Back to Journals » Cancer Management and Research » Volume 17
Long Non-Coding RNA TPTEP1 Exerts Tumor Suppressive Functions via Sequestering miR-4295 to Regulate Growth Arrest and DNA Damage-Inducible 45α Expression in Acute Myeloid Leukemia
Authors Liu X, Gui Y, Wang C, Xia K, Yang P
Received 13 July 2024
Accepted for publication 17 April 2025
Published 4 June 2025 Volume 2025:17 Pages 1059—1072
DOI https://doi.org/10.2147/CMAR.S486875
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Xueni Liu, Yonghui Gui, Chao Wang, Kang Xia, Peng Yang
Department of Blood Transfusion, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230001, People’s Republic of China
Correspondence: Peng Yang, Department of Blood Transfusion, The First Affiliated Hospital of Anhui Medical University, 218 JiXi Road, Hefei, 230001, Anhui, People’s Republic of China, Tel/Fax +86-55165908621, Email [email protected]
Introduction: Recent evidences show that long non-coding RNA (lncRNA) plays an essential role in physiology and pathophysiology. The purpose of this study was to determine the role and its potential underlying mechanisms of the lncRNA transmembrane phosphatase with tensin homology pseudogene 1 (TPTEP1) in acute myeloid leukemia (AML).
Methods: We detected the TPTEP1 and miR-4295 expression levels in AML cells. The vitro effects of TPTEP1 and miR-4295 on AML cells were analyzed. The correlation between miR-4295 and TPTEP1 or Growth arrest and DNA damage-inducible 45α (GADD45α) was confirmed by a luciferase reporter assay and RNA immunoprecipitation. The expression of GADD45α was investigated by Western blotting.
Results: TPTEP1 was down-regulated in AML cell lines and AML patients. Ectopic expression of TPTEP1 inhibited AML cells proliferation while promoted cells apoptosis. And we found that silencing miR-4295 produced the similar effect of TPTEP1 overexpression. TPTEP1 regulated the malignant behavior of AML cells by binding to miR-4295. In addition, overexpression of TPTEP up-regulated GADD45α, a direct target of miR-4295 which play a suppressive role in AML cells. Moreover, when AML cell lines were treated with a DNA methylation inhibitor, TPTEP1 expression was up-regulated.
Conclusion: This study reveals that the lncRNA TPTEP1 regulates the expression of GADD45α by sponging miR-4295 in AML cells, which may represent a novel therapeutic target for AML.
Keywords: acute myeloid leukemia, TPTEP1, miR-4295, GADD45α, methylation
Introduction
Acute myeloid leukemia (AML) is a heterogeneous malignancy characterized by uncontrolled proliferation of immature myeloid cells in peripheral blood and bone marrow.1,2 Currently, allogeneic hematopoietic stem cell transplantation and chemotherapy-based regimens still two common choices for the majority of AML patients. Despite significant progress in the therapeutic approaches of AML, the prognosis remains unideal. The 5-year survival for AML patients under the age of 60 years and older is no more than 50% and under 20%, respectively.2 A better understanding of AML’s molecular mechanisms and alternative therapeutic targets are urgently needed.
Long non-coding RNA (lncRNA) are RNA molecules over 200 nucleotides long that regulate gene expression.3,4 The abnormal lncRNA expression has been proven to be a key factor in the development of various tumors.5–7 The lncRNA transmembrane phosphatase with tensin homology pseudogene1 (TPTEP1) is involved in the progression of various human cancers, including hepatocellular and lung cancer. Cao et al8 identified that TPTEP1 inhibited NSCLC cells proliferation by impairing miR-328-5p. Dong et al9 found that hepatocellular carcinoma progression was enhanced via TPTEP1-mediated modulation of the miR-454-3p/DLG5 axis. Based on these data, we hypothesized that TPTEP1 expression significantly impacts the leukemogenesis of AML.
LncRNAs, as competitive endogenous RNA (ceRNAs), affect the expression of target genes through competitive binding specific microRNAs (miRNAs).10–12 Moreover, miRNAs play a critical role in regulating hematopoiesis, and their abnormal expression is associated with the pathogenesis of leukemia.13,14 Reference studies have found that miR-4295 functions as an oncomiR in glioma and osteosarcoma.15,16 Through bioinformatics analysis, we found that there may be an unstudied targeting relationship between miR-4295 and TPTEP1.
Growth arrest and DNA damage-inducible 45α (GADD45α), a downstream target gene of P53, is involved in cell cycle arrest, DNA demethylation, energy metabolism and tissue development.17,18 GADD45α is an important tumor suppressor, which is often expressed as genetic silencing in various cancer entities.18–20 Recent studies have shown that GADD45α was often down-regulated in chronic myeloid leukemia (CML) cells, up-regulation of GADD45α can enhance the sensitivity of CML cells to imatinib,21 while its biological role in AML cells still needs to be explored.
In the current study, we sought to define the role of TPTEP1/miR-4295/GADD45ɑ pathway in AML leukemogenesis and progression to fill some gaps in our knowledge of AML pathogenesis.
Materials and Methods
Patients and Normal Controls
Thirty-five newly diagnosed AML patients and 21 healthy controls were enrolled in this study (Supplementary Table 1). Peripheral whole blood samples from patients and healthy controls were collected in our hospital. The blood samples were subjected to erythrocyte lysis using erythrocyte lysis buffer (Qiagen, Hilden, Germany) to eliminate red blood cells. Subsequently, the samples were centrifuged to extract nucleated cells, which were then transferred to an Eppendorf tube and stored at −80 °C until further use.
Cell Culture and 5-Aza-2’-Deoxycytidine Treatment
Human AML cell lines SHI-1, THP-1, HL-60 and U937 were purchased from the China Center for Type Culture Collection (Shanghai, China). These four cell lines were cultured at 37°C with 5% CO2 in RPMI-1640 medium (Gibco, Carlsbad, USA) containing 10% fetal bovine serum (FBS, Gibco). The CD34+ hematopoietic stem and progenitor cell (CD34+) were kindly provided Dr. Qingsheng Li (First Affiliated Hospital, Anhui Medical University, Hefei, China).
AML cell lines were cultured in the same medium, treated with 5-aza-2’-deoxycytidine (5-AZA-CdR, a DNA methylation inhibitor) (Sigma, USA) at final concentration at 5 μM for 72 h. The cells were then harvested for isolation of total RNA or protein.
Bioinformatics Analysis
Expression patterns of TPTEP1 in the Cancer Genome Atlas cancer samples and survival charts of AML patients were obtained through Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) online database. The targeting relationships between TPTEP1 and miR-4295 were predicted with Starbase (https://rnasysu.com/encori/). TargetScan online software (http://www.targetscan.org) was performed to predict potential target genes of miR-4295.
Construction of Stable Cell Lines
To obtain cell lines stably overexpressing TPTEP1, U937 and HL-60 cells were infected with lentivirus carrying TPTEP1 gene (Lv-TPTEP1) or control lentivirus (Lv-control) (Genepharma, Shanghai, China) and screened by puromycin (Solarbio, Beijing, China). The efficiency of TPTEP1 overexpression was confirmed by qRT-PCR.
Cell Transfection and Ara-c Treatment
miRNA-4295 inhibitor (inhibitor), miR-4295 mimic, negative control (NC), pcDNA3.1-GADD45α were purchased from GenePharma (Shanghai, China). HL-60 and U973 cells were incubated in 6-well dishes (3.0×105/well), and then transfected with inhibitor, miR-4295 mimic, NC and pcDNA3.1-GADD45α at a final concentration of 50 nM singly or in combination using lipofectamine 3000 (Invitrogen, Carlsbad, USA). Protein assays, soft-agar colony formation, and qRT-PCR analyses were performed at 48 h after transfection. Cells were treated with 5 μg/mL Ara-c at 24 h after transfection. Ara-c treatment for 24 h, cell apoptosis was analyzed.
Cell Proliferation, Soft-Agar Colony Formation Assay, and Cell Apoptosis Analyses
Cell proliferation was assessed using the cell counting kit-8 (CCK-8) kit (Dojindo, Kumamoto, Japan). Briefly, 3.0×104 cells were inoculated into each well of 96-well plate and then treated at the indicate times. Cells were incubated in 10 μL CCK-8 solution. Absorbance at 450 nm was measured using a microtiter plate reader.
Cells were resuspended in RPMI-1640 medium containing 20% FBS with equal volume of 0.3% agar, and seeded in 6-well plates precasted 1.4 mL 1% agar at 5,000 cells per well. The colonies were counted and imaged 14 days later. Similarly, the cell apoptosis assays were performed in HL-60 and U937 cells by flow cytometry (BD Biosciences) using the Annexin V-FITC/PI Apoptosis Detection Kit (Beyotime, Nanjing, China).
Quantitative RT-PCR Assays
Total RNA was extracted using TRIZOL regents (Invitrogen, Santa Cruz, CA). cDNA was synthesized with random primers and Power SYBR Green PCR Master Mix. Level of miR-4295 was measured using a Hairpin-it miRNA qRT-PCR Quantitation Kit (GenePharma,Shanghai, China) according to manufacturer’s instructions. GAPDH and U6 were used as internal controls. The primer sequences are listed in Supplementary Table 2.
Luciferase Reporter Assay
Cells (1×105cells/well) were transfected with psi-CHECK2 vector containing TPTEP1-wt/mut or GADD45α-wt/mut 3’UTRs with 50 nM miR-4295 mimic or NC. The relative luciferase activity was measured 48 h post-transfection using the dual-luciferase reporter assay (Promega, Beijing, China)). The relative luciferase activity was evaluated 48 h after transfection using a Dual‐Luciferase® Reporter Assay System (Promega).
Western Blot Analysis
Cultured cells were harvested and lysed in buffer containing protease inhibitor (Roche, Mannheim, Germany). Equal amounts of protein (60 μg) were separated on 12% SDS-PAGE gels and then transferred onto PVDF membranes. Subsequently, the PVDF membranes were incubated with antibodies against GADD45α (1:1000, Cell Signaling Technology, Boston, USA), or β-actin (1:10000, Santa Cruz Biotechnology, Santa Cruz, CA). Subsequently, the membranes were extensively washed and incubated with HRP-conjugated secondary antibodies (1:1000; Sigma-Aldrich, St Louis, MO, USA). Signals were visualized with the enhanced chemiluminescence system (ComWin, Beijing, China). β-actin was used as an endogenous control.
RNA Immunoprecipitation (RIP) Assay
A RIP assay was performed with an RNA-Binding Protein Immunoprecipitation Kit according to the manufacturer’s protocol. The anti-Ago2 antibody used in this assay was purchased from Millipore. After RNA immunoprecipitation, the expression of TPTEP1 and miR-4295 was measured by qRT-PCR as previously described.
Statistical Analysis
All experiments were repeated at least three times, and all data were presented as the means ± SD. GraphPad Prism version 8.0 was used to analyze the date and draw the graphs. Student’s t test was used for comparisons between the two groups. All statistical tests were two-sided, and significance was assigned at P < 0.05.
Results
TPTEP1 Was Down-Regulated in AML and Was Associated With Poor Prognosis
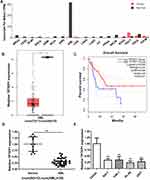
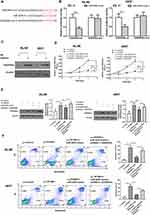
To investigate the role of TPTEP1 in AML, we examined its expression in various malignancies using the GEPIA database (Figure 1A). The expression levels of TPTEP1 were significantly lower in AML samples compared to healthy controls (Figure 1B). Moreover, GEPIA data exhibited that lower TPTEP1 expression was associated with poorer overall survival (OS) in AML patients after 5 years (Figure 1C). In addition, the relative expression levels of TPTEP1 in AML specimen and normal specimen were detected by qRT-PCR. Specifically, we found that the levels of TPTEP1 in 35 AML peripheral blood samples at the time of diagnosis were significantly lower than those in 12 healthy controls (Figure 1D). TPTEP1 expression was lower in AML cell lines than in hematopoietic stem cells, as observed in SHI-1, THP-1, HL-60, U937 and CD34+ cells (Figure 1E). These results indicate that TPTEP1 might play a critical role in the progress of AML.
Overexpression of TPTEP1 Inhibited AML Cells Proliferation and Promoted Cell Apoptosis
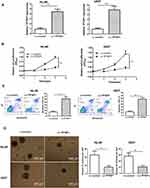
After evaluating TPTEP1 expression in AML, we further investigated its biological function by establishing TPTEP1 overexpression (Lv-TPTEP1) and control (Lv-control) cell lines through lentiviral infection in HL-60 and U937 cells. qRT-PCR results showed that the expression of TPTEP1 in AML cells was significantly increased after infection with Lv-TPTEP1 (Figure 2A). A CCK-8 proliferation assay showed that overexpression of TPTEP1 decreased cell growth in both HL-60 and U937 cells (Figure 2B). Additionally, overexpression of TPTEP1 significantly increased the proapoptotic effect of Ara-c (Figure 2C). Next, we tested whether the expression level of TPTEP1 is associated with clonogenicity in AML cells. The increase in TPTEP1 expression was coupled with attenuated colony formation of HL-60 and U937 cells (Figure 2D).
Down-Regulation of miR-4295 Inhibit AML Cells Proliferation and Induce Cell Apoptosis
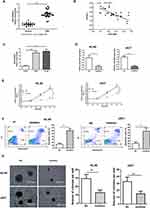
LncRNA may act as a ceRNA by sponging certain miRNAs to exert their biological functions.10–12 Thus, Starbase online database was applied to analyze the miRNA that potentially interacts with TPTEP1. Among the potential targets of TPTEP1, we focused on miR-4295, which has a complementary base sequence with TPTEP1. We found that miR-4295 level in 30 peripheral blood samples from AML patients at diagnosis time and two AML cell lines was highly expressed, but it was not in healthy controls and normal CD34+ cell (Figure 3A and C), and was negatively correlated with TPTEP1 level (Figure 3B). To further explore the biological role of miR-4295 in AML, cell proliferation was first examined using a loss-of-function approach. miR-4295 inhibitor or NC were transfected into in AML cells, respectively. Successful decreases in miR-4295 expression were detected using qRT-PCR (Figure 3D). The CCK-8 assay showed that inhibition of miR-4295 decreased the growth of AML cells (Figure 3E). Accordingly, miR-4295 suppression in both HL-60 and U937 cells promoted Ara-C-induced apoptosis of AML cells (Figure 3F). Furthermore, AML cells transfected with miR-4295 inhibitor formed smaller and fewer colonies, than NC transfected cells (Figure 3G). These data implied that miR-4295 might exert a carcinogenic role in AML.
TPTEP1 had a Negative Correlation With miR-4295
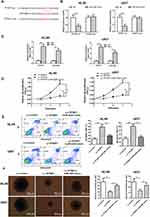
Starbase database was applied to analyze the miRNA that potentially interacts with TPTEP1. We focused on miR-4295, which has a complementary base sequence with TPTEP1 (Figure 4A). To further elucidate the interaction between TPTEP1 and miR-4295, the luciferase assays were performed. The results uncovered that merely the luciferase activity of TPTEP1-wt was lessened by miR-4295 (Figure 4B). Moreover, RIP assay showed that TPTEP1 and miR-4295 were preferentially enriched in the immunoprecipitates containing Ago2 relative to control IgG (Figure 4C). In the above experimental data, inhibition of miR-4295 suppressed proliferation and increased apoptosis, which was consistent with the effect of TPTEP1 overexpression. We next investigated whether there was a functional association between miR-4295 and TPTEP1. The CCK-8 and AnnexinV/PI assays showed that the enrichment of TPTEP1 reduced proliferation and increased apoptosis in AML cells, this effect was partially neutralized by miR-4295 mimic (Figure 4D and E). Clonal formation assay displayed similar results (Figure 4F). Taken together, these data support the notion that TPTEP1 might affect the cellular processes of AML cells in a miR-4295-participate pattern.
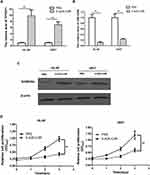
TPTEP1 Induced GADD45α up-Regulation by Inhibiting miR-4295
According to TargetScan analysis, we have identified GADD45α as a potential target mRNA of miR-4295 in AML. To verify this hypothesis, we cloned GADD45α wt/mut 3’-UTRs containing the miR-4295 binding site into the Psi-Check2 reporter (Figure 5A). miR-4295 inhibited luciferase activity in HL-60 and U937 cells transfected with the GADD45α wild-type 3’-UTR, but not with the GADD45α mutant 3’-UTR reporter (Figure 5B). Then, we assessed the effect of miR-4295 on GADD45α expression. The miR-4295 inhibitor was transfected into HL-60 and U937 cells, which express high levels of endogenous miR-4295 (Figure 3C). The knockdown of miR-4295 dramatically up-regulated the protein expression level of GADD45α in AML cell lines (Figure 5C). Furthermore, we transfected miR-4295 mimic into these AML cells, which were infected with Lv-TPTEP1. The Western blot assay showed that TPTEP1 upregulation significantly increased GADD45α protein expression in AML cells, and miR-4295 partially counteracted this effect (Figure 5E). Based on the above experimental results, we propose that TPTEP1 functions in AML cells through the miR-4295/GADD45α pathway. The results showed that the overexpression of TPTEP1 significantly decreased the proliferation of these AML cell lines, and miR-4295 could neutralize the suppressive effects. More importantly, GADD45α overexpression significantly decreased cell proliferation, while miR-4295 overexpression had opposite effect (Figure 5D). A parallel phenomenon was provided in a cell apoptosis assay (Figure 5F).
Up-Regulation of TPTEP1 Expression Mediated by 5-AZA-CdR
DNA methylation is an epigenetic modification that maintains gene stability and regulates gene expression. Specially, abnormal DNA promoter methylation is a crucial mechanism of gene function loss in carcinomas. It has been reported that DNA methylation regulates the position of TPTEP1 on chromosome 22q11.1.22 We hypothesized that hypermethylation might be responsible for the low expression of TPTEP1 in AML. We treated with 5-AZA-CdR in both HL60 and U937 cells for 72 h. The results showed that 5-AZA-CdR significantly increased the expression of TPTEP1 and GADD45α (Figure 6A and C), reduced the expression of miR-4295 (Figure 6B), simultaneously suppressed cell proliferation (Figure 6D) in AML cells.
In summary, these experimental findings support the idea that hypermethylation in AML suppresses TPTEP1 expression, elevates miR-4295 levels, and inhibits GADD45α expression to facilitate AML proliferation.
Discussion
Former work revealed that TPTEP1 could suppress AML cell proliferation by inactivating c-Jun N-terminal kinase (JNK)/c-JUN signaling pathway.23 However, little is known about the role of the TPTEP1/miR-4295/GADD45α axis in AML and the mechanism underlying TPTEP1 under-expression. In this study, we investigated the expression and role of TPTEP1 in AML based on multiple experiments. Our data showed a down-regulation of TPTEP in AML samples and AML cell lines. In addition, we demonstrated that ectopic expression of TPTEP1 increased GADD45α expression, a tumor suppressor, by competitively binding miR-4295, which resulted in the demotion of AML cell malignancy. We found that miR-4295 promotes proliferation and inhibits apoptosis by directly targeting GADD45α. We demonstrated the inverse relationship between miR-4295 and TPTEP1 expression, as well as miR-4295 and GADD45α expression in AML samples, establishing the significance of TPTEP1-miR-4295-GADD45α in malignant progression. LncRNA has presented as a key regulator of hematopoietic activity and its aberrant expression is related to the pathogenesis of leukemia.10,24 For instance, AML patients with high levels of 10 lncRNAs exhibit poorer clinical outcomes and shorter survival rates compared to those with low levels. The set includes DIRC3-AS1, AC004223.2, AC067735.1, AL355353.1, AL645608.1, AC025430.1, AF064858.2, AL645608.5, FP671120.3, and AC107398.3.25 HOTARM1 was decreased in AML-M3 patients compared with non-AML-M3 patients,26 whereas AML-M5 patients had higher levels of MALAT1 than other AML patients.27 Due to the limited number of de novo AML patients enrolled in the present study, we cannot conclude that TPTEP1 expression level is specific to AML subtypes.
High-throughput information stored in TCGA and GEPIA databases has been used to comprehensively analyze malignant mechanisms of tumors.28–32 Through GEPIA database, we found that TPTEP1 expression level in AML patients was significantly lower than that in healthy people, and its expression level was positively correlated with the overall survival rate of AML patients. Then, our data confirmed low expression level of TPTEP1 and indicated that TPTEP1 might play certain role in AML progression. Although, the role of miR-4295 in the progress of AML remains unclear. Recently, emerging data have reported that miR-4295 were abnormally elevated in multiple cancers, such as glioma, osteosarcoma, and promoted cancer cells proliferation, migration and invasion.15,16 Interestingly, we found that up-regulation of miR-4295 expression was a common event in both primary AML patients and AML cell lines. Through the analysis of miRNA-target interaction and expression profile, we found that miR-4295 not only had binding sites in GADD45α and TPTEP1, but also showed negative expression relationship with both GADD45α and TPTEP1. Therefore, we focused on the functional exploration of the role of TPTEP1-miR-4295-GADD45α in the malignancy of AML.
DNA methylation is one of the important epigenetic modifications means in gene expression regulation, involving covalent binding of methyl groups catalyzed by DNA methyltransferase to CpG cytosine.33,34 In many cancers, alteration of DNA methylation level leads to tumor suppressor gene abnormal silencing and protooncogenes transcription activation, contributes to oncology progression.35,36 For example, increased methylation upstream of the PRR34-AS1 promotor was observed in AML patients with poor overall survival,37 whereas aberrant expression and hypomethylation of MEG3 was associated with unfavorable outcomes in AML.38 We hypothesize that TPTEP1 also plays a tumor suppressor role in AML, as in non-small cell lung cancer,8 hepatocellular carcinoma,9 colorectal cancer and glioma.39,40 Hypermethylation may contribute to the diminution of TPTEP1, because TPTEP1 expression was significantly increased in AML cell lines treated with 5-AZA-CdR.
Taken together, TPTEP1 may act as a tumor suppressor, and hypermethylation of TPTEP1 leads to the derepression of miR-4295 expression, thus inhibits the expression of GADD45α and consequently contributes to AML development and progression. These findings may shed new light on basic medical research and clinical application for AML.
CRediT Authorship Contribution Statement
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Data Sharing Statement
The datasets upon which the conclusions of this study are available from the corresponding author on reasonable request.
Ethical Declarations
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Medical Ethics Committee and the Human Clinical Trial Committee of Anhui Medical University, with the approval number (20200928). All participants signed the Informed Consent Form.
Funding
This work was supported by Anhui Provincial Natural Science Foundation (#1808085MH273).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nepstad I, Hatfield KJ, Grønningsæter IS, Reikvam H. The PI3K-Akt-mTOR signaling pathway in human Acute Myeloid Leukemia (AML) cells. Int J mol Sci. 2020;21:2907. doi:10.3390/ijms21082907
2. Reville PK, Kadia TM. Maintenance therapy in AML. Front Oncol. 2020;10:619085. doi:10.3389/fonc.2020.619085
3. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi:10.1038/nrg.2015.10
4. Sanchez CA, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi:10.1111/cas.13642
5. Qin Y, Sun W, Zhang H, et al. LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer. Endocrine. 2018;59:555–564. doi:10.1007/s12020-017-1520-1
6. Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–787. doi:10.4161/rna.28828
7. Zhu D, Yu Y, Wang W, et al.. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 2019;8:6064–6081. doi:10.1002/cam4.2494
8. Cao F, Wang Z, Feng Y, et al. lncRNA TPTEP1 competitively sponges miR-328-5p to inhibit the proliferation of non‑small cell lung cancer cells. Oncol Rep. 2020;43:1606–1618. doi:10.3892/or.2020.7522
9. Dong Y, Wang Q, Sun J, Liu H, Wang H. Long non-coding RNA TPTEP1 exerts inhibitory effects on hepatocellular carcinoma by impairing microRNA-454-3p-mediated DLG5 downregulation. Dig Liver Dis. 2022;54:268–279. doi:10.1016/j.dld.2021.04.014
10. Dahariya S, i P, Kumar S, et al. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. doi:10.1016/j.molimm.2019.04.011
11. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinf. 2017;15:177–186. doi:10.1016/j.gpb.2016.12.005
12. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi:10.4161/rna.20481
13. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi:10.1038/nrg1379
14. Memari F, Joneidi Z, Taheri B, et al. Epigenetics and Epi-miRNAs: potential markers/therapeutics in leukemia. Biomed Pharmacother. 2018;106:1668–1677. doi:10.1016/j.biopha.2018.07.133
15. Li X, Zheng J, Diao H, Liu Y. RUNX3 is down-regulated in glioma by Myc-regulated miR-4295. J Cell Mol Med. 2016;20:518–525. doi:10.1111/jcmm.12736
16. Cheng JP, Huang B, Duan JH, Yi KJ, Zhuang ZL. miR-4295 promotes cell proliferation, migration and invasion of osteosarcoma through targeting interferon regulatory factor. Oncol Lett. 2020;20:260. doi:10.3892/ol.2020.12123
17. Amanullah A, Azam N, Balliet A, et al.. Cell signalling: cell survival and a Gadd45-factor deficiency. Nature. 2003;424(741). doi:10.1038/424741b
18. Hoffman B, Liebermann DA. Gadd45 in modulation of solid tumors and leukemia. Adv Exp Med Biol. 2013;793:21–33. doi:10.1007/978-1-4614-8289-5_2
19. Hoffman B, Liebermann DA. Gadd45 modulation of intrinsic and extrinsic stress responses in myeloid cells. J Cell Physiol. 2009;218:26–31. doi:10.1002/jcp.21582
20. Mancini M, Leo E, Aluigi M, et al.. Gadd45a transcriptional induction elicited by the Aurora kinase inhibitor MK-0457 in Bcr-Abl-expressing cells is driven by Oct-1 transcription factor. Leuk Res. 2012;36:1028–1034. doi:10.1016/j.leukres.2012.03.025
21. Liu X, Wang C, Cheng Y, Yang P. Upregulation of GADD45α by Lentivirus vector enhances the drug sensitivity of imatinib-resistant K562IR cells. Acta Universitatis Med Anhui. 2021;56:1920–1924. doi:10.19405/j.cnki.issn1000-1492.2021.12.014
22. Liang Q, Ding J, Xu R, Xu Z, Zheng S. The novel human endogenous retrovirus-related gene, psiTPTE22-HERV, is silenced by DNA methylation in cancers. Int, J Cancer. 2010;127:1833–1843. doi:10.1002/ijc.25213
23. Li L, Zhao W. The mutual regulatory loop between TPTEP1 and miR-1303 in leukemogenesis of acute myeloid leukemia. Cancer Cell Int. 2021;21:260. doi:10.1186/s12935-021-01966-0
24. Bhat AA, Younes SN, Raza SS, et al.. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. mol Cancer. 2020;19:57. doi:10.1186/s12943-020-01175-9
25. Tang P, Xie M, Wei Y, et al. A 10-long non-coding rna-based expression signature as a potential biomarker for prognosis of acute myeloid leukemia. Med Sci Monit. 2019;25:4999–5004. doi:10.12659/MSM.917182
26. Díaz-beyá M, Brunet S, Nomdedéu J, et al.. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6:31613–31627. doi:10.18632/oncotarget.5148
27. Huang JL, Liu W, Tian LH, et al.. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol Rep. 2017;38:1353–1362. doi:10.3892/or.2017.5802
28. Chen C, Liang C, Wang S, et al.. Expression patterns of immune checkpoints in acute myeloid leukemia. J Hematol Oncol. 2020;13:28. doi:10.1186/s13045-020-00853-x
29. Giordano TJ. The cancer genome atlas research network: a sight to behold. Endocr Pathol. 2014;25:362–365. doi:10.1007/s12022-014-9345-4
30. Li F, Guo P, Dong K, Guo P, Wang H, Lv X. Identification of key biomarkers and potential molecular mechanisms in renal cell carcinoma by bioinformatics analysis. J Comput Biol. 2019;26:1278–1295. doi:10.1089/cmb.2019.0145
31. Liu XS, Zhou LM, Yuan LL, et al.. NPM1 is a prognostic biomarker involved in immune infiltration of lung adenocarcinoma and associated With m6A modification and glycolysis. Front Immunol. 2021;12:724741. doi:10.3389/fimmu.2021.724741
32. Zimta AA, Tomuleasa C, Sahnoune I, Calin GA, Berindan-neagoe I. Long non-coding RNAs in myeloid malignancies. Front Oncol. 2019;9:1048. doi:10.3389/fonc.2019.01048
33. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi:10.1016/j.cell.2012.06.013
34. Skvortsova K, Stirzaker C, Taberlay P. The DNA methylation landscape in cancer. Essays Biochem. 2019;63:797–811. doi:10.1042/EBC20190037
35. Klutstein M, Nejman D, Greenfield R, Cedar H. DNA Methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi:10.1158/0008-5472.CAN-15-3278
36. Koch A, Joosten SC, Feng Z, et al.. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459–466. doi:10.1038/s41571-018-0004-4
37. Nan FY, Gu Y, Xu ZJ, et al.. Abnormal expression and methylation of PRR34-AS1 are associated with adverse outcomes in acute myeloid leukemia. Cancer Med. 2021;10:5283–5296. doi:10.1002/cam4.4085
38. Sellers ZP, Bolkun L, Kloczko J, et al.. Increased methylation upstream of the MEG3 promotor is observed in acute myeloid leukemia patients with better overall survival. Clin Clin Epigenet. 2019;11:50. doi:10.1186/s13148-019-0643-z
39. Fattahi F, Kiani J, Alemrajabi M, et al.. Overexpression of DDIT4 and TPTEP1 are associated with metastasis and advanced stages in colorectal cancer patients: a study utilizing bioinformatics prediction and experimental validation. Cancer Cell Int. 2021;21(1):303. doi:10.1186/s12935-021-02002-x
40. Tang T, Wang LX, Yang ML, Zhang RM. lncRNA TPTEP1 inhibits stemness and radioresistance of glioma through miR‑106a‑5p‑mediated P38 MAPK signaling. Mol Med Rep. 2020;22:4857–4867. doi:10.3892/mmr.2020.11542
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.