Back to Journals » International Journal of Nanomedicine » Volume 19
Liver Extracellular Vesicles and Particles Enriched β-Sitosterol Effectively Promote Liver Regeneration in Mice
Authors Gongye X, Xia P, Ma T, Chai Y, Chen Z, Zhu Y, Qu C, Liu J, Guo WW, Zhang M, Liu Y, Tian M, Yuan Y
Received 22 February 2024
Accepted for publication 31 July 2024
Published 8 August 2024 Volume 2024:19 Pages 8117—8137
DOI https://doi.org/10.2147/IJN.S465346
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Farooq A. Shiekh
Xiangdong Gongye,1,2,* Peng Xia,1,2,* Tianyin Ma,1,2,* Yibo Chai,1,2 Zhang Chen,1,2 Yimin Zhu,1,2 Chengming Qu,1,2 Jie Liu,1,2 Wing Wa Guo,1,2 Minghe Zhang,1,2 Yingyi Liu,1,2 Ming Tian,1,2 Yufeng Yuan1– 3
1Department of Hepatobiliary & Pancreatic Surgery, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China; 2Clinical Medicine Research Center for Minimally Invasive Procedure of Hepatobiliary & Pancreatic Diseases of Hubei Province, Hubei, People’s Republic of China; 3Taikang Center for Life and Medical Sciences of Wuhan University, Wuhan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ming Tian; Yufeng Yuan, Email [email protected]; [email protected]
Background: The liver’s regenerative capacity allows it to repair itself after injury. Extracellular vesicles and particles (EVPs) in the liver’s interstitial space are crucial for signal transduction, metabolism, and immune regulation. Understanding the role and mechanism of liver-derived EVPs in regeneration is significant, particularly after partial hepatectomy, where the mechanisms remain unclear.
Methods: A 70% hepatectomy model was established in mice, and EVPs were isolated and characterized using electron microscopy, nanocharacterization, and Western blot analysis. Combined metabolomic and transcriptomic analyses revealed β-sitosterol enrichment in EVPs and activation of the Hedgehog signaling pathway during regeneration. The role of β-sitosterol in EVPs on the Hedgehog pathway and its targets were identified using qRT-PCR, Western blot analysis. The regulation of carnitine synthesis by this pathway was determined using a dual luciferase assay. The effect of a β-sitosterol diet on liver regeneration was verified in mice.
Results: After 70% hepatectomy, the liver successfully regenerated without liver failure or death. At 24 hours post-surgery, tissue staining showed transient regeneration-associated steatosis (TRAS), with increased Ki67 positivity at 48 hours. EVPs displayed a spherical lipid bilayer structure with particle sizes of 70– 130 nm. CD9, CD63, and CD81 in liver-derived EVPs were confirmed. Transcriptomic and metabolomic analyses showed EVPs supplementation significantly promoted carnitine synthesis and fatty acid oxidation. Tissue staining confirmed accelerated TRAS resolution and enhanced liver regeneration with EVP supplementation. Mass spectrometry identified β-sitosterol in EVPs, which binds to Smo protein, activating the Hedgehog pathway. This led to the nuclear transport of Gli3, stimulating Setd5 transcription and inducing carnitine synthesis, thereby accelerating fatty acid oxidation. Mice with increased β-sitosterol intake showed faster TRAS resolution and liver regeneration compared to controls.
Conclusion: Liver-derived EVPs promote regeneration after partial hepatectomy. β-sitosterol from EVPs accelerates fatty acid oxidation and promotes liver regeneration by activating Hedgehog signaling pathway.
Keywords: liver regeneration, extracellular vesicles and particles, β-sitosterol, carnitine synthesis, hedgehog signaling pathway
Graphical Abstract:

Introduction
The liver is a key solid organ in mammals that plays an indispensable role in the digestive and immune systems.1,2 The liver is also unique in that it possesses the ability to repair damage and regenerate precisely to meet the body’s needs.3,4 However, in the presence of persistent liver damage caused by harmful factors from various sources, the regenerative capacity often fails to prevent acute or chronic liver function failure. Liver transplantation still remains the primary choice for treating end-stage liver disease; nevertheless, the limited donor source and rejection-related reactions after transplantation often restrict the promotion of transplantation therapy.5,6 While cell therapies and other alternative bioengineering strategies are currently available,7 the problem of insufficient cell survival and differentiation rate cannot still be solved.8,9 Furthermore, liver regeneration is not always beneficial, and the potential risk of tumorigenesis cannot be ignored.10–12 Therefore, the development of a method that promotes normal regenerative repair of the liver is a hot topic in the field of liver research.
Liver regeneration is initiated after partial hepatectomy due to the reduction in liver volume and loss of hepatocytes. Notably, no new lobes are formed during this process. At the tissue level, the volume and number of hepatic lobules formed by the proliferation of various cell types in the postoperative residual liver tissues continue to increase. At the molecular level, signaling pathways mediated by cytokines, chemokines, enzyme-linked/G protein-coupled receptors, integrins and environmental stress exhibit interrelated regulatory effects.13,14 These pathways intertwine to form a complex regulatory network that maintains the homeostasis of the hepatic microenvironment.15
Tissue-derived extracellular vesicles and particles (EVPs) encapsulate a wide range of proteins, nucleic acid molecules, and metabolites and mediate a variety of intercellular information exchange processes.16–18 EVPs have attracted much attention owing to their unique physical, chemical, and biological properties, as well as their good application prospects in the diagnosis, treatment, and prognosis monitoring of various diseases. Increasingly more studies have shown their roles in maintaining body homeostasis and regulating different pathophysiological activities.16,19,20 For instance, iron encapsulated in EV is transported from hepatocytes to hematopoietic stem cells, leading to disrupted iron distribution in the liver. Iron-deficient hepatocytes exhibit pathological features of increased lipid synthesis and insulin resistance via the HIF2α (hypoxia inducible factor 2α)-ATF4 (activating transcription factor 4) pathway.21 Debanjali Dasgupta et al reported that upon activation of IRE1A (inositol-requiring enzyme 1 alpha), mouse hepatocytes release exosomes enriched with inflammatory ceramides. These inflammatory exosomes recruit monocyte-derived macrophages to the liver, contributing to inflammatory injury responses. Developing strategies to block these pathways could potentially slow the progression of inflammation in patients with non-alcoholic fatty liver disease, thereby treating the disease and promoting liver regeneration.22 Moreover, following partial hepatectomy, apart from the initial mechanical signals triggered by drastic changes in intrahepatic blood flow pressure (shear stress due to portal vein blood flow through a narrower outflow tract), various chemical signal transmissions between cells and tissues require a reliable carrier to overcome the complex physical and chemical barriers of the internal environment.23
The application of mesenchymal stem cells and hepatocyte-derived vesicles for liver diseases, including liver regeneration regulation and repair, has been widely discussed. A study has demonstrated the involvement of EVPs carrying miRNAs in the repair and regeneration processes of polytrauma and surgical invasive injuries.24 Engineered modified EVPs have also been demonstrated to contribute to the regenerative repair of soft tissues, such as esophageal fistulas.25 The role of naturally existing tissue-derived vesicles represents an immediate homeostatic response. A previous study has reported the therapeutic efficacy of exogenous liver tissue vesicles using a model of carbon tetrachloride-mediated chemical hepatic necrosis.26 EVPs have also been identified to have a protective effect during hepatic ischemia-reperfusion injury by ameliorating local inflammation.27 However, the specific signaling mechanisms of these vesicles in promoting regeneration and repair have not been thoroughly studied to date. Therefore, further studies investigating EVPs in liver tissues are required to explore their therapeutic potential and functional mechanism.
The aim of this study was to reveal the regulatory mechanism of metabolites in EVPs for liver regeneration. Accordingly, we established a regeneration model after 70% hepatectomy in mice, isolated EVPs from freshly resected liver tissues, and then characterized their physicochemical properties, including morphology, particle size, and immune molecular composition. Furthermore, we confirmed the biological effects of EVPs on hepatocyte uptake and regeneration both in vivo and in vitro and also conducted time-course experiments to trace the changes that occurred during liver regeneration with or without exogenous liver tissue-derived EVPs (LT-EVPs) supplementation. Subsequently, we analyzed the metabolite profile in LT-EVPs and identified the role of β-sitosterol in enhancing liver regeneration by activating Hedgehog signaling to upregulate carnitine synthesis, thereby enhancing fatty acid oxidation.
Materials and Methods
The Hepatectomy Using Mice Model
Six-week-old male C57BL/6 mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The animals were placed in an animal house and kept in a standard feeding environment for more than one week prior to preparation for liver partial resection. Animal experiments were performed in accordance with all regulations set by the Chinese Council on Animal Care, and the study protocol was approved by Experimental Animal Welfare Ethics Committee, Zhongnan Hospital of Wuhan University (Approval NO. ZN2022032). As for the diet groups, we added β-sitosterol in the feed for mice and controlled the intake levels to 20 mg/kg per day.28,29 The 6-week-old mice were initiated on this dietary pattern after entering the animal laboratory. Partial hepatectomy was performed at 1 week later, and the same diet was continued postoperatively. On the day of surgery, after the mice were anesthetized with isoflurane gas, a transverse incision was made under the xiphoid process to open the abdominal cavity and extrude the resection part of the liver. The root of the liver lobe was ligated with saline-soaked 5–0 braided thread, and the liver lobe and excess thread were removed after confirming the tightness of the knot. Following the removal of the middle lobe and left bipartite lobe, the abdominal cavity was closed and sutured. The procedure for the sham operation group involved opening the abdominal cavity, dissociating the hepatic round ligament, and then closing the abdomen. EVPs (20 µg of EVPs protein per gram of body weight) were injected through the tail veins of mice, while the control group received an injection of the same volume of PBS. Animals were sacrificed at different time points postoperatively according to each group’s time point requirements (24 h, 48 h, and 72 h). The set number of mice at each time point was 3. The weight of the regenerated liver was measured and recorded in each group. The livers were partially frozen and partially fixed in paraformaldehyde.
The Biochemical Examination of Mice Serum
Blood was obtained from mice via apical puncture and was cooled on ice. Plasma was extracted after centrifugation at 500 g per minute for 10 min at room temperature (25°C). Plasma biochemical parameters were measured using an automatic biochemical analyzer (BK-280, Shandong Broco, China).
Tissue Histological Analysis
For immunohistochemistry, the tissues were fixed, embedded, and sectioned. Xylene and ethanol were used to deparaffinize and rehydrate the paraffin-embedded tissues. For antigen retrieval, slides were steamed for 15 minutes in Tris-ethylenediaminetetraacetic acid (EDTA) buffer (pH 9.0; Abcam, Cambridge, MA, USA). Then, slides were incubated in 0.3% H2O2-methanol (Aladdin, Shanghai, China) for 10 minutes. Slides were blocked with 4% donkey serum in Tris-buffered saline with Tween 20 (TBST) for 1 hour. Subsequently, slides were incubated with the primary antibody overnight at 4°C, followed by incubation with the biotinylated secondary antibody for 60 minutes. Slides were then incubated for 30 minutes with VECTASTAIN Elite ABC Reagent (Vector Labs, Burlingame, CA, USA) and reacted with the peroxidase substrate solution 3.3′-diaminobenzidine (DAB, Vector Labs) until the desired stain intensity developed. The dehydrated slides (processed through 70% ethanol-xylene substitute) were air-dried and mounted with neutral tree gum (Solaibao, Beijing, China). For Oil Red, tissue samples were fixed in 4% paraformaldehyde for 24–48 hours at room temperature and washed in phosphate-buffered saline (PBS) to remove excess fixative. Fixed tissues were embedded in OCT compound, frozen at −20°C, cut into 10 μm thick sections using a cryostat, and mounted on glass slides. For staining, slides were first dehydrated in 60% isopropanol for 5 minutes, then immersed in an Oil Red O working solution (prepared by diluting a 0.5% Oil Red O stock solution in isopropanol with distilled water at a 3:2 ratio and filtering before use) for 10–15 minutes at room temperature. After staining, slides were briefly rinsed in 60% isopropanol to remove excess stain, then counterstained with hematoxylin for 1–2 minutes. Slides were rinsed in running tap water for 5 minutes to remove excess hematoxylin, then mounted with an aqueous mounting medium to avoid dissolving the Oil Red O stain. Stained sections were examined under a light microscope, where lipid droplets appeared red and nuclei were stained blue. For Nile Red, tissue was washed in 1 × PBS prior to staining with Nile Red solution (7385–67-3, Solarbio, Beijing, China) diluted 1:100 with PBS for 20 min in the dark. Samples were then washed twice with PBS, and stained with DAPI. Images were acquired by IF microscopy.
For histological analysis, areas of interest were selected from 6 to 10 randomly chosen microscopic fields (×20 and ×63 magnification) per mouse (distributed over three non-consecutive tissue sections) or per well of the cell culture chamber. The analysis was performed using ImageJ software (version 1.47v; NIH, Bethesda, MD).
Western Blotting Analysis
Briefly, total proteins were extracted using Radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, Shanghai, China). The total protein concentrations in the supernatants were quantified using the Bicinchoninic Acid (BCA) protein assay (Beyotime Biotechnology). Proteins were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using gels of varying concentrations (10%, 12.5%, and 15%) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked for 1–2 hours with 5% nonfat dry milk (Biosharp, Hefei, Anhui, China) and then incubated with the various primary antibodies overnight at 4°C. Subsequently, the membranes were incubated with the appropriate secondary antibodies. Protein bands were detected using an enhanced chemiluminescence (ECL) detection reagent (GE Healthcare, Amersham, Buckinghamshire, UK). All antibodies used in this study are listed in the Supplementary Materials.
RNA Extraction, Quantitative Real‐time Polymerase Chain Reaction (qRT‐PCR) and RNA-Seq
Total RNA was extracted from cells or tissue samples using the TRIzol, following the manufacturer’s protocol. The concentration and purity of the extracted RNA were assessed using a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
For quantitative real-time polymerase chain reaction (qRT-PCR), 1 µg of total RNA was reverse transcribed into cDNA using the HiScript II Q RT SuperMix (Vazyme, Nanjing, Jiangsu, China) according to the manufacturer’s instructions. qRT-PCR was performed using the 2× ChamQ Universal SYBR qPCR Master Mix (Vazyme). The qRT-PCR conditions were set as follows: initial denaturation at 95°C for 40s, followed by 40 cycles of denaturation at 95°C for 10s, annealing at 57°C for 25s, and extension at 72°C for 20s. Relative gene expression levels were calculated using the 2^(-ΔΔCt) method. The primers used for the target genes and reference gene are listed in Supplementary Materials.
For sequencing, the amplified PCR products were purified and sequenced by Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Differentially expressed genes (DEGs) were defined as genes with a P value < 0.05 and an absolute log2 (fold change) > 1.
Liquid Chromatography-Mass Spectrometry (LC/MS) and Untargeted Metabolomics
The samples were separated using an ACQUITY UPLC® HSS T3 column (2.1 × 100 mm, 1.8 µm; Waters, Milford, MA, USA). The column temperature was set to 25°C, with a flow rate of 0.3 mL/min and an injection volume of 2 μL. The mobile phase composition was: A - water with 25 mmol/L ammonium acetate and 25 mmol/L ammonia; B - acetonitrile. The gradient elution program was as follows: 0–1 min, 95% B; 1–14 min, B linearly changed from 95% to 65%; 14–16 min, B linearly changed from 65% to 40%; 16–18 min, B maintained at 40%; 18–18.1 min, B linearly changed from 40% to 95%; and 18.1–23 min, B maintained at 95%.
Briefly, buffer S1 nuclease (Takara Biotechnology, Dalian, Liaoning, China), alkaline phosphatase (Takara Biotechnology), and phosphodiesterase I (Sigma-Aldrich) were added to 1 μg of RNA. The mixture was incubated at 37°C until the RNA was completely digested into nucleosides. The mixture was then extracted with chloroform (Sigma-Aldrich), and the resulting aqueous layer was collected for analysis using LC-MS/MS.
The sample extracts were analyzed using an Ultra Performance Liquid Chromatography (UPLC)-MS/MS system (UPLC, ExionLC™ AD, Applied Biosystems 6500 Triple Quadrupole; Foster City, CA, USA). The analytical conditions were as follows: LC Column: Waters ACQUITY UPLC HSS T3 C18 (1.8 μm; Waters, Milford, MA, USA); Solvent System: Water (2 mmol/L NH4HCO3): methanol (2 mmol/L NH4HCO3); Gradient Program: 95:5 V/V at 0 min, 95:5 V/V at 1 min, 5:95 V/V at 9 min, 5:95 V/V at 11 min, 95:5 V/V at 11.1 min, 95:5 V/V at 14 min; Flow Rate: 0.30 mL/min; Temperature: 40°C; Injection Volume: 10 μL.
The effluent was alternatively connected to an electrospray ionization-triple quadrupole-linear ion trap. A specific set of multiple reaction monitoring transitions was monitored for each period according to the metabolites eluted within this period.
Cell Culture and Proliferation
As a normal mouse liver cell line, NCTC-1469 is considered an excellent material for an in vitro study of mouse liver.30 The NCTC-1469 mouse normal liver cell line and its complete medium were purchased from Procell Inc (Wuhan). Cell culture conditions were as follows: 37°C with 5% CO2.
CCK8 Cell Proliferation: Cells added PBS or EVPs (active protein concentration 50 ug/mL) for 24 hours were reseeded into 96-well plates at a density of 20 cells/μL, with each well containing 100 μL of cell suspension (totaling 2000 cells per well). A blank control group was also prepared by adding 100 μL of cell-free medium to specific wells to eliminate any medium effects on the results. Additionally, 100 μL of PBS was added to the outermost wells of the 96-well plate, and the plate was placed closest to the water pan in the CO₂ incubator to prevent liquid evaporation from affecting the experiment. After allowing the cells to adhere for 5–6 hours, 10 μL of CCK8 reagent was added to each well, and the plate was gently tapped to mix the reagent. The plate was incubated in the CO₂ incubator for 1 hour, after which the absorbance at 450 nm was measured using a microplate reader. To prevent air bubbles from affecting the absorbance readings, any bubbles in the wells were carefully punctured with a syringe needle before measurement. Absorbance measurements were taken every 24 hours. The absorbance difference between the experimental and blank control wells was used to construct a time-course line graph.
EdU immunofluorescence: After inoculating the cells with cell-crawling tablets, the cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton-X100 for 10 min, and treated with 10% sheep serum at 37°C for 1 h. The cells were incubated with primary antibody at 4°C overnight, cleaned with phosphate buffer solution (PBS) on the next day, incubated with fluorescent secondary antibody for 1 h, and subsequently re-stained with DAPI. They were then photographed using a confocal microscope.
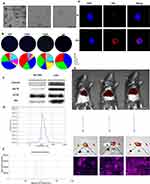
EVPs Separation and Characterization
The liver tissues were processed as soon as possible after dissection to isolate the EVPs or stored in liquid nitrogen. The livers were cut into relatively large pieces and pretreated with PBS to purify any damaged organelles or cellular debris generated when cutting the liver tissues.26 Add 2 mL of type II collagenase solution (prepared by dissolving 100 mg of dry powder in 10 mL MEM medium) to the tissue sample and resuspend thoroughly. Incubate the mixture in a shaking incubator set to 37°C and 150 rpm, mixing and pipetting the solution every 10 minutes to ensure even digestion until no large tissue fragments remain. Perform sequential gradient centrifugation at room temperature using the following settings: 300 g for 5 minutes, 2000 g for 15 minutes, and 10000 g for 30 minutes. After each centrifugation step, carefully decant the supernatant. Filter the supernatant through a 0.22 μm pore filter to remove smaller cellular debris. Transfer the filtrate to ultracentrifuge tubes and centrifuge at 100000 g for 2 hours. Discard the supernatant to obtain the crude EVPs pellet. Resuspend the pellet in pre-chilled PBS and centrifuge again at 100000 g for 90 minutes. Finally, discard the supernatant and resuspend the pellet in PBS to obtain the purified EVPs preparation. According to the MISEV guidelines, we characterized the morphology of isolated EVPs using transmission electron microscopy (TEM). We analyzed EVPs surface markers through automated fluorescence detection and validated these markers using Western blotting. Additionally, we performed comprehensive characterization of EVPs by analyzing their particle size distribution and Zeta potential distribution.
Subsequently, we added lipophilic DiR dye to the EVPs suspension at a working concentration of 10 µM. The mixture was incubated overnight at 4°C. Subsequently, free dye was removed by ultracentrifugation at 100000 g for 1 hour, and resuspend to obtain fluorescently labeled EVPs preparations.
In Vitro EVPs Uptake Assay: Add fluorescent EVPs to the culture medium containing cell coverslips at a concentration of 4 µg/µL. Collect the cell coverslips at 4-hour and 24-hour time points, and wash with PBS to remove dead cells. Fix the cells with paraformaldehyde and stain the nuclei. Observe and capture images using a confocal microscope.
In Vivo EVPs Uptake Assay: Inject 50µL fluorescent EVPs into mice via the tail vein. Use an in vivo imaging system to capture the distribution of EVPs within the mice. After 48 hours, euthanize the mice and collect the heart, liver, spleen, lungs, and kidneys. Capture images of the organ-level EVP fluorescence distribution. Finally, preserve liver tissue in paraformaldehyde protected from light, and capture images of tissue sections to observe the distribution of EVP fluorescence within the tissue.
Dual Luciferase Assay
One day before transfection, seed 0.5–2×10^5 cells per well in a 24-well plate with 500 μL of antibiotic-free medium, ensuring that cell confluence reaches 30–50% at the time of transfection. The cells were divided into two groups: Group A (control group) with an empty vector, pSetd5 (mouse)-Fluc-hRluc-Neo (firefly luciferase plasmid), and Renilla luciferase plasmid, and Group B (experimental group) with pECMV-Gli3-m-FLAG, pSetd5 (mouse)-Fluc-hRluc-Neo, and Renilla luciferase plasmid. For each well, dilute the plasmids (pECMV-Gli3-m-FLAG or empty vector, pSetd5 (mouse)-Fluc-hRluc-Neo, and pRL-TK) in 50 μL of Opti-MEM reduced-serum medium to the desired concentration and mix gently. Vortex Lipofectamine 2000 gently before use, then dilute 1 μL of Lipofectamine 2000 in 50 μL of Opti-MEM reduced-serum medium and incubate at room temperature for 5 minutes. Combine the diluted plasmid solutions with the diluted Lipofectamine 2000 to a total volume of 100 μL, mix gently, and incubate at room temperature for 20 minutes. Add 100 μL of the transfection complex to each well, gently swirl the plate to ensure even distribution, and incubate the cells in a CO2 incubator at 37°C. After 4–6 hours, replace the medium. Collect cells according to the protocol provided by Promega’s Dual-Luciferase® Reporter Assay System. Measure the firefly luciferase and Renilla luciferase activities in both experimental and control groups, using the ratio of the two to determine the transcriptional activation of the Setd5 gene by Gli3 protein.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8.0 and SPSS 26.0 software. Results are presented as mean ± standard deviation (SD). Differences between groups were compared using the t-test.
Results
Liver Regeneration Was Closely Related to Fatty Acid Oxidation
A 70% hepatectomy model was first established in mice, which were fed with normal diet postoperatively. These mice were sacrificed, and samples were obtained and weighed at 1–3 days after surgery. The liver-to-body weight ratio was successfully restored to near normal levels over a period of time. Interestingly, significant hepatic steatosis was observed at 24 h after liver biopsy (Figure 1A). By reviewing the literature, we identified this phenomenon as transient regeneration-associated steatosis (TRAS), defined as a transient steatosis caused by fatty acid accumulation from the catabolism of peripheral adipose tissues into the liver through circulation.31–33 Hematoxylin and eosin (H&E) staining (Figure 1A) revealed that there was no significant inflammatory infiltration and necrosis during liver regeneration.
Oil Red, Nile Red, and Ki67 staining was performed to verify the authenticity of steatosis and regeneration processes. Based on the most significant steatosis at 24 h (Figure 1B and C) and the most significant Ki67 at 48 h (Figure 1D), we proposed a hypothesis as to whether TRAS depletion was in response to the liver regeneration process (ie, whether enhancing lipid combustion would fuel liver regeneration).34
To identify intrinsic molecular changes during regeneration, we chose 24 h as the main study time point and conducted combined transcriptomic and untargeted metabolomic analysis, with the static normal liver as the control. We found significant differences in metabolism-related genes. (p<0.05) For instance, the most significantly upregulated gene was Atp1a3 (Figure 2A), a catalytic component of active enzyme that catalyzes the hydrolysis of ATP to provide energy.35 Therefore, we performed a gene set variation analysis (GSVA) based on metabolic pathways, with the normal liver as the blank control, and observed a significant upregulation of DNA replication (Figure 2B), further validating liver regeneration at the genetic level.
Differential metabolites during liver regeneration were identified through untargeted metabolomic analysis. The corresponding functions of these differential metabolites were also analyzed by KEGG (Figure 2C) enrichment. The 48-h analysis results compared to the normal liver are presented in Supplementary Figure 1.
By analyzing the results, we concluded that metabolism and regeneration processes were inseparable, suggesting the important role of metabolic reprogramming. The untargeted metabolomic results also indicated the significance of the carnitine synthesis pathway (Figure 2D). Carnitine is closely related to energy metabolism in organisms and promotes fatty acid oxidation.36 Numerous studies have confirmed that enhancing fatty acid oxidation can effectively promote liver regeneration.31,34,37–39
While metabolic reprogramming is often regarded as one of the hallmark features of tumors, regenerating normal hepatocytes also undergo metabolic reprogramming.40 A recent study has reported for the first time that alterations in cardiomyocyte energy metabolism could regenerate the heart.41 EVs have been reported to regulate the metabolism of lipids.42 Therefore, we set the goal of further research on the function of LT-EVPs.
Isolation and Characterization of LT-EVPs
LT-EVPs exist in the vast extracellular environment in the liver and play an important role in regulating the metabolic state and inflammatory response of the liver.43,44 Based on previous studies,26,45–47 it is reasonable to assume that LT-EVPs can mediate the repair of liver homeostasis after liver resection. Morphological analysis using transmission electron microscopy at different magnifications showed that EVPs were spherical lipid-bilayer structures (Figure 3A). The capture rates of CD9, CD63, and CD81 on EVPs by the fully automated fluorescence detection and analysis system indicated the prevalence of markers (Figure 3B). By comparing Western blots, we found that hepatic EVPs in vivo were highly enriched for the canonical markers CD63, Hsp70, and Alix, but not for the intracellular marker calnexin (Figure 3C). Additionally, the main components of EVPs were characterized by nanoparticle size analysis (Figure 3D) and zeta potential analysis (Figure 3E) with a particle size of 123.4 nm (88.9%) and a zeta potential of −30.2 mV. Overall, we revealed the typical physicochemical characteristics of EVPs from their molecular shape, size distribution, zeta potential, and composition of tagged proteins.48
The uptake of EVPs into the hepatocytes was confirmed by in vitro localization assays (Figure 3F). The in vivo results indicated that they could be taken up by the liver tissues through the body’s circulation (Figure 3G). Furthermore, the tracer experiment results indicated the good targeting properties of tissue-derived EVPs based on the homing property,49,50 which has the natural advantages to be developed as a delivery system.
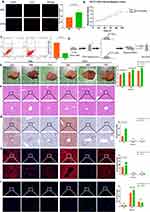
LT-EVPs Promoted Liver Regeneration in vivo and Vitro
We hypothesized that EVPs within liver tissues might contribute to the promotion of liver regeneration. Therefore, we investigated the effects of EVPs on NCTC-1469 cells in vitro. EdU fluorescence staining revealed that the addition of EVPs significantly increased the DNA replication activity and promoted hepatocyte proliferation (Figure 4A, p=0.0048). Additionally, the CCK-8 cell proliferation curve confirmed the improvement in cell proliferation ability (Figure 4B). We also found that EVPs effectively reduced the hepatocyte apoptosis rate (Figure 4C); however, no significant change in the cell cycle occurred (results not shown). The in vitro antiapoptotic effect of EVPs might be attributed to the enhancement of energy metabolism in cells, which is consistent with the findings of previous studies.51,52 The effect of EVPs on liver regeneration after liver resection was investigated in mice (Figure 4D), with no significant difference between EVP and sham group (Supplementary Figure 2, p>0.05).
Interestingly, the liver regeneration rate was significantly increased in the EVP administration group (Figure 4E), and there was no effect on liver inflammatory expression (Figure 4F) and liver injury enzyme levels (Supplementary Figure 3). Notably, the non-estesterified fatty acid (NEFA) levels in the blood of the treated group significantly decreased at 24 h (Supplementary Figure 3, p<0.05), suggesting that NEFA uptake and utilization in the regenerated liver were enhanced.31 The expression of genes involved in fatty acid β-oxidation and carnitine synthesis during regeneration was also more significantly enhanced in the treated group (Supplementary Figure 4, p<0.05). Through Oil Red staining (Figure 4G) and Nile Red staining (Figure 4H), we confirmed the enhanced TRAS dissipation rate within the treated groups. Additionally, the Ki67 fluorescence index of the treated group was significantly enhanced at 48 h (Figure 4I). Overall, these results suggest that exogenous EVPs supplementation increased the NEFA uptake and utilization in the liver postoperatively and, on this basis, facilitated complete regeneration.
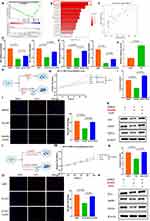
LT-EVPs Promoted Hedgehog Signaling Pathway Activation and Setd5 Transcription for Carnitine Synthesis
Similarly, we compared the transcriptomic and untargeted metabolomic changes at 24 h and 48 h between the two groups. Molecular changes at 24 h were further analyzed in detail and explored, as the earliest and most significant changes occurred at this time node. We examined considerably different molecules at the RNA level, and the results are presented as a volcano plot in Supplementary Figure 5A. Interestingly, the molecule with the largest fold change in upregulation was Cyp2d11, which is present in liver microsomes. This enzyme is involved in the NADPH-dependent electron transport pathway and oxidizes various structurally unrelated compounds such as steroids, fatty acids, and exogenous substances.53 A conserved molecule such as Scd3 also plays a key role in fatty acid metabolism in mammals.54
Using GSVA, we focused on the significant upregulation of the Hedgehog signaling pathway (Supplementary Figure 5B), which was also found on the GSEA (Figure 5A). KEGG analysis revealed significant differences in cell proliferation and energy metabolism, particularly oxidative phosphorylation (Supplementary Figure 5C). The enrichment of biological processes (Supplementary Figure 5D), cellular components (Supplementary Figure 5E), and molecular functions (Supplementary Figure 5F) by GO of differentially expressed genes also showed that cell functions such as energy metabolism, division, and proliferation were significantly enhanced.
Metabolites showing significant differences are presented in a ring heat map in Supplementary Figure 5G, which is more convenient for analyzing changes in related metabolic processes. Interestingly, SMPDB analysis revealed that the most significant changes in metabolic pathways occurred in carnitine synthesis (Figure 5B), suggesting the enhanced effect of exogenous EVP supplementation on metabolic reprogramming.
The 48-h analysis revealed functional differences in bile secretion and xenobiotic metabolism in the treated groups, suggesting a faster recovery of normal liver function (Supplementary Figure 6). Additionally, carnitine synthesis still showed a significant difference at this point.
Gli3 Promoted Downstream Gene Setd5 Transcription and Enhanced Carnitine Synthesis
The Hedgehog signaling pathway is a morphogenetic signaling pathway that controls the fate of progenitor cells and tissue construction during embryogenesis, and reactivation occurs during various types of liver injury in adults.55 One of its prominent roles in mature tissues is the promotion of regeneration.23 Gli3 is the terminal signal node of the Hedgehog signaling pathway. When this pathway is activated, Gli3 is transported from the cytoplasm to the nucleus as a transcription factor to promote the transcription of downstream genes. Findings of previous studies have supported the positive significance of Hedgehog signaling pathway activation for liver regeneration after liver resection;56–58 nonetheless, its relationship with hepatic metabolic reprogramming remains unclear.
Carnitine synthesis appears to be important throughout the liver regeneration process. Hence, we hypothesized that Hedgehog signaling pathway could regulate carnitine synthesis. As a methyltransferase, Setd5 is the initial rate-limiting enzyme in the carnitine synthesis pathway.59 By combining the normal mouse liver sequencing data (GSE233598) from the GEO database with the sequencing data generated during the experiment, we found that Setd5 expression was significantly positively correlated with Gli3 expression through gene correlation analysis (Figure 5C). Subsequently, we verified the hypothesized regulatory relationship between Gli3 as a transcription factor and Setd5 gene transcription in NCTC-1469 cell lines. Using pharmacological modulation, we observed that Gant61, which is widely recognized as a Gli inhibitor,60 inhibited Gli3 RNA expression in hepatocytes and that the inhibitory effect was partially reversed by exogenous EVP supplementation; similar results were obtained from the gene interference experiments (Figure 5D). The RNA expression level of Setd5 also exhibited the same trend (Figure 5E). We verified this relationship using a dual luciferase assay (Figure 5F) and further designed a cell proliferation-related experimental protocol for drug interference (Figure 5G). Based on the CCK-8 cell proliferation curve (Figure 5H), carnitine ELISA detection (Figure 5I) and EdU fluorescence detection (Figure 5J), we confirmed that Gant61 inhibited carnitine synthesis and reduced the energy supply to inhibit hepatocyte proliferation. Additionally, Western blot analysis confirmed the regulatory relationship between Gli3 and Setd5 at the protein level (Figure 5K). The small interference RNA regulation experiments also supported the same conclusion (Figure 5L–P). In this regard, we identified the role of Gli3 as a transcription factor regulating the transcription of Setd5, a key enzyme for carnitine synthesis.
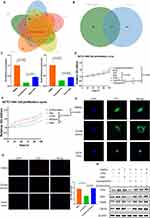
β-sitosterol in EVPs activated Hedgehog signaling through a non-canonical pathway
Based on the above results, we next investigated the specific molecules involved in the activation of Hedgehog signaling by EVPs. A recently published report showed that palmitic acid carried by extracellular vesicles reprogramed the hepatic metabolic state.61 In the absence of Hedgehog ligands, Ptch1 inhibited the activity of the seven-fold transmembrane protein Smoothened (Smo) by reducing its accessibility to obtaining sterols from the inner membrane leaflet.23 By reviewing the literature, we found that sterols could activate the Hedgehog signaling pathway.23,62 Under the guidance of this idea, we analyzed the metabolites within EVPs of independent individual origin. Based on mass spectrometry data, we screened the enriched metabolites in LT-EVPs and found β-sitosterol (Figure 6A). The metabolites are detailed in Supplementary Table 1. From SwissTarget and TargetNet databases were used to predict the potential targets of β-sitosterol, and Smo was identified as a potential target (Figure 6B, Supplementary Table 2). This is consistent with previous studies; nevertheless, the mechanism of action in the liver has not been elucidated.63,64 We employed computer simulation of molecular docking to predict the binding sites of sitosterol to Smo. The score (6.68163538) of this combination mode in the PLANET scoring model was within the range of the benchmark score of the training set, indicating the rationality of the combination.65 Notably, sitosterol was not detected in the regenerated liver tissues from mass spectrometry assay (results not shown). Combined with single-cell database analysis,66 Smo was found to be predominantly present in hepatocytes (Supplementary Figure 7), suggesting that β-sitosterol in EVPs reprograms liver metabolism by regulating hepatocyte function.
Next, we further validated sitosterol for the Hedgehog signaling pathway and the role of liver cell proliferation. We first identified cyclopamine, a known inhibitor of Smo, as a means of interference and verified its inhibitory effect on Gli3 and Setd5 RNA expression levels, which could be partially offset by β-sitosterol (Figure 6C). In the in vitro cell proliferation assay, CCK-8 cell proliferation curve assay was used to verify the effects of SAG hydrochloride, an activator, and cyclopamine, an inhibitor. β-sitosterol had a similar effect, promoted the proliferation as SAG, and partially offset cyclopamine’s inhibition of proliferation effect (Figure 6D). Furthermore, we found that increasing the β-sitosterol content in EVPs could enhance the cell proliferation effect (Figure 6E). Protein nuclear translocation of Gli3 was also inhibited by cyclopamine, while it could be recovered by β-sitosterol (Figure 6F). EdU fluorescence detection confirmed the regulatory relationship with the proliferation effect (Figure 6G). Additionally, Western blot analysis confirmed the regulatory relationship at the protein level (Figure 6H). Taken together, we demonstrated that β-sitosterol in LT-EVPs could activate the Hedgehog signaling pathway by binding to Smo.
Perioperative Sitosterol Diet Contributed to Postoperative Liver Regeneration in Mice
Consistent with the hypothesis, β-sitosterol diet significantly enhanced the liver regeneration rate (Figure 7A and F, p<0.05). H&E staining revealed that the regenerating liver had no obvious inflammatory infiltration and necrotic foci, which was consistent with the experimental results described above. Moreover, lipid droplets dissipated more rapidly in the liver of mice fed with β-sitosterol (Figure 7B). This phenomenon was also confirmed by Oil Red staining (Figure 7C) and Nile Red dyeing (Figure 7D). The Ki67 staining results verified the enhanced liver regeneration rate in the β-sitosterol diet group (Figure 7E). The results for the serum indexes of mice, including liver function enzymology, suggested secure sitosterol dosing; however, free fatty acids showed no obvious difference with the index of previous LT-EVP dosing. This result might be associated with the more abundant content in the EVPs (Supplementary Figure 8).
Discussion
Our present study revealed that liver regeneration after partial hepatectomy was closely related to lipid oxidation and carnitine synthesis. In in vivo and in vitro experimental models, exogenous EVPs effectively promoted liver regeneration. In liver sections, the accelerated dissipation of regrowth-associated steatosis was observed to provide energetic support for liver regeneration.34 For the first time, we confirmed the function of LT-EVPs; that is, they could accelerate liver regeneration after partial hepatectomy.
We investigated how EVPs mediated the acceleration of regeneration and observed that EVPs could be taken up by the hepatocytes after injection. EVPs could stimulate hepatocyte proliferation and reduce the proportion of apoptosis, suggesting that EVPs might play an important role in tissue regeneration after liver resection. Based on a series of untargeted metabolomic analyses, the carnitine synthesis pathway was observed to be consistently enriched as the top pathway in the mouse liver regeneration model. Recent studies have also shown that carnitine can significantly promote regeneration after hepatectomy.37 Functional studies in vivo confirmed that EVPs enhanced fatty acid metabolism after hepatectomy while inducing the acceleration of regeneration. β-sitosterol in EVPs is mainly absorbed by the digestive tract.67,68 In support of our findings, increasing the β-sitosterol content of the diet could promote liver regeneration. These findings collectively suggest that sitosterol is a key beneficial metabolite in hepatic EVPs and contributes to liver regeneration. Significant gaps remain in the study of β-sitosterol regarding liver regeneration post-hepatectomy. Our literature review identified only one mechanistic study focusing on the treatment of hepatic fibrosis using the Chinese herbal preparation Xiao Chai Hu Tang.69 However, this study lacked comprehensive experimental validation. Metabolic reprogramming seems to be the cornerstone for normal liver regeneration; thus, how to correctly distinguish proliferative from neoplastic metabolic reprogramming will also be an interesting topic in the future.
We investigated the acceleration effects of EVPs on liver regeneration in terms of mechanism and found that the Hedgehog signaling pathway was significantly activated and carnitine synthesis-related genes were transcripted after exogenous EVP supplementation. This effect was confirmed through in vitro and in vivo experiments. The Hedgehog signaling pathway plays a main role in the regeneration and repair of mature tissues; therefore, Hedgehog signaling pathway activation by EVPs at least partially explains its positive significance on liver tissue regeneration. We further demonstrated the direct interaction between β-sitosterol in EVPs and the Hedgehog signaling pathway in the liver. We identified Smo as the relevant surface receptor on hepatocytes and further demonstrated the activation of Smo protein by sitosterol. The Smo inhibitor effectively abolished the pro-proliferative effect of sitosterol on hepatocytes. Our findings suggest that EVPs carry sitosterol, which is taken up by the hepatocytes after partial liver resection and binds to Smo to activate the Hedgehog signaling pathway, thereby enhancing hepatic energy supply for regeneration.
We also explored the effect of perioperative dietary supplementation of β-sitosterol on postoperative liver regeneration. The administration of β-sitosterol, a dietary phytosterol, can promote liver regeneration, which has great implications for clinical translation. Previous reports have proposed improved perioperative dietary strategies for the benefit of liver surgery results.70 Therefore, the promotion of β-sitosterol diet during the perioperative period of liver disease has a good prospect. However, this study has certain limitations. Firstly, we have not identified the specific mechanism that promotes the formation of TRAS, which is related to the intrahepatic accumulation of NEFA and involves long-distance communication between the liver and peripheral adipose tissue during liver regeneration.71,72 Secondly, the limited time span of the sample prevented observation of multiple waves of liver fat accumulation and proliferation. This limitation caused difficulties in the kinetic study of liver regeneration.73–75 Further investigation is required to overcome these limitations, such as the use of organoids and organ-on-a-chips. These technologies allow for continuous observation of changes during liver regeneration.76–78 Finally, it is important to note that the liver of patients in clinical practice may not be normal, and concomitant diseases can also impair the liver’s ability to regenerate and repair after trauma. Our group’s future research will focus on verifying whether this method is also beneficial for the liver in different pathological states.
In conclusion, our study revealed that β-sitosterol was a hepato-regenerative component that could bind to the Smo protein on hepatocytes to activate the Hedgehog signaling pathway and promote carnitine synthesis, thereby enhancing liver regeneration. Additionally, EVPs encapsulated metabolites to regulate the metabolic state of the liver and stimulate its regeneration, which could be an effective drug delivery strategy. Therefore, they offer good development prospects for the prevention and treatment of insufficient liver regeneration in the future.
Conclusion
Our in vitro and in vivo experiments have confirmed that LT-EVPs can promote liver regeneration following liver resection. The β-sitosterol contained in these EVPs plays a crucial role in this process. β-sitosterol activates the Hedgehog signaling pathway, enhances carnitine synthesis in the liver, and improves the energy supply to hepatocytes, thereby supporting liver regeneration.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
Animal experiments were performed in accordance with all regulations set by the Chinese Council on Animal Care, and the study protocol was approved by Experimental Animal Welfare Ethics Committee, Zhongnan Hospital of Wuhan University. (Approval No. WP20230052).
Acknowledgments
We would like to express our special thanks to the Institute of Model Animals of Wuhan University for providing technical advice and other help, as well as the noble life dedication of animals for scientific inquiry.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Research Fund of the Health Commission of Hubei Province (grant no. WJ2021M255); Cancer Research and Translational Platform Project of Zhongnan Hospital of Wuhan University (grant no. ZLYNXM202004); Grant from the Key Research; Development Program of Hubei Province, China (grant no. 2021BCA114); Fundamental Research Funds for the Central Universities(grant no.413000411) and Outstanding Doctoral (Postdoctoral) Program of Zhongnan Hospital of Wuhan University(grant no. ZNYB2021021). Dr. Peng Xia was granted the following funding: The research fund from medical Sci-Tech innovation platform of Zhongnan Hospital, Wuhan University (No. PTXM2024029), The China Postdoctoral Science Foundation (No. 2023M742698) and The China National Postdoctoral Program for Innovative Talents (BX20240271)
Disclosure
The authors declare that they have no competing interests.
References
1. Stanger BZ. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol. 2015;77:179–200. doi:10.1146/annurev-physiol-021113-170255
2. Kubes P, Jenne C. Immune Responses in the Liver. Annu Rev Immunol. 2018;36(1):247–277. doi:10.1146/annurev-immunol-051116-052415
3. Power C, Rasko JE. Whither prometheus’ liver? Greek myth and the science of regeneration. Ann Intern Med. 2008;149(6):421–426. doi:10.7326/0003-4819-149-6-200809160-00009
4. Hess S, et al. In vivo partial reprogramming by bacteria promotes adult liver organ growth without fibrosis and tumorigenesis. Cell Rep Med. 2022;3(11):100820. doi:10.1016/j.xcrm.2022.100820
5. Lieber SR, Schiano TD, Rhodes R. Should living donor liver transplantation be an option when deceased donation is not? J Hepatol. 2018;68(5):1076–1082. doi:10.1016/j.jhep.2017.10.024
6. Mathurin P. Early liver transplantation for acute alcoholic hepatitis: we can’t say no. J Hepatol. 2021;75(3):718–722. doi:10.1016/j.jhep.2021.05.019
7. Ma H, Wang C, Liang S, et al. ROCK inhibition enhanced hepatocyte liver engraftment by retaining membrane CD59 and attenuating complement activation. Mol Ther. 2023;31(6):1846–1856. doi:10.1016/j.ymthe.2023.02.018
8. Viswanathan P, Gupta P, Kapoor S, Gupta S. Thalidomide promotes transplanted cell engraftment in the rat liver by modulating inflammation and endothelial integrity. J Hepatol. 2016;65(6):1171–1178. doi:10.1016/j.jhep.2016.07.008
9. Forbes SJ, Gupta S, Dhawan A. Cell therapy for liver disease: from liver transplantation to cell factory. J Hepatol. 2015;62(1):S157–169. doi:10.1016/j.jhep.2015.02.040
10. Herranz-Iturbide M, López-Luque J, Gonzalez-Sanchez E, et al. NADPH oxidase 4 (Nox4) deletion accelerates liver regeneration in mice. Redox Biol. 2021;40:101841. doi:10.1016/j.redox.2020.101841
11. Li H, Zhang L. Liver regeneration microenvironment of hepatocellular carcinoma for prevention and therapy. Oncotarget. 2017;8(1):1805–1813. doi:10.18632/oncotarget.12101
12. Zhang H, Xia T, Xia Z, et al. KIF18A inactivates hepatic stellate cells and alleviates liver fibrosis through the TTC3/Akt/mTOR pathway. Cell Mol Life Sci. 2024;81(96). doi:10.1007/s00018-024-05114-5
13. Liu T, Li J, Li Q, et al. Environmental eustress promotes liver regeneration through the sympathetic regulation of type 1 innate lymphoid cells to increase IL-22 in mice. Hepatology. 2023;78(1):136–149. doi:10.1097/HEP.0000000000000239
14. Huppert SS, Schwartz RE. Multiple Facets of Cellular Homeostasis and Regeneration of the Mammalian Liver. Annu Rev Physiol. 2023;85(1):469–493. doi:10.1146/annurev-physiol-032822-094134
15. de Haan LR, van Golen RF, Heger M. Molecular Pathways Governing the Termination of Liver Regeneration. Pharmacol Rev. 2024;76(3):500–558. doi:10.1124/pharmrev.123.000955
16. Li SR, Man Q-W, Gao X, et al. Tissue-derived extracellular vesicles in cancers and non-cancer diseases: present and future. J Extracell Vesicles. 2021;10(14):e12175. doi:10.1002/jev2.12175
17. Wang J, Li L, Zhang Z, et al. Extracellular vesicles mediate the communication of adipose tissue with brain and promote cognitive impairment associated with insulin resistance. Cell Metab. 2022;34(9):1264–1279e1268. doi:10.1016/j.cmet.2022.08.004
18. Gongye X, Tian M, Xia P, et al. Multi-omics analysis revealed the role of extracellular vesicles in hepatobiliary & pancreatic tumor. J Control Release. 2022;350:11–25. doi:10.1016/j.jconrel.2022.08.010
19. Liao Z, Li S, Lu S, et al. Metformin facilitates mesenchymal stem cell-derived extracellular nanovesicles release and optimizes therapeutic efficacy in intervertebral disc degeneration. Biomaterials. 2021;274:120850. doi:10.1016/j.biomaterials.2021.120850
20. Tian M, Ticer T, Wang Q, et al. Adipose-Derived Biogenic Nanoparticles for Suppression of Inflammation. Small. 2020;16(10):e1904064. doi:10.1002/smll.201904064
21. Gao H, Jin Z, Bandyopadhyay G, et al. Aberrant iron distribution via hepatocyte-stellate cell axis drives liver lipogenesis and fibrosis. Cell Metab. 2022;34(8):1201–1213e1205. doi:10.1016/j.cmet.2022.07.006
22. Dasgupta D, Nakao Y, Mauer AS, et al. IRE1A Stimulates Hepatocyte-Derived Extracellular Vesicles That Promote Inflammation in Mice With Steatohepatitis. Gastroenterology. 2020;159(4):1487–1503e1417. doi:10.1053/j.gastro.2020.06.031
23. Zhang Y, Beachy PA. Cellular and molecular mechanisms of Hedgehog signalling. Nat Rev Mol Cell Biol. 2023;24(9):668–687. doi:10.1038/s41580-023-00591-1
24. Groven RVM, Greven J, Mert Ü, et al. Circulating miRNA expression in extracellular vesicles is associated with specific injuries after multiple trauma and surgical invasiveness. Front Immunol. 2023;14:1273612. doi:10.3389/fimmu.2023.1273612
25. Silva AKA, Perretta S, Perrod G, et al. Thermoresponsive Gel Embedded with Adipose Stem-Cell-Derived Extracellular Vesicles Promotes Esophageal Fistula Healing in a Thermo-Actuated Delivery Strategy. ACS Nano. 2018;12(10):9800–9814. doi:10.1021/acsnano.8b00117
26. Lee J, Kim SR, Lee C, et al. Extracellular vesicles from in vivo liver tissue accelerate recovery of liver necrosis induced by carbon tetrachloride. J Extracell Vesicles. 2021;10(10):e12133. doi:10.1002/jev2.12133
27. Calleri A, Roggio D, Navarro-Tableros V, et al. Protective Effects of Human Liver Stem Cell-Derived Extracellular Vesicles in a Mouse Model of Hepatic Ischemia-Reperfusion Injury. Stem Cell Rev Rep. 2021;17(2):459–470. doi:10.1007/s12015-020-10078-7
28. Abo-Zaid OA, Moawed FS, Ismail ES, Farrag MA. beta-sitosterol attenuates high- fat diet-induced hepatic steatosis in rats by modulating lipid metabolism, inflammation and ER stress pathway. BMC Pharmacol Toxicol. 2023;24(1):31. doi:10.1186/s40360-023-00671-0
29. Feng S, Dai Z, Liu A, et al. beta-Sitosterol and stigmasterol ameliorate dextran sulfate sodium-induced colitis in mice fed a high fat Western-style diet. Food Funct. 2017;8(11):4179–4186. doi:10.1039/c7fo00375g
30. Liu Y, Xu W, Zhai T, You J, Chen Y. Silibinin ameliorates hepatic lipid accumulation and oxidative stress in mice with non-alcoholic steatohepatitis by regulating CFLAR-JNK pathway. Acta Pharm Sin B. 2019;9(4):745–757. doi:10.1016/j.apsb.2019.02.006
31. Qu X, Wen Y, Jiao J, et al. PARK7 deficiency inhibits fatty acid beta-oxidation via PTEN to delay liver regeneration after hepatectomy. Clin Transl Med. 2022;12(9):e1061. doi:10.1002/ctm2.1061
32. Gazit V, Weymann A, Hartman E, et al. Liver regeneration is impaired in lipodystrophic fatty liver dystrophy mice. Hepatology. 2010;52(6):2109–2117. doi:10.1002/hep.23920
33. Walldorf J, Hillebrand C, Aurich H, et al. Propranolol impairs liver regeneration after partial hepatectomy in C57Bl/6-mice by transient attenuation of hepatic lipid accumulation and increased apoptosis. Scand J Gastroenterol. 2010;45(4):468–476. doi:10.3109/00365520903583848
34. Kachaylo E, Tschuor C, Calo N, et al. PTEN Down-Regulation Promotes beta-Oxidation to Fuel Hypertrophic Liver Growth After Hepatectomy in Mice. Hepatology. 2017;66(3):908–921. doi:10.1002/hep.29226
35. You Y, Muraoka S, Jedrychowski MP, et al. Human neural cell type-specific extracellular vesicle proteome defines disease-related molecules associated with activated astrocytes in Alzheimer’s disease brain. J Extracell Vesicles. 2022;11(1):e12183. doi:10.1002/jev2.12183
36. Crentsil V. Mechanistic contribution of carnitine deficiency to geriatric frailty. Ageing Res Rev. 2010;9(3):265–268. doi:10.1016/j.arr.2010.02.005
37. Zhou X, et al. L-carnitine promotes liver regeneration after hepatectomy by enhancing lipid metabolism. J Transl Med. 2023;21(1):487. doi:10.1186/s12967-023-04317-x
38. Kumar S, Verma AK, Rani R, et al. Hepatic Deficiency of Augmenter of Liver Regeneration Predisposes to Nonalcoholic Steatohepatitis and Fibrosis. Hepatology. 2020;72(5):1586–1604. doi:10.1002/hep.31167
39. Kohjima M, Tsai T-H, Tackett BC, et al. Delayed liver regeneration after partial hepatectomy in adipose differentiation related protein-null mice. J Hepatol. 2013;59(6):1246–1254. doi:10.1016/j.jhep.2013.07.025
40. Hall Z, Chiarugi D, Charidemou E, et al. Lipid Remodeling in Hepatocyte Proliferation and Hepatocellular Carcinoma. Hepatology. 2021;73(3):1028–1044. doi:10.1002/hep.31391
41. Li X, Wu F, Günther S, et al. Inhibition of fatty acid oxidation enables heart regeneration in adult mice. Nature. 2023;622(7983):619–626. doi:10.1038/s41586-023-06585-5
42. Gao H, Zeng Y, Huang X, et al. Extracellular vesicles from organoid-derived human retinal progenitor cells prevent lipid overload-induced retinal pigment epithelium injury by regulating fatty acid metabolism. J Extracell Vesicles. 2024;13(1):e12401. doi:10.1002/jev2.12401
43. He Y, et al. Neutrophil-to-hepatocyte communication via LDLR-dependent miR-223-enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. J Clin Invest. 2021;131(3). doi:10.1172/JCI141513
44. Zhao Y, Zhao M-F, Jiang S, et al. Liver governs adipose remodelling via extracellular vesicles in response to lipid overload. Nat Commun. 2020;11(1):719. doi:10.1038/s41467-020-14450-6
45. Driscoll J, Wehrkamp C, Ota Y, et al. Biological Nanotherapeutics for Liver Disease. Hepatology. 2021;74(5):2863–2875. doi:10.1002/hep.31847
46. Povero D, Pinatel EM, Leszczynska A, et al. Human induced pluripotent stem cell-derived extracellular vesicles reduce hepatic stellate cell activation and liver fibrosis. JCI Insight. 2019;5(14). doi:10.1172/jci.insight.125652
47. Borrelli DA, Yankson K, Shukla N, et al. Extracellular vesicle therapeutics for liver disease. J Control Release. 2018;273:86–98. doi:10.1016/j.jconrel.2018.01.022
48. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi:10.1080/20013078.2018.1535750
49. Al faruque H, Choi ES, Kim JH, Kim E. Enhanced effect of autologous EVs delivering paclitaxel in pancreatic cancer. J Control Release. 2022;347:330–346. doi:10.1016/j.jconrel.2022.05.012
50. Pirisinu M, Pham TC, Zhang DX, et al. Extracellular vesicles as natural therapeutic agents and innate drug delivery systems for cancer treatment: recent advances, current obstacles, and challenges for clinical translation. Semin Cancer Biol. 2022;80:340–355. doi:10.1016/j.semcancer.2020.08.007
51. Xue S, Zhou X, Sang W, et al. Cartilage-targeting peptide-modified dual-drug delivery nanoplatform with NIR laser response for osteoarthritis therapy. Bioact Mater. 2021;6(8):2372–2389. doi:10.1016/j.bioactmat.2021.01.017
52. Lee TY, Liu M-S, Huang L-J, et al. Bioenergetic failure correlates with autophagy and apoptosis in rat liver following silver nanoparticle intraperitoneal administration. Part Fibre Toxicol. 2013;10(1). doi:10.1186/1743-8977-10-40
53. Wong G, Itakura T, Kawajiri K, Skow L, Negishi M. Gene family of male-specific testosterone 16 alpha-hydroxylase (C-P-450(16 alpha)) in mice. Organization, differential regulation, and chromosome localization. J Biol Chem. 1989;264(5):2920–2927. doi:10.1016/S0021-9258(19)81700-3
54. Bai Y, McCoy JG, Levin EJ, et al. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015;524(7564):252–256. doi:10.1038/nature14549
55. Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54(2):366–373. doi:10.1016/j.jhep.2010.10.003
56. Papa S, Bubici C. Feeding the Hedgehog: a new meaning for JNK signalling in liver regeneration. J Hepatol. 2018;69(3):572–574. doi:10.1016/j.jhep.2018.05.026
57. Langiewicz M, Graf R, Humar B, Clavien PA. JNK1 induces hedgehog signaling from stellate cells to accelerate liver regeneration in mice. J Hepatol. 2018;69(3):666–675. doi:10.1016/j.jhep.2018.04.017
58. Langiewicz M, Schlegel A, Saponara E, et al. Hedgehog pathway mediates early acceleration of liver regeneration induced by a novel two-staged hepatectomy in mice. J Hepatol. 2017;66(3):560–570. doi:10.1016/j.jhep.2016.10.014
59. Kerner J, Hoppel C. Genetic disorders of carnitine metabolism and their nutritional management. Annu Rev Nutr. 1998;18(1):179–206. doi:10.1146/annurev.nutr.18.1.179
60. Zubcic V, Rinčić N, Kurtović M, et al. GANT61 and Lithium Chloride Inhibit the Growth of Head and Neck Cancer Cell Lines Through the Regulation of GLI3 Processing by GSK3beta. Int J Mol Sci. 2020;21(17):6410. doi:10.3390/ijms21176410
61. Wang G, Li J, Bojmar L, et al. Tumour extracellular vesicles and particles induce liver metabolic dysfunction. Nature. 2023;618(7964):374–382. doi:10.1038/s41586-023-06114-4
62. Deshpande I, Liang J, Hedeen D, et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 2019;571(7764):284–288. doi:10.1038/s41586-019-1355-4
63. Che Omar MT. Data analysis of molecular dynamics simulation trajectories of beta-sitosterol, sonidegib and cholesterol in smoothened protein with the CHARMM36 force field. Data Brief. 2020;33:106350. doi:10.1016/j.dib.2020.106350
64. Truong VL, Bak MJ, Lee C, Jun M, Jeong WS. Hair Regenerative Mechanisms of Red Ginseng Oil and Its Major Components in the Testosterone-Induced Delay of Anagen Entry in C57BL/6 Mice. Molecules. 2017;22(9):1505. doi:10.3390/molecules22091505
65. Zhang X, et al. PLANET: a Multi-objective Graph Neural Network Model for Protein-Ligand Binding Affinity Prediction. J Chem Inf Model. 2023. doi:10.1021/acs.jcim.3c00253
66. Tabula Muris C, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi:10.1038/s41586-018-0590-4
67. Panayotis N, Freund PA, Marvaldi L, et al. beta-sitosterol reduces anxiety and synergizes with established anxiolytic drugs in mice. Cell Rep Med. 2021;2(5):100281. doi:10.1016/j.xcrm.2021.100281
68. Kim HJ, Fan X, Gabbi C, et al. Liver X receptor beta (LXRbeta): a link between beta-sitosterol and amyotrophic lateral sclerosis-Parkinson’s dementia. Proc Natl Acad Sci U S A. 2008;105(6):2094–2099. doi:10.1073/pnas.0711599105
69. Qiang R, Zhang Y, Wang Y. Mechanisms of Xiaochaihu Decoction on Treating Hepatic Fibrosis Explored by Network Pharmacology. Dis Markers. 2022;2022:8925637. doi:10.1155/2022/8925637
70. Cornide-Petronio ME, Alvarez-Mercado AI, Jimenez-Castro MB, Peralta C. Current Knowledge about the Effect of Nutritional Status, Supplemented Nutrition Diet, and Gut Microbiota on Hepatic Ischemia-Reperfusion and Regeneration in Liver Surgery. Nutrients. 2020;12(2):284. doi:10.3390/nu12020284
71. Abshagen K, Degenhardt B, Liebig M, et al. Liver-specific Repin1 deficiency impairs transient hepatic steatosis in liver regeneration. Sci Rep. 2018;8(1):16858. doi:10.1038/s41598-018-35325-3
72. Rudnick DA, Davidson NO. Functional Relationships between Lipid Metabolism and Liver Regeneration. Int J Hepatol. 2012;2012:549241. doi:10.1155/2012/549241
73. Chen T, Oh S, Gregory S, Shen X, Diehl AM. Single-cell omics analysis reveals functional diversification of hepatocytes during liver regeneration. JCI Insight. 2020;5(22). doi:10.1172/jci.insight.141024
74. Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18(1):40–55. doi:10.1038/s41575-020-0342-4
75. Zou Y, Bao Q, Kumar S, et al. Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS One. 2012;7(2):e30675. doi:10.1371/journal.pone.0030675
76. Fahrner R, Groger M, Settmacher U, Mosig AS. Functional integration of natural killer cells in a microfluidically perfused liver on-a-chip model. BMC Res Notes. 2023;16(1):285. doi:10.1186/s13104-023-06575-w
77. Yin Y, Sichler A, Ecker J, et al. Gut microbiota promote liver regeneration through hepatic membrane phospholipid biosynthesis. J Hepatol. 2023;78(4):820–835. doi:10.1016/j.jhep.2022.12.028
78. Danoy M, et al. Multi-omics analysis of hiPSCs-derived HLCs matured on-chip revealed patterns typical of liver regeneration. Biotechnol Bioeng. 2021;118(10):3716–3732. doi:10.1002/bit.27667
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.