Back to Journals » Journal of Inflammation Research » Volume 17
Important Role of Mitochondrial Dysfunction in Immune Triggering and Inflammatory Response in Rheumatoid Arthritis
Authors Li P , Zhou M, Wang J, Tian J, Zhang L, Wei Y, Yang F, Xu Y, Wang G
Received 6 October 2024
Accepted for publication 15 December 2024
Published 27 December 2024 Volume 2024:17 Pages 11631—11657
DOI https://doi.org/10.2147/JIR.S499473
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Adam Bachstetter
Pingshun Li,1,2 Mengru Zhou,2 Jia Wang,2 Jiexiang Tian,2 Lihuan Zhang,2 Yong Wei,2 Fang Yang,2 Yali Xu,1 Gang Wang2
1College of Integrative Chinese and Western Medicine, Gansu University of Chinese Medicine, Lanzhou, 730000, People’s Republic of China; 2Department of Rheumatology and Bone Disease, Affiliated Hospital of Gansu University of Chinese Medicine, Lanzhou, 730000, People’s Republic of China
Correspondence: Gang Wang, Email [email protected]
Abstract: Rheumatoid arthritis (RA) is an inflammatory autoimmune disease, primarily characterized by chronic symmetric synovial inflammation and erosive bone destruction.Mitochondria, the primary site of cellular energy production, play a crucial role in energy metabolism and possess homeostatic regulation capabilities. Mitochondrial function influences the differentiation, activation, and survival of both immune and non-immune cells involved in RA pathogenesis. If the organism experiences hypoxia, genetic predisposition, and oxidative stress, it leads to mitochondrial dysfunction, which further affects immune cell energy metabolism, synovial cell proliferation, apoptosis, and inflammatory signaling, causing the onset and progression of RA; and, mitochondrial regulation is becoming increasingly important in the treatment of RA.In this review, we examine the structure and function of mitochondria, analyze the potential causes of mitochondrial dysfunction in RA, and focus on the mechanisms by which mitochondrial dysfunction triggers chronic inflammation and immune disorders in RA. We also explore the effects of mitochondrial dysfunction on RA immune cells and osteoblasts, emphasizing its key role in the immune response and inflammatory processes in RA. Furthermore, we discuss potential biological processes that regulate mitochondrial homeostasis, which are of great importance for the prevention and treatment of RA.
Keywords: mitochondria, rheumatoid arthritis, immune triggers, inflammatory response, osteoblasts, synovial cells
Introduction
Mitochondria are intracellular organelles present in almost all eukaryotic organisms and are involved in various cellular processes, primarily playing a critical role in energy production. They also regulate calcium homeostasis, produce reactive oxygen species(ROS), and are involved in cellular proliferation and metabolism.1,2 Mitochondria are particularly important for immune cells, eg, T cells, macrophages, and neutrophils, which serve as the sentinel cells of the immune response and require a large supply of energy for their rapid response and metabolic reprogramming processes.3 Interest in these organelles has grown with recent findings linking mitochondria to various pathologies, including immune disorders, inflammation, cancer, aging, and neurodegenerative diseases.4 Dysfunctional mitochondria are unable to meet the energy demands of high-energy tissues such as the heart, brain, and skeletal muscles, leading to a wide range of clinical manifestations.5
RA is a systemic autoimmune disease characterized by synovial inflammation and hyperplasia, the production of autoantibodies, including rheumatoid factor (RF) and anti-citrullinated protein antibodies (ACPA), cartilage destruction, vascular opacification, and bone erosion.6 In RA, autoimmune tissue destruction manifests as synovitis, a joint inflammation triggered and sustained by complex interactions among various dendritic cell (DC) subtypes, T cells, macrophages, B cells, neutrophils, fibroblasts, and osteoclasts.7 Due to the persistent activation and incomplete clearance of specific autoantigens in RA patients, this ongoing immune cell activation and abnormal immune response result in recurrent chronic synovitis in the joints. This process leads to the formation of vascular opacities, which invade the cartilage-bone junction, causing bone erosion and cartilage degeneration.8,9
Studies have shown that mitochondrial dysfunction plays a significant role in the development of RA. Structurally and functionally intact mitochondria are essential for the normal survival of osteoblasts, synoviocytes, and immune cells. Once mitochondrial function is compromised, it affects the survival, activation, and differentiation of both immune and non-immune cells involved in RA pathogenesis, ultimately contributing to the development of RA.10,11 In this review, we examine the structure and function of mitochondria, analyze the potential causes of mitochondrial dysfunction in RA, and focus on the mechanisms by which mitochondrial dysfunction triggers chronic inflammation and immune disorders in RA. We also review the effects of mitochondrial dysfunction on immune cells and osteoblasts in RA, highlighting the physiological functions and pathological roles of mitochondria in RA, to promote the understanding of mitochondrial mechanisms and emphasize their role in RA pathogenesis.
Mitochondrial Structure
Mitochondria are intracellular organelles primarily responsible for energy conversion and supply within the cell.12 They function mainly through three key systems: the respiratory chain, the tricarboxylic acid cycle, and fatty acid oxidation. Additionally, mitochondria play critical roles in regulating apoptosis, cell signaling, and maintaining cellular metabolic homeostasis.13,14
Bilayer Membrane Structure
Mitochondria are enclosed by two phospholipid membranes: the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM), which create two distinct spaces within the organelle, the matrix and the intermembrane space (IMS).15 These membranes differ significantly in lipid composition, transmembrane protein characteristics, permeability, and shape, which reflect the organelle’s symbiotic origin.
The OMM closely resembles eukaryotic membranes in lipid composition, while the IMM is similar to bisphosphatidylglycerol-rich bacterial membranes.16 The IMM has a high protein-to-lipid ratio and forms densely stacked invaginations in the matrix called cristae.17 These cristae increase the surface area and the capacity for ATP production, as the oxidative phosphorylation (OXPHOS) machinery is embedded within them, along with other essential proteins. Mitochondrial cristae morphology undergoes significant changes in the face of mitochondrial stress or energy demand, and optimizes mitochondrial function mainly by adjusting the number and structure of cristae to protect cells from damage. For example, cold stimulation promotes the transport of MICOS complex subunit MIC19 protein to mitochondria in brown adipose tissue, which in turn promotes the formation of mitochondrial cristae and improves mitochondrial function; the mechanism of this is mainly related to the activation of the unfolded protein response (UPR) pathway.18 The portion of the IMM that does not extend into the matrix but runs parallel to the OMM is known as the inner boundary membrane (IBM). The cristae and IBM are connected by narrow tubular or slit-like structures called cristae junctions (CJ) (Figure 1).19
Additionally, the OMM and IMM differ significantly in permeability. The IMM is far less permeable than the OMM. The IMM is mainly characterized by the presence of voltage-dependent anion channels (VDAC) and α-helical transport proteins, such as protein translocases and various metabolite and ion carriers, which facilitate the passage of ions and small molecules.20 In contrast, the OMM contains channel-forming proteins such as β-barrel transmembrane hydrophilic pores, which allow the passage of precursor proteins, small hydrophilic metabolites (eg, O2 and CO2), and ions. This selective permeability enables the formation of electrochemical gradients across the mitochondrial membrane, which are essential for ATP production. Additionally, it tightly regulates the concentration of other ions, such as calcium, which plays a crucial role in intracellular signaling.21,22
Matrix and Intermembrane Space
Mitochondrial function is not only closely related to its double-membrane structure, but the mitochondrial matrix also plays an important role
The interior of the mitochondrion consists of the matrix and the intermembrane space.23 The matrix contains various soluble enzymes and mitochondrial DNA (mtDNA), while the intermembrane space lies between the inner and outer membranes. Mitochondrial DNA shares typical characteristics with bacterial DNA: it is a circular, double-stranded molecule containing 16,569 base pairs (bp) and lacks introns, primarily exhibiting cis-trans arrangements.24 Structurally, each gene is adjacent to the next, except for substitution loops or non-coding regions in the D-loop, although some genes may overlap.
Unlike nuclear DNA, mtDNA exists in multiple copies within a cell, ranging from approximately 100 to 10,000 copies, depending on the energy demands of the tissue.25 Additionally, mtDNA genetic codons differ slightly from nuclear DNA, using distinct codons for tryptophan and methionine, and it includes only two termination codons. Over the course of evolution, most mitochondrial genes were lost or transferred to nuclear DNA, leaving mtDNA with only 37 genes: 11 messenger RNAs (mRNAs) encoding 13 proteins, 2 ribosomal RNAs (rRNA, 12S and 16S), and 22 tRNAs.26
Mitochondrial Function
Energy Metabolism
Mitochondria serve as the intracellular “energy factories” and are the primary site of oxidative phosphorylation within the cell.27 The mitochondrial respiratory chain forms the core structure of oxidative phosphorylation, consisting of four enzyme complexes: nicotinamide adenine dinucleotide ubiquinone reductase (NADH dehydrogenase) (Complex I, CI), succinate ubiquinone oxidoreductase (Complex II, CII), ubiquinone cytochrome oxidoreductase (Complex III, CIII), and cytochrome c oxidase (Complex IV, CIV). These complexes are accompanied by two mobile electron carriers, ubiquinone (CoQ) and cytochrome c (Cyt c), which are embedded in the inner mitochondrial membrane, creating a continuous reaction system.28,29 Through this system, mitochondria oxidize and break down nutrients such as sugars, fats, and amino acids, releasing significant amounts of energy.30 This process involves the electron transport chain and ATP synthase. The electron transport chain transfers electrons and protons from substrates to oxygen, creating a proton gradient that is harnessed by ATP synthase to generate ATP, ATP as a mediator of energy storage and energy conversion, which powers various cellular functions.31 Therefore, mitochondria are the central site of oxidative metabolism in eukaryotes and are essential for maintaining normal cellular physiological functions (Figure 2).32
Apoptosis and Autophagy
Mitochondria play a crucial role in both apoptosis and autophagy.33 When cells are severely damaged or exposed to specific signals, mitochondria promote the oligomerization of pro-apoptotic proteins BAX and BAK at the outer mitochondrial membrane (OMM) by regulating transmembrane potential and bioenergetic exhaustion.34 This process induces the release of pro-apoptotic proteins, including cytochrome c, from the intermembrane space (IMS) into the cytosol.35 In the cytosol, cytochrome c binds to and activates apoptotic protease-activating factor 1 (Apaf-1) and proteasome 9, forming a complex known as the “apoptosome”. The apoptosome then cleaves and activates caspase-3, leading to DNA fragmentation, degradation of cytoskeletal and nuclear proteins, protein cross-linking, and the formation of apoptotic vesicles. This mechanism is essential for maintaining internal environmental stability by preventing excessive proliferation of abnormal cells and the development of malignant cells.36 Mitochondria also participate in autophagy, a process that preserves cellular homeostasis by removing damaged or surplus organelles and proteins.37
Regulation of Calcium Ion Homeostasis
Mitochondria have the capacity to rapidly take up calcium ions, which pass through the OMM via VDAC, known for their high conductivity and low anion selectivity.38 In contrast, calcium ion passage through the IMM is more tightly regulated. It involves the mitochondrial calcium uniporter (MCU), which translocates Ca2+ into the matrix, and the Na+/Ca2+ exchanger, predominantly found in excitable cells such as muscle and brain tissue. In other cell types, the H+/Ca2+ exchanger releases calcium ions from the matrix into the IMS.39 This process allows mitochondria to act as a buffer for intracellular calcium ion storage.
ROS Production and Scavenging
Mitochondria are a major source of intracellular ROS and also possess mechanisms for scavenging ROS.40 ROS play a role in cellular signaling and immune responses, but excessive ROS can result in oxidative stress and cellular damage.41
Synthesizing Proteins and Nucleic Acids
Mitochondria are a key site for the synthesis of essential biomolecules, including proteins, nucleic acids, and lipids, which are critical for normal cellular function and metabolism.42
Immune and Inflammatory Regulation
Mitochondria play a crucial role in regulating immune responses and inflammatory processes by activating immune signaling pathways, serving as a platform for innate immune signaling, and participating in both apoptotic and necrotic pathways.43 Mitochondrial dysfunction is closely linked to the development and progression of various immune and inflammation-related diseases.44
Other Functions
Mitochondria also participate in several other physiological processes, such as cholesterol and hormone synthesis in the liver, blood glucose homeostasis, iron-sulfur cluster synthesis, pyrimidine synthesis, regulation of the intracellular environment, and muscle metabolism (Figure 3).45,46
Causes of Mitochondrial Dysfunction in RA
Hypoxia
RA is primarily characterized by chronic synovitis, synovial cell proliferation, and the formation of vascular opacification. Under normal physiological conditions, a layer of endothelial cells lines the interior of synovial blood vessels, where leukocyte migration is minimal, the vasculature remains stable, and blood perfusion and oxygen supply are maintained at normal levels.47 In chronic synovitis associated with RA, unstable vasculature interacts abnormally with endothelial cells and surrounding tissues. As vascular opacification forms, endothelial cells become activated, lose polarity, and detach, protruding into the vessel lumen. This leads to vascular dysfunction, severely impairing the transport of nutrients and oxygen through the synovial membrane, resulting in hypoxia.48 The severity of synovitis correlates with the degree of oxygen deprivation in the synovial membrane.49 Biniecka et al cultured primary rheumatoid arthritis synovial fibroblasts (RASF) under hypoxic conditions and observed mitochondrial function. The study demonstrated that hypoxia induced mitochondrial dysfunction, promoting glycolysis, pannus formation, and neovascularization.50 Furthermore, hypoxia leads to significant alterations in mitochondrial structure, genome, and dynamics, reducing ATP synthesis, increasing ROS production, and accelerating the accumulation of mtDNA mutations.51,52
Mutations in Mitochondrial DNA (mtDNA)
The high mutation rate of mtDNA can be attributed to its lack of protection by histones and chromatin structures, making it more vulnerable to environmental damage. Additionally, the activity of DNA repair enzymes is less robust in mitochondria compared to the nucleus.53 Mutations in genes encoding mitochondrial proteins often result in altered peptide sequences, leading to mitochondrial dysfunction.When the mutation occurs, it may affect the function of the mitochondrial respiratory chain complex, so that electron transport and oxidative phosphorylation processes are impaired, and ATP cannot be synthesized efficiently, thus affecting organismal function.This dysfunction can cause cellular damage, such as increased ROS production, immune cell activation, and autoantibody production, all of which can exacerbate the progression of RA.54
Inflammatory environments have been shown to promote mtDNA mutations. LC Harty et al discovered that higher levels of TNF-α or interferon gamma (IFN-γ) in the synovial membrane were associated with an increased frequency of mtDNA mutations. In in vitro experiments, treating RA FLS with TNF-α also resulted in a higher frequency of mtDNA mutations, indicating that inflammation can induce mtDNA mutations. This suggests a vicious cycle between mitochondrial dysfunction and inflammation.55 Additionally, hypoxia is a key factor in inducing mtDNA mutations. M. Biniecka et al found a greater number of mtDNA mutations in RA fibroblasts cultured under hypoxic conditions in vitro.56
Oxidative Stress
Excessive ROS production in mitochondria increases the risk of mtDNA mutations, impairs ATP synthesis, and contributes to mitochondrial dysfunction.57 In RA patients, ROS levels may significantly rise due to environmental stressors (eg, pathogenic factors). The accumulation of ROS leads to oxidative stress, which causes DNA damage. This damage results in the production of incorrect polypeptides encoded by mtDNA, further damaging the mitochondria and leading to cellular dysfunction and disease.58
Mechanisms Associated with Mitochondrial Dysfunction Triggering Chronic Inflammation and Immune Disorders in RA
In recent years, mitochondria have become a central focus in immunology for two key reasons. First, there is increasing evidence that metabolic reprogramming of immune cells is crucial for the proper regulation of immune activation. Regulating mitochondrial metabolic pathways, such as respiration, β-oxidation, and OXPHOS, can significantly alter immune cell properties and shape immune responses.59 Second, mitochondria have been shown to play a direct role in activating the immune system, as the release of various mitochondrial products can directly trigger immune and inflammatory responses, ultimately contributing to the pathogenesis of RA.60
MtDNA Release
The innate immune system, as the first line of defense, protects against numerous injuries and microbial invasions. Pattern recognition receptors (PRRs) in cells detect specific pathogen-associated molecular patterns (PAMPs) to recognize pathogenic infections.61 PRRs are broadly categorized into four types: NOD-like receptors (NLRs), Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and C-type lectin receptors (CLRs).62
The nucleus of the cell contains all the DNA that makes up the human genome and is called “nuclear DNA”. The mitochondria in a cell are encoded by “mitochondrial DNA”, which makes up only 0.1% of nuclear DNA and is inherited exclusively from the mother’s side of the family. However, in a recent study, researchers compared the mitochondrial and nuclear DNA of thousands of people and found that mitochondrial DNA can be transferred in trace amounts to the nucleus and that changes in mitochondrial DNA are determined by nuclear DNA. This finding tells us that there is a subtle relationship between the mitochondria and the nucleus in our cells that is critical to maintaining cellular health.63
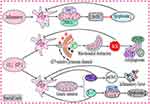
mtDNA is located in the mitochondrial matrix near the electron transport chain, which is the primary source of ROS. Due to this proximity, mtDNA is particularly vulnerable to oxidative stress. When mitochondria experience damage or stress, they release danger-associated molecular patterns (DAMPs) such as mtDNA, succinate, N-formyl peptides, ROS, cardiolipin, and ATP, all of which activate multiple immune response pathways.64 When mtDNA is present in the cytoplasm or extracellular environment, it acts as a potent agonist for PRRs, initiating a range of signaling pathways. These pathways lead to the upregulation of type I interferons, pro-inflammatory chemokines, and cytokines, contributing to chronic inflammation and immune dysregulation in RA (Figure 4).65
The cGAS-STING Pathway
The structure and signaling mechanisms of the cGAS-STING pathway have been extensively studied in recent years. Structurally, cGAS contains three dsDNA-binding sites and can recognize both self and non-self DNA in a sequence-independent manner.66 cGAS monomers interact with mtDNA that is over 45 base pairs in length, forming a stable network of ladder-like dimers that activate cGAS.67 Upon activation, cGAS catalyzes the conversion of GTP and ATP into 2′3′-cyclic GMP-AMP (cGAMP), a second messenger that binds to the endoplasmic reticulum (ER)-resident protein, stimulator of interferon genes (STING), causing a conformational change in its C-terminal tail.68 This conformational shift recruits TANK-binding kina se 1 (TBK1) to STING, leading to the activation of the NF-κB and IRF3 pathways. This, in turn, triggers the transcription of hundreds of interferon-stimulated genes (ISGs), which exert potent antiviral effects.69
TLR9 Pathway
TLR9 is a type I integral membrane protein with an N-terminal ligand-recognizing domain, a single transmembrane helix, and a C-terminal cytoplasmic signaling domain. The N-terminal domain exhibits a characteristic horseshoe shape formed by leucine-rich repeat (LRR) motifs.70 TLR9 is predominantly localized in plasma cell-like dendritic cells, monocytes, B-cells, and macrophages. It is situated in the ER and translocates to intracellular compartments upon stimulation by hypomethylated CpG DNA.71 TLR9 was the first receptor identified to recognize unmethylated CpG DNA, a hallmark of both bacterial DNA and mtDNA, making mtDNA a potent agonist for TLR9.72 When bound to mtDNA, TLR9 recruits myeloid differentiation factor 88 (MyD88), forming a signaling complex with interleukin-1 (IL-1) receptor-associated kinase (IRAK) and tumor necrosis factor receptor-associated factor 6 (TRAF6), as well as IRF7. This complex also includes transforming growth factor-β-activated kinase 1 (TAK1), which interacts with NF-κB inhibitor IκB (IκKBα) or TAK1-mitogen-activated protein kinase 1 (MAPK1). Together, these pathways promote the expression of pro-inflammatory genes by activating transcription factors such as IRF7, NF-κB, and activator protein-1 (AP-1).73
Inflammatory
Vesicles mtDNA plays a key role in activating inflammatory vesicles, such as NLRP3 and AIM2, both of which promote the secretion of pro-inflammatory cytokines.74 NLRP3 is a PRR located on the cell membrane that responds to a variety of pathogen-derived and endogenous substances. Upon encountering DAMPs, NLRP3 undergoes a conformational change, exposing its central nucleotide-binding and oligomerization domains, which promotes its oligomerization and activation.75 AIM2 is another cellular receptor that recognizes dsDNA and triggers an inflammatory vesicle cascade. Extracellular dsDNA released by necrotic cells is phagocytosed and recognized intracellularly by AIM2, which activates inflammatory vesicles and drives the secretion of cytokines such as IL-18 and IL-1β.76 As a result, cytoplasmic mtDNA serves as an ideal endogenous agonist, triggering the activation of NLRP3 and AIM2 vesicles. This activation leads to caspase-1 cleavage, and the resulting active caspases promote the maturation and secretion of pro-inflammatory cytokines, including IL-1, IL-6, IL-18, and TNF-α.77
ROS Generation
ROS are by-products of cellular aerobic respiration, generated when electrons are transferred from highly reducing compounds to molecular oxygen. ROS encompass several types, including superoxide anion (O2-), hydrogen peroxide (H2O2), hydroxyl radical (HO-), and nitric oxide (NO), which also falls under the category of ROS.78 Mitochondria are the primary site of ROS generation. Upon exposure to various stimuli such as pathogens, radiation, or photoactivation, electrons leak from the electron transport chain (ETC) complexes in the cytoplasm, cell membrane, or other cellular compartments like peroxisomes and the endoplasmic reticulum (ER). These leaked electrons interact with molecular oxygen to form superoxide anions, which are subsequently converted to H2O2 through the action of mitochondria-specific superoxide dismutase (SOD).79,80
Mitochondrial ROS (mt-ROS) play a critical role in the formation and activation of the NLRP3 inflammasome,81 a cytoplasmic multiprotein complex composed of NLRP3, ASC, and caspase-1, serving as a key mediator of innate immunity.82
mt-ROS promote NLRP3 activation through several mechanisms (Figure 5): (1) promoting NF-κB transcription, which increases the naturally low levels of NLRP3 sensors, (2) oxidizing mtDNA synthesized via the TLR pathway, which directly binds to and activates NLRP3, and (3) enhancing mt-ROS production due to impaired autophagy proteins (eg, beclin-1), further driving NLRP3 activation. NLRP3 is an intracellular sensor that, upon stimulation by mt-ROS, forms an inflammasome through interactions between a single NLRP3 pyrin domain unit and the ASC pyrin domain. ASC then interacts with pro-caspase-1 through its CARD domain, forming an activated inflammasome. This activation triggers caspase-1 and leads to the release of pro-inflammatory factors such as IL-1β and IL-18, thereby contributing to the inflammatory response.40,83
Mitochondrial Dynamics
Mitochondrial dynamics refer to the equilibrium state in which mitochondria continuously divide and fuse within the cell, maintaining their morphology, number, size, and structure.84 Fusion allows for the mixing of mitochondrial contents, helping to preserve the functional integrity of mitochondria, while division enables mitochondria to adapt to changes in intracellular energy demands and to remove damaged mitochondria through autophagy in response to cellular injury.85 This dynamic balance is crucial for the energy supply, metabolic regulation, and overall health of the cell. Changes in mitochondrial dynamics can manifest as varying morphological forms within the cytoplasm, such as dots, fragments, or elongated structures.86
Mitochondrial dynamics are closely tied to essential cellular functions, including cell proliferation, metabolism, and migration, and are regulated by various enzymes and proteins.87 Mitochondrial fission is primarily controlled by dynamin-related protein 1 (Drp1), which is recruited to the outer mitochondrial membrane upon stimulation. Drp1 interacts with other proteins and hydrolyzes GTP, leading to the contraction of both the inner and outer mitochondrial membranes, ultimately triggering fission. Conversely, mitochondrial fusion is mediated by mitofusin 1 and 2 (Mfn1/2) for the outer membrane and optic atrophy 1 (Opa1) for the inner membrane, which are responsible for promoting membrane fusion.88 These regulatory mechanisms ensure that mitochondria can quickly respond and maintain cellular homeostasis under various physiological and pathological conditions (Figure 6).
 |
Figure 6 Mitochondrial dynamics. Abbreviations: Opa1, outer membrane and optic atrophy; Mfn1/2, mitofusin 1 and 2; Drp, dynamin-related protein; FIS1, Fission 1 (mitochondrial outer membrane) Homolog. |
Normal mitochondrial dynamics are essential for maintaining proper cellular function, while dysregulation of these dynamics is closely linked to the development of various diseases, including cancer, autoimmune disorders, and neurodegenerative conditions.89 In RA, dysregulated mitochondrial dynamics may contribute to disease progression by influencing processes such as mitochondrial ROS production, mtDNA release, and apoptosis, all of which play roles in regulating immune and inflammatory responses. For instance, total glucosides of Paeonia lactiflora (TGP), a commonly used drug for RA treatment, have been shown to reduce oxidative stress and inflammation caused by mt-ROS by correcting mitochondrial dynamics and improving mitochondrial function. This effect may be mediated by the AMPK pathway.90
Mitochondrial fission and fusion are critical for mitochondrial function, with dynamin-related protein 1 (DNM1L) serving as a key regulator of mitochondrial fission. Upregulation of DNM1L expression has been associated with shorter mitochondrial length and increased RA severity in the ST of RA patients. This is primarily because upregulated DNM1L expression and mitochondrial fission promote cell survival, LC3B-associated autophagy in FLS, and ROS production, all of which contribute to inflammation by regulating the AKT/IKK/NFKBIA/NF-κB signaling pathway. Inhibition of DNM1L in FLS leads to mitochondrial depolarization, mitochondrial elongation, decreased cell viability, reduced production of ROS, IL-8, and COX-2, and increased apoptosis.91
Mitochondrial Autophagy
Mitochondrial autophagy, or mitophagy, is a crucial quality control process that allows cells to selectively encapsulate and degrade damaged or excess mitochondria, helping to regulate mitochondrial numbers and maintain energy metabolism.92 Mitophagy involves four main steps (Figure 7): (1) Mitochondrial damage, often indicated by membrane potential depolarization due to reactive oxygen species (ROS), nutrient deprivation, cellular senescence, or external stressors, which is a prerequisite for autophagy; (2) Encapsulation of mitochondria by autophagosomes: a double-membrane structure forms around the damaged mitochondrion and elongates, eventually enclosing it in an autophagosome; (3) Fusion of the mitochondrial autophagosome with a lysosome; and (4) Degradation of mitochondria by lysosomal enzymes, where lysosomal hydrolases break down the encapsulated mitochondrion.93
 |
Figure 7 Mitochondrial autophagy process. Abbreviations: MMP, mitochondrial membrane potential; LC3, Microtubule-associated protein 1 light chain 3; PINK1, PTEN induced putative kinase 1. |
The mechanisms underlying mitochondrial autophagy can be broadly categorized into two pathways: ubiquitin-dependent and non-ubiquitin-dependent pathways (Figure 8).94 In the ubiquitin-dependent pathway, mitochondrial surface proteins undergo ubiquitination to induce autophagy. The most extensively studied pathway in this category is the PINK1/Parkin pathway, which plays a key role in clearing damaged mitochondria in mammals. In healthy mitochondria, PINK1 levels are kept low because PINK1 is continuously translocated to the inner mitochondrial membrane and degraded by proteases. However, when mitochondrial membrane potential (MMP, ΔΨm) is compromised, the translocation of PINK1 is disrupted, leading to its accumulation on the outer mitochondrial membrane. This accumulation of PINK1 recruits Parkin, an E3 ubiquitin ligase, to the mitochondria. Upon mitochondrial damage, PINK1 phosphorylates Parkin, activating it to ubiquitinate outer mitochondrial membrane proteins, thereby recruiting downstream receptor proteins and initiating ubiquitin-dependent mitophagy.95
In addition to the PINK1/Parkin pathway, other ubiquitin-dependent pathways also promote mitochondrial autophagy without relying on Parkin. For instance, PINK1 can directly recruit autophagy receptors such as OPTN and NDP52 to promote autophagy through ubiquitin phosphorylation. Furthermore, several other E3 ubiquitin ligases, including SMURF1, MUL1, and Gp78, can also participate in the ubiquitination of mitochondrial proteins, thereby inducing mitochondrial autophagy.96
Unlike PINK1/Parkin-mediated pathways, certain proteins located on the outer mitochondrial membrane contain LIR (LC3-interacting region) motifs and serve as autophagy receptors. These proteins can directly bind to LC3 without the need for ubiquitination, initiating mitochondrial autophagy. In mammals, key receptors involved in this non-ubiquitin-dependent pathway include Nip3-like protein X (NIX)/BCL2-interacting protein 3-like (BNIP3L), BCL2-interacting protein 3 (BNIP3), and FUN14 domain-containing protein 1 (FUNDC1).97,98
Mitochondrial autophagy selectively removes damaged or dysfunctional mitochondria. By clearing these damaged mitochondria, the process helps maintain mitochondrial quality and function, reduces ROS production, and limits the release of mtDNA, thereby inhibiting inflammatory responses. When mitochondrial autophagy is impaired, mitochondrial dysfunction can occur, leading to excess ROS production and accumulation, which triggers immune responses and oxidative cellular damage, contributing to the onset of RA. For example, the traditional Chinese herbal formula Gui Zhi Shao Yao Zhi Mu Tang (GSZG), used to treat RA-associated bone destruction, has been shown to inhibit bone erosion by regulating the ROS/NF-κB axis through the PINK1/Parkin-mediated autophagy pathway, thus reducing osteoclast precursor production.99 Additionally, various bioinformatics approaches have identified mitochondrial autophagy-related genes, such as ATG5, GSK3A, MFN1, NRAS, RELA, RRAS2, SLC25A46, and TSCA, as potential biomarkers for RA diagnosis and progression.100
Mitochondrial Biogenesis
Mitochondrial biogenesis is the process by which new mitochondria are generated through the growth and division of pre-existing mitochondria, producing large amounts of ATP. This process is coordinated by both mitochondrial DNA (mtDNA) and the nuclear genome.101 The primary regulator of mitochondrial biogenesis is peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α).102 PGC-1α activates nuclear respiratory factor 1 and 2 (Nrf1/2) and estrogen-related receptor-α, which, in turn, stimulate mitochondrial transcription factor A (TFAM), promoting mtDNA replication and transcription, thereby driving mitochondrial biogenesis.103 PGC-1α itself is regulated by various pathways, including the AMP-dependent protein kinase (AMPK) and cAMP-response element-binding protein (CREB) pathways. The sirtuin 1 (SIRT1)/PGC-1α signaling pathway also plays a significant role in regulating mitochondrial biogenesis by acting on PGC-1α to promote mtDNA replication and transcription.104
Proper mitochondrial biogenesis can increase both the number and functionality of mitochondria, boosting cellular energy metabolism and enhancing immune and anti-inflammatory responses.105 However, when mitochondrial biogenesis is abnormally activated, it can lead to a substantial increase in mitochondrial numbers and excessive mtDNA release, which may result in the overproduction and accumulation of ROS. This, in turn, can trigger immune responses and cause inflammatory damage.
Mitochondrial Antiviral Signaling Proteins
Mitochondrial antiviral signaling protein (MAVS) is a junction protein located on the outer mitochondrial membrane, playing a critical role in the innate immune system. MAVS is also known by other names, such as IFN-β promoter stimulator (IPS-1), caspase activation and recruitment domain (CARD)-containing adaptor inducing IFN-β (Cardif), and virus-induced signaling adapter (VISA).106 In the resting state, MAVS remains inactive. However, during viral infections, the interferon gene stimulator protein (STING), also known as MITA, MPYS, or ERIS, acts as a crucial intermediary in the recognition of DNA viruses, with the MAVS-STING interaction being key to mitochondria-mediated antiviral innate immune responses.107 When RNA viruses are detected, RIG-I recognizes viral double-stranded RNA, activating the RIG-I/MAVS antiviral pathway.108
Recent research on MAVS’ downstream signaling pathways has shown that it mediates viral activation of NF-κB and IRF3(Figure 9):109 (1) MAVS strongly activates IRF3 through the Toll-like receptor (TLR) and RIG-I-like receptor (RLR) families. During interferon (IFN) induction, MAVS recruits TANK-binding kinase 1 (TBK1) and IKKϵ, which phosphorylate interferon regulatory factors 3 and 7 (IRF3/IRF7). IRF3/IRF7 are essential for the production of IFN. (2) In the resting state, NF-κB is sequestered in the cytoplasm by members of the inhibitory κB (IκB) family. Upon MAVS activation, IκB kinases (IKKα, IKKβ, and IKKγ) are recruited. IKKγ, also known as the NF-κB essential modulator (NEMO), induces the phosphorylation of IκB. This phosphorylation causes IκB degradation, allowing NF-κB to translocate to the nucleus and bind to enhancers/promoters of target genes, thereby triggering a pro-inflammatory response.
NLRP3 Inflammasome
The NLRP3 inflammasome is a multiprotein complex composed of NLRP3, ASC, and caspase-1. Mitochondrial dysfunction can activate the NLRP3 inflammasome, leading to the activation of caspase-1 and the subsequent release of inflammatory cytokines, such as IL-1β and IL-18, which are central to the inflammatory response (Figure 4).110
Ferulic Acid
Ferulic acid, also known as fumaric acid, is a significant modulator of innate immunity. Fumaric acid hydratase (FH) is a metabolic enzyme in the tricarboxylic acid (TCA) cycle that catalyzes the reversible conversion of fumarate to malate. When mitochondrial dysfunction occurs, FH activity decreases, leading to the abnormal accumulation of fumaric acid. Studies have shown that increased intracellular levels of fumaric acid induce remodeling of the mitochondrial network and the formation of mitochondria-derived vesicles, which release mtDNA into the cytoplasm. This triggers the activation of the cGAS-STING-TBK1 pathway, initiating immune responses and inflammatory reactions.111
LKB1-AMPK Signaling Pathway
The LKB1-AMPK signaling pathway plays a crucial role in regulating cellular metabolism, proliferation, and survival in response to changing nutrient and energy demands. LKB1-AMPK signaling promotes ATP-generating catabolic pathways and allows for metabolic flexibility in T cells under energetic stress. Through metabolic reprogramming, LKB1 and AMPK influence T cell differentiation and function.112
Activation of LKB1 is linked to changes in mitochondrial metabolism and fitness, as well as increased mevalonate metabolism under specific conditions. By regulating various downstream targets, AMPK signaling inhibits glycolysis, glutaminolysis, and fatty acid synthesis while promoting catabolic processes like mitophagy and autophagy. AMPK also drives mitochondrial biogenesis by facilitating mitochondrial apoptosis and regulating mitochondrial dynamics, thereby impacting immune and inflammatory responses.113
Role of the Mitochondrial Bilayer Membrane Structure in the Regulation of Inflammation
The mitochondrial bilayer membrane structure provides dual-layered control over cellular processes. The OMM serves as a platform for signaling molecules, such as MAVS in the RIG-I pathway, and NLRP3 inflammasomes.114 Mitochondrial stress and injury result in the release of mitochondrial damage-associated molecular patterns (mtDAMPs) from their respective PRRs into the cytoplasm or extracellular space. These mtDAMPs include mtDNA, ATP, formyl peptides, cardiolipin, cytochrome c, succinate, and mitochondrial transcription factor A (TFAM). Once released, mtDAMPs activate NLRP3 inflammasomes, upregulate MyD88 and NF-κB, and increase the production of cytokines like TNFα, IL-1, IL-6, and IL-10, playing a significant role in regulating cellular stress and inflammation.115
Other Mechanisms
Mitochondria also influence immune and inflammatory responses by regulating cellular metabolism, calcium homeostasis, endoplasmic reticulum stress, and other signaling pathways.116
Impact of Mitochondrial Dysfunction on RA Immune Cells
Innate immune cells, including T cells, macrophages, and dendritic cells (DCs), play a crucial role in regulating the immune-inflammatory response in RA. Abnormal activation of these cells can lead to the overproduction of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-17, exacerbating the disease.117 The energy required for the survival and function of immune cells is primarily derived from glucose, fatty acids, and amino acids. These molecules fuel the tricarboxylic acid (TCA) cycle via glucose metabolism, gluconeogenesis, and glutaminolysis, respectively. Mitochondria are central to energy production in immune cells, with oxidative phosphorylation providing a stable ATP supply under normal conditions. Research has shown that disruptions in immune cell metabolism, particularly mitochondrial dysfunction, are key contributors to the pathogenesis of RA.
T Cells
Under normal physiological conditions, primary T cells transform into effector T cells after recognizing antigens presented by antigen-presenting cells (APCs), initiating proliferation. Different activation states of T cells require distinct metabolic patterns to meet their functional needs. Effector T cells, including pro-inflammatory T cells found in inflammatory tissue lesions, rely heavily on rapid glucose uptake and glycolysis to meet their energy demands.118 In short, T cells require substantial energy to exert their effects (Figure 10).
In RA patients, the synovial cavity presents a relatively hypoxic microenvironment, with significantly reduced glucose concentrations. This forces tissue-resident cells to seek alternative energy sources, such as lactate.119 Research has shown that under hypoxic and low-glucose conditions, CD8+ T cells in RA convert glutamine to lactate to meet their energy needs.120 Thus, both CD4+ and CD8+ T cells in RA patients may exhibit mitochondrial defects that compel them to rely on lactate for energy. Persistent hypoxia and low glucose levels can lead to lactate accumulation. Studies suggest that in RA synovitis, this accumulation upregulates lactate transporter protein(LTP) expression in CD4+ and CD8+ T cells, supporting the hypothesis that synovial T cells consume lactate. Lactate consumption has been linked to the persistence of T cells in inflamed tissues, with lactate-consuming T cells producing higher levels of IL-17, further exacerbating the inflammatory response.121
As RA progresses, T cells shift their energy metabolism beyond mitochondria and lactate utilization, diverting carbon into the pentose phosphate pathway (PPP) to produce NADPH and nucleotides. This process is regulated by two key rate-limiting enzymes: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) and glucose-6-phosphate dehydrogenase (G6PD).122 PFKFB3, a crucial rate-limiting enzyme in glycolysis, is suppressed in RA T cells, leading to decreased glycolysis and reduced T cell activation.123 In contrast, G6PD is overexpressed in RA T cells, resulting in elevated NADPH levels and increased reduced glutathione (GSH). Glutathione’s antioxidant properties impair the signaling functions of intracellular ROS, which, in turn, prevents activation of the ATM kinase, a cell cycle checkpoint regulator. This leads to the overproliferation of T cells, favoring inflammatory Th1 and Th17 differentiation, further worsening the inflammatory response.124
B Cells
A key feature of RA is the production of autoantibodies, including RF and ACPA, which are primarily produced by B cells. Studies on B cell biology show that naïve B cells are metabolically quiescent, primarily storing energy and synthesizing precursors. This relatively inactive state is lifted when B cells are activated, allowing them to use stored energy for growth and proliferation.125 While the role of mitochondria in B cell development and differentiation is well-established, there is limited data on how mitochondrial regulation of B cell function contributes to RA pathogenesis.
Experimental evidence provides indirect insights into this relationship. First, the effects of mitochondria on B cells may involve the peroxisome proliferator-activated receptor γ (PPARγ), a ligand-activated transcription factor involved in adipogenesis and lipid metabolism, which regulates B cell function and plays a critical role in antibody responses.126 Second, B-cell activation factor (BAFF), also known as B lymphocyte stimulator (BLyS), plays a role in B cell activation, proliferation, and differentiation.127 BAFF-induced signaling regulates B cell metabolism by regulating glucose metabolism and mitochondrial activity. Treatment with BAFF increases B cell size, cellular protein content, and mitochondrial membrane potential, making BAFF-treated B cells more prone to proliferation.128
Macrophages
Macrophages play a crucial role in the pathogenesis of RA and are key cells in bone immunity. These cells can be polarized into two types: pro-inflammatory M1 macrophages and anti-inflammatory M2 macrophages, depending on environmental signals. Macrophages interact with various cell types, including T cells, B cells, and fibroblast-like synoviocytes, leading to the production of numerous inflammatory cytokines, which contribute to bone and cartilage destruction.129 In RA, M1 macrophages are characterized by increased mitochondrial activity, which increases their energy demands. To meet these needs, macrophages shift their glucose metabolism from oxidative phosphorylation to glycolysis, driving the production of large amounts of ROS and ATP.130 This increased mitochondrial activity is associated with the formation of mitochondria-associated membranes (MAMs), structures that connect mitochondria with other organelles, such as the endoplasmic reticulum (ER).131 The formation of MAMs allows mitochondria to exchange calcium with the ER, regulating various cellular signaling pathways. This interaction promotes the production of pro-inflammatory cytokines, including CCL18, TNFα, IL-6, and IL-1β, and histone modifications, which are closely linked to the progression of RA.132
Effects of Mitochondrial Dysfunction on RA Osteoblasts
Fibroblast-Like Synoviocytes (Figure 11)
Fibroblast-like synoviocytes (FLS) are the primary cell type in the synovial membrane, comprising a lining layer and a sublining layer. Their main function is to produce lubricin and hyaluronic acid, essential for joint lubrication.133 During the progression of RA, mitochondrial dysfunction, primarily involving abnormalities in mitochondrial fission and autophagy, plays a critical role in FLS metabolism and apoptosis. FLS not only secrete inflammatory cytokines and chemokines that drive inflammation but also promote the production of matrix metalloproteinases, which degrade cartilage and stimulate osteoclast differentiation, leading to bone erosion.134
In RA, mitochondria in FLS exhibit stress-related phenotypes, such as fragmentation and elevated expression of the mitochondrial fission GTPase dynamin-related protein 1 (Drp1).91 DNM1L, a key regulator of mitochondrial fission, is highly expressed in FLS. Silencing DNM1L expression increases mitochondrial length and induces mitochondrial depolarization. Inhibiting mitochondrial fission also decreases the ratio of LC3B-II to LC3B-I, promotes FLS apoptosis, and reduces inflammation in RA synoviocytes.135
The synovial environment in RA patients is characterized by hypoxia, which increases mitochondrial membrane potential by activating mitochondrial ATP-sensitive potassium channels. This leads to mitochondrial dysfunction in FLS and is associated with elevated ROS production from a dysfunctional electron transport chain. The increased ROS levels further upregulate key proteins in autophagy, such as ATG5 and LC3, promoting the formation of autophagic vesicles.136 Moreover, hypoxia-induced mitochondrial dysfunction in RA FLS can cause mutations in the mitochondrial genome, resulting in increased secretion of pro-inflammatory cytokines, including IL-17 and TNF-α, and disruption of mitochondrial homeostasis.137 TNF-α, in particular, acts as a mitochondrial stressor, reducing mitochondrial autophagy and leading to the release of mtDNA, which further drives the production of additional pro-inflammatory factors.138 Mitochondria are key regulators of programmed cell death, with the balance between pro-apoptotic Bax and anti-apoptotic Bcl-2 proteins playing a key role in FLS survival. IL-17 has been shown to increase the expression of the anti-apoptotic protein Bcl-2 in RA FLS, promoting their survival. Consequently, IL-17 is considered a significant factor in rescuing RA FLS from apoptosis, contributing to the persistence of inflammation.139,140
Osteoclasts
Osteoclasts are derived from osteoclast precursor cells, which originate from the hematopoietic lineage of monocytes and macrophages. Their primary role in bone homeostasis is to secrete substances that dissolve inorganic bone components and degrade bone matrix proteins, promoting bone resorption. In RA, osteoclasts are the key effector cells responsible for bone destruction.141 The activity and function of osteoclasts are closely linked to mitochondrial health, as alterations in mitochondrial function can either inhibit or overactivate osteoclasts, disrupting bone tissue homeostasis. The influence of mitochondria on osteoclast activity can be understood through the following mechanisms (Table 1):
 |
Table 1 Effects of Mitochondrial Dysfunction on RA Osteoblasts |
(1) Oxidative phosphorylation energy supply mode: Oxidative phosphorylation is the primary energy source for osteoclast proliferation and differentiation. Studies by Lemma et al142 revealed that cellular oxygen consumption and the expression of respiratory chain-related proteins significantly increased during osteoclast proliferation and differentiation. Further, research by Kim et al143 demonstrated that inhibiting the mitochondrial oxidative phosphorylation pathway suppressed osteoclast differentiation, highlighting the critical role of mitochondrial oxidative phosphorylation in supporting osteoclast development and function.
(2) ROS production-clearance: Mitochondria are the primary site for both the production and clearance of ROS. During oxidative phosphorylation, a large amount of ROS is generated as a byproduct of energy production. Accumulation of ROS can lead to oxidative stress, causing severe damage to cell structures and functions. To counter this, mitochondria possess their own antioxidant system, which includes enzymatic components such as peroxidase, Mn-SOD, and non-enzymatic systems like cytochrome and reduced coenzyme Q10.148 These systems maintain a dynamic balance between ROS production and scavenging, which plays a crucial role in regulating osteogenesis and osteoclast differentiation. Arai et al153 found that elevated intracellular ROS levels inhibited osteoblast mineralization, while ROS also acted as signaling molecules promoting osteoclast differentiation. Studies by Fraser et al and Baek et al144,145 showed that H2O2 promoted the proliferation and differentiation of osteoclasts. Evidence of H2O2 activity was found in the cranial bones of mice, in bone marrow cells, and in human bone marrow cells. Additionally, a study by Srinivasan et al154 demonstrated that adding antioxidants to mouse cells significantly reduced the number of differentiated and mature osteoclasts. This suggests that intracellular ROS influences osteoclast differentiation through pathways such as RANK/RANKL signaling, as NF-κB activity was found to be elevated under hypoxic conditions. Ishii et al155 discovered that ROS were involved in the transcription of PGC-1β, which positively regulates ROS production, thereby promoting osteoclastogenesis. Furthermore, Kim et al156 found that knocking down the endoplasmic reticulum transmembrane protein 64 in mice reduced intracellular calcium oscillations, decreased ROS levels, and inhibited bone resorption, suggesting that calcium signaling may also be a pathway through which ROS regulate osteoclast differentiation.
(3) Mitochondrial dynamics pathway: Mitochondrial dynamics involve several key processes, including mitochondrial biogenesis, fusion, and autophagy. Mitochondrial biogenesis replenishes new mitochondria, while autophagy removes senescent and damaged mitochondria. Mitochondrial fusion and fission provide energy and support signaling pathways essential for cellular functions. Lemma et al142 observed that osteoblast differentiation is associated with changes such as increased mitochondrial size, number, and elevated expression of electron transport chain complexes. This suggests a potential link between mitochondrial dynamics and osteoblast differentiation, possibly mediated by PGC-1β. PGC-1β, a member of the peroxisome proliferator-activated receptor gamma coactivator 1 family, plays a key role in regulating transcription during mitochondrial biogenesis.
(4) Mitochondrial calcium unidirectional transporter (MCU): The MCU is the primary calcium transporter within mitochondria and is crucial for maintaining mitochondrial structure and function.157 Research by Yinbo Wang146 demonstrated that MCU promotes osteoclast differentiation, playing distinct roles at different stages. During the early stage of osteoclast differentiation, MCU increases mitochondrial calcium levels, supporting ATP production needed for differentiation. In the later stages, MCU induces ROS production, which further promotes osteoclast differentiation.
(5) Mitochondrial Ferroptosis: Ferroptosis, or iron-dependent cell death, is a non-apoptotic form of necrotic cell death driven by the accumulation of intracellular iron and toxic lipid peroxides.158 Iron overload is a critical factor in ferroptosis. Osteoclast differentiation is closely tied to the OPG/RANK/RANKL signaling pathway, with factors such as RANK, RANKL, TNF, and IL-6 driving macrophage-to-osteoclast differentiation.159 Li et al147 found that iron overload induces the expression of RANKL and IL-6, which, in turn, promotes osteoclast differentiation. This suggests that mitochondrial ferroptosis may be mediated by the OPG/RANK/RANKL pathway during the osteoclast differentiation process.
Osteoblasts
Osteoblasts are specialized cells responsible for bone formation during bone repair and reconstruction. They primarily differentiate from bone mesenchymal stem cells (BMSCs) in the bone marrow. Abnormalities in osteoblast differentiation and function play a key role in the development of bone metabolic diseases,160 which manifest as decreased bone mass and sparse trabeculae, leading to increased bone fragility and a higher risk of fractures. Mitochondria serve as the energy source for cellular functions, producing large amounts of ATP through oxidative phosphorylation to support osteoblast metabolism. Additionally, mitochondria influence osteoclastogenesis by participating in osteogenesis-related signaling pathways. The regulation of osteoblast function by mitochondria is primarily mediated through the following mechanisms (Table 1):
(1) Regulation of oxidative phosphorylation: Osteoblast differentiation and mineralization require substantial energy to synthesize high levels of collagen and bone matrix proteins. Mitochondrial oxidative phosphorylation is crucial for maintaining osteoblast function and is also involved in regulating osteogenic signaling pathways that affect osteoblast proliferation and differentiation. β-catenin, a key component in this process, enters the nucleus and binds to transcription factors such as LEF/TCF, regulating the expression of osteogenic genes like Runx2 and osteocalcin.161 These genes are essential for osteogenesis. β-catenin maintains its activity through acetylation, a process dependent on the mitochondria-produced acetyl donor, acetyl-CoA. Acetyl-CoA is generated in the tricarboxylic acid cycle and exported from the mitochondria as citrate. Shares et al148 found that stimulating mitochondrial oxidative phosphorylation with galactose increased acetyl-CoA levels, upregulating β-catenin acetylation and promoting osteogenic differentiation. Conversely, inhibiting ATP-citrate lyase (ACLY) with SB204990 reduced β-catenin acetylation levels, reversing the effects of galactose-stimulated mitochondrial oxidative phosphorylation on osteoblast differentiation. These findings suggest that mitochondrial oxidative phosphorylation supports osteoblast differentiation by promoting β-catenin acetylation.
(2) Mitochondrial biogenesis: Mitochondrial biogenesis is the process by which mature mitochondria generate new mitochondria, co-regulated by mtDNA and nuclear DNA (nDNA). This process is essential for repairing mitochondrial structures and maintaining their function. The key regulator of mitochondrial biogenesis is peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), which acts as an intermediary regulator. PGC-1α is activated by upstream signaling molecules such as AMPK, SIRT1, and SIRT3. Once activated, PGC-1α influences downstream signaling molecules, including NRF-1 and NRF-2, which upregulate mitochondrial biogenesis, reduce ROS production, maintain mitochondrial quality, and promote the proliferation and differentiation of osteoclasts.149,150
(3) Mitochondrial dynamics: Mitochondrial dynamics, encompassing processes like fission and fusion, play a vital role in osteoblast activity by influencing energy production, ROS accumulation, and signal transduction. Forni et al151 found that changes in mitochondrial dynamics are critical for the differentiation of MSCs into osteoblasts, adipocytes, and chondrocytes. Increased levels of Mnf2 and increased mitochondrial fusion were shown to promote osteogenic differentiation. Similarly, Wan et al162 demonstrated that regulating mitochondrial dynamics, including accelerated mitochondrial production, helps maintain the energy balance required during osteoblast proliferation and differentiation.
(4) Mitochondrial autophagy: Dysregulated mitochondrial autophagy disrupts the functional integrity of mitochondria, leading to reduced ATP production and excessive oxidative stress due to ROS accumulation. This impairs the survival and bone-forming abilities of BMSCs, directly affecting osteoblast proliferation and differentiation.163
Conclusion
In recent years, significant progress has been made in understanding the role of mitochondria in immune cell function and the pathogenesis of autoimmune diseases like RA. Various factors, including hypoxia, oxidative stress, and mtDNA mutations, can lead to mitochondrial dysfunction, which in turn affects RA through multiple mechanisms: (1) Direct Effects: Mitochondrial dysfunction directly influences the differentiation and function of synoviocytes, osteoclasts, and osteoblasts, disrupting bone metabolic homeostasis and contributing to RA pathogenesis. (2) Indirect Effects: Mitochondrial components, such as mtDNA, ROS, and the processes of mitochondrial dynamics, autophagy, and biogenesis, play key roles in immune regulation. These dysfunctions can trigger the release of mitochondrial antiviral signaling proteins (MAVS), NLRP3 inflammasomes, and signaling pathways such as LKB1-AMPK, leading to immune dysregulation, which promotes the development of RA and amplifies inflammatory responses.
However, the effects of mitochondrial dysfunction on other immune cells, such as neutrophils and B cells, require further exploration. Neutrophils, a source of self-antigens that drive inflammation and tissue damage.It has been found that mitochondrial stress induces the rapid release of mitochondrial protein-derived molecules (eg, N-formyl peptide (mtNFP) and N-formylmethionine (fMet)), both of which act through the Formyl Peptide Receptor-1 (FPR1), which is abundantly expressed on neutrophils, and which, in combination with homologous ligands, can trigger the release of antigens and activation processes in neutrophils and play a role in inducing inflammation in RA.164,165 Consequently,Mitochondrial dysregulation in neutrophils significantly impacts the progression of RA.166
Currently, the regulatory mechanism of mitochondria on neutrophils and B cells is mainly related to metabolic reorganization; when mitochondria are dysfunctional, the metabolic mode of neutrophils and B cells tends to favor glycolysis rather than oxidative phosphorylation, leading to abnormal activation of neutrophils and B cells, and inducing inflammatory responses in RA.152 However, other data on mitochondrial regulation of neutrophils and B cells to induce RA are limited and deserve further study.
In clinical settings, antirheumatic drugs that regulate mitochondrial homeostasis by regulating mitochondrial biosynthesis, dynamics, and autophagy have shown promising results in treating RA. For example, methotrexate (MTX) inhibits the mitochondrial folate pathway through competitive inhibition of dihydrofolate reductase (DHFR), affecting mitochondrial biogenesis and exerting antiproliferative effects.167 Additionally, leflunomide inhibits mitochondrial dihydroorotate dehydrogenase (DHODH), reducing pyrimidine synthesis and increasing mitochondrial fusion by promoting Mfn1/2 expression.168 This review examines mitochondrial structure and function in the context of RA, analyzes the potential causes and effects of mitochondrial dysfunction, and highlights how this dysfunction influences immune cells and osteoblasts, contributing to RA pathogenesis. Thus, identifying targeted biological processes (eg, mitochondrial biogenesis, dynamics and mitochondrial autophagy) to regulate mitochondrial homeostasis, holds great promise for improving RA prevention and treatment strategies.
Data Sharing Statement
The data are included in the article as table.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Supported by the National Natural Science Foundation of China Regional projects (No. 81860858, 81960832),the Gansu Provincial Natural Science Foundation Youth Fund Program (No.22JR11RA133,21JR11RA158,23JRRA1584,24JRRA548),The Second Affiliated Hospital of Gansu University of Traditional Chinese Medicine Youth Program (No. ZX-62000004-2022-092),Gansu Province Famous Traditional Chinese Medicine Practitioners Inheritance Studio Construction Project (National TCM Regulation and Caixin Letter [2021] No. 50) and 2023 Central Finance Transfer Payment for Local Projects for Capacity Enhancement of Chinese Medicine Counseling and Evidence Construction(No. 2101704).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Monzel AS, Enríquez JA, Picard M. Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nat Metab. 2023;5(4):546–562. doi:10.1038/s42255-023-00783-1
2. Akbari M, Kirkwood TBL, Bohr VA. Mitochondria in the signaling pathways that control longevity and health span. Ageing Res Rev. 2019;54:100940. doi:10.1016/j.arr.2019.100940
3. He L, Maheshwari A. Mitochondria in Early Life. Curr Pediatr Rev. 2023;19(4):395–416. doi:10.2174/1573396319666221221110728
4. Tiwari-Heckler S, Robson SC, Longhi MS. Mitochondria Drive Immune Responses in Critical Disease. Cells. 2022;11(24):4113. doi:10.3390/cells11244113
5. Disha B, Mathew RP, Dalal AB, et al. Mitochondria in biology and medicine-2023. Mitochondrion. 2024;76:101853. doi:10.1016/j.mito.2024.101853
6. Lin YJ, Anzaghe M, Schülke S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells. 2020;9(4):880. doi:10.3390/cells9040880
7. Jang S, Kwon EJ, Lee JJ. Rheumatoid Arthritis: pathogenic Roles of Diverse Immune Cells. Int J Mol Sci. 2022;23(2):905. doi:10.3390/ijms23020905
8. Radu AF, Bungau SG. Management of Rheumatoid Arthritis: an Overview. Cells. 2021;10(11):2857. doi:10.3390/cells10112857
9. Díaz-González F, Hernández-Hernández MV. Rheumatoid arthritis. Med Clin. 2023;161(12):533–542. doi:10.1016/j.medcli.2023.07.014
10. Wang X, Fan D, Cao X, et al. The Role of Reactive Oxygen Species in the Rheumatoid Arthritis-Associated Synovial Microenvironment. Antioxidants. 2022;11(6):1153. doi:10.3390/antiox11061153
11. Jávor P, Mácsai A, Butt E, et al. Mitochondrial Dysfunction Affects the Synovium of Patients with Rheumatoid Arthritis and Osteoarthritis Differently. Int J Mol Sci. 2022;23(14):7553. doi:10.3390/ijms23147553
12. O’Sullivan JDB, Bullen A, Mann ZF. Mitochondrial form and function in hair cells. Hear Res. 2023;428:108660. doi:10.1016/j.heares.2022.108660
13. Zhou QY, Ren C, Li JY, et al. The crosstalk between mitochondrial quality control and metal-dependent cell death. Cell Death Dis. 2024;15(4):299. doi:10.1038/s41419-024-06691-w
14. Kane MS, Benavides GA, Osuma E, et al. The interplay between sex, time of day, fasting status, and their impact on cardiac mitochondrial structure, function, and dynamics. Sci Rep. 2023;13(1): 21638. doi:10.1038/s41598-023-49018-z
15. Hinton A, Claypool SM, Neikirk K, et al. Mitochondrial Structure and Function in Human Heart Failure. Circ Res. 2024;135(2):372–396. doi:10.1161/CIRCRESAHA.124.323800
16. Iovine JC, Claypool SM, Alder NN. Mitochondrial compartmentalization: emerging themes in structure and function. Trends Biochem Sci. 2021;46(11):902–917. doi:10.1016/j.tibs.2021.06.003
17. Huang C, Deng K, Wu M. Mitochondrial cristae in health and disease. Int J Biol Macromol. 2023;235:123755. doi:10.1016/j.ijbiomac.2023.123755
18. Caron C, Bertolin G. Cristae shaping and dynamics in mitochondrial function. J Cell Sci. 2024;137(1):jcs260986. doi:10.1242/jcs.260986
19. Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793(1):5–19. doi:10.1016/j.bbamcr.2008.06.013
20. Busch KB. Inner mitochondrial membrane compartmentalization: dynamics across scales. Int J Biochem Cell Biol. 2020;120:105694. doi:10.1016/j.biocel.2020.105694
21. Kadam A, Jadiya P, Tomar D. Post-translational modifications and protein quality control of mitochondrial channels and transporters. Front Cell Dev Biol. 2023;11:1196466. doi:10.3389/fcell.2023.1196466
22. McStay GP. Complex formation and turnover of mitochondrial transporters and ion channels. J Bioenerg Biomembr. 2017;49(1):101–111. doi:10.1007/s10863-016-9648-x
23. He XD, Zhang F, Huang Y, et al. Potential Indicators of Mitochondrial Structure and Function. Curr Pharm Des. 2022;28(21):1738–1744. doi:10.2174/1381612828666220520161200.
24. He B, Yu H, Liu S, et al. Mitochondrial cristae architecture protects against mtDNA release and inflammation. Cell Rep. 2022;41(10):111774. doi:10.1016/j.celrep.2022.111774
25. Pereira CV, Gitschlag BL, Patel MR. Cellular mechanisms of mtDNA heteroplasmy dynamics. Crit Rev Biochem Mol Biol. 2021;56(5):510–525. doi:10.1080/10409238.2021.1934812
26. Newman LE, Shadel GS. Mitochondrial DNA Release in Innate Immune Signaling. Annu Rev Biochem. 2023;92:299–332. doi:10.1146/annurev-biochem-032620-104401
27. Suh J, Lee YS. Mitochondria as secretory organelles and therapeutic cargos. Exp Mol Med. 2024;56(1):66–85.
28. Guan S, Zhao L, Peng R. Mitochondrial Respiratory Chain Supercomplexes: from Structure to Function. Int J Mol Sci. 2022;23(22):13880. doi:10.3390/ijms232213880
29. Song N, Mei S, Wang X, et al. Focusing on mitochondria in the brain: from biology to therapeutics. Transl Neurodegener. 2024;13(1):23. doi:10.1186/s40035-024-00409-w
30. Jenkins BC, Neikirk K, Katti P, et al. Mitochondria in disease: changes in shapes and dynamics. Trends Biochem Sci. 2024;49(4):346–360. doi:10.1016/j.tibs.2024.01.011
31. Trinchese G, Cimmino F, Catapano A, et al. Mitochondria: the gatekeepers between metabolism and immunity. Front Immunol. 2024;15:1334006. doi:10.3389/fimmu.2024.1334006
32. Kim KH, Lee CB. Socialized mitochondria: mitonuclear crosstalk in stress. Exp Mol Med. 2024;56(5):1033–1042. doi:10.1038/s12276-024-01211-4
33. Luo Y, Ma J, Lu W. The Significance of Mitochondrial Dysfunction in Cancer. Int J Mol Sci. 2020;21(16):5598. doi:10.3390/ijms21165598
34. Zhang S, Rao S, Yang M, et al. Role of Mitochondrial Pathways in Cell Apoptosis during He-Patic Ischemia/Reperfusion Injury. Int J Mol Sci. 2022;23(4):2357. doi:10.3390/ijms23042357
35. Wang H, Zhang C, Li M, et al. Antimicrobial Peptides Mediate Apoptosis by Changing Mitochondrial Membrane Permeability. Int J Mol Sci. 2022;23(21):12732. doi:10.3390/ijms232112732
36. Vringer E, Tait SWG. Mitochondria and cell death-associated inflammation. Cell Death Differ. 2023;30(2):304–312. doi:10.1038/s41418-022-01094-w
37. Li A, Gao M, Liu B, et al. Mitochondrial autophagy: molecular mechanisms and implications for cardiovascular disease. Cell Death Dis. 2022;13(5):444. doi:10.1038/s41419-022-04906-6
38. Garbincius JF, Elrod JW. Mitochondrial calcium exchange in physiology and disease. Physiol Rev. 2022;102(2):893–992. doi:10.1152/physrev.00041.2020
39. Tamarit J, Britti E, Delaspre F, et al. Mitochondrial iron and calcium homeostasis in Friedreich ataxia. Iubmb Life. 2021;73(3):543–553. doi:10.1002/iub.2457
40. Harrington JS, Ryter SW, Plataki M, et al. Mitochondria in health, disease, and aging. Physiol Rev. 2023;103(4):2349–2422. doi:10.1152/physrev.00058.2021
41. Palma FR, Gantner BN, Sakiyama MJ, et al. ROS production by mitochondria: function or dysfunction? Oncogene. 2024;43(5):295–303. doi:10.1038/s41388-023-02907-z
42. Baker ZN, Forny P, Pagliarini DJ. Mitochondrial proteome research: the road ahead. Nat Rev Mol Cell Biol. 2024;25(1):65–82. doi:10.1038/s41580-023-00650-7
43. Cortés M, Brischetto A, Martinez-Campanario MC, et al. Inflammatory macrophages reprogram to immunosuppression by reducing mitochondrial translation. Nat Commun. 2023;14(1):7471.
44. Murphy MP, O’Neill LAJ. A break in mitochondrial endosymbiosis as a basis for inflammatory diseases. Nature. 2024;626(7998):271–279. doi:10.1038/s41586-023-06866-z
45. Zong Y, Li H, Liao P, et al. Mitochondrial dysfunction: mechanisms and advances in therapy. Signal Transduct Target Ther. 2024;9(1):124. doi:10.1038/s41392-024-01839-8
46. Koludarova L, Battersby BJ. Mitochondrial protein synthesis quality control. Hum Mol Genet. 2024;33(R1):R53–R60. doi:10.1093/hmg/ddae012
47. Sies H, Mailloux RJ, Jakob U. Fundamentals of redox regulation in biology. Nat Rev Mol Cell Biol. 2024;25(9):701–719. doi:10.1038/s41580-024-00754-8
48. Hua P, Liang R, Tu Y, et al. Reactive oxygen species and nitric oxide scavenging nanoparticles alleviating rheumatoid arthritis through adjusting the seeds and growing soils. Acta Pharm Sin B. 2023;13(12):5016–5029. doi:10.1016/j.apsb.2023.07.021
49. Alivernini S, Firestein GS, McInnes IB. The pathogenesis of rheumatoid arthritis. Immunity. 2022;55(12):2255–2270. doi:10.1016/j.immuni.2022.11.009
50. Biniecka M, Canavan M, McGarry T, et al. Dysregulated bioenergetics: a key regulator of joint inflammation. Ann Rheum Dis. 2016;75(12):2192–2200. doi:10.1136/annrheumdis-2015-208476
51. Vega RB, Horton JL, Kelly DP. Maintaining ancient organelles: mitochondrial biogenesis and maturation. Circ Res. 2015;116(11):1820–1834. doi:10.1161/CIRCRESAHA.116.305420
52. Nakahira K, Hisata S, Choi AM. The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxid Redox Signal. 2015;23(17):1329–1350. doi:10.1089/ars.2015.6407
53. Kopinski PK, Singh LN, Zhang S, et al. Mitochondrial DNA variation and cancer. Nat Rev Cancer. 2021;21(7):431–445. doi:10.1038/s41568-021-00358-w
54. Du J, Yu S, Wang D, et al. Germline and somatic mtDNA mutation spectrum of rheumatoid arthritis patients in the Taizhou area, China. Rheumatology. 2020;59(10):2982–2991. doi:10.1093/rheumatology/keaa063
55. Harty LC, Biniecka M, O’Sullivan J, et al. Mitochondrial mutagenesis correlates with the local inflammatory environment in arthritis. Ann Rheum Dis. 2012;71(4):582–588. doi:10.1136/annrheumdis-2011-200245
56. Biniecka M, Fox E, Gao W, et al. Hypoxia induces mitochondrial mutagenesis and dysfunction in inflammatory arthritis. Arthritis Rheum. 2011;63(8):2172–2182. doi:10.1002/art.30395
57. Jiang G, Jiang T, Chen J, et al. Mitochondrial dysfunction and oxidative stress in diabetic wound. J Biochem Mol Toxicol. 2023;37(7):e23407. doi:10.1002/jbt.23407
58. Wang DK, Zheng HL, Zhou WS, et al. Mitochondrial Dysfunction in Oxidative Stress-Mediated Intervertebral Disc Degeneration. Orthop Surg. 2022;14(8):1569–1582. doi:10.1111/os.13302
59. Harapas CR, Idiiatullina E, Al-Azab M, et al. Organellar homeostasis and innate immune sensing. Nat Rev Immunol. 2022;22(9):535–549. doi:10.1038/s41577-022-00682-8
60. Weyand CM, Goronzy JJ. The immunology of rheumatoid arthritis. Nat Immunol. 2021;22(1):10–18. doi:10.1038/s41590-020-00816-x
61. Denning NL, Aziz M, Gurien SD, et al. DAMPs and NETs in Sepsis. Front Immunol. 2019;10:2536. doi:10.3389/fimmu.2019.02536
62. Walker KA, Basisty N, Wilson DM, et al. Connecting aging biology and inflammation in the omics era. J Clin Invest. 2022;132(14):e158448. doi:10.1172/JCI158448
63. Wei WTuna S, Keogh MJ, Keogh MJ, et al. Germline selection shapes human mitochondrial DNA diversity.Science. Science (New York, N.Y.). 2019;364(6442):eaau6520. doi:10.1126/science.aau6520
64. Bryant JD, Lei Y, VanPortfliet JJ, et al. Assessing Mitochondrial DNA Release into the Cytosol and Subsequent Activation of Innate Immune-related Pathways in Mammalian Cells. Curr Protoc. 2022;2(2):e372. doi:10.1002/cpz1
65. Faas MM, de Vos P. Mitochondrial function in immune cells in health and disease. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165845. doi:10.1002/cpz1.372
66. Chung KW, Dhillon P, Huang S, et al. Mitochondrial Damage and Activation of the STING Pathway Lead to Renal Inflammation and Fibrosis. Cell Metab. 2019;30(4):784–799. doi:10.1016/j.cmet.2019.08.003
67. Yu CH, Davidson S, Harapas CR, et al. TDP-43 Triggers Mitochondrial DNA Release via mPTP to Activate cGAS/STING in ALS. Cell. 2020;183(3):636–649.e18. doi:10.1016/j.cell.2020.09.020
68. Chen R, Du J, Zhu H, et al. The role of cGAS-STING signalling in liver diseases. Jhep Rep. 2021;3(5):
69. Kim J, Kim HS, Chung JH. Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway. Exp Mol Med. 2023;55(3):510–519. doi:10.1038/s12276-023-00965-7
70. Tripathi A, Bartosh A, Whitehead C, et al. Activation of cell-free mtDNA-TLR9 signaling mediates chronic stress-induced social behavior deficits. Mol Psychiatry. 2023;28(9):3806–3815. doi:10.1038/s41380-023-02189-7
71. Saber MM, Monir N, Awad AS, et al. TLR9: a friend or a foe. Life Sci. 2022;307:120874. doi:10.1016/j.lfs.2022.120874
72. Xu Y, Shen J, Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 2020;16(1):3–17. doi:10.1080/15548627.2019.1603547
73. Ward GA, Dalton RP R, Meyer BS, et al. Oxidized Mitochondrial DNA Engages TLR9 to Activate the NLRP3 Inflammasome in Myelodysplastic Syndromes. Int J Mol Sci. 2023;24(4):3896. doi:10.3390/ijms24043896
74. Liao S, Luo J, Kadier T, et al. Mitochondrial DNA Release Contributes to Intestinal Ischemia/Reperfusion Injury. Front Pharmacol. 2022;13:854994. doi:10.3389/fphar.2022.854994
75. Austad SN, Ballinger S, Buford TW, et al. Targeting whole body metabolism and mitochondrial bioenergetics in the drug development for Alzheimer’s disease. Acta Pharm Sin B. 2022;12(2):511–531. doi:10.1016/j.apsb.2021.06.014
76. Han W, Cui J, Sun G, et al. Nano-sized microplastics exposure induces skin cell senescence via triggering the mitochondrial localization of GSDMD. Environ Pollut. 2024;349:123874. doi:10.1016/j.envpol.2024.124123
77. Pezone A, Olivieri F, Napoli MV, et al. Inflammation and DNA damage: cause, effect or both. Nat Rev Rheumatol. 2023;19(4):200–211. doi:10.1038/s41584-022-00905-1
78. Zhang B, Pan C, Feng C, et al. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep. 2022;27(1):45–52. doi:10.1080/13510002.2022.2046423
79. Antonucci S, Di Lisa F, Kaludercic N. Mitochondrial reactive oxygen species in physiology and disease. Cell Calcium. 2021;94:102344. doi:10.1016/j.ceca.2020.102344
80. Kasai S, Shimizu S, Tatara Y, et al. Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology. Biomolecules. 2020;10(2):320. doi:10.3390/biom10020320
81. Shekhova E, Silverman N. Mitochondrial reactive oxygen species as major effectors of antimicrobial immunity. PLoS Pathog. 2020;16(5):e1008470. doi:10.1371/journal.ppat.1008470
82. Hadrava Vanova K, Kraus M, Neuzil J, et al. Mitochondrial complex II and reactive oxygen species in disease and therapy. Redox Rep. 2020;25(1):26–32. doi:10.1080/13510002.2020.1752002
83. Mukherjee A, Ghosh KK, Chakrabortty S, et al. Mitochondrial Reactive Oxygen Species in Infection and Immunity. Biomolecules. 2024;14(6):670. doi:10.3390/biom14060670
84. Quintana-Cabrera R, Scorrano L. Determinants and outcomes of mitochondrial dynamics. Mol Cell. 2023;83(6):857–876. doi:10.1016/j.molcel.2023.02.012
85. Zhen W, Zhao H, Li Y. Mitochondrial dynamics in health and disease: mechanisms and potential targets. Signal Transduct Target Ther. 2023;8(1):333. doi:10.1038/s41392-023-01547-9
86. Yapa NMB, Lisnyak V, Reljic B, et al. Mitochondrial dynamics in health and disease. FEBS Lett. 2021;595(8):1184–1204. doi:10.1002/1873-3468.14077
87. Whitley BN, Engelhart EA, Hoppins S. Mitochondrial dynamics and their potential as a therapeutic target. Mitochondrion. 2019;49:269–283. doi:10.1016/j.mito.2019.06.002
88. Zacharioudakis E, Gavathiotis E. Mitochondrial dynamics proteins as emerging drug targets. Trends Pharmacol Sci. 2023;44(2):112–127. doi:10.1016/j.tips.2022.11.004
89. Chan DC. Mitochondrial Dynamics and Its Involvement in Disease. Annu Rev Pathol. 2020;15:235–259. doi:10.1146/annurev-pathmechdis-012419-032711
90. Wang M, Li Q, Zhang Y, et al. Total Glucosides of Peony Protect Cardiomyocytes against Oxidative Stress and Inflammation by Reversing Mitochondrial Dynamics and Bioenergetics. Oxid Med Cell Longev. 2020;2020:6632413. doi:10.1155/2020/6632413
91. Wang X, Chen Z, Fan X, et al. Inhibition of DNM1L and mitochondrial fission attenuates inflammatory response in fibroblast-like synoviocytes of rheumatoid arthritis. J Cell Mol Med. 2020;24(2):1516–1528. doi:10.1111/jcmm.14837
92. Onishi M, Yamano K, Sato M, et al. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. doi:10.15252/embj.2020104705
93. Tang Y, Xu W, Liu Y, et al. Autophagy protects mitochondrial health in heart failure. Heart Fail Rev. 2024;29(1):113–123. doi:10.1007/s10741-023-10354-x
94. Kim JS, Chapman WC, Lin Y. Mitochondrial Autophagy in Ischemic Aged Livers. Cells. 2022;11(24):4083. doi:10.3390/cells11244083
95. Yan C, Shi Y, Yuan L, et al. Mitochondrial quality control and its role in osteoporosis. Front Endocrinol. 2023;14:1077058. doi:10.3389/fendo.2023.1077058
96. Goodall EA, Kraus F, Harper JW. Mechanisms underlying ubiquitin-driven selective mitochondrial and bacterial autophagy. Mol Cell. 2022;82(8):1501–1513. doi:10.1016/j.molcel.2022.03.012
97. Abate M, Festa A, Falco M, et al. Mitochondria as playmakers of apoptosis, autophagy and senescence. Semin Cell Dev Biol. 2020;98:139–153. doi:10.1016/j.semcdb.2019.05.022
98. Towers CG, Wodetzki DK, Thorburn J, et al. Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy. Dev Cell. 2021;56(14):2029–2042.e5. doi:10.1016/j.devcel.2021.06.003
99. Yao H, Xiang L, Huang Y, et al. Guizhi Shaoyao Zhimu granules attenuate bone destruction in mice with collagen-induced arthritis by promoting mitophagy of osteoclast precursors to inhibit osteoclastogenesis. Phytomedicine. 2023;118:154967. doi:10.1016/j.phymed.2023.154967
100. In II, Deng W. Construction and validation of a diagnostic model for rheumatoid arthritis based on mitochondrial autophagy-related genes. Heliyon. 2024;10(3):e24818. doi:10.1016/j.heliyon.2024.e24818
101. Popov LD. Mitochondrial biogenesis: an update. J Cell Mol Med. 2020;24(9):4892–4899. doi:10.1111/jcmm.15194
102. Liu L, Li Y, Chen G, et al. Crosstalk between mitochondrial biogenesis and mitophagy to maintain mitochondrial homeostasis. J Biomed Sci. 2023;30(1):86. doi:10.1186/s12929-023-00975-7
103. Zhao T, Zhang J, Lei H, et al. NRF1-mediated mitochondrial biogenesis antagonizes innate antiviral immunity. EMBO J. 2023;42(16):e113258. doi:10.15252/embj.2022113258
104. Halling JF, Pilegaard H. PGC-1α-mediated regulation of mitochondrial function and physiological implications. Appl Physiol Nutr Metab. 2020;45(9):927–936. doi:10.1139/apnm-2020-0005
105. Blanco FJ, Fernández-Moreno M. Mitochondrial biogenesis: a potential therapeutic target for osteoarthritis. Osteoarthritis Cartilage. 2020;28(8):1003–1006. doi:10.1016/j.joca.2020.03.018
106. Yasukawa K, Koshiba T. Mitochondrial reactive zones in antiviral innate immunity. Biochim Biophys Acta Gen Subj. 2021;1865(3):129839. doi:10.1016/j.bbagen.2020.129839
107. Weng GX, Ling T, Hou W, et al. Mitochondrial DUT-M potentiates RLR-mediated antiviral signaling by enhancing VISA and TRAF2 association. Mol Immunol. 2021;132:117–125. doi:10.1016/j.molimm.2021.01.023
108. Li X, Hou P, Ma W, et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol Immunol. 2022;19(1):67–78. doi:10.1038/s41423-023-01023-y
109. Wu M, Pei Z, Long G, et al. Mitochondrial antiviral signaling protein: a potential therapeutic target in renal disease. Front Immunol. 2023;14:1266461. doi:10.3389/fimmu.2023.1266461
110. Fu J, Wu H. Structural Mechanisms of NLRP3 Inflammasome Assembly and Activation. Annu Rev Immunol. 2023;41:301–316. doi:10.1146/annurev-immunol-081022-021207
111. Zecchini V, Paupe V, Herranz-Montoya I, et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature. 2023;615(7952):499–506. doi:10.1038/s41586-023-05770-w
112. Chou WC, Rampanelli E, Li X, et al. Impact of intracellular innate immune receptors on immunometabolism. Cell Mol Immunol. 2022;19(3):337–351. doi:10.1038/s41423-021-00780-y
113. Saravia J, Raynor JL, Chapman NM, et al. Signaling networks in immunometabolism. Cell Res. 2020;30(4):328–342. doi:10.1038/s41422-020-0301-1
114. Zhu M, Barbas AS, Lin L, et al. Mitochondria Released by Apoptotic Cell Death Initiate Innate Immune Responses. Immunohorizons. 2018;2(11):384–397. doi:10.4049/immunohorizons.1800063
115. Zhong Z, Lemasters JJ. A Unifying Hypothesis Linking Hepatic Adaptations for Ethanol Metabolism to the Proinflammatory and Profibrotic Events of Alcoholic Liver Disease. Alcohol Clin Exp Res. 2018;42(11):2072–2089. doi:10.1111/acer.13877
116. Cui L, Weiyao J, Chenghong S, et al. Rheumatoid arthritis and mitochondrial homeostasis: the crossroads of metabolism and immunity. Front Med. 2022;9:1017650. doi:10.3389/fmed.2022.1017650
117. Gravallese EM, Firestein GS, Longo DL. Rheumatoid Arthritis - Common Origins, Divergent Mechanisms. N Engl J Med. 2023;388(6):529–542. doi:10.1056/NEJMra2103726
118. Rambold AS, Pearce EL. Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends Immunol. 2018;39(1):6–18. doi:10.1016/j.it.2017.08.006
119. Hu Z, Zhao TV, Huang T, et al. The transcription factor RFX5 coordinates antigen-presenting function and resistance to nutrient stress in synovial macrophages. Nat Metab. 2022;4(6):759–774. doi:10.1038/s42255-022-00585-x
120. Souto-Carneiro MM, Klika KD, Abreu MT, et al. Effect of Increased Lactate Dehydrogenase A Activity and Aerobic Glycolysis on the Proinflammatory Profile of Autoimmune CD8+ T Cells in Rheumatoid Arthritis. Arthritis Rheumatol. 2020;72(12):2050–2064. doi:10.1002/art.41420
121. Haas R, Smith J, Rocher-Ros V, et al. Lactate Regulates Metabolic and Pro-inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol. 2015;13(7):e1002202. doi:10.1371/journal.pbio.1002202
122. Yang Z, Shen Y, Oishi H, et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci Transl Med. 2016;8(331):331ra38. doi:10.1126/scitranslmed.aad7151
123. Yang Z, Fujii H, Mohan SV, et al. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210(10):2119–2134. doi:10.1084/jem.20130252
124. Pucino V, Certo M, Bulusu V, et al. Lactate Buildup at the Site of Chronic Inflammation Promotes Disease by Inducing CD4+ T Cell Metabolic Rewiring. Cell Metab. 2019;30(6):1055–1074.e8. doi:10.1016/j.cmet.2019.10.004
125. Farmer JR, Allard-Chamard H, Sun N, et al. Induction of metabolic quiescence defines the transitional to follicular B cell switch. Sci Signal. 2019;12(604):eaaw5573. doi:10.1126/scisignal.aaw5573
126. Ramon S, Bancos S, Thatcher TH, et al. Peroxisome proliferator-activated receptor γ B cell-specific-deficient mice have an impaired antibody response. J Immunol. 2012;189(10):4740–4747. doi:10.4049/jimmunol.1200956
127. Smulski CR, Eibel H. BAFF and BAFF-Receptor in B Cell Selection and Survival. Front Immunol. 2018;9:2285. doi:10.3389/fimmu.2018.02285
128. Patke A, Mecklenbräuker I, Erdjument-Bromage H, et al. BAFF controls B cell metabolic fitness through a PKC beta- and Akt-dependent mechanism. J Exp Med. 2006;203(11):2551–2562. doi:10.1084/jem.20060990
129. Zhang Z, Zhang R, Li L, et al. Macrophage migration inhibitory factor (MIF) inhibitor, Z-590 suppresses cartilage destruction in adjuvant-induced arthritis via inhibition of macrophage inflammatory activation. Immunopharmacol Immunotoxicol. 2018;40(2):149–157. doi:10.1080/08923973.2018.1424896
130. Weyand CM, Zeisbrich M, Goronzy JJ. Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Curr Opin Immunol. 2017;46:112–120. doi:10.1016/j.coi.2017.04.010
131. Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8(11):656–664. doi:10.1038/nrrheum.2012.153
132. Sun P, Liu Y, Deng X, et al. An inhibitor of cathepsin K, icariin suppresses cartilage and bone degradation in mice of collagen-induced arthritis. Phytomedicine. 2013;20(11):975–979. doi:10.1016/j.phymed.2013.04.019
133. Cush JJ. Rheumatoid Arthritis: early Diagnosis and Treatment. Rheum Dis Clin North Am. 2022;48(2):537–547. doi:10.1016/j.rdc.2022.02.010
134. Di Matteo A, Bathon JM, Emery P. Rheumatoid arthritis. Lancet. 2023;402(10416):2019–2033. doi:10.1016/S0140-6736(23)01525-8
135. Brillo V, Chieregato L, Leanza L, et al. Mitochondrial Dynamics, ROS, and Cell Signaling: a Blended Overview. Life. 2021;11(4):332. doi:10.3390/life11040332
136. Wu J, Feng Z, Chen L, et al. TNF antagonist sensitizes synovial fibroblasts to ferroptotic cell death in collagen-induced arthritis mouse models. Nat Commun. 2022;13(1):676. doi:10.1038/s41467-021-27948-4
137. Fearon U, Canavan M, Biniecka M, et al. Hypoxia, mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat Rev Rheumatol. 2016;12(7):385–397. doi:10.1038/nrrheum.2016.69
138. Willemsen J, Neuhoff MT, Hoyler T, et al. TNF leads to mtDNA release and cGAS/STING-dependent interferon responses that support inflammatory arthritis. Cell Rep. 2021;37(6):109977. doi:10.1016/j.celrep.2021.109977
139. Lee SY, Kwok SK, Son HJ, et al. IL-17-mediated Bcl-2 expression regulates survival of fibroblast-like synoviocytes in rheumatoid arthritis through STAT3 activation. Arthritis Res Ther. 2013;15(1):R31. doi:10.1186/ar4179
140. Kim EK, Kwon JE, Lee SY, et al. IL-17-mediated mitochondrial dysfunction impairs apoptosis in rheumatoid arthritis synovial fibroblasts through activation of autophagy. Cell Death Dis. 2017;8(1):e2565. doi:10.1038/cddis.2016.490
141. Zhang T, Wang L, Duan X, et al. Sirtuins mediate mitochondrial quality control mechanisms: a novel therapeutic target for osteoporosis. Front Endocrinol. 2024;14:1281213. doi:10.3389/fendo.2023.1281213
142. Lemma S, Sboarina M, Porporato PE, et al. Energy metabolism in osteoclast formation and activity. Int J Biochem Cell Biol. 2016;79:168–180. doi:10.1016/j.biocel.2016.08.034
143. Kim JM, Jeong D, Kang HK, et al. Osteoclast precursors display dynamic metabolic shifts toward accelerated glucose metabolism at an early stage of RANKL-stimulated osteoclast differentiation. Cell Physiol Biochem. 2007;20(6):935–946. doi:10.1159/000110454
144. Fraser JH, Helfrich MH, Wallace HM, et al. Hydrogen peroxide, but not superoxide, stimulates bone resorption in mouse calvariae. Bone. 1996;19(3):223–226. doi:10.1016/8756-3282(96)00177-9
145. Baek KH, Oh KW, Lee WY, et al. Association of oxidative stress with postmenopausal osteoporosis and the effects of hydrogen peroxide on osteoclast formation in human bone marrow cell cultures. Calcif Tissue Int. 2010;87(3):226–235. doi:10.1007/s00223-010-9393-9
146. Bertero E, Popoiu TA, Maack C. Mitochondrial calcium in cardiac ischemia/reperfusion injury and cardioprotection. Basic Res Cardiol. 2024;119(4):569–585. doi:10.1007/s00395-024-01060-2
147. Li Y, Bai B, Zhang Y. Bone abnormalities in young male rats with iron intervention and possible mechanisms. Chem Biol Interact. 2018;279:21–26. doi:10.1016/j.cbi.2017.11.005
148. Shares BH, Busch M, White N, et al. Active mitochondria support osteogenic differentiation by stimulating β-catenin acetylation. J Biol Chem. 2018;293(41):16019–16027. doi:10.1074/jbc.RA118.004102
149. Wang Y, Zhao X, Lotz M, et al. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor γ coactivator 1α. Arthritis Rheumatol. 2015;67(8):2141–2153. doi:10.1002/art.39182
150. Wang J, Li J, Song D, et al. AMPK: implications in osteoarthritis and therapeutic targets.American. J Transl Res. 2020;12(12):7670–7681. doi:10.1002/art.39182
151. Forni MF, Peloggia J, Trudeau K, et al. Murine mesenchymal stem cell commitment to differentiation is regulated by mitochondrial dynamics. Stem Cells. 2016;34(3):743–755. doi:10.1002/stem.2248
152. Wang Y, McLean AS. The Role of Mitochondria in the Immune Response in Critical Illness. Crit Care. 2022;26(1):80. doi:10.1186/s13054-022-03908-2
153. Arai M, Shibata Y, Pugdee K, et al. Effects of reactive oxygen species (ROS) on antioxidant system and osteoblastic differentiation in MC3T3-E1 cells. IUBMB Life. 2007;59(1):27–33. doi:10.1080/15216540601156188
154. Srinivasan S, Koenigstein A, Joseph J, et al. Role of mitochondrial reactive oxygen species in osteoclast differentiation. Ann N Y Acad Sci. 2010;1192(1):245–252. doi:10.1111/j.1749-6632.2009.05377.x
155. Ishii KA, Fumoto T, Iwai K, et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat Med. 2009;15(3):259–266. doi:10.1038/nm.1910
156. Kim H, Kim T, Jeong BC, et al. Tmem64 modulates calcium signaling during RANKL-mediated osteoclast differentiation. Cell Metab. 2013;17(2):249–260. doi:10.1016/j.cmet.2013.01.002
157. Rossini M, Filadi R. Sarcoplasmic Reticulum-Mitochondria Kissing in Cardiomyocytes: ca2+, ATP, and Undisclosed Secrets. Front Cell Dev Biol. 2020;8:532. doi:10.3389/fcell.2020.00532
158. Liu P, Feng Y, Li H, et al. Ferrostatin-1 alleviates lipopolysaccharide-induced acute lung injury via inhibiting ferroptosis. Cell Mol Biol Lett. 2020;25:10. doi:10.1186/s11658-020-00205-0
159. Yokota K, Sato K, Miyazaki T, et al. Characterization and Function of Tumor Necrosis Factor and Interleukin-6-Induced Osteoclasts in Rheumatoid Arthritis. Arthritis Rheumatol. 2021;73(7):1145–1154.doi. doi:10.1002/art.41666
160. Zhu YS, Mo TT, Jiang C, et al. Osteonectin bidirectionally regulates osteoblast mineralization. J Orthop Surg Res. 2023;18(1):761. doi:10.1186/s13018-023-04250-1
161. Doumpas N, Lampart F, Robinson MD, et al. TCF /LEF dependent and independent transcriptional regulation of Wnt/βcatenin target genes, The EMBO. THE EMBO Journal. 2019;38(2):e98873. doi:10.15252/embj.201798873
162. Wan MC, Tang XY, Li J, et al. Upregulation of mitochondrial dynamics is responsible for osteogenic differentiation of mesenchymal stem cells cultured on self-mineralized collagen membranes. Acta Biomater. 2021;136:137–146. doi:10.1016/j.actbio.2021.09.039
163. Lee SY, An HJ, Kim JM, et al. PINK1 deficiency impairs osteoblast differentiation through aberrant mitochondrial homeostasis. Stem Cell Res Ther. 2021;12(1):589. doi:10.1186/s13287-021-02656-4
164. Banoth B, Cassel SL. Mitochondria in innate immune signaling. Transl Res. 2018;202:52–68. doi:10.1016/j.trsl.2018.07.014
165. Kuley R, Duvvuri B, Wallin JJ, et al. Mitochondrial N-formyl methionine peptides contribute to exaggerated neutrophil activation in patients with COVID-19. Virulence. 2023;14(1):2218077. doi:10.1080/21505594.2023.2218077
166. Injarabian L, Devin A, Ransac S, et al. Neutrophil Metabolic Shift during their Lifecycle: impact on their Survival and Activation. Int J Mol Sci. 2019;21(1):287. doi:10.3390/ijms21010287
167. Bedoui Y, Guillot X, Sélambarom J, et al. Methotrexate an Old Drug with New Tricks. Int J Mol Sci. 2019;20(20):5023. doi:10.3390/ijms20205023
168. Yu M, Nguyen ND, Huang Y, et al. Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. JCI Insight. 2019;5(16):e126915. doi:10.1172/jci.insight.126915
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Network Pharmacology Analysis and Experimental Validation to Investigate the Mechanism of Total Flavonoids of Rhizoma Drynariae in Treating Rheumatoid Arthritis
Chen G, Luo J, Liu Y, Yu X, Liu X, Tao Q
Drug Design, Development and Therapy 2022, 16:1743-1766
Published Date: 8 June 2022