genetically predicted levels of these cells and susceptibility to PHN. Several potential biological mechanisms may underlie this novel association: (1) Enhanced Neuroinflammatory Response: Previous studies have demonstrated that B cell-mediated autoimmune responses can contribute to neuropathic pain development,49 although specific investigations of CD27+IgD-CD38dim B cell in PHN are lacking. (2) Sustained Immune Activation: Research has shown that gut dysbiosis and immune system activation contribute to the development and progression of neuropathy,50 suggesting potential mechanisms for chronic pain states.3) Neuroimmune Modulation:Evidence indicates that immune cells can modulate neural function through various mediators, including serotonin and other immune modulators,51 which may influence pain sensitization.While these mechanistic hypotheses are supported by indirect evidence, further experimental validation is necessary to elucidate the precise role of CD27 on IgD- CD38dim B cell in the pathogenesis of PHN.
FSC-A (Forward Scatter-Area) on myeloid Dendritic Cells (mDCs) serves as a critical parameter for identifying and characterizing mDCs based on their size and granularity. Myeloid dendritic cells are professional antigen-presenting cells that bridge innate and adaptive immunity through antigen presentation and cytokine production. Although no direct evidence has previously confirmed an association between FSC-A on myeloid Dendritic Cell and PHN, our MR analyses demonstrated a positive causal relationship between genetically predicted levels of these cells and susceptibility to PHN. Two potential biological mechanisms may underpin this novel association: (1) Enhanced Antigen Presentation and T Cell Activation: During viral infections like VZV, the enhanced antigen presentation capabilities of dendritic cells contribute to sustained immune responses and potential tissue damage.52 The size and complexity of mDCs, indicated by FSC-A, may reflect their activation status and antigen-presenting capacity.(2) Cytokine-Mediated Neuroinflammation: Activated mDCs secrete pro-inflammatory cytokines, including interleukin-12 (IL-12), tumor necrosis factor-alpha (TNF-α), and interleukin-1 beta (IL-1β),53 which can sensitize nociceptors and contribute to neuropathic pain development.54 The physical attributes of mDCs, as indicated by FSC-A, may correlate with their cytokine-producing capacity. Further experimental validation is required to clarify the specific role of FSC-A in mDCs regarding PHN pathogenesis.
BAFF-R (B-cell activating factor receptor) expression on naive-mature and transitional B cells represents a crucial marker for B cell survival and maturation. BAFF-R signaling is essential for B cell homeostasis and function through the regulation of B cell survival, differentiation, and activation. Although no direct evidence has previously confirmed an association between BAFF-R expressing B cells and PHN, this MR analyse demonstrated a negative causal relationship between genetically predicted levels of these cells and susceptibility to PHN. Potential biological mechanisms may underpin this novel association:BAFF-R signaling in naive-mature and transitional B cells promotes the production of regulatory B cells (Bregs), which secrete anti-inflammatory cytokines such as interleukin-10 (IL-10) and Transforming growth factor beta (TGF-β).55 These immunomodulatory factors can suppress neuroinflammation and potentially alleviate neuropathic pain development.56 BAFF-R expressing B cells play a crucial role in maintaining B cell tolerance and preventing autoimmune responses.57 The reduced levels of these cells might lead to dysregulated immune responses, potentially contributing to neural tissue damage and chronic pain states.
CD123 expression on plasmacytoid Dendritic Cells (pDCs) and CD62L+ pDCs, along with CD25 on activated regulatory T cells (Tregs), represents crucial markers for these immunoregulatory cell populations. These cells play essential roles in immune homeostasis and inflammation regulation through various mechanisms including cytokine production and immune response modulation. This MR analyse demonstrated a negative causal relationship between genetically predicted levels of these cells and susceptibility to PHN. pDCs, characterized by CD123 expression, produce type I interferons which can modulate microglial activation states.58 Through interaction with microglia, pDCs may suppress excessive neuroinflammation and reduce neuropathic pain development. The CD62L+ subset of pDCs shows enhanced capacity for tissue trafficking and immune regulation.59 CD25+ Tregs secrete anti-inflammatory cytokines (IL-10, TGF-β) that can directly influence microglial phenotype, promoting an M2-like anti-inflammatory state.60 This interaction between Tregs and microglia creates an immunosuppressive microenvironment that may protect against chronic pain development. Both pDCs and Tregs can infiltrate the nervous system and interact with resident microglia, forming a regulatory network that maintains immune homeostasis.61 This cellular interplay may prevent excessive microglial activation and subsequent neuroinflammation associated with PHN. The interaction between these immune cells and microglia appears crucial in PHN pathogenesis. Microglia, as central nervous system resident immune cells, respond to signals from both pDCs and Tregs, potentially shifting their phenotype from pro-inflammatory to anti-inflammatory states.62 This modulation of microglial function through immune cell interactions may represent a key mechanism in preventing or alleviating PHN.
CD33+ HLA-DR+ CD14- cells represent a subset of myeloid cells with immunoregulatory properties. These cells are characterized by the expression of CD33 (Siglec-3) and HLA-DR, while lacking CD14, suggesting their potential role in immune modulation and antigen presentation. Although no direct evidence has previously confirmed an association between CD33+ HLA-DR+ CD14- cells and PHN, our MR analyses demonstrated a negative causal relationship between genetically predicted levels of these cells and susceptibility to PHN. The protective mechanism may operate through Immunomodulatory Function: CD33+ myeloid cells can suppress excessive inflammatory responses through the production of anti-inflammatory mediators and the regulation of T cell responses.63 The engagement of CD33 (Siglec-3) inhibitory receptors on these cells leads to reduced production of pro-inflammatory cytokines and promotes the resolution of inflammation,64 potentially limiting the development of chronic pain states like PHN. Further experimental validation is required to fully elucidate the protective role of CD33+ HLA-DR+ CD14- cells in PHN pathogenesis.
Advantages and Limitations
This MR demonstrates multiple strengths. Firstly, it represents the most comprehensive and methodical investigation to date into the causal association between Immune cells and PHN, encompassing an analysis of 731 immune cells. Secondly, a rigorous MR analysis was employed to mitigate inherent limitations such as reverse causality and confounding variables. To enhance the reliability of the findings, a variety of techniques were utilized to uphold the assumptions of MR and minimize potential biases. The strength of the findings is evidenced by the consistent directionality of the three MR estimates and sensitivity analysis. Additionally, the validity of the results was reinforced through a MVMR analysis. Furthermore, colocalization analyses were conducted to illustrate how modifications at key loci within the GWAS signaling pathway can impact PHN through the regulation of Immune cells expression at both the gene and protein levels.
There are several limitations to the current study. One limitation is the restricted number of SNPs available for identifying SNPs of interest at the genome-wide level. To mitigate this limitation, a more lenient threshold for MR analysis was employed, a practice commonly observed in other studies. Nonetheless, all selected SNPs exhibited F-statistic values exceeding 10, suggesting the robustness of our IVs. Additionally, to minimize the impact of ethnic diversity, only individuals of European descent from the FinnGen were included in the MR analysis. Hence, additional research is warranted to investigate the applicability of our findings to diverse databases and populations. Another constraint of this study is the reliance on sample size for the accuracy of MR estimation. Therefore, it is imperative to increase the sample size to ensure the credibility of our outcomes. Furthermore, while MR analysis offers valuable insights into causality, it is essential to underscore the importance of validating our results through rigorous RCTs and fundamental research prior to clinical application.
Conclusion
In summary, the present MR study has identified eight immune cells populations with genetic susceptibility that are causally linked to the development of PHN. Notably, CD27-expressing IgD- CD38dim B cells have been highlighted as a particularly promising therapeutic target, meriting further exploration. The identification of these immune cells signatures holds substantial implications for the early detection, prevention, and management of PHN. These findings not only contribute to the design of future clinical trials but also offer new therapeutic perspectives. Moreover, the comprehensive analysis of genomic and proteomic data provides a robust framework for exploring the underlying etiology and molecular mechanisms of PHN, thereby potentially aiding in the development of targeted immunotherapeutic strategies. Our findings constitute a substantial advancement in elucidating the immunological basis of PHN and may lay the groundwork for personalized therapeutic approaches informed by immune cells profiles.
Data Sharing Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Ethics Approval and Consent to Participate
In accordance with the Measures for Ethical Review of Life Sciences and Medical Research Involving Humans (2023) issued by the National Health Commission of the People’s Republic of China and affiliated governmental bodies, research projects are exempt from ethical review procedures when they exclusively utilize either legally obtained public data or employ non-interventional observational methods to collect data on public behavior. This exemption policy is explicitly detailed in Article 32 of the measures (https://www.gov.cn/zhengce/zhengceku/2023-02/28/content_5743658.htm) and further clarified in the official interpretation document (https://www.gov.cn/zhengce/2023-02/28/content_5743660.htm).
Acknowledgments
We would like to express our gratitude to the GWAS Catalog and FinnGen for generously providing GWAS data, and acknowledge the financial support from the Science and Technology Foundation of Guizhou Provincial Health Commission (Grant No. gzwkj2024-422). Additionally, We extend our thanks to the drawing services offered by www.figdraw.com.
Funding
This research was funded by a grant from the Science and Technology Foundation of Guizhou Provincial Health Commission (Grant No. gzwkj2024-422).
Disclosure
The authors declare no conflicts of interest.
References
1. Rice ASC, Dworkin RH, McCarthy TD, et al. EMA401, an orally administered highly selective angiotensin II type 2 receptor antagonist, as a novel treatment for postherpetic neuralgia: a randomised, double-blind, placebo-controlled Phase 2 clinical trial. Lancet. 2014;383(9929):1637–1647. doi:10.1016/S0140-6736(13)62337-5
2. Lee SH, Lee JY, Yeon H, et al. Pain changes and new neurologic sign in post-herpetic neuralgia: a clue in the diagnosis of malignancy-a case report. Ann Palliat Med. 2022;11(8):2773–2777. doi:10.21037/apm-21-2567
3. Mizukami A, Sato K, Adachi K, et al. Impact of herpes zoster and post-herpetic neuralgia on health-related quality of life in Japanese adults aged 60 years or older: results from a prospective, observational cohort study. Clin Drug Investig. 2018;38(1):29–37. doi:10.1007/s40261-017-0581-5
4. Wang Y, Jia T. Causal links between blood inflammation markers and postherpetic neuralgia risk: insights from a two-sample Mendelian randomization study. Front Neurol. 2024;15:1411541. doi:10.3389/fneur.2024.1411541
5. Truini A, Galeotti F, Haanpaa M, et al. Pathophysiology of pain in postherpetic neuralgia: a clinical and neurophysiological study. Pain. 2008;140(3):405–410. doi:10.1016/j.pain.2008.08.018
6. Miller YI, Navia-Pelaez JM, Corr M, Yaksh TL. Lipid rafts in glial cells: role in neuroinflammation and pain processing. J Lipid Res. 2020;61(5):655–666. doi:10.1194/jlr.TR119000468
7. Chen Y, Zhou Y, Li XC, et al. Neuronal GRK2 regulates microglial activation and contributes to electroacupuncture analgesia on inflammatory pain in mice. Biol Res. 2022;55:5.
8. Zhu D, Peng T, Zhang Z, et al. Mesenchymal stem cells overexpressing XIST induce macrophage M2 polarization and improve neural stem cell homeostatic microenvironment, alleviating spinal cord injury. J Tissue Eng. 2024;15:20417314231219280. doi:10.1177/20417314231219280
9. Oaklander AL, Stocks EA, Mouton PR. Number of Langerhans immune cells in painful and non-painful human skin after shingles. Arch Dermatol Res. 2003;294(12):529–535. doi:10.1007/s00403-002-0362-7
10. Shepherd AJ, Mickle AD, Golden JP, et al. Macrophage angiotensin II type 2 receptor triggers neuropathic pain. Proc Natl Acad Sci U S A. 2018;115(34):E8057–e8066. doi:10.1073/pnas.1721815115
11. Peng Q, Guo X, Luo Y, et al. Dynamic immune landscape and VZV-specific T cell responses in patients with herpes zoster and postherpetic neuralgia. Front Immunol. 2022;13:887892. doi:10.3389/fimmu.2022.887892
12. Zuccolo L, Holmes MV. Commentary: Mendelian randomization-inspired causal inference in the absence of genetic data. Int J Epidemiol. 2017;46(3):962–965. doi:10.1093/ije/dyw327
13. Zheng Q, Wang D, Lin R, et al. Mendelian randomization analysis suggests no associations of human herpes viruses with amyotrophic lateral sclerosis. Front Neurosci. 2023;17:1299122. doi:10.3389/fnins.2023.1299122
14. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi:10.1136/bmj.k601
15. Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. doi:10.1136/bmj.n2233
16. Richmond RC, Davey Smith G. Mendelian Randomization: concepts and Scope. Cold Spring Harb Perspect Med. 2022;12(1):a040501. doi:10.1101/cshperspect.a040501
17. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi:10.1002/gepi.21758
18. Boef AG, Dekkers OM, le Cessie S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol. 2015;44(2):496–511. doi:10.1093/ije/dyv071
19. Orrù V, Steri M, Sidore C, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52(10):1036–1045. doi:10.1038/s41588-020-0684-4
20. Sidore C, Busonero F, Maschio A, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47(11):1272–1281. doi:10.1038/ng.3368
21. Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518. doi:10.1038/s41586-022-05473-8
22. Yang J, Yan B, Zhao B, et al. Assessing the causal effects of human serum metabolites on 5 major psychiatric disorders. Schizophr Bull. 2020;46(4):804–813. doi:10.1093/schbul/sbz138
23. Choi KW, Chen C-Y, Stein MB, et al. Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample Mendelian randomization study. JAMA Psychiatry. 2019;76(4):399–408. doi:10.1001/jamapsychiatry.2018.4175
24. Lv X, Hu Z, Liang F, et al. Causal relationship between ischemic stroke and its subtypes and frozen shoulder: a two-sample Mendelian randomization analysis. Front Neurol. 2023;14:1178051. doi:10.3389/fneur.2023.1178051
25. Gill D, Brewer CF, Monori G, et al. Effects of genetically determined iron status on risk of venous thromboembolism and carotid atherosclerotic disease: a Mendelian randomization study. J Am Heart Assoc. 2019;8(15):e012994. doi:10.1161/JAHA.119.012994
26. Zhang F, Deng S, Zhang J, et al. Causality between heart failure and epigenetic age: a bidirectional Mendelian randomization study. ESC Heart Fail. 2023;10(5):2903–2913. doi:10.1002/ehf2.14446
27. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880–1906. doi:10.1002/sim.6835
28. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–1739. doi:10.1093/ije/dyx034
29. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi:10.1007/s10654-015-0011-z
30. Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178(7):1177–1184. doi:10.1093/aje/kwt084
31. Ren Z, Simons P, Wesselius A, Stehouwer CDA, Brouwers M. Relationship between NAFLD and coronary artery disease: a Mendelian randomization study. Hepatology. 2023;77(1):230–238. doi:10.1002/hep.32534
32. Chen M, Xie CR, Shi YZ, Tang TC, Zheng H. Gut microbiota and major depressive disorder: a bidirectional Mendelian randomization. J Affect Disord. 2022;316:187–193. doi:10.1016/j.jad.2022.08.012
33. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi:10.1007/s10654-017-0255-x
34. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi:10.1093/ije/dyv080
35. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi:10.1038/s41588-018-0099-7
36. Flatby HM, Ravi A, Damås JK, Solligård E, Rogne T. Circulating levels of micronutrients and risk of infections: a Mendelian randomization study. BMC Med. 2023;21(1):84. doi:10.1186/s12916-023-02780-3
37. Mbutiwi FIN, Dessy T, Sylvestre MP. Mendelian randomization: a review of methods for the prevention, assessment, and discussion of pleiotropy in studies using the fat mass and obesity-associated gene as an instrument for adiposity. Front Genet. 2022;13:803238. doi:10.3389/fgene.2022.803238
38. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. doi:10.1371/journal.pgen.1007081
39. Wu F, Huang Y, Hu J and Shao Z. (2020). Mendelian randomization study of inflammatory bowel disease and bone mineral density. BMC Med, 18(1), 10.1186/s12916-020-01778-5
40. Patil A, Vyshnavi S, Raja T, et al. A Randomized clinical trial comparing the efficacy of ultrasound-guided erector spinae block and paravertebral block in preventing postherpetic neuralgia in patients with zoster-associated pain. J Anaesthesiol Clin Pharmacol. 2024;40(3):510–515. doi:10.4103/joacp.joacp_82_23
41. Sanderson E. Multivariable Mendelian randomization and mediation. Cold Spring Harb Perspect Med. 2021;11(2):a038984. doi:10.1101/cshperspect.a038984
42. Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251–260. doi:10.1093/aje/kwu283
43. Xiao S, Guo J, Zhang W, et al. A six-microRNA signature nomogram for preoperative prediction of tumor deposits in colorectal cancer. Int J Gen Med. 2022;15:675–687. doi:10.2147/IJGM.S346790
44. Grant AJ, Burgess S. Pleiotropy robust methods for multivariable Mendelian randomization. Stat Med. 2021;40(26):5813–5830. doi:10.1002/sim.9156
45. Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51(5):768–769. doi:10.1038/s41588-019-0404-0
46. Foley CN, Staley JR, Breen PG, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12(1):764. doi:10.1038/s41467-020-20885-8
47. Yun Z, Guo Z, Li X, et al. Genetically predicted 486 blood metabolites in relation to risk of colorectal cancer: a Mendelian randomization study. Cancer Med. 2023;12(12):13784–13799. doi:10.1002/cam4.6022
48. Larbi A, Pawelec G, Wong SC, et al. Impact of age on T cell signaling: a general defect or specific alterations? Ageing Res Rev. 2011;10(3):370–378. doi:10.1016/j.arr.2010.09.008
49. Boakye PA, Tang SJ, Smith PA. Mediators of neuropathic pain; focus on spinal microglia, CSF-1, BDNF, CCL21, TNF-α, Wnt ligands, and interleukin 1β. Front Pain Res. 2021;2:698157.
50. Mázala-de-Oliveira T, Jannini de Sá YAP, Carvalho VF. Impact of gut-peripheral nervous system axis on the development of diabetic neuropathy. Mem Inst Oswaldo Cruz. 2023;118:e220197. doi:10.1590/0074-02760220197
51. Wan M, Ding L, Wang D, Han J, Gao P. Serotonin: a potent immune cell modulator in autoimmune diseases. Front Immunol. 2020;11:186.
52. Steain M, Sutherland JP, Rodriguez M, et al. Analysis of T cell responses during active varicella-zoster virus reactivation in human ganglia. J Virol. 2014;88(5):2704–2716. doi:10.1128/JVI.03445-13
53. Segura E, Amigorena S. Inflammatory dendritic cells in mice and humans. Trends Immunol. 2013;34(9):440–445. doi:10.1016/j.it.2013.06.001
54. Grace PM, Hutchinson MR, Maier SF, Watkins LR. Pathological pain and the neuroimmune interface. Nat Rev Immunol. 2014;14(4):217–231. doi:10.1038/nri3621
55. Yang M, Sun L, Wang S, et al. Novel function of B cell-activating factor in the induction of IL-10-producing regulatory B cells. J Immunol. 2010;184(7):3321–3325. doi:10.4049/jimmunol.0902551
56. Rubtsova K, Rubtsov AV, Thurman JM, et al. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J Clin Invest. 2017;127(4):1392–1404. doi:10.1172/JCI91250
57. Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9(7):491–502. doi:10.1038/nri2572
58. Isaksson M, Ardesjö B, Rönnblom L, et al. Plasmacytoid DC promote priming of autoimmune Th17 cells and EAE. Eur J Immunol. 2009;39(10):2925–2935. doi:10.1002/eji.200839179
59. Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15(8):471–485. doi:10.1038/nri3865
60. Liesz A, Suri-Payer E, Veltkamp C, et al. Regulatory T cells are key cerebroprotective immunomodulators in acute experimental stroke. Nat Med. 2009;15(2):192–199. doi:10.1038/nm.1927
61. Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci. 2017;20(2):136–144. doi:10.1038/nn.4475
62. Masuda T, Sankowski R, Staszewski O, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566(7744):388–392. doi:10.1038/s41586-019-0924-x
63. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi:10.1038/nri2056
64. Duan S, Paulson JC. Siglecs as immune cell checkpoints in disease. Annu Rev Immunol. 2020;38(1):365–395. doi:10.1146/annurev-immunol-102419-035900
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.