Back to Journals » Cancer Management and Research » Volume 17
Gut Microbiota–Tumor Microenvironment Interactions: Mechanisms and Clinical Implications for Immune Checkpoint Inhibitor Efficacy in Cancer
Authors Said SS, Ibrahim WN
Received 6 September 2024
Accepted for publication 21 November 2024
Published 25 January 2025 Volume 2025:17 Pages 171—192
DOI https://doi.org/10.2147/CMAR.S405590
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Sawsan Sudqi Said, Wisam Nabeel Ibrahim
Department of Biomedical Sciences, College of Health Sciences, QU Health, Qatar University, Doha, Qatar
Correspondence: Wisam Nabeel Ibrahim, Department of Biomedical sciences, College of Health sciences, QU Health, Qatar University, Qatar, Email [email protected]
Abstract: Cancer immunotherapy has transformed cancer treatment in recent years, with immune checkpoint inhibitors (ICIs) emerging as a key therapeutic approach. ICIs work by inhibiting the mechanisms that allow tumors to evade immune detection. Although ICIs have shown promising results, especially in solid tumors, patient responses vary widely due to multiple intrinsic and extrinsic factors within the tumor microenvironment. Emerging evidence suggests that the gut microbiota plays a pivotal role in modulating immune responses at the tumor site and may even influence treatment outcomes in cancer patients receiving ICIs. This review explores the complex interactions between the gut microbiota and the tumor microenvironment, examining how these interactions could impact the effectiveness of ICI therapy. Furthermore, we discuss how dysbiosis, an imbalance in gut microbiota composition, may contribute to resistance to ICIs, and highlight microbiota-targeted strategies to potentially overcome this challenge. Additionally, we review recent studies investigating the diagnostic potential of microbiota profiles in cancer patients, considering how microbial markers might aid in early detection and stratification of patient responses to ICIs. By integrating insights from recent preclinical and clinical studies, we aim to shed light on the potential of microbiome modulation as an adjunct to cancer immunotherapy and as a diagnostic tool, paving the way for personalized therapeutic approaches that optimize patient outcomes.
Keywords: ICIs, immunotherapy, gut microbiota, microbiome, tumor microenvironment
Introduction
The growing resistance of cancer to traditional therapies has led scientists to explore new treatment approaches. In recent decades, immunotherapy has emerged as a groundbreaking method, with several drugs showing significant success, particularly when combined with other therapies. Notably, patients with advanced metastatic non-small cell lung cancer (NSCLC) treated with immune checkpoint inhibitors (ICIs) have shown markedly improved survival outcomes compared to those receiving chemotherapy.1 Despite these advancements, immunotherapy still faces challenges in terms of efficacy for certain cancers.
A promising avenue for enhancing immunotherapy involves the human microbiome. Emerging evidence suggests that incorporating the microbiome can boost the effectiveness of immunotherapy and improve cancer outcomes.2 As a result, research has focused on understanding the impact of the gut microbiome on human health and its potential to optimize cancer therapies. Recent studies have identified associations between specific gut microbiota and treatment success, particularly with ICIs targeting PD-1, PD-L1, and CTLA-4.3 The gut microbiome plays a critical role in modulating the immune system, including its response to cancer therapies. It enhances immune function through mechanisms such as releasing short-chain fatty acids, interacting with toll-like receptors, and producing IgA to prevent bacterial invasion.4 These effects have spurred interest in reprogramming gut microbiota to improve immune responses across various contexts.
While the microbiome is being leveraged to enhance cancer therapies, changes in microbiome composition during prolonged treatment have been observed. The relationship between the gut microbiome, immune system, and cancer is complex, requiring a better understanding of how these interactions affect drug resistance and treatment outcomes.
A deeper understanding of these mechanisms could transform cancer treatment strategies. This review summarizes the latest findings on the role of microbiota in modulating immune checkpoint inhibitors in cancer treatment.
Gut Microbiota: Structure, Diversity, and Health
The human body hosts trillions of microbiomes, including bacteria, fungi, viruses, and archaea, residing in or on various surfaces. Colonization of gut microbiomes begins early in life, with the mode of delivery influencing an infant’s gut microbiome diversity. Infants born via caesarian section tend to harbor more opportunistic microbes, such as Staphylococcus, Haemophilus, Enterobacter cancerogenus, Hormaechei, and Veillonella species, compared to those delivered vaginally.5 The adult gut contains between 10¹³ and 1014 microbiomes, comprising over 1000 microbial species and more than 7000 bacterial strains.4 Gut microbiome composition varies based on factors like age, race, sex, geography, and lifestyle, including diet, prebiotics, and probiotics. Among these, bacteria are the most common commensal organisms, primarily colonizing the gastrointestinal tract, where their metabolites regulate physiological processes like mental health, inflammation, and immunity.
Imbalance in the microbiome (dysbiosis) is linked to diseases such as diabetes, asthma, allergies, autism, cardiovascular, neurological, and cancer-related conditions.6 Dysbiosis reflects changes in microbial diversity and abundance. Early microbiome research relied on culturing techniques, but many microbes are unculturable. Modern approaches such as 16S rRNA sequencing, metagenomics, transcriptomics, and proteomics have allowed researchers to differentiate between healthy and diseased microbiomes.7
16S rRNA sequences are mapped and clustered into operational taxonomic units (OTUs) based on sequence similarities. Next-generation sequencing offers diversity indices that mathematically describe the complexity of microbiome samples and their differences. In 1960, Whittaker introduced the concept of “gamma diversity” (total species diversity within a landscape), which comprises “alpha diversity” (within-sample diversity) and “beta diversity” (differences between samples).8 High alpha diversity indicates a sample with numerous abundant species, while low beta diversity suggests that two samples share most species.
Early classifications of healthy microbiomes were based on their resilience to stress and ability to recover from disturbances.6 Recent research has highlighted the role of dysbiotic microbiota in cancer therapy and cardiotoxicity.9 In 2017, the interaction between microbiota and cancer treatments gained attention, particularly in the context of immunotherapy. Routy et al demonstrated that lung and kidney cancer patients with primary resistance to PD-1 inhibitors had lower levels of Akkermansia muciniphila, a mucus-degrading bacterium, compared to responders.10 Restoring immune responses through fecal transplants or oral supplementation of Akkermansia muciniphila has shown promise in mouse models of epithelial tumors.11 Additionally, studies in melanoma patients have revealed a healthier microbiota profile in responders to PD-1 inhibitors compared to non-responders.
Gut Microbiota-Immune System Crosstalk
Gut microbiota play a crucial role in digestion by producing metabolites that nourish gastrointestinal (GI) epithelial cells and protect against pathogens. The immune system in the intestine acts as a protective barrier, preventing gut microbes from crossing the epithelial layer while avoiding inappropriate immune responses to commensal bacteria in the intestinal lumen. This balance helps reduce the risk of diseases like inflammatory bowel disease and celiac disease, as well as other immune-related disorders. The gut immune system maintains a delicate equilibrium between pro-inflammatory and anti-inflammatory responses, and this regulation is essential for immune homeostasis in conditions such as obesity, cancer, and Parkinson’s disease.5 The interaction between gut microbiota and the host’s immune system is a complex and dynamic process that either maintains physiological stability or contributes to disease development. The immune system consists of two main components: the innate and adaptive systems, which work together to neutralize immunological threats without triggering excessive immune responses. However, in certain chronic diseases and inflammatory conditions, the immune system may become overactive, leading to adverse outcomes and complicating treatment.
Initially, innate immune cells—such as dendritic cells, natural killer cells, macrophages, and polymorphonuclear cells—provide the first line of defense against pathogens. These cells release chemokines and cytokines, signaling adaptive immune cells like B cells, T cells, and regulatory T cells to join in eliminating the invading agent.
Neonatal Gut Microbiota Shapes the Immune System
Gut microbiota begin to form during delivery or shortly after birth, with the composition depending on the method of delivery. In vaginal births, a newborn’s microbiota closely resembles the mother’s vaginal and fecal microbiota, while cesarean section deliveries result in microbiota acquired from the immediate environment and skin. Newborns also receive secretory immunoglobulin A (SIgA) from their mother, which is critical for developing both their immune system and microbiota.12 SIgA neutralizes toxins, binds to M cells in the gut epithelium, and coats non-invasive pathogens, facilitating the activation of B and T cells. The proximal small intestine has the highest concentration of SIgA-coated microbiota, followed by the distal small intestine and colon. This process helps the immune system recognize and eliminate harmful pathogens, promoting overall health.
During intrauterine development, maternal FOXp3+ CD4+ regulatory T cells suppress the fetal innate immune response to maternal antigens. However, delays in microbiota colonization and low microbial diversity in newborns can affect the maturation of adaptive immune cells. A Swedish study on 65 children found that early gut colonization significantly influenced T cell development. The presence of Bifidobacterium in the neonatal gut during the first week of life was linked to increased production of cytokines like IL-5, IL-6, IL-13, TNF, and memory CD45RO+CD4+ T cells by 36 months.13 By age three, gut microbiota such as Enterococcus, Staphylococcus aureus, and Clostridium were associated with IL-13, IL-5, and TNF production. Additionally, at 4 and 8 months, the presence of CD27+ memory B cells was linked to both Bifidobacterium and E. coli.13 The composition of infant gut microbiota has changed over time. Previously, it was dominated by Clostridium difficile with low diversity, but recent studies show an increased presence of E. coli, Bacteroides, coagulase-negative Staphylococcus, and S. aureus in the infant gut lumen.
Gut Microbiota-Innate Immune System Axis
The intestinal epithelium, consisting of multiple structural layers, works with gut microbiota as the first line of defense against pathogens and inflammatory disorders. This barrier is composed of a pre-epithelial mucus layer, the epithelial layer (including M cells, Paneth cells, epithelial cells, and mucipare cells), and a post-epithelial layer with immune cells like dendritic cells, lymphocytes, and macrophages.14 The mucus layer contains tight junction proteins and antimicrobial peptides (AMPs), which help protect against bacterial invasion. Paneth cells, predominantly located in the small intestine, are essential for host-microbiota interactions. These specialized epithelial cells express receptors like Toll-like receptors (TLRs), RIG-I-like receptors, and NOD-like receptors, which recognize bacterial patterns and activate protective responses.14,15 Paneth cells also produce antimicrobial peptides, such as defensins, that not only defend against pathogens but also help shape the composition of the microbiota.14,16
The gut’s innate immune defense involves a variety of strategies to maintain homeostasis. The mucus layer, AMPs, and secretory IgA (SIgA) play key roles in keeping commensal microorganisms balanced while ensuring a protective immune response. Intestinal epithelial cells (IECs) also regulate immune responses through the MyD88 and NF-kB pathways in response to commensals, while TLR-2 signaling maintains epithelial integrity16 (Figure 1C). IECs secrete signals that regulate innate lymphoid cells (ILCs), which in turn produce cytokines associated with different immune responses (Th1, Th2, and Th17) (Figure 1B). Despite lacking antigen recognition, ILCs release a range of cytokines, such as IFN-γ, IL-5, IL-13, IL-17A, and IL-22, which help maintain gut immune homeostasis.
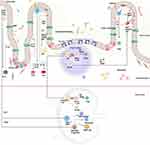 |
Figure 1 The interaction between commensal bacteria and intestinal epithelial cells influences the innate and adaptive immune systems’ ability to maintain homeostasis and prevent inflammation. In response to microbiota signals, IECs interact with their immediate environment, identify microbial signals, and mediate the gut immune response in a regulatory process. (A) Tolerogenic function is achieved through the production of thymic stromal lymphopoietin (TSLP), TGF-beta, and RA by stimulating DCs. Activated DCs then travel to Mesenteric lymph nodes and Peyer’s patch, where they interact with naïve T cells, leading to Treg development and differentiation of naive B cells into Ig A+ secreting plasma cells. (B) Furthermore, the regulatory function of IECs that produce semaphorin 7A SEMA7A has been established by activating IL10-producing macrophages and T regs. (C) IECs secreted cytokines, such as thymic stromal lymphopoietin (TSLP), which causes basophil progenitor cells to differentiate into basophils, and interleukin-25 (IL-25), which promotes the formation of monocytes, mast cells, basophils, and macrophages from type 2 multipotent progenitor cells. Moreover, IEC-derived cytokines govern the development of innate lymphoid cells into three groups: ILC1, ILC2, and ILC3. Created in BioRender. Nabeel, W. (2025) https://BioRender.com/ h28g657. |
In response to commensal microbe signals, IECs secrete immunoregulatory molecules like TGF-beta, thymic stromal lymphopoietin (TSLP), and retinoic acids. These signals activate dendritic cells (DCs) and macrophages, which promote a tolerogenic immune environment by producing IL-10 and retinoic acids.5 CD103+ DCs, a dominant subset of intestinal macrophages, present antigens to T cells in secondary lymphoid organs, like Peyer’s patches and mesenteric lymph nodes. Activated T cells secrete TGF-beta, promoting the development of regulatory T cells that help prevent excessive inflammation. Additionally, intestinal macrophages remove pathogens and commensal bacteria that penetrate the epithelial barrier.16
When invasive pathogens or pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharides cross the epithelial barrier, they trigger goblet cells to release mucin and reinforce the mucus layer. PAMPs are recognized by TLRs on innate immune cells like neutrophils, DCs, and macrophages, initiating an immune response.5,14 Gut microbiota also influence gut-associated lymphoid tissue (GALT), stimulating innate immune cells, such as DCs and natural killer cells, in response to pathogens. Certain bacteria, including Lactobacilli and Clostridium, help regulate the inflammatory response by promoting regulatory T cell activity.14 Normally, the innate immune system efficiently clears pathogens, but if compromised, pathogens can spread to systemic circulation, potentially leading to sepsis.
Several factors, such as antibiotics, NSAIDs, and aging, can disrupt the balance of gut microbiota, leading to the overgrowth of pathogenic organisms like Clostridium difficile, which can cause infection and diarrhea. C. difficile toxins activate innate immune cells via TLR4, TLR5, and NOD1, triggering the release of pro-inflammatory cytokines like IL-12, IL-18, and IFN-γ, which contribute to infection severity.17 Recent studies have shown that Bifidobacterium longum BB536 can enhance innate immunity by increasing the expression of activation markers (CD86 and HLA-DR) on dendritic cells and promoting the release of pro-inflammatory cytokines like IFN-α1 and IFN-β.18
Gut Microbiota-Adaptive Immune System Axis
Bacterial invasion of the epithelial layers and subsequent entry into mesenteric lymph nodes (MLNs) or systemic circulation triggers the adaptive immune system, which consists of humoral and cellular components. Antigen-presenting cells (APCs), such as dendritic cells, macrophages, and neutrophils, present bacterial antigens to T lymphocytes, particularly intraepithelial lymphocytes (IELs) and lamina propria lymphocytes. Notably, intraepithelial γδ T cells play a vital role in the adaptive immune response by secreting cytokines and antimicrobial compounds, serving as a defense mechanism against numerous infections. These γδ T cells, found in mucosal barriers like the lungs, skin, and gut, are crucial for maintaining immune balance.19,20
Invasive pathogens in the bloodstream correlate with a reduction in γδ T lymphocytes within epithelial layers, increasing vulnerability to infections. Studies show that the loss or insufficiency of mucosal T lymphocytes adversely impacts outcomes in enteric infections. Additionally, some gut microbiota and their metabolites stimulate γδ T cells. Antibiotic use can disrupt gut microbiota, causing hypoxia in the intestinal tract and altering its composition. This shift often results in a decrease in Clostridium and an increase in Desulfovibrio, leading to the activation of γδ T cells.20 γδ T cells exhibit dual roles as both innate and adaptive immune cells. In conditions such as inflammatory bowel disease (IBD), these cells release pro-inflammatory cytokines like IL-17A, TNF-α, and IFN-γ, contributing to the disease’s pathology.20
Commensal microbiota plays a significant role in T cell activation. Microbial antigens stimulate dendritic cells (DCs) to release IL-6 via TLR/MyD88-dependent and independent pathways, activating T cells.21 MyD88 serves as a key adaptor for innate immune receptors recognizing microbial signals. Its deficiency impacts microbiome composition, as shown in studies linking MyD88 signaling to the prevention of type 1 diabetes by modifying gut microbiota.22,23
Moreover, specific subtypes of T cells are activated by certain bacteria more than others, with segmented filamentous bacteria (SFB) identified as major inducers of intestinal Th17 cells. Innate IL-23 production can modify Th17 responses within the gut, further emphasizing the complex relationship between microbiota and the immune system.21
Additionally, polysaccharides from Bacteroides fragilis activate IL-10-producing Foxp3+ regulatory T cells through the TLR2 pathway.
Recent research has also explored γδ T cells’ role in immune responses against lung cancer and their interactions with microbiota. Lung cancer progression has been linked to the activation of γδ T cells and certain microbiota. Inhibiting γδ T cells, microbiota, or their metabolites can hinder lung cancer development.19 Lung microbiota, such as Propionibacterium acnes, Aneurinibacillus aneurinilyticus, and others, contribute to lung cancer by stimulating neutrophils and alveolar macrophages to release IL-1B and IL-23, which in turn activate γδ T cells and promote inflammation.19 T cell activation is also influenced by microbial composition. For example, the activation of CD8+ cytotoxic T cells is initiated by APCs recognizing microbial antigens, with cytokines from CD4+ T cells further stimulating cytotoxic responses. This underscores the importance of understanding the interplay between gut microbiota and systemic immunity.17
Follicular helper T (Tfh) cells, which play a crucial role in regulating microbiota composition, also interact with gut microbes. Research has found that Tfh differentiation is influenced by microbiota, with specific deficiencies, such as in PD-1 or P2RX7, linked to altered microbiota composition.24 SFB has been shown to promote Tfh differentiation in Peyer’s patches, influencing effector CD4+ T helper cell responses by limiting IL-2 access. Tfh cells are essential for supporting B cell functions, such as memory B cell development, affinity maturation, and germinal center formation.
In conclusion, the interaction between microbiota and T lymphocytes shapes both the innate and adaptive immune systems. This complex relationship highlights the profound impact that gut microbiota can have on overall immune function and health. Figure 1 demonstrated the interactions of gut microbiota with the innate and adaptive immune systems.
Dysbiosis Associated Cancer
Initial research aimed to identify strains of healthy microbiota such as Prevotella copri, Ruminococcus obeum, Bacteroides finegoldii, and Lachnobacterium bovis.6 The microbiome’s composition in different body sites can reflect transitions from healthy to diseased states and the transmission of microbiota between these sites. Dysbiosis of gut microbiota can lead to pathological issues and trigger immune responses to disease treatments. Colorectal cancer (CRC), the third most common cancer globally, has garnered significant attention, with an expected incidence of 2.2 million cases by 2030.25,26
Studies have shown that patients with CRC who develop metastasis often present with altered intestinal microbiota, including strains like Fusobacterium, Selenomonas, Prevotella, and Bacteroides.27 The gut microbiota in CRC patients differs significantly from that of healthy individuals, and various factors beyond microbiota—including inflammation, bacterial metabolites, virulence factors, and genotoxicity—may influence CRC treatments.26 While CRC results from multiple factors, including genetic and environmental influences, the microbiota is increasingly recognized as an environmental contributor to cancer development. Limited data exist on the relationship between microbiota and aging, but a study in rats found that while gut microbiota diversity decreases with age, Firmicutes increase and Bacteroides decline. Ruminococcus was shown to negatively correlate with pro-inflammatory cytokine IFN-γ levels, indicating a positive role in gut health.28 Microbiome diversity correlates with tumor stage, suggesting potential microbial markers for cancer progression. For example, Fusobacterium and Parvimonas are more abundant in grade 3 CRC tumors, while Fusicatenibacter, Blautia, Intestimonas, and Romboutsia are enriched in grade 2 tumors.29 Metagenomic sequencing of fecal samples has revealed differences in microbiota diversity between young and old CRC patients. Younger patients had a greater diversity of bacteria, with species like Flavonifractor plautii, Fusobacterium, and Odoribacter more prominent, while older patients had less diversity, dominated by Streptococcus, Fusobacterium, and Gemella.25 Tumor location and stage also affect microbiota composition. In older patients, Fusobacterium was more prevalent in early-stage (stage 0-III) CRC tumors on the left side, while Christensenellaceae was abundant in late-stage (stage IV) tumors on the right side.
Functional and metabolic analyses of microbiota are crucial in understanding CRC prognosis. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses have highlighted distinct bacterial metabolites in young and old CRC patients. KEGG analysis showed higher short-chain fatty acid production in controls compared to CRC patients, while GO analysis found higher DNA and RNA binding activity in younger CRC patients, suggesting increased cellular invasion and proliferation.25
Extracellular vesicles (EVs) derived from gut microbiota have been proposed as potential biomarkers for CRC diagnosis. A study involving 116 control patients and 91 CRC patients analyzed EVs from urine samples, finding significant differences between the two groups. CRC patients showed elevated levels of Proteobacteria, Actinobacteria, and Firmicutes, while Bacteroides and Verrucomicrobia were reduced compared to the control group. Staphylococcus and Acinetobacter were more enriched at the genus level in CRC patients.30 Further analysis revealed no significant differences in alpha or beta diversity based on CRC location (proximal/distal) or between early and late-stage CRC groups.30
In hepatocellular carcinoma (HCC), early-stage patients showed an abundance of Acinetobacter, while butyrate-producing bacteria and liposaccharide-producing bacteria were elevated compared to cirrhosis patients.31 Similarly, Sellimonas abundance is associated with a higher risk of estrogen receptor-positive breast cancer, while Alphaproteobacteria abundance correlates with a lower risk of prostate cancer.32
Pancreatic cancer is associated with reduced microbiota diversity and a high prevalence of Proteobacteria. Studies have shown that the pancreatic microbiota differs significantly from that of the stomach and duodenum. In pancreatic ductal adenocarcinoma (PDAC), distinct bacterial compositions within the tumor microenvironment (TME) have been found to reduce anti-tumor immunity and promote tumor progression. The predominant microbiota in non-tumorous pancreatic tissue consists of Proteobacteria and Firmicutes. In PDAC, Lactobacillus, Bacteroides, and Peptoniphilus were significantly elevated, leading to reduced effector T cells and unfavorable immune responses. Some bacterial metabolites, like tryptophan produced by Lactobacillus, may exert immunosuppressive effects by inhibiting intertumoral macrophages.33,34
Microbiome-Based Diagnostic Applications in Cancer
The gut microbiome, particularly through analysis of fecal and plasma samples, is emerging as a promising noninvasive tool for cancer diagnosis. Fecal microbiome analysis has been extensively explored for colorectal cancer (CRC) detection, with studies identifying specific bacterial markers linked to CRC. For instance, a gene marker from Lachnoclostrium spp. demonstrated a sensitivity of 48% and specificity of 79% for detecting colorectal adenomas.35 When combined with other bacterial species such as Fusobacterium nucleatum, Clostridium hathewayi, and Bacteroides clarus, sensitivity and specificity for CRC detection improved to 94% and 81%, respectively.36 Similarly, Clostridium symbiosum showed promise as a CRC biomarker, yielding a sensitivity of 80% and specificity of 55%, outperforming the well-known cancer marker F. nucleatum.37 This model achieved AUROC scores of 0.74 for early-stage CRC and 0.76 across all stages—higher than traditional fecal immunochemical tests (FIT). Notably, combining C. symbiosum with FIT further increased AUROC scores to 0.80 and 0.87, suggesting that fecal microbial markers can complement existing diagnostic methods.
Meta-analyses of fecal metagenomes across diverse CRC populations have also highlighted a core set of 29 enriched bacterial species, including F. nucleatum and Parvimonas micra, consistently differentiating CRC patients from controls with AUROC scores ranging from 0.71 to 0.92.38,39 Additional findings indicated elevated trimethylamine synthesis, potentially from Lachnospiraceae species and H. hathewayi, pointing to microbial pathway markers.40 While these fecal microbiome signatures are robust across populations, it remains unclear whether they play a causative role in CRC or reflect downstream effects.
Microbial markers in the fecal microbiome have been studied in other cancers as well. A 16S rRNA analysis in lung cancer patients revealed significant shifts in 24 bacterial genera. A model based on nine of these genera, including Bacteroides and Clostridium, predicted lung cancer with an AUROC score of 0.76.41 These preliminary results highlight the potential of gut microbiota as diagnostic markers beyond CRC, although further validation across larger populations is required.
In addition to the gut microbiome, plasma cell-free DNA (cfDNA) is gaining interest as a diagnostic tool for cancer. Originally developed for prenatal testing, cfDNA assays—such as the FDA-approved cobas EGFR Mutation Test and Guardant360 CDx—now aid in cancer diagnostics, particularly for detecting mutations in non-small cell lung cancer (NSCLC).42,43 FoundationOne Liquid CDx, approved in 2020, expands this approach by screening over 300 genes associated with multiple cancers, including NSCLC, prostate, ovarian, and breast cancer.44
More recently, researchers have investigated microbial DNA signatures in plasma cfDNA as potential cancer biomarkers. Next-generation sequencing of plasma from breast cancer patients, for example, revealed distinct microbial reads, with Acinetobacter spp. prevalent in controls and Pseudomonas spp. and Sphingomonas spp. in patients.45 A diagnostic model trained on these microbial signatures successfully distinguished among 20 different cancer types and healthy controls, achieving AUROC scores consistently above 0.85. This accuracy was maintained for early-stage cancers (stages I and II), with AUROC scores exceeding 0.80. Fungal DNA signatures in plasma also demonstrated diagnostic potential, with combined bacterial and fungal data achieving an AUROC score of 0.92. These findings underscore the potential of plasma-based microbial cfDNA assays as an early-stage diagnostic tool, although further research is necessary to refine and validate these approaches.
Although these diagnostics show potential for detecting cancer experimentally, these methods are likely to complement, rather than replace, conventional techniques such as imaging and tissue biopsies in the near term. As our understanding of the microbiome’s complex interactions with cancer progresses, however, such diagnostics could evolve into standalone approaches, particularly valuable for early detection and reducing the need for invasive procedures.
Effects of Gut Dysbiosis on Cancer Progression
Introduction to Gut Microbiota and Dysbiosis
The human gut microbiota is a complex ecosystem comprising diverse microorganisms, including bacteria, fungi, archaea, and viruses, predominantly made up of bacterial groups like Firmicutes and Bacteroidetes. This microbiota is integral in regulating metabolism, immune function, and maintaining overall balance in the intestinal tract.46 The gut microbiota interacts closely with the host’s mucosal immune system to promote tolerance to beneficial microbes and control harmful pathogens through mechanisms such as producing inhibitory metabolites, modulating immune responses, maintaining a protective mucus barrier, and competitively using nutrients.47 This equilibrium is crucial for preventing pathological infections and ensuring overall health.
However, an imbalance in this microbial community, known as gut dysbiosis, has been linked to the pathogenesis and progression of various diseases, including cancer.48 Dysbiosis disrupts fundamental processes within the host, contributing to cancer development through alterations in metabolic pathways, chronic inflammation, and compromised intestinal barrier function. For example, dysbiosis can lead to the production of carcinogenic compounds and reduce protective metabolites like short-chain fatty acids (SCFAs), which are known for their anti-inflammatory and anti-cancer properties.49 Furthermore, dysbiosis can trigger chronic inflammation, a known cancer risk factor, and compromise the intestinal barrier, allowing toxic substances to enter the bloodstream and promote cancer progression.50 Additionally, dysbiosis can weaken immune surveillance against tumors, as seen in colorectal cancer, where specific pathogenic bacteria such as Fusobacterium nucleatum create a pro-tumorigenic environment.51 Emerging research also indicates a potential link between dysbiosis and breast cancer through modulation of hormone-driven pathways.52 The intricate mechanisms by which gut dysbiosis influences cancer progression intersect with multiple hallmarks of cancer across various types as shown in Figure 2.
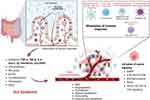 |
Figure 2 Mechanisms Linking Gut Dysbiosis to Cancer Progression; This illustration depicts the complex interplay between gut dysbiosis and cancer progression, highlighting how an imbalance in gut microbiota influences various cancer hallmarks. Gut dysbiosis alters microbial metabolite production, leading to enhanced signaling pathways that promote cancer cell proliferation and inhibit Tumor suppressor functions. Dysbiosis increases levels of pro-angiogenic factors such as VEGF, facilitating Tumor vascularization mediated by inflammatory cytokines (eg, TNF-α, IL-6) and microbial metabolites. Inflammatory responses triggered by gut dysbiosis degrade the extracellular matrix, aiding cancer cell invasion and metastasis. Gut dysbiosis affects nutrient absorption and metabolite availability, influencing cellular energy production and survival pathways in cancer cells. Dysbiosis also leads to an immunosuppressive Tumor microenvironment, achieved through modulation of T-cell function and immune checkpoint proteins, inhibition of natural killer cells and dendritic cells with recruitment of myeloid-derived suppressor cells (MDSCs). Created in BioRender. Nabeel, W. (2025) https://BioRender.com/ y49m093. |
Sustaining Proliferative Signaling and Evading Growth Suppressors
In normal physiology, cellular growth signals are tightly regulated. Cancer cells, however, often acquire the ability to sustain these signals autonomously, bypassing typical regulatory mechanisms.53 Gut dysbiosis can exacerbate this by altering the production of microbial metabolites that interact with signaling pathways. For instance, bacterial enzymes can convert primary bile acids into secondary bile acids, which act as signaling molecules that promote proliferation in liver and colorectal cancer cells.54 Certain pathogenic strains produce toxins like colibactin that induce DNA damage, disrupting cell cycle regulation and linking them to colorectal cancer.55 Moreover, dysbiosis can lead to decreased levels of SCFAs like butyrate, which are crucial for promoting apoptosis in cancer cells.56 Lower SCFA levels reduce apoptotic signaling, supporting cancer cell survival. Additionally, dysbiosis can influence cancer cell immortality by favoring bacteria that produce metabolites enhancing telomerase expression, thus extending cancer cell lifespan.57
Studies in germ-free mouse models have demonstrated that the gut microbiome significantly impacts cancer onset and progression. For instance, overexpression of the enzyme SQLE has been shown to enhance colorectal cancer cell proliferation by promoting cell cycle progression and inhibiting apoptosis, with contributions from pathogenic bacteria.58 Alterations in the microbiome associated with different stages of gastric cancer have highlighted specific bacteria as key players in disease progression. For example, infection with Helicobacter pylori initiates an inflammatory response, leading to the loss of acid-secreting cells and facilitating the colonization of other bacterial species, thus contributing to gastric carcinogenesis.59 Non-H. pylori bacteria produce microbial metabolites like N-nitroso compounds and lactate, promoting cancer through mechanisms such as inflammation, immune modulation, DNA damage, and epithelial-mesenchymal transition (EMT).60 Additionally, bacteria like H. pylori affect key cellular pathways, such as the AKT pathway, leading to deregulation of tumor suppressors and alterations in proliferative and survival pathways.61 Similarly, interactions with cellular structures like E-cadherin can disrupt cell junctions and activate oncogenic signaling pathways like β-catenin.62 Some bacteria, such as Salmonella enterica, can influence cell survival and proliferation by interacting with pathways like MAPK and AKT.63 Pathogenic infections leading to dysbiosis can cause DNA damage and genomic instability, thereby initiating and advancing tumors in susceptible cells. Bacterial toxins like colibactin and cytolethal distending toxin (CDT) cause DNA breaks, leading to mutations and tumor development.64 Additionally, pathogens like Shigella flexneri can disrupt DNA damage response and repair pathways, increasing mutation risks during DNA repair.65
Inducing Angiogenesis
Tumor growth necessitates a blood supply, which is achieved through angiogenesis, the formation of new blood vessels. Dysbiosis can contribute to increased levels of pro-angiogenic factors such as vascular endothelial growth factor (VEGF), facilitating tumor vascularization. Studies have shown that certain gut bacteria can stimulate VEGF production, promoting angiogenesis in cancers like pancreatic cancer.66 The relationship between gut dysbiosis and angiogenesis involves complex interactions where microbial imbalance influences inflammatory responses and biochemical signaling to support new blood vessel growth, which is essential for tumor development. Elevated levels of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-1β, associated with dysbiosis, can increase VEGF expression and enhance angiogenesis in cancers like colorectal cancer.67 Additionally, microbial metabolites such as secondary bile acids can induce angiogenesis, impacting liver cancers like hepatocellular carcinoma by promoting liver inflammation and tumor vascularization.68 Dysbiosis also impairs intestinal barrier function, allowing substances like lipopolysaccharides (LPS) to enter the bloodstream and trigger systemic inflammation and angiogenesis, particularly relevant to pancreatic cancer progression through PI3K/AKT signaling.69,70 The altered immune landscape due to dysbiosis, marked by reduced immune surveillance and increased immunosuppression, supports an angiogenic environment that aids malignancies like gastric cancer.71 These insights suggest potential therapeutic approaches, including probiotics, prebiotics, dietary interventions, and targeted therapies aimed at modulating the gut microbiota, reducing inflammation, and inhibiting angiogenic pathways in cancer treatment.
Activating Invasion and Metastasis
The ability of cancer cells to invade surrounding tissues and metastasize to distant sites is a critical hallmark of cancer.53 This complex process involves several steps: detachment from the primary tumor, acquisition of an invasive phenotype through epithelial-mesenchymal transition (EMT), local invasion, intravasation into the circulatory system, systemic transportation, extravasation, and colonization in secondary organs.72 Microbial imbalance in the gut can promote inflammation, facilitating extracellular matrix degradation and cancer cell spread. For example, Fusobacterium nucleatum has been associated with enhanced metastatic potential in colorectal cancer by binding to cancer cells and promoting invasion.73 Circulating tumor cells (CTCs) typically arise from epithelial tumor cells that have undergone EMT, marked by loss of adhesion and polarity, cytoskeletal reorganization, acquisition of tumor stem cell properties, and resistance to therapy. EMT is regulated by transcription factors such as Snail 1, Slug, ZEB1, Twist, and FOXC2, and influenced by tumor microenvironment signaling pathways like WNT, Notch, Hedgehog, TGFβ, FGF, EGF, and HGF.74,75 Tumor hypoxia further activates signaling pathways like PI3K, WNT/β-catenin, and MAPK, crucial for regulating EMT.76 Gut dysbiosis significantly impacts this process by altering the tumor microenvironment, supporting systemic inflammation, and modulating immune responses, thus creating conditions favorable for tumor cell invasion and metastasis. Dysbiosis can also affect the production of cytokines and growth factors, influencing cancer cell metastatic potential.77 Before arriving at the premetastatic niche, VEGFR1-positive hematopoietic progenitor cells migrate from the bone marrow into the circulation and establish in secondary organs, where they adhere to fibronectin produced by fibroblasts. This process is mediated by integrin VLA-4 and influenced by chemokine SDF-1 binding to the CXCR4 receptor on breast cancer cells, particularly associated with bone metastasis.78 Gut dysbiosis enhances systemic inflammation, increasing pro-inflammatory cytokines and stimulating overproduction of growth factors like VEGF-A, FGF1, EGF, and PDGF, which further support angiogenesis and metastasis. Metabolic byproducts of dysbiosis can also modulate angiogenic factor expression through signaling pathways within cancer cells. Additionally, exosomes released by cancer and immune cells, carrying proangiogenic molecules, play a role in tumor angiogenesis and metastasis, with gut dysbiosis potentially altering their composition and function.79,80 This intricate interaction between gut microbiome dysregulation and the tumor microenvironment underscores the systemic effects of gut health on cancer progression and metastasis, highlighting the need for a balanced gut microbiome in cancer prevention and treatment.
Deregulating Cellular Energetics
Cancer cells often exhibit altered metabolism, known as the Warburg effect, where they preferentially use glycolysis over oxidative phosphorylation for energy production, even under normoxic conditions. Gut dysbiosis impacts cancer cell metabolism, particularly in colorectal and pancreatic cancers, by affecting nutrient absorption and metabolite availability. In colorectal cancer, dysbiosis disrupts normal fermentation processes, impacting SCFA production like butyrate, which can be utilized by hypoxic tumor cells to support survival and proliferation.81 This process is mediated through the activation of the butyrate receptor GPR109A, which modulates the NF-κB pathway and influences inflammatory responses and cell survival.82 Dysbiosis also affects the synthesis of amino acids and the conversion of primary bile acids into secondary bile acids. These secondary bile acids have been implicated in DNA damage and the activation of carcinogenic signaling pathways, particularly through the FXR and TGR5 receptors in the liver and colorectal cancers.83,84
In pancreatic cancer, dysbiosis exacerbates the reliance of cancer cells on alternative energy sources, such as amino acids and lipids. This metabolic reprogramming, often referred to as the Warburg effect, illustrates the cancer cells’ preference for glycolysis, which results in the accumulation of glycolytic intermediates used as building blocks and an increased production of lactic acid.70,85 Lactic acid modulates the tumor microenvironment by suppressing immune surveillance and promoting angiogenesis through the activation of HIF-1α, which enhances the expression of vascular endothelial growth factor (VEGF).
Hypoxia, a common feature in solid tumors like pancreatic adenocarcinoma, activates hypoxia-inducible factors (HIFs) that upregulate glucose transporters (such as GLUT1) and glycolytic enzymes (including HK2 and PFKFB3), boosting glucose uptake and metabolism.86,87 HIFs also play a role in stimulating angiogenesis by upregulating VEGF, contributing to the tumor’s growth and metastatic potential.88 Dysbiosis can aggravate these hypoxic responses by promoting systemic inflammation through the release of microbial metabolites that mimic host signaling molecules, further influencing metabolic adaptations in cancer cells.
Avoiding Immune Destruction
The immune system plays a crucial role in identifying and eliminating cancer cells. However, gut dysbiosis can lead to an immunosuppressive tumor microenvironment that diminishes the efficacy of immune surveillance. For example, certain bacteria can modulate T-cell function or influence the expression of checkpoint proteins, which cancer cells exploit to evade immune detection. This intricate interplay between gut microbiota and immune responses significantly influences cancer progression, especially in colorectal cancer, hepatocellular carcinoma, and melanoma.
In colorectal cancer, the gut microbiota’s role is highlighted through its ability to modulate the immune landscape via cytokine signaling and T helper cell differentiation. Transplantation of microbiota from CRC patients into mice has shown to induce colorectal tumorigenesis, mediated through the elevation of pro-inflammatory cytokines such as interleukin-17 (IL-17) and interferon-gamma (IFN-γ), which enhance Th1 and Th17 cell infiltration in the colon.89 These conditions promote inflammatory and oncogenic pathways in the intestinal epithelium.
Specific pathogens such as Fusobacterium nucleatum facilitate CRC progression by recruiting myeloid-derived suppressor cells (MDSCs) and inhibiting natural killer (NK) cells and T cells through mechanisms involving the engagement of the TIGIT receptor, which inhibits NK cell cytotoxicity.90 Research reveals that Peptostreptococcus anaerobius is significantly more abundant in stool samples and colon biopsies from patients with colorectal cancer (CRC) and advanced adenoma than in those from healthy controls.91 This bacterium predominantly colonizes the colon and stimulates cell proliferation by interacting with TLR2 and TLR4 receptors on colon cells, leading to increased reactive oxidative species which enhance cholesterol synthesis, driving cell proliferation.
Hepatocellular Carcinoma (HCC): In HCC, the gut-liver axis is critical, with gut microbiota-derived metabolites and endotoxins like lipopolysaccharide (LPS) exacerbating liver pathology. LPS activates the NF-κB signaling pathway in hepatic stellate and Kupffer cells, leading to inflammation and fibrogenesis.92 Dysbiosis also affects bile acid metabolism, increasing the production of oncogenic secondary bile acids.93 Additionally, the decrease in beneficial microbes such as Akkermansia muciniphila and the presence of specific bacteria like Clostridium species are linked to increased MDSC infiltration and reduced levels of hepatic CXCR6+ natural killer T cells, respectively, which collectively dampen antitumor immunity.94,95
Melanoma: In melanoma, gut microbiota composition can significantly alter the efficacy of immunotherapies. Certain bacteria affect the responsiveness to checkpoint inhibitors by influencing the immune checkpoint axis, altering the expression of CTLA-4 and PD-1 on T cells.96 Skin microbiota also play a role by producing antimicrobial peptides and modulating cytokine profiles, such as increasing IL-17 production, which can affect immune surveillance and influence melanoma progression.97
These examples underscore the profound impact of gut microbiota on immune system function across various cancers. By modulating cytokine production, influencing immune cell differentiation, and interacting with metabolic pathways, gut microbiota can shape the tumor microenvironment, promoting immune evasion and cancer progression. Targeting microbial communities to restore a balanced gut microbiome offers a promising strategy for enhancing immune response and improving outcomes in cancer therapy.
Gut Microbiota Translocation
Introduction to Gut-Liver Axis and Disease
Previous research has highlighted the significant role of gut microbiota in diseases such as colorectal cancer, liver diseases, and particularly hepatocellular carcinoma (HCC). The liver is intricately connected to the gut via the portal vein, serving as a primary pathway for substances absorbed in the gut to reach the liver. Thus, any increase in intestinal permeability can facilitate the translocation of gut microbiota and their metabolites to the liver, potentially influencing liver health and disease progression.
In patients with hepatocellular carcinoma, metabolites from gut microbiota such as lipopolysaccharides (LPS) have been shown to trigger inflammation and liver injury, which in turn promote hepatocarcinogenesis.88 Additionally, dysregulation of bile acids by gut microbiota has been identified as a contributing factor to liver cirrhosis and cancer. Recent research in HCC has noted an increase in LPS-producing microbiota in patients with liver cirrhosis-HCC, while butyrate-producing microbiomes are decreased.88 Bile acids are crucial for maintaining liver health, and alterations in bile-acid-associated microbiota can significantly impact liver metabolism. For instance, Lithocholic acid, a secondary bile acid derived from the dehydroxylation of chenodeoxycholic acid (CDCA), has been found to be hepatotoxic and increases the risk of developing HCC.98
Gut Microbiota and Colorectal Cancer
A recent nested case-control study has further explored the connection between gut microbiota and colorectal cancer (CRC) through the analysis of circulating biomarkers related to gut microbiome activity. This study involved 261 CRC patients and 261 controls, monitored over a follow-up period of up to 15 years. Biomarkers such as LPS-binding protein (LBP), soluble CD14 (sCD14), and endotoxin core antibody (EndoCAb) immunoglobulin M (IgM) were measured to assess their association with CRC incidence. The results indicated a positive association between sCD14 levels and the incidence of CRC, suggesting that sCD14 could be a marker of microbial translocation and subsequent immune activation.99 These biomarkers are initiated primarily through the translocation of microbial lipopolysaccharides, which result in gut barrier dysfunction. Factors such as alcohol consumption and a high-fat diet are known to compromise gut barrier integrity. Upon reaching the gut barrier, LPS can bind to the cell surface Toll-like receptor 4 (TLR4) via the MD2-TLR4 complex, activating the production of inflammatory cytokines that stimulate the immune response. However, the EndoCAb was found to have a low association with CRC incidence, indicating variable impacts of different biomarkers.99 Thus, this dysfunction of the gut barrier alters the balance between commensal and deleterious bacteria in the gut microbiota, leading to the activation of inflammatory cytokines and prolonged inflammation that targets the epithelial colorectal cells, potentially culminating in CRC. The study underscores the importance of maintaining gut barrier integrity to prevent the adverse effects of microbial translocation on liver and colorectal health. This highlights the need for targeted interventions that could modulate gut microbiota to prevent or mitigate the progression of liver-related diseases and colorectal cancer.
Clinical Value of Gut Microbiota in Cancer Prognosis and Treatment
The gut microbiome is a stable, diverse component of the human body that is closely linked to individual health. Factors such as genetics, diet, environment, medications, and smoking can cause variations in the gut microbiome, distinguishing drug responders from non-responders.100,101 The gut microbiome interacts with the immune system, influencing the differentiation of lymphocytes, NK cells, and Tregs, which suggests its potential as a predictor of immunotherapy efficacy. Stool samples provide easy access to gut microbiome data, enabling clinicians to gather baseline information before treatment. Several studies have shown that the baseline gut microbiome composition can serve as a promising predictor of immunotherapy response.
Specific gut bacteria have been associated with improved outcomes in cancer treatment. For example, the presence of Akkermansia muciniphila has been linked to enhanced efficacy of PD-1 blockade in lung, liver, and kidney cancer patients.93,102,103 This bacterium is thought to modulate the immune system by boosting the recruitment and activation of immune cells, particularly CD8+ T cells, which are essential for antitumor immunity.104 Similarly, Bifidobacterium has been associated with better outcomes in melanoma patients receiving anti-PD-L1 therapy.105 Bifidobacterium spp. promotes the maturation and function of dendritic cells, leading to a stronger T-cell response against tumors.106 Beyond specific bacterial species, the overall diversity of the gut microbiome is a significant predictor of cancer prognosis. A more diverse microbiome supports a balanced immune system, reduces chronic inflammation, and promotes effective antitumor responses. Patients with higher gut microbiome diversity have shown better progression-free survival (PFS) and overall survival (OS) in cancers such as melanoma and non-small cell lung cancer (NSCLC).107–109
While immunotherapy activates the immune response, it can also lead to immune-related adverse events (irAEs). Predicting these adverse events, particularly severe ones, is crucial for preventing complications and optimizing treatment. Interestingly, molecular markers like PD-L1 expression levels may not reliably predict irAEs, as a Phase 3 clinical trial found similar irAE rates in NSCLC patients regardless of PD-L1 expression.110 However, the gut microbiome plays a more substantial role in predicting irAEs.
The gut microbiome can act as both a risk and protective factor for irAEs. Checkpoint inhibitor colitis (CIC) is one of the most common irAEs. Faecalibacterium prausnitzii is considered a risk factor for CIC, while Bacteroides fragilis, with its anti-inflammatory properties, acts as a protective factor in the gastrointestinal tract.111,112 Enrichment of Bacteroidetes, which promotes Treg differentiation, has been linked to resistance against CIC development.112 In liver cancer, reduced gut microbiome diversity and abundance were associated with severe immunotherapy-related colitis, suggesting that the microbiome can predict both the occurrence and severity of irAEs.113
In metastatic melanoma patients treated with Ipilimumab, a higher abundance of Faecalibacterium and other Firmicutes correlated with improved responses (longer PFS and OS) but also increased colitis risk, with fewer Tregs in peripheral blood.114 This indicates that specific bacteria can predict both immunotherapy efficacy and the likelihood of irAEs. The gut microbiome may also predict other irAEs such as diarrhea and skin toxicity. Prevotellamassilia timonensis, a biomarker for severe colitis in liver cancer patients, has also been linked to severe diarrhea.115 In advanced NSCLC patients, reduced gut microbiome diversity was associated with higher rates of skin toxicity during immunotherapy.116 A prospective cohort study (NCT03688347) found that bacteria such as Bifidobacterium and Desulfovibrio could predict overall irAE risk rather than specific irAE types.117 Neoadjuvant immunotherapy is becoming an increasingly popular treatment option for cancer patients. In esophageal squamous cell carcinoma (ESCC) patients receiving neoadjuvant camrelizumab and chemotherapy, taxonomic features of the gut microbiome were found to predict both pathological responses and severe adverse events (grade ≥3)118 expanding the potential applications of microbiome biomarkers in cancer prognosis.
Gut Microbiota Disparities in Treatment Outcomes
Interactions With Immune Checkpoint Inhibitors (ICIs)
The interaction of cancer cells with the immune system primarily occurs through T cells, which identify cancer antigens presented by Major Histocompatibility Complex class I (MHC I) as in demonstrated in Figure 3A. While immune checkpoints and co-stimulatory molecules limit the immune response to prevent autoimmunity, they also allow tumors to evade immune destruction as shown in Figure 3B. Tumor-infiltrating lymphocytes interact with Tumor antigens via the PD-1/PD-L1 signaling pathway. The combination of PD-L1 and PD-1 sends inhibitory signals that limit T cell growth and activation, allowing Tumor immune evasion and distant metastases as shown in Figure 3C. Anti-PD-1 or anti-PD-L1 therapies, on the other hand, may minimize Tumor cell evasion while enhancing immune responses; inhibiting immunological checkpoints between T cells and malignant cells increases the number of activated T cells capable of recognizing and killing Tumor cells (Figure 3D).119 These therapies have shown significant success in treating diseases such as lung cancer120 and malignant melanoma,121 despite challenges related to immune resistance at various treatment stages.100
Microbiota’s Role in Immune Modulation
Research has increasingly focused on the role of gut microbiota in cancer development and modulating immune responses during infections and cancer. Animal studies have examined the impact of ICIs on the translocation of specific gut microbiomes to secondary lymph nodes and tumors, exploring tumor-draining lymph nodes (TDLNs), primary tumors, and mesenteric lymph nodes (MLNs).122 For example, in studies involving melanoma treated with a combination of anti-PD-1 and anti-CTLA4, a higher abundance of bacteria was observed in the MLNs of treated groups compared to untreated groups, highlighting the significant influence of ICIs on gut microbiota dynamics.122 These studies suggest that ICIs may enhance the translocation of microbiota via dendritic cells, which are recruited into secondary lymphoid organs in association with CCR7, activating a potent anti-tumor immune response, especially involving CD8+ T cells.
Influence on Lymphocyte Relocation and Immunotherapy Efficacy
Recent research has demonstrated that the gut microbiota substantially affects the efficacy of immunotherapy by influencing the relocation of lymphocytes within the ileal lumen. For instance, a study showed that the commensal microbiome preserves immune cells in the ileal lamina propria and MLNs, where high endothelial venules express adhesion molecules like Mucosal Addressin Cell Adhesion Molecule-1 (MAdCAM-1).123 These molecules interact with α4β7 integrin on lymphocytes, including regulatory T cells (Tregs) and T helper cells, enhancing tumor control while avoiding disruptions by ICIs.
Conversely, antibiotics and dysbiosis can disrupt MAdCAM-1 interactions, forcing Tregs and T helper cells to migrate to tumor microenvironments, potentially accelerating tumor growth. For instance, a particular strain of Enterocloster, including E. clostridioformis, has been shown to down-regulate MAdCAM-1, influencing T cell positioning and activity.123
Case Studies and Clinical Outcomes
Fecal microbiota transplantation (FMT) combined with anti-PD-1 therapy has been observed to reverse immunological resistance to PD-1 and improve the tumor microenvironment (TME) in melanoma research.124 Patients responding to treatment exhibited a higher presence of specific T cell subsets, and the modification of the gut microbiome through FMT influenced responses to PD-1 blockade.
Additionally, a comprehensive study assessed the contribution of gut microbiota in patients with non-small cell lung cancer and metastatic colorectal cancer. The presence of butyrate-producing bacteria correlated with improved progression-free survival, highlighting the potential of targeting gut microbiota to enhance the outcomes of immunotherapy.125
Ongoing Research
A cohort study involving 175 metastatic melanoma patients receiving ICI therapy is currently underway to profile the gut microbiota and its correlation with treatment outcomes.126 This study is examining the microbiota’s association with monotherapy, combination therapy, and ICI-related side effects like colitis. Initial findings suggest that specific microbial patterns, such as the abundance of SCFA producers or certain Clostridia species, may correlate with progression-free survival and immune-related adverse effects. These investigations (demonstrated in Table 1) underline the critical role of gut microbiota in modulating immune responses and influencing the efficacy of cancer therapies, paving the way for novel therapeutic strategies that integrate microbiome modulation with conventional treatments to enhance patient outcomes.
 |
Table 1 Microbiomes Modulating the Impact of Immune Checkpoints in Cancer |
Mechanisms Underlying the Interaction Between Gut Microbiota and ICI’s Efficacy
Microbiota-Driven Activation of Innate Immunity: Mechanistic Insights for Immune Checkpoint Inhibitors
The gut microbiota plays a pivotal role in augmenting the response to immune checkpoint inhibitors (ICIs) by activating various components of the innate immune system. For instance, Bifidobacterium enhances the anti-cancer efficacy of ICIs by stimulating dendritic cells, which increases CD8+ T cell activity and helps overcome PD-L1 resistance.3 Similarly, Bacteroides fragilis not only prompts dendritic cell maturation and IL-12 activation, enhancing anti-CTLA-4 antibody efficacy, but also stimulates macrophage polarization to M1, bolstering innate immunity.3 Furthermore, the microbiota induces interferon gene stimulators and IFN signaling, which facilitates the transition from innate to adaptive immunity, converting mononuclear phagocytic cells into anti-tumor macrophages and activating the NKs-DCs axis.127,128
Microbiota-Driven Activation of Adaptive Immunity: Mechanistic Insights for Immune Checkpoint Inhibitors
The gut microbiota significantly influences the efficacy of ICIs across various cancer types by modulating adaptive immune responses. In melanoma, the presence of microbes such as those from the Clostridiales, Ruminococcaceae, and Faecalibacterium families enriches the tumor microenvironment (TME), enhancing antigen presentation to dendritic cells and boosting effector CD4+ and CD8+ T cell responses.3 Clinical trials involving fecal microbiota transplantation (FMT) have shown that the enrichment of Firmicutes and Actinobacteria can amplify PD-1 inhibition to enhance anti-tumor responses.124
Microbiota-Driven Metabolic Modulation of ICIs
Gut microbiota produces a myriad of metabolites that influence the immune response, particularly during ICI treatment. Metabolites such as short-chain fatty acids (SCFAs), inosine, and trimethylamine N-oxide (TMAO) have shown significant interactions with ICIs, boosting anti-tumor immune responses.3 For instance, Clostridium and Enterococcus-produced TMAO can cause ER stress leading to gasdermin-E-mediated pyroptosis, thereby enhancing CD8+ T cell functionality in the anti-tumor response.129,130 Additionally, SCFAs like propionic acid, produced by A. muciniphila, disrupt tumor cell proliferation and enhance ICI effects by inhibiting apoptosis.
Translocation and Immune Cell Recruitment in the TME
Recent studies have explored how the translocation of specific microbiota influences immune cell dynamics within the TME. For instance, Lactobacillus reuteri has been found to migrate to tumor sites, releasing metabolites like indole-3-aldehyde (I3A) from dietary tryptophan, which modulates the immune response by activating the aryl hydrocarbon receptor (AhR) on CD8+ T cells and promoting TC1 differentiation.131–133 This activation facilitates the production of IFNγ by CD8+ T cells, enhancing the anti-tumor immune response and potentially prolonging progression-free survival (PFS) in melanoma patients under ICI therapy.134–136
Influence of Gut Microbiota on Tumor Immunogenicity and ICI Efficacy
Gut microbiota can provoke tumor immunogenicity, enhancing ICI efficacy by producing tumor-cross antigens that activate anti-tumor immunity. An illustrative example is Bifidobacterium breve, which expresses an antigen similar to a tumor antigen found in melanoma cells, leading to T cell-mediated tumor cell destruction.137 Additionally, microbiota-derived metabolites like inosine have been shown to sensitize tumor cells to T-cell cytotoxicity, highlighting a novel mechanism through which microbiota can influence the efficacy of ICIs.138 The interactions between gut microbiota and ICIs illustrate a complex network involving innate and adaptive immunity, metabolic modulation, and direct impacts on the TME. This intricate relationship underscores the potential of leveraging gut microbiota in enhancing the efficacy of cancer immunotherapies, offering promising avenues for future research and therapeutic strategies.
Microbiome-Based Therapeutics to Improve Immune Checkpoint Inhibitors Efficacy
Fecal Microbial Transplantation (FMT)
Intensive chemotherapy often disrupts gut microbiota composition, leading to dysbiosis. Fecal microbial transplantation (FMT) has emerged as a novel approach to restore the gut microbiota during disease treatment. In acute myeloid leukemia (AML) patients, microbiota dysbiosis due to chemotherapy and antibiotics has been observed. A Phase II clinical study revealed that autologous fecal microbiota transfer (AFMT) is a safe and effective method for re-establishing gut microbiota, compared to traditional FMT.139 Ongoing studies, including a randomized clinical trial (NCT04758507) with advanced renal cell carcinoma patients, are exploring the significant role of gut microbiota in modulating the effectiveness of immune checkpoint inhibitors.2 Further research using FMT capsules in gastrointestinal cancer aims to counteract resistance to ICIs, particularly anti-PD-1 treatments. Recent trials, such as one involving advanced melanoma patients (NCT03772899), reported a 65% response rate using FMT in combination with PD-1 inhibitors like nivolumab or pembrolizumab, achieving up to a 20% complete response rate.140 Another phase II trial (ChiCTR2100046768) documented improved survival in treating refractory microsatellite stable metastatic colorectal cancer using a combination of FMT with anti-PD-1 inhibitors, highlighting the importance of specific bacterial populations such as Proteobacteria and Lachnospiraceae in responders.141
Dietary Interventions
Diet plays a crucial role in modulating tumor immunity. For instance, a high salt diet has been found to promote the translocation of Bifidobacterium into the tumor microenvironment (TME), enhancing NK cell activity and anti-tumor immunity.3 A nested case-control study linked dietary metabolites like choline, betaine, and phenylacetylglutamine (PAGIn) with lethal prostate cancer, suggesting that microbiota-related metabolites produced from specific diets influence cancer progression.142 Additionally, the traditional herbal remedy Xiao-Chai-Hu-Tang (XCHT) has been studied for its antidepressant properties and potential to influence cancer treatment. A xenografted mouse model of colorectal cancer treated with XCHT under chronic restraint stress showed increased longevity and modified levels of specific gut bacteria, which may suppress TLR4/MyD88/NF-κB signaling involved in tumor growth.143
Probiotics and Prebiotics
Probiotics are used to support disease treatment by restoring microbiota balance. Recent studies highlight the beneficial role of Clostridium butyricum in post-gastrectomy gastric cancer patients. This probiotic promotes gut microbiota regeneration, enhancing immune function and reducing inflammation through metabolites like butyric acid, which also helps in regenerating host intestinal epithelial cells.144
Prebiotics, indigestible food ingredients that promote the growth of beneficial gut bacteria, have also shown promise. In a Phase I clinical trial (NCT05303493), the prebiotic camu camu influenced the growth of health-promoting gut bacteria like Ruminococcus bromii in advanced melanoma patients experiencing immune-related adverse events (irAEs) from ICIs. This intervention appeared to minimize irAEs and enhance the immune response.145,146
Future Directions
This review has underscored the complex interplay between gut microbiota, its metabolites, and immune checkpoint inhibitors. The dynamic interactions suggest the need for further research to uncover the underlying mechanisms of this modulatory effect and investigate distinct microbiota compositions that may improve immunotherapy outcomes. Personalized microbiota modification methods, advanced spatial multi-omics tools, and targeted metabolomics analysis are crucial for optimizing therapeutic efficacy and understanding individual variability in response to cancer treatments. Moreover, monitoring the microbiota throughout treatment could provide insights into how microbiome composition affects treatment efficacy and patient outcomes, paving the way for personalized medicine in cancer management.
Acknowledgment
The publication of this article was supported by Qatar University.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Ruiz-Patiño A, Arrieta O, Cardona AF. et al. Immunotherapy at any line of treatment improves survival in patients with advanced metastatic non-small cell lung cancer (NSCLC) compared with chemotherapy (Quijote-CLICaP). Thorac Cancer. 2020;11(2):353–361. doi:10.1111/1759-7714.13272
2. Li X, Zhang S, Guo G, et al. Gut microbiome in modulating immune checkpoint inhibitors. EBioMedicine. 2022;82:104163. doi:10.1016/j.ebiom.2022.104163
3. Lu Y, Yuan X, Wang M, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. 2022;15(1):47. doi:10.1186/s13045-022-01273-9
4. Rebersek M. Gut microbiome and its role in colorectal cancer. BMC Cancer. 2021;21(1):1325. doi:10.1186/s12885-021-09054-2
5. Campbell C, Kandalgaonkar MR, Golonka RM, et al. Crosstalk between Gut Microbiota and Host Immunity: impact on Inflammation and Immunotherapy. Biomedicines. 2023;11(2). doi:10.3390/biomedicines11020294.
6. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8(1):51. doi:10.1186/s13073-016-0307-y
7. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31(1):69–75. doi:10.1097/MOG.0000000000000139
8. Finotello F, Mastrorilli E, Di Camillo B. Measuring the diversity of the human microbiota with targeted next-generation sequencing. Brief Bioinform. 2018;19(4):679–692. doi:10.1093/bib/bbw119
9. Kunika NF, Rangrez AY, Rangrez AY. Exploring the Involvement of Gut Microbiota in Cancer Therapy-Induced Cardiotoxicity. Int J mol Sci. 2023;24(8):7261. doi:10.3390/ijms24087261
10. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi:10.1126/science.aan3706
11. Santoni M, Piva F, Conti A, et al. Re: gut Microbiome Influences Efficacy of PD-1-based Immunotherapy Against Epithelial Tumors. Eur Urol. 2018;74(4):521–522. doi:10.1016/j.eururo.2018.05.033
12. Pabst O, Cerovic V, Hornef M. Secretory IgA in the Coordination of Establishment and Maintenance of the Microbiota. Trends Immunol. 2016;37(5):287–296. doi:10.1016/j.it.2016.03.002
13. Rabe H, Lundell A-C, Sjöberg F, et al. Neonatal gut colonization by Bifidobacterium is associated with higher childhood cytokine responses. Gut Microbes. 2020;12(1):1–14. doi:10.1080/19490976.2020.1847628
14. Giorgetti G, Brandimarte G, Fabiocchi F, et al. Interactions between Innate Immunity, Microbiota, and Probiotics. J Immunol Res. 2015;2015:501361. doi:10.1155/2015/501361
15. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi:10.1016/j.cell.2004.07.002
16. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi:10.1038/nri3608
17. Wang C, Li Q, Ren J. Microbiota-Immune Interaction in the Pathogenesis of Gut-Derived Infection. Front Immunol. 2019;10:1873. doi:10.3389/fimmu.2019.01873
18. Li Y, Arai S, Kato K, et al. The Potential Immunomodulatory Effect of Bifidobacterium longum subsp. longum BB536 on Healthy Adults through Plasmacytoid Dendritic Cell Activation in the Peripheral Blood. Nutrients. 2023;16(1):42. doi:10.3390/nu16010042
19. Jin C, Lagoudas GK, Zhao C, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell. 2019;176(5):998–1013.e16. doi:10.1016/j.cell.2018.12.040
20. Li Y, Wang Y, Shi F, et al. Phospholipid metabolites of the gut microbiota promote hypoxia-induced intestinal injury via CD1d-dependent γδ T cells. Gut Microbes. 2022;14(1):2096994. doi:10.1080/19490976.2022.2096994
21. Feng T, Elson CO. Adaptive immunity in the host–microbiota dialog. Mucosal Immunol. 2011;4(1):15–21. doi:10.1038/mi.2010.60
22. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi:10.1038/s41422-020-0332-7
23. Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109–1113. doi:10.1038/nature07336
24. Proietti M, Cornacchione V, Rezzonico Jost T, et al. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer’s patches to promote host-microbiota mutualism. Immunity. 2014;41(5):789–801. doi:10.1016/j.immuni.2014.10.010
25. Yang Y, Du L, Shi D, et al. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12(1):6757. doi:10.1038/s41467-021-27112-y
26. Cheng Y, Ling Z, Li L. The Intestinal Microbiota and Colorectal Cancer. Front Immunol. 2020;11:615056. doi:10.3389/fimmu.2020.615056
27. Kroemer G, Zitvogel L. Cancer immunotherapy in 2017: the breakthrough of the microbiota. Nat Rev Immunol. 2018;18(2):87–88. doi:10.1038/nri.2018.4
28. Meng C, Feng S, Hao Z, et al. Changes in gut microbiota composition with age and correlations with gut inflammation in rats. PLoS One. 2022;17(3):e0265430. doi:10.1371/journal.pone.0265430
29. Kneis B, Wirtz S, Weber K, et al. Colon Cancer Microbiome Landscaping: differences in Right- and Left-Sided Colon Cancer and a Tumor Microbiome-Ileal Microbiome Association. Int J mol Sci. 2023;24(4):3265. doi:10.3390/ijms24043265
30. Yoon H, Kim N-E, Park J, et al. Analysis of the gut microbiome using extracellular vesicles in the urine of patients with colorectal cancer. Korean J Intern Med. 2023;38(1):27–38. doi:10.3904/kjim.2022.112
31. Kang Y, Cai Y, Yang Y. The Gut Microbiome and Hepatocellular Carcinoma: implications for Early Diagnostic Biomarkers and Novel Therapies. Liver Cancer. 2022;11(2):113–125. doi:10.1159/000521358
32. Wei Z, Yang B, Tang T, et al. Gut microbiota and risk of five common cancers: a univariable and multivariable Mendelian randomization study. Cancer Med. 2023;12(9):10393–10405. doi:10.1002/cam4.5772
33. Nakano S, Kawamoto Y, Komatsu Y, et al. Analysis of the Pancreatic Cancer Microbiome Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration-Derived Samples. Pancreas. 2022;51(4):351–357. doi:10.1097/MPA.0000000000002028
34. Abe S, Masuda A, Matsumoto T, et al. Impact of intratumoral microbiome on tumor immunity and prognosis in human pancreatic ductal adenocarcinoma. J Gastroenterol. 2024;59(3):250–262. doi:10.1007/s00535-023-02069-5
35. Liang JQ, Li T, Nakatsu G, et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2020;69(7):1248–1257. doi:10.1136/gutjnl-2019-318532
36. Liang JQ, Wong SH, Szeto CH, et al. Fecal microbial DNA markers serve for screening colorectal neoplasm in asymptomatic subjects. J Gastroenterol Hepatol. 2021;36(4):1035–1043. doi:10.1111/jgh.15171
37. Xie YH, Gao Q-Y, Cai G-X, et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: test and Validation Studies. EBioMedicine. 2017;25:32–40. doi:10.1016/j.ebiom.2017.10.005
38. Löwenmark T, Li X, Löfgren-Burström A, et al. Parvimonas micra is associated with tumour immune profiles in molecular subtypes of colorectal cancer. Cancer Immunol Immunother. 2022;71(10):2565–2575. doi:10.1007/s00262-022-03179-4
39. Kunzmann AT, Proença MA, Jordao HW, et al. Fusobacterium nucleatum tumor DNA levels are associated with survival in colorectal cancer patients. Eur J Clin Microbiol Infect Dis. 2019;38(10):1891–1899. doi:10.1007/s10096-019-03649-1
40. Hulme H, Meikle LM, Strittmatter N, et al. Microbiome-derived carnitine mimics as previously unknown mediators of gut-brain axis communication. Sci Adv. 2020;6(11):eaax6328. doi:10.1126/sciadv.aax6328
41. Jiang H, Zeng W, Zhang X, et al. Gut microbiota and its metabolites in non-small cell lung cancer and brain metastasis: from alteration to potential microbial markers and drug targets. Front Cell Infect Microbiol. 2023;13:1211855. doi:10.3389/fcimb.2023.1211855
42. Malapelle U, Sirera R, Jantus-Lewintre E, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn. 2017;17(3):209–215. doi:10.1080/14737159.2017.1288568
43. Bauml JM, Li BT, Velcheti V, et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer. 2022;166:270–278. doi:10.1016/j.lungcan.2021.10.007
44. Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15(9):e0237802. doi:10.1371/journal.pone.0237802
45. Huang YF, Chen Y-J, Fan T-C, et al. Analysis of microbial sequences in plasma cell-free DNA for early-onset breast cancer patients and healthy females. BMC Med Genomics. 2018;11(Suppl 1):16. doi:10.1186/s12920-018-0329-y
46. Bull MJ, Plummer NT. Part 1: the Human Gut Microbiome in Health and Disease. Integr Med (Encinitas). 2014;13(6):17–22.
47. Khan I, Bai Y, Zha L, et al. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front Cell Infect Microbiol. 2021;11:716299. doi:10.3389/fcimb.2021.716299
48. Madhogaria B, Bhowmik P, Kundu A. Correlation between human gut microbiome and diseases. Infect Med (Beijing). 2022;1(3):180–191. doi:10.1016/j.imj.2022.08.004
49. Tan C, Wu Q, Wang H, et al. Dysbiosis of Gut Microbiota and Short-Chain Fatty Acids in Acute Ischemic Stroke and the Subsequent Risk for Poor Functional Outcomes. JPEN J Parenter Enteral Nutr. 2021;45(3):518–529. doi:10.1002/jpen.1861
50. Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J. 2014;20(3):181–189. doi:10.1097/PPO.0000000000000048
51. Brennan CA, Clay SL, Lavoie SL, et al. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes. 2021;13(1):1987780. doi:10.1080/19490976.2021.1987780
52. Ruo SW, Alkayyali T, Win M, et al. Role of Gut Microbiota Dysbiosis in Breast Cancer and Novel Approaches in Prevention, Diagnosis, and Treatment. Cureus. 2021;13(8):e17472. doi:10.7759/cureus.17472
53. Hanahan D. Hallmarks of Cancer: new Dimensions. Cancer Discov. 2022;12(1):31–46. doi:10.1158/2159-8290.CD-21-1059
54. Yang R, Qian L. Research on Gut Microbiota-Derived Secondary Bile Acids in Cancer Progression. Integr Cancer Ther. 2022;21:15347354221114100. doi:10.1177/15347354221114100
55. Chat H, Dalmasso G, Godfraind C, et al. Cytotoxic necrotizing factor 1 hinders colon tumorigenesis induced by colibactin-producing Escherichia coli in Apc Min/+ mice. Gut Microbes. 2023;15(1):2229569. doi:10.1080/19490976.2023.2229569
56. Li Q, Cao L, Tian Y, et al. Butyrate Suppresses the Proliferation of Colorectal Cancer Cells via Targeting Pyruvate Kinase M2 and Metabolic Reprogramming. mol Cell Proteomics. 2018;17(8):1531–1545. doi:10.1074/mcp.RA118.000752
57. Al-Daghri NM, Abdi S, Sabico S, et al. Gut-Derived Endotoxin and Telomere Length Attrition in Adults with and without Type 2 Diabetes. Biomolecules. 2021;11(11):1693. doi:10.3390/biom11111693
58. He X, Lan H, Jin K, et al. Cholesterol in colorectal cancer: an essential but tumorigenic precursor? Front Oncol. 2023;13:1276654. doi:10.3389/fonc.2023.1276654
59. Wang M, Yang G, Tian Y, et al. The role of the gut microbiota in gastric cancer: the immunoregulation and immunotherapy. Front Immunol. 2023;14:1183331. doi:10.3389/fimmu.2023.1183331
60. Li Q, Yu H. The role of non-H. pylori bacteria in the development of gastric cancer. Am J Cancer Res. 2020;10(8):2271–2281.
61. Wei J, Nagy TA, Vilgelm A, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139(4):1333–1343. doi:10.1053/j.gastro.2010.06.018
62. Prasad SK, Bhat S, Shashank D, et al. Bacteria-Mediated Oncogenesis and the Underlying Molecular Intricacies: what We Know So Far. Front Oncol. 2022;12:836004. doi:10.3389/fonc.2022.836004
63. Wu LH, Pangilinan CR, Lee CH. Downregulation of AKT/mTOR signaling pathway for Salmonella-mediated autophagy in human anaplastic thyroid cancer. J Cancer. 2022;13(11):3268–3279. doi:10.7150/jca.75163
64. Lai YR, Chang Y-F, Ma J, et al. From DNA Damage to Cancer Progression: potential Effects of Cytolethal Distending Toxin. Front Immunol. 2021;12:760451. doi:10.3389/fimmu.2021.760451
65. Bergounioux J, Elisee R, Prunier A-L, et al. Calpain activation by the Shigella flexneri effector VirA regulates key steps in the formation and life of the bacterium’s epithelial niche. Cell Host Microbe. 2012;11(3):240–252. doi:10.1016/j.chom.2012.01.013
66. Sevcikova A, Mladosievicova B, Mego M, et al. Exploring the Role of the Gut and Intratumoral Microbiomes in Tumor Progression and Metastasis. Int J mol Sci. 2023;24(24):17199. doi:10.3390/ijms242417199
67. Bhat AA, Nisar S, Singh M, et al. Cytokine- and chemokine-induced inflammatory colorectal tumor microenvironment: emerging avenue for targeted therapy. Cancer Commun. 2022;42(8):689–715. doi:10.1002/cac2.12295
68. Chen JL, Wang L, Li R, et al. High expression of endothelial progenitor cell-induced angiogenic markers is associated with bile acid levels in HCC. Oncol Lett. 2020;20(3):2729–2738. doi:10.3892/ol.2020.11815
69. Massoumi RL, Teper Y, Ako S, et al. Direct Effects of Lipopolysaccharide on Human Pancreatic Cancer Cells. Pancreas. 2021;50(4):524–528. doi:10.1097/MPA.0000000000001790
70. Sun Y, Wu C, Ma J, et al. Toll-like receptor 4 promotes angiogenesis in pancreatic cancer via PI3K/AKT signaling. Exp Cell Res. 2016;347(2):274–282. doi:10.1016/j.yexcr.2016.07.009
71. Majewski M, Mertowska P, Mertowski S, et al. Microbiota and the Immune System-Actors in the Gastric Cancer Story. Cancers (Basel). 2022;14(15):3832. doi:10.3390/cancers14153832
72. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
73. Xu C, Fan L, Lin Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13(1):1980347. doi:10.1080/19490976.2021.1980347
74. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev mol Cell Biol. 2019;20(2):69–84. doi:10.1038/s41580-018-0080-4
75. Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
76. Abou Khouzam R, Zaarour RF, Brodaczewska K, et al. The Effect of Hypoxia and Hypoxia-Associated Pathways in the Regulation of Antitumor Response: friends or Foes? Frontiers in Immunology. 2022;13:828875. doi:10.3389/fimmu.2022.828875
77. Biragyn A, Ferrucci L. Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 2018;19(6):e295–e304. doi:10.1016/S1470-2045(18)30095-0
78. Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–827. doi:10.1038/nature04186
79. Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol. 2007;28(7):299–307. doi:10.1016/j.it.2007.05.007
80. Mishra S, Amatya SB, Salmi S, et al. Microbiota and Extracellular Vesicles in Anti-PD-1/PD-L1 Therapy. Cancers (Basel). 2022;14(20):5121. doi:10.3390/cancers14205121
81. Huang JT, Mao YQ. The impact of the microbiome in cancer: targeting metabolism of cancer cells and host. Front Oncol. 2022;12:1029033. doi:10.3389/fonc.2022.1029033
82. Thangaraju M, Cresci GA, Liu K, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69(7):2826–2832. doi:10.1158/0008-5472.CAN-08-4466
83. Caliceti C, Punzo A, Silla A, et al. New Insights into Bile Acids Related Signaling Pathways in the Onset of Colorectal Cancer. Nutrients. 2022;14(14):2964. doi:10.3390/nu14142964
84. Wang X, Fu X, Van Ness C, et al. Bile Acid Receptors and Liver Cancer. Curr Pathobiol Rep. 2013;1(1):29–35. doi:10.1007/s40139-012-0003-6
85. Aykut B, Pushalkar S, Chen R, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. doi:10.1038/s41586-019-1608-2
86. Leung SWS, Shi Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol Sin. 2022;43(2):251–259. doi:10.1038/s41401-021-00647-y
87. Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):126. doi:10.1186/s13046-020-01629-4
88. Zheng R, Wang G, Pang Z, et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020;9(12):4232–4250. doi:10.1002/cam4.3045
89. Wong SH, Zhao L, Zhang X, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology. 2017;153(6):1621–1633.e6. doi:10.1053/j.gastro.2017.08.022
90. Wu J, Li Q, Fu X. Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Transl Oncol. 2019;12(6):846–851. doi:10.1016/j.tranon.2019.03.003
91. Tsoi H, Chu ESH, Zhang X, et al. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152(6):1419–1433.e5. doi:10.1053/j.gastro.2017.01.009
92. Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. doi:10.1038/s41575-018-0011-z
93. Pellegrino A, Coppola G, Santopaolo F, et al. Role of Akkermansia in Human Diseases: from Causation to Therapeutic Properties. Nutrients. 2023;15(8):1815. doi:10.3390/nu15081815
94. Ma C, Han M, Heinrich B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391):6391. doi:10.1126/science.aan5931
95. Behary J, Amorim N, Jiang X-T, et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat Commun. 2021;12(1):187. doi:10.1038/s41467-020-20422-7
96. Mekadim C, Skalnikova HK, Cizkova J, et al. Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol. 2022;22(1):63. doi:10.1186/s12866-022-02458-5
97. Ibrahim WN, Doolaanea AA, Bin Abdull Rasad MSB. Effect of shRNA Mediated Silencing of YB-1 Protein on the Expression of Matrix Collagenases in Malignant Melanoma Cell In Vitro. Cells. 2018;7(1):7. doi:10.3390/cells7010007
98. Lee PC, Wu C-J, Hung Y-W, et al. Gut microbiota and metabolites associate with outcomes of immune checkpoint inhibitor-treated unresectable hepatocellular carcinoma. J Immunother Cancer. 2022;10(6):e004779. doi:10.1136/jitc-2022-004779
99. Shi M, Zong X, Hur J, et al. Circulating markers of microbial translocation and host response to bacteria with risk of colorectal cancer: a prospective, nested case-control study in men. EBioMedicine. 2023;91:104566. doi:10.1016/j.ebiom.2023.104566
100. Said SS, Ibrahim WN. Cancer Resistance to Immunotherapy: comprehensive Insights with Future Perspectives. Pharmaceutics. 2023;15(4):1143. doi:10.3390/pharmaceutics15041143
101. Said SS, Ibrahim WN. Breaking Barriers: the Promise and Challenges of Immune Checkpoint Inhibitors in Triple-Negative Breast Cancer. Biomedicines. 2024;12(2):369. doi:10.3390/biomedicines12020369
102. Wu Z, Zhang S, Li L, et al. The gut microbiota modulates responses to anti-PD-1 and chemotherapy combination therapy and related adverse events in patients with advanced solid tumors. Front Oncol. 2022;12:887383. doi:10.3389/fonc.2022.887383
103. Lan X, Ma J, Huang Z, et al. Akkermansia muciniphila might improve anti-PD-1 therapy against HCC by changing host bile acid metabolism. J Gene Med. 2024;26(1):e3639. doi:10.1002/jgm.3639
104. Luo ZW, Xia K, Liu Y-W, et al. Extracellular Vesicles from Akkermansia muciniphila Elicit Antitumor Immunity Against Prostate Cancer via Modulation of CD8(+) T Cells and Macrophages. Int J Nanomedicine. 2021;16:2949–2963. doi:10.2147/IJN.S304515
105. Procaccianti G, Roggiani S, Conti G, et al. Bifidobacterium in anticancer immunochemotherapy: friend or foe? Microbiome Res Rep. 2023;2(3):24. doi:10.20517/mrr.2023.23
106. Gavzy SJ, Kensiski A, Lee ZL, et al. Bifidobacterium mechanisms of immune modulation and tolerance. Gut Microbes. 2023;15(2):2291164. doi:10.1080/19490976.2023.2291164
107. Guo C, Kong L, Xiao L, et al. The impact of the gut microbiome on tumor immunotherapy: from mechanism to application strategies. Cell Biosci. 2023;13(1):188. doi:10.1186/s13578-023-01135-y
108. Zhang M, Liu J, Xia Q. Role of gut microbiome in cancer immunotherapy: from predictive biomarker to therapeutic target. Exp Hematol Oncol. 2023;12(1):84. doi:10.1186/s40164-023-00442-x
109. Zhang H, Xu Z. Gut-lung axis: role of the gut microbiota in non-small cell lung cancer immunotherapy. Front Oncol. 2023;13:1257515. doi:10.3389/fonc.2023.1257515
110. Gao W, Liu Q, Zhou Y, et al. The Predictive Model Construction for Immune-Related Adverse Events in Non-Small Cell Lung Cancer Patients Receiving Immunotherapy. Technol Cancer Res Treat. 2023;22:15330338231206705. doi:10.1177/15330338231206705
111. Tang L, Wang J, Lin N, et al. Immune Checkpoint Inhibitor-Associated Colitis: from Mechanism to Management. Front Immunol. 2021;12:800879. doi:10.3389/fimmu.2021.800879
112. He Q, Niu M, Bi J, et al. Protective effects of a new generation of probiotic Bacteroides fragilis against colitis in vivo and in vitro. Sci Rep. 2023;13(1):15842. doi:10.1038/s41598-023-42481-8
113. Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol. 2020;72(2):230–238. doi:10.1016/j.jhep.2019.08.016
114. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. doi:10.1093/annonc/mdx108
115. Pei B, Ma X, Wu N, et al. Effect of microbiome group on immune checkpoint inhibitors in treatment of gastrointestinal tumors. Chin J Cancer Res. 2023;35(3):252–265. doi:10.21147/j.issn.1000-9604.2023.03.05
116. Liu T, Xiong Q, Li L, et al. Intestinal microbiota predicts lung cancer patients at risk of immune-related diarrhea. Immunotherapy. 2019;11(5):385–396. doi:10.2217/imt-2018-0144
117. Chau J, Yadav M, Liu B, et al. Prospective correlation between the patient microbiome with response to and development of immune-mediated adverse effects to immunotherapy in lung cancer. BMC Cancer. 2021;21(1):808. doi:10.1186/s12885-021-08530-z
118. Xu L, Qi Y, Jiang Y, et al. Crosstalk between the gut microbiome and clinical response in locally advanced thoracic esophageal squamous cell carcinoma during neoadjuvant camrelizumab and chemotherapy. Ann Transl Med. 2022;10(6):325. doi:10.21037/atm-22-1165
119. Wang J, Yang H-R, Wang D-J, et al. Association between the gut microbiota and patient responses to cancer immune checkpoint inhibitors. Oncol Lett. 2020;20(6):342. doi:10.3892/ol.2020.12205
120. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi:10.1056/NEJMoa1501824
121. Robert C, Schachter J, Long GV, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med. 2015;372(26):2521–2532. doi:10.1056/NEJMoa1503093
122. Choi Y, Lichterman JN, Coughlin LA, et al. Immune checkpoint blockade induces gut microbiota translocation that augments extraintestinal antitumor immunity. Sci Immunol. 2023;8(81):eabo2003. doi:10.1126/sciimmunol.abo2003
123. Fidelle M, Rauber C, Alves Costa Silva C, et al. A microbiota-modulated checkpoint directs immunosuppressive intestinal T cells into cancers. Science. 2023;380(6649):eabo2296. doi:10.1126/science.abo2296
124. Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371(6529):595–602. doi:10.1126/science.abf3363
125. Martini G, Ciardiello D, Dallio M, et al. Gut microbiota correlates with antitumor activity in patients with mCRC and NSCLC treated with cetuximab plus avelumab. Int J Cancer. 2022;151(3):473–480. doi:10.1002/ijc.34033
126. Björk JR, Bolte LA, Maltez Thomas A, et al. Longitudinal gut microbiome changes in immune checkpoint blockade-treated advanced melanoma. Nat Med. 2024;30(3):785–796. doi:10.1038/s41591-024-02803-3
127. Lam KC, Araya RE, Huang A, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338–5356.e21. doi:10.1016/j.cell.2021.09.019
128. Jin Y, Dong H, Xia L, et al. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J Thorac Oncol. 2019;14(8):1378–1389. doi:10.1016/j.jtho.2019.04.007
129. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi:10.1126/science.aac4255
130. Mager LF, Burkhard R, Pett N, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369(6510):1481–1489. doi:10.1126/science.abc3421
131. Kang X, Lau HC, Yu J. Modulating gut microbiome in cancer immunotherapy: harnessing microbes to enhance treatment efficacy. Cell Rep Med. 2024;5(4):101478. doi:10.1016/j.xcrm.2024.101478
132. Joachim L, Göttert S, Sax A, et al. The microbial metabolite desaminotyrosine enhances T-cell priming and cancer immunotherapy with immune checkpoint inhibitors. EBioMedicine. 2023;97:104834. doi:10.1016/j.ebiom.2023.104834
133. Bender MJ, McPherson AC, Phelps CM, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. 2023;186(9):1846–1862.e26. doi:10.1016/j.cell.2023.03.011
134. Fong W, Li Q, Ji F, et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut. 2023;72(12):2272–2285. doi:10.1136/gutjnl-2023-329543
135. Kawanabe-Matsuda H, Takeda K, Nakamura M, et al. Dietary Lactobacillus -Derived Exopolysaccharide Enhances Immune-Checkpoint Blockade Therapy. Cancer Discov. 2022;12(5):1336–1355. doi:10.1158/2159-8290.CD-21-0929
136. Griffin ME, Espinosa J, Becker JL, et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science. 2021;373(6558):1040–1046. doi:10.1126/science.abc9113
137. Bessell CA, Isser A, Havel JJ, et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight. 2020;5(8). doi:10.1172/jci.insight.135597.
138. Zhang L, Jiang L, Yu L, et al. Inhibition of UBA6 by inosine augments tumour immunogenicity and responses. Nat Commun. 2022;13(1):5413. doi:10.1038/s41467-022-33116-z
139. Malard F, Vekhoff A, Lapusan S, et al. Gut microbiota diversity after autologous fecal microbiota transfer in acute myeloid leukemia patients. Nat Commun. 2021;12(1):3084. doi:10.1038/s41467-021-23376-6
140. Routy B, Lenehan JG, Miller WH, et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med. 2023;29(8):2121–2132. doi:10.1038/s41591-023-02453-x
141. Zhao W, Lei J, Ke S, et al. Fecal microbiota transplantation plus tislelizumab and fruquintinib in refractory microsatellite stable metastatic colorectal cancer: an open-label, single-arm, phase II trial (RENMIN-215). EClinicalMedicine. 2023;66:102315. doi:10.1016/j.eclinm.2023.102315
142. Reichard CA, Naelitz BD, Wang Z, et al. Gut Microbiome-Dependent Metabolic Pathways and Risk of Lethal Prostate Cancer: prospective Analysis of a PLCO Cancer Screening Trial Cohort. Cancer Epidemiol Biomarkers Prev. 2022;31(1):192–199. doi:10.1158/1055-9965.EPI-21-0766
143. Shao S, Jia R, Zhao L, et al. Xiao-Chai-Hu-Tang ameliorates tumor growth in cancer comorbid depressive symptoms via modulating gut microbiota-mediated TLR4/MyD88/NF-κB signaling pathway. Phytomedicine. 2021;88:153606. doi:10.1016/j.phymed.2021.153606
144. Cao W, Zheng C, Xu X, et al. Clostridium butyricum potentially improves inflammation and immunity through alteration of the microbiota and metabolism of gastric cancer patients after gastrectomy. Front Immunol. 2022;13:1076245. doi:10.3389/fimmu.2022.1076245
145. Pang SA, Elkrief A, Capella MP, et al. Two Cases of Durable and Deep Responses to Immune Checkpoint Inhibition-Refractory Metastatic Melanoma after Addition of Camu Camu Prebiotic. Curr Oncol. 2023;30(9):7852–7859. doi:10.3390/curroncol30090570
146. Villemin C, Six A, Neville BA, et al. The heightened importance of the microbiome in cancer immunotherapy. Trends Immunol. 2023;44(1):44–59. doi:10.1016/j.it.2022.11.002
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Overexpression of TREM1 is Associated with the Immune-Suppressive Microenvironment and Unfavorable Prognosis in Pan-Cancer
Zhou X, Lin K, Fu L, Liu F, Lin H, Chen Y, Zhuang B, Liang H, Deng Q, Wang Z, Chen W, Luo J, Cao J, Li P
Journal of Inflammation Research 2023, 16:1375-1391
Published Date: 27 March 2023
Exploring the Correlation Between GPR176, a Potential Target Gene of Gastric Cancer, and Immune Cell Infiltration
Gu X, Shen H, Xiang Z, Li X, Zhang Y, Zhang R, Su F, Wang Z
Pharmacogenomics and Personalized Medicine 2023, 16:519-535
Published Date: 1 June 2023
Integrating Bulk and Single-Cell RNA Sequencing Reveals Heterogeneity, Tumor Microenvironment, and Immunotherapeutic Efficacy Based on Sialylation-Related Genes in Bladder Cancer
Tan Z, Chen X, Zuo J, Fu S, Wang J, Wang H
Journal of Inflammation Research 2023, 16:3399-3417
Published Date: 14 August 2023
NOTCH1 Mutations Predict Superior Outcomes of Immune Checkpoint Blockade in Non-Small Cell Lung Cancer
Huang Q, Cao H, Yao Q, Zhou X, Li H, Bai Q, Hu H
ImmunoTargets and Therapy 2023, 12:165-173
Published Date: 5 December 2023
The Role of Type 2 Diabetes Mellitus–Related Risk Factors and Drugs in Hepatocellular Carcinoma
Mai Y, Meng L, Deng G, Qin Y
Journal of Hepatocellular Carcinoma 2024, 11:159-171
Published Date: 19 January 2024


