Back to Journals » Infection and Drug Resistance » Volume 18
Major Predominant Serotypes and Virulence Genes and Antibiotic Resistance Characteristics of Klebsiella pneumoniae Clinical Isolates in Middle and East China
Authors Pan WK, Chen SN, Yang MJ, Tao LP, Wang MQ, Zhang XW, Xu YH, Yan J, Qin JF, Sun AH
Received 23 October 2024
Accepted for publication 26 February 2025
Published 13 March 2025 Volume 2025:18 Pages 1451—1464
DOI https://doi.org/10.2147/IDR.S502323
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wang-Kai Pan,1,* Sui-Ning Chen,1,* Mei-Juan Yang,2,* Liang-Ping Tao,3 Mei-Qi Wang,4 Xin-Wei Zhang,5 Yin-Hai Xu,6 Jie Yan,7 Jiang-Feng Qin,1 Ai-Hua Sun1
1School of Basic Medical Sciences and Forensic Medicine, Hangzhou Medical College, Hangzhou, Zhejiang, 310053, People’s Republic of China; 2The First Hospital of Putian City, Putian, Fujian, 351199, People’s Republic of China; 3The First People’s Hospital of Chuzhou City, Chuzhou, Anhui, 239001, People’s Republic of China; 4The People’s Hospital of Xiuning County, Xiuning, Anhui, 245499, People’s Republic of China; 5The First Affiliated Hospital of Henan University, Kaifeng, Henan, 475001, People’s Republic of China; 6The Affiliated Hospital of Xuzhou Medical University, Xuzhou, Jiangsu, 221002, People’s Republic of China; 7Zhejiang Provincial Society for Microbiology, Hangzhou, Zhejiang, 310058, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ai-Hua Sun; Jiang-Feng Qin, Email [email protected]; [email protected]
Background: Klebsiella pneumoniae is a common opportunistic pathogen. Predominant serotypes, virulence genes, and resistance characteristics of K. pneumoniae isolates from patients in different regions of China require further investigation.
Methods: K. pneumoniae isolates from patients and healthy individuals in middle and east China were identified using an auto-bacterial detector. Major serotypes and virulence genes in the isolates were detected by polymerase chain reaction, while drug resistance of the isolates was determined using broth microdilution assays.
Results: Respiratory K. pneumoniae infection was observed in 70.0% of the patients. Of the K. pneumoniae isolates from patients, 42.3% were hypervirulent K (hvKp) serotypes, of which 30.1% and 37.0% belonged to K1 and K2 serotypes with 78.6– 87.8% positive rates of rmpA and rmpA2 virulence genes. The isolates from healthy individuals had fewer hvKp serotypes and rmpA/rmpA2 genes (7.2% and 22.9%/26.5%). Resistance rates (38.6– 79.5%) of the isolates from healthy individuals against 14 antibiotics were higher than those from patients (16.4– 40.8%). The isolates from patients were sensitive to amikacin (83.6%) and polymyxin-B (93.9%) but presented 20.3% and 26.6% resistance rates to imipenem and meropenem, respectively. The isolates from patients with urinary infections exhibited higher resistances (42.1– 52%) to cefoxitin, cefuroxime, ceftriaxone, ciprofloxacin, and levofloxacin than those from patients with respiratory or blood infections (22.4– 39.3%). In the isolates from patients, the K47 and K64 serotypes exhibited multiple drug resistance (65– 90%) against 14 antibiotics but all the hvKp serotypes displayed much lower antibiotic resistance (1.9– 26.0%).
Conclusion: K1/K2 were the major predominant hvKp serotypes with rmpA/rmpA2 virulence genes and carbapenem-resistant K. pneumoniae strains were prevalent in patients from middle and east China. The hvKp serotypes have low antibiotic resistance, but K. pneumoniae isolates from patients with urinary infections resist the cephalosporin/quinolone antibiotics for treatment of bacterial urinary infections. Amikacin and polymyxin-B can be used to treat drug-resistant K. pneumoniae infections.
Keywords: Klebsiella pneumonia, extraintestinal infection, capsular serotypes, virulence genes, antibiotic resistance, multicenter study
Introduction
Klebsiella pneumoniae, a member of the Gram-negative Enterobacteriaceae family, is a typical component of the healthy intestinal microbiota. However, this bacterium can act as an opportunistic pathogen that causes various extraintestinal infectious diseases, including pneumonia, urinary infection, liver abscesses, sepsis, and meningitis, due to bacterial translocation.1–3 Although K. pneumoniae rarely causes intraintestinal infections, the emergence of strains with enhanced antibiotic resistance, including carbapenem resistance and production of multiple extended-spectrum β-lactamases, has become a significant global public health concern.4,5
Based on the diversity of capsular polysaccharide-based animal toxicity, K. pneumoniae strains can be classified into hypovirulent classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp).6,7 Due to the lack of rapid and convenient methods for identifying hvKp in clinical practice, the string test has been widely used in clinical laboratories. However, recent studies have reported that the string test can only be used to identify hypermucoviscous K. pneumoniae (hmKp), as hypermucoviscosity and hypervirulence are the two complementary but distinct phenotypes of K. pneumoniae species.8,9 Numerous previous studies have demonstrated that geographic differences in the prevalent hvKp serotypes and both virulence genes and drug resistance in different K. pneumoniae strains played important roles in the pathogenicity and prevalence of this bacterial pathogen.10–15 Therefore, investigating the epidemiological characteristics of hvKp capsular serotypes, virulence genes, and antibiotic resistance of K. pneumoniae isolates from patients in different countries and regions has significant clinical importance.
All K. pneumoniae strains possessed capsules composed of different capsular polysaccharides. Based on the variations in K antigens within the CPS molecules, K. pneumoniae strains can be classified into numerous capsular serotypes.16 Notably, the capsule is a key virulence factor of K. pneumoniae, enabling it to resist macrophage phagocytosis, block exogenous harmful substances, and mediate bacterial adhesion to host cells. Some capsular serotypes of K. pneumoniae have also been shown to be closely related to drug resistance.16–18 For example, K1, K2, K5, K20, K54, and K57 have been identified as the major hvKp serotypes in many countries in the world including China,16–20 whereas K47 and K64 serotypes are more prevalent in the multidrug-resistant (MDR) K. pneumoniae strains.18,21
K. pneumoniae can also produce various virulence factors. Previous studies have revealed that the products of the rmpA and rmpA2 genes promote CPS synthesis of K. pneumoniae. Inactivation of either the rmpA or rmpA2 gene leads to a significant decrease in the virulence of K. pneumoniae strains.21,22 The wcaG gene product catalyzes the synthesis of fucose, a virulent component in the CPS of K. pneumoniae, which enables the bacterium to resist phagocytosis by host macrophages.23 Adhesion and subsequent colonization of host cells play critical roles in the process of bacterial infection. Type I and III pili of K. pneumoniae have been confirmed to mediate the adhesion and colonization of host epithelial cells.24 The fimH and mrkD genes are responsible for encoding type I and III pilins of K. pneumoniae.24,25 In addition to the major virulence factors mentioned above, K. pneumoniae can also produce some other virulence-related factors, such as yersiniabactin, colibactin, aerobactin, and salmochelin responsible for iron ion uptake and subsequent transport from hosts.15 However, the characteristic correlations between these major virulence genes and capsular serotypes in K. pneumoniae clinical isolates remain poorly understood.
Drug resistance in pathogenic bacteria, including K. pneumoniae, has always been a significant medical challenge.26–28 Antibiotics are commonly used to treat various K. pneumoniae-infected diseases. However, the World Health Organization (WHO) reported that the antibiotic resistance of K. pneumoniae clinical isolates from different countries and regions has been continuously increasing in recent years.29 In China, antibiotic-resistant K. pneumoniae strains are becoming increasingly prevalent.30 Previous views suggested that the virulence of K. pneumoniae is negatively correlated with drug resistance. Nevertheless, recent studies have found that some K. pneumoniae plasmids can carry both multidrug resistance and virulence genes, resulting in the emergence of multidrug-resistant hvKp (MDR-hvKp).31,32 Therefore, the correlation between antibiotic resistance and virulence in K. pneumoniae clinical isolates is an interesting research topic.
In the present study, 710 K. pneumoniae isolates were obtained from six hospitals in different regions of middle and east China between 2022 and 2024 to determine the major predominant hvKp serotypes (K1, K2, K5, K20, K54, and K57) and MDR-related serotypes (K47 and K64),16–20,31–33 virulence genes (rmpA, rmpA2, wcaG, fimH, and mrkD),21–25 and resistance to commonly used antibiotics.26–30 This study aimed to identify the common K. pneumoniae-infected disease types, major predominant hvKp serotypes and virulence genes, and the correlation between serotypes and antibiotic resistance in K. pneumoniae isolates from patients in middle and east China. These results will help improve the clinical therapy and control of K. pneumoniae-infected diseases.
Materials and Methods
Ethics Statement
This study was conducted in accordance with the Declaration of Helsinki, and informed consent was obtained from all patients and healthy individuals for routine physical examinations to collect clinical specimens to isolate K. pneumoniae strains. The study protocol was approved by the Ethics Committee of Hangzhou Medical College (permission No. 2021–111).
Separation and Identification of K. pneumoniae Isolates
Sputum, bronchoalveolar lavage fluid, urine, urinary tract secretions, and peripheral blood samples from 627 patients, and stool samples from 83 healthy individuals for routine physical examinations were collected from six hospitals in Zhejiang (n = 194), Jiangsu (n = 164), Anhui (n = 169), and Henan (n = 100) provincial regions in middle and east China between 2022 and 2024 to isolate K. pneumoniae strains. Each sample was streaked for inoculation on a Columbia blood agar plate (BioMérieux, France) and cultured at 37°C for 16–18 h. Colonies on the plates were selected for bacterial enrichment by cultivation on Columbia blood agar plates and subsequent bacterial identification using a VITEK2-Compact Auto-Bacterial Detector (bioMérieux).
String Test
Each of the 710 K. pneumoniae isolates was inoculated onto Columbia blood agar plates and cultured at 37°C with 5% CO2 for 14–16 h. A bacterial inoculation loop was used to stretch a single colony of K. pneumoniae in the plates. A colonial mucoviscous string length ≥5mm was considered as the hypermucoviscous phenotype K. pneumoniae (hmKp).34
Determination of Major Capsular Serotypes in K. pneumoniae Isolates
Polymerase chain reaction (PCR) was used to determine the capsular serotypes (K1, K2, K5, K20, K47, K54, K57, and K64) in K. pneumoniae isolates.35 Briefly, genomic DNAs of the 710 K. pneumoniae isolates were extracted using a Bacterial Genomic DNA MiniPrep Kit (Axygen, USA) and then quantified using ultraviolet spectrophotometry.36 Using 100 ng of each genomic DNA as a template, separate PCRs were performed using a High-Fidelity Ex-Taq PCR Kit (TaKaRa-Bio, China) and different pairs of serotyping primers (Table 1). The PCRs were initiated by incubation at 95 °C for 3 min for DNA denaturation, followed by 35 cycles at 95 °C for 30s, 53–55°C for 30s and 72°C for 30–60 s to amplify the target gene fragments, followed by incubation at 72°C for 7 min for extension. For serotyping of each K. pneumoniae isolate, three independent PCRs were repeated for detection. The PCR products were examined by 1.5% agarose gel electrophoresis at 110V for 40 min.
 |
Table 1 Primers Used in the PCRs of This Study |
Detection of Major Virulence Genes in K. pneumoniae Isolates by PCR
PCR was also used to detect the major virulence genes of K. pneumoniae (rmpA, rmpA2, wcaG, fimH, and mrkD).37 Genomic DNAs of the 710 K. pneumoniae isolates were extracted using a Bacterial Genomic DNA MiniPrep Kit (Axygen, USA) and then quantified using ultraviolet spectrophotometry as previously described.36 Subsequently, separate PCRs were performed to detect the five virulence genes from the DNA templates as described earlier. The primers used in the PCR for detecting the virulence genes of K. pneumoniae are shown in Table 1. The PCR products were examined by 1.5% agarose gel electrophoresis.
Drug Susceptibility Test
Broth microdilution assays in vitro were performed to determine the minimum inhibitory concentration (MIC) of ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, cefoxitin, cefuroxime, ceftazidime, ceftriaxone, cefotaxime, cefoperazone/sulbactam, meropenem, imipenem, amikacin, ciprofloxacin, levofloxacin, and polymyxin-B in 710 K. pneumoniae isolates as previously reported.38 The obtained results were interpreted and determined as sensitive, intermediate, and resistant according to the 2021 Clinical and Laboratory Standards Institute (CLSI) M100-Ed31.38 The sensitive, intermediate, or resistant result indicated that the MIC of an antibiotic was below, close to, or above its clinically common blood drug concentration. Staphylococcus aureus ATCC25923 and Escherichia coli ATCC25922 were used as controls.
Data Analysis
The obtained data were statistically analyzed using SPSS software (version 29.0). The chi-squared test (χ2-test) and Fisher’s exact probability test were used to determine significant differences. Statistical significance was defined as p<0.05.
Results
Types and Distribution of K. pneumoniae-Infected Diseases in Patients
Among the 627 K. pneumoniae-infected patients, 73.0% (458/627), 16.3% (102/627), and 10.7% (67/627) of the patients were diagnosed with respiratory, urinary, and blood infections, respectively, and respiratory infection was the most common disease type (Figure 1A). The percentage of patients with respiratory or blood infections from the Jiangsu provincial region was significantly lower or higher (56.7% or 27.4%) than those from the Zhejiang, Anhui and Henan provincial regions (70.1–87.6% or 1.8–7.0%) (Figure 1B). Additionally, the percentage of patients with urinary infections from the Zhejiang provincial region was significantly higher (23.7%) than that from the other three provincial regions (10.6–15.8%) (Figure 1B). These data suggest that respiratory infections are a major K. pneumoniae-infected disease in patients from different provincial regions of middle and east China. However, there are regional or geographic differences in the predominant disease types caused by K. pneumoniae.
Predominant Capsular Serotypes in the K. pneumoniae Isolates
Among the 627 K. pneumoniae isolates from patients with respiratory, urinary, or blood infections, 12.9% (81/627), 15.6% (98/627), 3.7% (23/627), 4.2% (26/627), 2.4% (15/627), and 3.5% (22/627) were determined by PCR as K1, K2, K5, K20, K54, and K57 hvKp serotypes (42.3% in total from 265/627), respectively. In contrast, 4.6% (29/627) and 3.2% (20/627) of the isolates (7.8% in total) were identified as K47 and K64 MDR-related serotypes (7.8% in total from 49/627) (Figure 2A and B). However, in the 83 K. pneumoniae isolates from the intestines of healthy individuals, only 1.2% (1/83), 2.4% (2/83), 1.2% (1/83), 1.2% (1/83), and 1.2% (1/83) of the isolates (7.2% in total from 6/83) were determined by PCR as K1, K2, K20, K54, and K57 hvKp serotypes, respectively (Figure 2B). Notably, the K5 hvKp serotype was undetectable in the isolates from the healthy individuals. Moreover, 4.8% (4/83) and 26.5% (22/83) of the 83 K. pneumoniae isolates from healthy individuals (31.3% in total from 26/83) were identified as the K47 and K64 MDR-related serotypes, respectively (Figure 2B). These data suggest that K1 and K2 are the major predominant hvKp serotypes prevalent in patients in middle and east China, whereas the K47 and K64 MDR-related serotypes are more common in K. pneumoniae isolates from healthy individuals.
Distribution Differences in Serotypes of the K. pneumoniae Isolates from Different Patients and Provincial Regions
Only the positive rate of the K2 hvKp serotype in K. pneumoniae isolates from patients with respiratory infections was significantly higher than that in patients with urinary or blood infections. However, neither the K1, K5, K20, K54, and K57 hvKp serotypes nor the K47 and K64 MDR-related serotypes in K. pneumoniae isolates from patients with different diseases displayed significant differences (Table 2). Moreover, there was no significant difference in the distribution of hvKp serotypes among K. pneumoniae isolates from patients in the different provincial regions (Table 3). However, the positive rate of K47 or K64 MDR-related serotypes in the isolates from patients in the Henan or Zhejiang provincial regions (8.3% or 15.0%) was significantly higher than that of patients in the other three provincial regions (0–4.3%) (Table 3). These data suggest that the K2 hvKp serotype is more common in K. pneumoniae isolates from patients with respiratory infections and that the prevalence of K. pneumoniae K47 and K64 MDR-related serotypes in patients has significant regional differences in middle and east China.
 |
Table 2 Capsular Serotypes of K. pneumoniae Isolates from Different Patients |
 |
Table 3 Capsular Serotypes in K. pneumoniae Isolates from Patients in Different Regions |
Predominant Virulence Genes in the K. pneumoniae Isolates
In the 627 K. pneumoniae isolates from patients, 30.3% (190/627) of the isolates were positive in the string test, which was significantly higher than that (10.8%, 9/83) in the 83 K. pneumoniae isolates from the intestines of healthy individuals (p<0.01). The positive rates of the rmpA, rmpA2, and wcaG virulence genes in the 627 K. pneumoniae isolates from patients were 47.5% (298/627), 42.7% (268/627), and 20.1% (126/627), respectively, which were significantly higher than those of 22.9% (19/83), 26.5% (22/83), and 6.0% (5/83) of the 83 K. pneumoniae isolates from healthy individuals (Figure 3A and B). Furthermore, the rmpA and rmpA2 virulence genes demonstrated significantly higher positive detection rates than the wcaG virulence gene in the K. pneumoniae isolates from patients (p < 0.01). However, the 94.9% (595/627) and 94.9% (595/627) positive rates of the fimH and mrkD virulence genes in the 627 K. pneumoniae isolates from patients showed no significant differences compared to the 95.2% (79/83) and 92.8% (77/83) of the 83 K. pneumoniae isolates from healthy individuals (Figure 3B). These data suggest that the rmpA, rmpA2, and wcaG, but not fimH and mrkD genes, play important roles in the virulence of K. pneumoniae isolates from patients in middle and east China
Distribution Differences in Virulence Genes of the K. pneumoniae Isolates from Different Patients and Regions
The rmpA and rmpA2 virulence genes were more common in the 627 K. pneumoniae isolates from patients with respiratory infections (52.0% and 46.5%, respectively) than in those from the patients with urinary or blood infections (34.3% and 32.4%, or 37.3% and 32.8%, respectively) (Table 4). Moreover, the positive rates (30.3% and 30.0%) of the wcaG virulence gene in K. pneumoniae isolates from patients in the Anhui and Henan provincial regions were significantly higher than those from the patients in the Zhejiang and Jiangsu provincial regions (14.4% and 10.4%, respectively) (Table 5). These data suggest that the rmpA and rmpA2 virulence genes play important roles in the pathogenesis of K. pneumoniae-caused respiratory infections and that the distribution of wcaG virulence genes in K. pneumoniae isolates has regional or geographic differences in middle and east China.
 |
Table 4 Virulence Genes in K. pneumoniae Isolates from Different Patients |
 |
Table 5 Virulence Genes in the K. pneumoniae Isolates from Patients in Different Regions |
Correlation of hvKp Serotypes With Virulence Genes in the K. pneumoniae Isolates
In the 265 hvKp serotypes from 627 K. pneumoniae isolates, both fimH and mrkD virulence genes displayed similar high positive rates (92.3–98.0%) in the six different hvKp serotypes (Table 6). However, the positive rates (78.6–87.8%) of rmpA and rmpA2 genes in the K1 and K2 hvKp serotypes were significantly higher than those (39.1–60.9%) in the other four hvKp serotypes (Table 6). In addition, the wcaG gene in the K1 and K54 hvKp serotypes also had significantly higher positive rates (63.0% and 73.3%) than those (11.5%–18.2%) in the other four hvKp serotypes (Table 6). These data suggest that the rmpA and rmpA2 genes are related to the virulence of the predominant K1 and K2 hvKp serotypes in K. pneumoniae isolates from patients in middle and east China.
 |
Table 6 Correlation of hvKp Serotypes with Virulence Genes from Patients |
Extensive Antibiotic Resistance of the K. pneumoniae Isolates
The 627 K. pneumoniaeisolates from patients exhibited a much higher resistance rate (85.3%) against ampicillin but presented significantly higher sensitivity rates to amikacin (83.6%) and polymyxin-B (93.9%) among the 16 tested antibiotics (Figure 4 and Table S1). In particular, the resistance rates of the isolates against meropenem and imipenem, the two commonly used carbapenem antibiotics, reached 26.6% and 20.3%, respectively. Furthermore, the isolates exhibited relatively higher levels of resistance against the other 11 antibiotics (26.5–40.8%) belonging to either β-lactams or quinolones. Interestingly, except for ampicillin and polymyxin-B, the resistance rates (38.6–79.5%) of the 83 K. pneumoniae isolates from the intestines of healthy individuals against the other 14 tested antibiotics were significantly higher than those (16.4–40.8%) of the K. pneumoniae isolates from different clinical samples of the patients (Figure 4 and Table S1). These data suggest that K. pneumoniae isolates prevalent in patients in middle and east China exhibit extensive and relatively high resistance to the most commonly used antibiotics, including over 20% resistance rates against imipenem and meropenem, but display high sensitivity to amikacin and polymyxin-B.
Diversity of Antibiotic Resistance in the K. pneumoniae Isolates From Different Patients and Provincial Regions
Except for the much higher resistance rates (83.3%, 83.6%, and 86.0%) against ampicillin in the 627 K. pneumoniae isolates from the patients with different diseases, the resistance rates (42.1–55.9%) of the isolates from patients with urinary infections to cefoxitin, cefuroxime, ceftriaxone, ciprofloxacin, and levofloxacin were significantly higher than those (22.4–39.3%) from patients with respiratory or blood infections (Figure 5 and Table S2). However, there were no significant differences in the resistance rates against any of the 16 tested antibiotics among the isolates from patients in different provincial regions (Figure 6 and Table S3). These data suggest that K. pneumoniae isolates from patients with urinary infections in middle and east China exhibit selective resistance to some specific antibiotics.
 |
Figure 5 Antibiotic resistance of K. pneumoniae isolates from different patients. The abbreviations of 16 tested antibiotics were the same as shown in Figure 4. *indicates p<0.01 vs the resistance rates in the isolates against all the other tested antibiotics. ▲indicates p<0.05 vs the resistance rates in the isolates from patients with respiratory or blood infections. |
 |
Figure 6 Antibiotic resistance of K. pneumoniae isolates from patients in different regions. Antibiotic resistance of K. pneumoniae isolates from different patients. The abbreviations of 16 tested antibiotics were the same as shown in Figure 4. *indicates p<0.01 vs the resistance rates in the isolates against all the other 15 antibiotics. |
Correlation of Serotypes With Antibiotic Resistance in the K. pneumoniae Isolates
Except for ampicillin and polymyxin-B, the K47 and K64 MDR-related serotypes in the 627 K. pneumoniae isolates from patients displayed significantly higher resistance rates (65.0–90.0%) to the other 14 tested antibiotics (Figure 7 and Table S4). In contrast, the 265 K. pneumoniae isolates belonging to six different major hvKp serotypes showed much lower resistance rates (1.9–26.0%) to these 14 antibiotics. The 313 non hvKp/MDR-related classical K. pneumoniae (cKp) isolates displayed significantly higher resistance rates (20.8–53.0%) than those of the hvKp isolates (Figure 7 and Table S4). These data suggest that the K47 and K64 MDR-related serotypes in K. pneumoniae isolates from patients possess multiple resistances to the commonly used antibiotics and the hvKp serotypes, including K1 and K2 prevailing in patients in Middle and East China, are not related to antibiotic resistance.
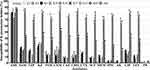 |
Figure 7 Correlation between K. pneumoniae serotypes in patients with antibiotic resistance. Antibiotic resistance of K. pneumoniae isolates from different patients. The abbreviations of 16 tested antibiotics were the same as shown in Figure 4. *indicates p<0.05 vs the resistance rates in the cKp isolates. ▲indicates p<0.01 vs the resistance rates in the K1, K2, K5, K20, K54, and K57 hvKp isolates, as well as in the K47 and K64 MDR-related isolates. |
Discussion
K. pneumoniae is a common opportunistic bacterial pathogen that can cause various diseases in nearly all human tissues and organs.1–3 However, respiratory infection, including bronchitis and pneumonia, is the most common K. pneumoniae-infected disease.39 In the present study, 627 K. pneumoniae-infected patients from four provincial regions (Zhejiang, Jiangsu, Anhui, and Henan) in Middle and East China were clinically diagnosed with respiratory, urinary, or blood infection, with over 70% of the patients suffering from K. pneumoniae respiratory infection. These data indicate that the respiratory tract is the most susceptible site for K. pneumoniae isolates in middle and east China.
K. pneumoniae is a typical Gram-negative bacterium with a polysaccharide capsule. In addition to lipopolysaccharide endotoxin, the capsule is another key virulence factor of K. pneumoniae.24,40 Some special capsular serotypes of K. pneumoniae, such as K1, K2, K5, K20, K54, and K57, have been confirmed as the major hvKp serotypes.16–20 Previous studies have reported that K1 and K2 are the most common hvKp capsular serotypes in Asia,7,41,42 but are rare in Europe and North America.43,44 The present study also revealed that K1 and K2 are the predominant hvKp serotypes in the patients in Middle and East China. rmpA, rmpA2, and wcaG have been reported as important virulence genes of K. pneumoniae.21–23 The present study found that the rmpA and rmpA2 genes displayed significantly higher positive rates (47.5% and 42.7%) than that the wcaG gene (20.1%) in the K. pneumoniae isolates from patients. However, all the three virulence genes had significantly higher carrying rates in the isolates from patients than those from healthy individuals. The positive rates of both rmpA and rmpA2 genes in the K. pneumoniae isolates from patients in middle and east China were also notably higher than those in northern Japan (22.2% and 19.8%) and New York of America (4.16% and 2.81%), but similar to those in eastern India (48.4% and 57.5%).45–47 In particular, the K1 and K2 hvKp serotypes in the K. pneumoniae isolates from patients in this study exhibited significantly higher positive rates (78.6–87.8%) for rmpA and rmpA2 genes than the K5, K20, K54, and K57 hvKp serotypes (39.1–60.9%). These data indicate that K1, K2, rmpA, and rmpA2 are the predominant hvKp serotypes and virulence genes in K. pneumoniae isolates from patients in middle and east China, respectively, and that both rmpA and rmpA2 genes are closely related to the virulence of K1 and K2 hvKp serotypes. In addition, K. pneumoniae isolates from patients with respiratory infections had the significantly higher proportions of rmpA and rmpA2 genes (52.0% and 46.5%, respectively) than those from patients with urinary or blood infections (32.4–37.3%), suggesting a possible reason for the higher incidence rate of K. pneumoniae respiratory infections in this study.
Pili composed of pilins are surface structures in many bacteria that act as important virulence factors by mediating bacterial adhesion and colonization in hosts.48 Most K. pneumoniae strains have type I pilin-encoding fimH gene and type III pilin-encoding mrkD gene.49 Our study showed that 92.8–95.2% of the K. pneumoniae isolates from patients and healthy individuals were positive for fimH and mrkD genes. A recent study also reported that the positive rates of the two pilin-encoding genes in the K. pneumoniae isolates from a central European region were as high as 91.7% and 96.3%, respectively.50 Therefore, fimH and mrkD genes are required for adhesion and colonization of K. pneumoniae in healthy individuals and patients, but not suitable for determining the virulence of K. pneumoniae isolates.
In clinical practice, β-lactam and quinolone antibiotics are usually selected to treat K. pneumoniae-infected diseases.51 β-lactam antibiotics can be classified into penicillins, cephalosporins, and carbapenems based on their structural differences in β-lactam rings and their side chains in molecules.52 Among the 16 antibiotics used in the drug susceptibility test of this study, 14 are the commonly used types of penicillins, cephalosporins, carbapenems, and quinolones, while the other two belong to aminoglycoside (amikacin) or polymyxin antibiotics. A previous study reported that K. pneumoniae is naturally resistant to ampicillin.4 Our drug susceptibility test also showed that the ampicillin resistance rate was as high as over 85% in the 627 K. pneumoniae isolates from patients. More importantly, the six common cephalosporin antibiotics (cefazolin, cefoxitin, cefuroxime, ceftazidime, ceftriaxone, and cefotaxime) tested in this study had 30.3–41.3% resistance rates in the 627 isolates, which were similar to the resistance rates of cephalosporin antibiotics (20.8%–42.1%) in the K. pneumoniae isolates from patients in southwest China but notably lower than those in many other different countries of Asia (65.4%–87.2%) and southern Europe (50%–60%).4,5,30 Compared to penicillin and cephalosporin antibiotics, carbapenem antibiotics exhibit many advantages, such as a wider antibacterial spectrum, strong antibacterial activity, and high β-lactase stability.53 However, the K. pneumoniae isolates from patients in middle and east China also exhibited 26.6% and 20.3% resistance rates against meropenem and imipenem, the two commonly used carbapenem antibiotics, which were obviously higher than those in southwest and north China (7.0%–14.1%) but remarkably lower than those in several other countries of Asia (30.8%–89.0%) and southern Europe (50%–60%).4,5,30,54 Fortunately, the 627 K. pneumoniae isolates from patients in this study displayed high sensitivity to amikacin (83.6%) and polymyxin-B (93.9%). These data indicate that the K. pneumoniae isolates prevailing in middle and east China have higher resistance to most cephalosporin antibiotics and ampicillin cannot be used to treat K. pneumoniae-infected diseases. In addition, these data also indicate the prevalence of carbapenem-resistant K. pneumoniae strains in patients in Middle and East China.
The continuous and widespread use of antibiotics can induce bacteria to develop antibiotic resistance.55 Except for ampicillin and polymyxin-B, the 83 K. pneumoniae isolates from the intestines of the healthy individuals in this study presented much higher resistance rates against the other 14 tested antibiotics than the K. pneumoniae isolates from the patients, which may be due to intestinal bacteria, including K. pneumoniae, often stimulated by oral antibiotics. Moreover, the K. pneumoniae isolates from patients with urinary infections showed significantly higher resistance rates (42.1–55.9%) against cefoxitin, cefuroxime, ceftriaxone, ciprofloxacin, and ciprofloxacin than those (22.4–39.3%) from the patients with respiratory or blood infections. These five antibiotics are commonly used to treat bacterial urinary infections in clinical practice.56,57 Previous studies reported that K47 and K64 are the major MDR-related serotypes, and hvKp serotypes had higher resistance rates against many different antibiotics.15,58–60 In the present study, except for ampicillin and polymyxin-B, the K47 and K64 serotypes in the K. pneumoniae isolates from patients presented much higher resistance against the other 14 tested antibiotics; however, the hvKp serotypes in the isolates were unexpectedly found to have very low resistance to these antibiotics compared to the non-hvKp/MDR-related K. pneumoniae isolates. K. pneumoniae strains have been reported to evolve in two different directions: high toxicity-low drug resistance and low toxicity-high drug resistance.61,62 Therefore, the correlation between hvKp serotypes and antibiotic resistance in K. pneumoniae isolates from different countries and regions requires further investigation. In addition, compared to K. pneumoniae isolates from the intestines of healthy individuals containing much fewer hvKp serotypes and rmpA/rmpA2/wcaG virulence genes but exhibiting significantly higher antibiotic resistance, K. pneumoniae isolates from different patients containing significantly more hvKp serotypes and virulence genes but displaying relatively lower antibiotic resistance seem to be exogenous strains. However, no differences were found in the disease types between K. pneumoniae-infected inpatients and outpatients and in the major hvKp serotypes, virulence genes, and antibiotic resistance characteristics between the K. pneumoniae isolates from the two different types of patients.
Conclusion
K. pneumoniae infection is a common disease in China. This study revealed the major predominant hvKp serotypes, virulence genes, and antibiotic resistance characteristics of K. pneumoniae isolates from patients in middle and east China. Respiratory infections are the main K. pneumoniae-induced diseases. K1 and K2 were the major predominant hvKp serotypes in K. pneumoniae isolates from the patients. rmpA and rmpA2 are the main virulence-determining genes that are closely related to the K1 and K2 hvKp serotypes. The K. pneumoniae isolates exhibited extensive and relatively high resistance to the most commonly used antibiotics, including over 20% resistance rates against meropenem and imipenem. K. pneumoniae isolates from patients with urinary infections displayed significantly higher resistance to cefoxitin, cefuroxime, ceftriaxone, ciprofloxacin, and levofloxacin than those from patients with respiratory or blood infections. In addition to ampicillin and polymyxin-B, the K47 and K64 MDR-related serotypes in K. pneumoniae isolates exhibited high resistance to all the other tested 14 β-lactam and quinolone antibiotics. However, the major hvKp serotypes in the isolates showed much lower resistance rates to these 14 antibiotics, indicating that the high virulence of K. pneumoniae is not related to the antibiotic resistance.
Acknowledgments
This work was supported by a grant from the Innovation and Entrepreneurship Training Program for College Students of Zhejiang Province (No. S202413023093) and a grant from the Scientific Research Special Project of Putian College for the First Hospital of Putian City (No. 2024084).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112(27):e3574–3581. doi:10.1073/pnas.1501049112
2. Lapp Z, Han JH, Wiens J, et al. Patient and microbial genomic factors associated with carbapenem-resistant Klebsiella pneumoniae extraintestinal colonization and infection. mSystems. 2021;6(2):e00177–00221. doi:10.1128/mSystems.00177-21
3. Du Q, Xu Q, Pan F, et al. Association between intestinal colonization and extraintestinal infection with carbapenem-resistant Klebsiella pneumoniae in children. Microbiol Spectr. 2023;11(2):e0408822. doi:10.1128/spectrum.04088-22
4. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
5. Effah CY, Sun T, Liu S, et al. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1–9. doi:10.1186/s12941-019-0343-8
6. Dai P, Hu D. The making of hypervirulent Klebsiella pneumoniae. J Clin Lab Anal. 2022;36(12):e24743. doi:10.1002/jcla.24743
7. Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniaeinfection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. doi:10.1128/AAC.01127-16
8. Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111–1123. doi:10.1080/21505594.2017.1317412
9. Harada S, Doi Y. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol. 2018;56(9):e00959–01018. doi:10.1128/JCM.00959-18
10. Wu W, Jiang Y, Zhou W, et al. Genomic characteristics of carbapenem-resistant Klebsiella pneumoniae isolated from neonatal patients in Southwest China during 2017–2021. Infect Drug Resist. 2023;16:6725–6733. doi:10.2147/IDR.S426565
11. Le MN, Kayama S, Wyres KL, et al. Genomic epidemiology and temperature dependency of hypermucoviscous Klebsiella pneumoniae in Japan. Microb Genom. 2022;8(5):mgen000827. doi:10.1099/mgen.0.000827
12. Singh S, Pathak A, Rahman M, et al. Genetic characterisation of colistin resistant Klebsiella pneumoniae clinical isolates from North India. Front Cell Infect Microbiol. 2021;11:666030. doi:10.3389/fcimb.2021.666030
13. Kaye KS, Gupta V, Mulgirigama A, et al. Prevalence, regional distribution, and trends of antimicrobial resistance among female outpatients with urine Klebsiella spp. isolates: a multicenter evaluation in the United States between 2011 and 2019. Antimicrob Resist Infect Control. 2024;13(1):21. doi:10.1186/s13756-024-01372-x
14. Budia-Silva M, Kostyanev T, Ayala-Montaño S, et al. International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nat Commun. 2024;15(1):5092. doi:10.1038/s41467-024-49349-z
15. Kocsis B. Hypervirulent Klebsiella pneumoniae: an update on epidemiology, detection and antibiotic resistance. Acta Microbiol Immunol Hung. 2023;70(4):278–287. doi:10.1556/030.2023.02186
16. Dong N, Yang X, Chan EW, et al. Klebsiella species: taxonomy, hypervirulence and multidrug resistance. eBioMedicine. 2022;79:103998. doi:10.1016/j.ebiom.2022.103998
17. Fang CT, Chuang YP, Shun CT, et al. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
18. Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812–1820. doi:10.3201/eid2011.140206
19. Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi:10.1016/j.jinf.2015.07.010
20. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–00019. doi:10.1128/CMR.00001-19
21. Tang HL, Chiang MK, Liou WJ, et al. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis. 2010;29(6):689–698. doi:10.1007/s10096-010-0915-1
22. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–00818. doi:10.1128/JCM.00776-18
23. Turton JF, Perry C, Elgohari S, et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(5):541–547. doi:10.1099/jmm.0.015198-0
24. Li B, Zhao Y, Liu C, et al. Molecular Pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–1081. doi:10.2217/fmb.14.48
25. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
26. Ferri M, Ranucci E, Romagnoli P, et al. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–2876. doi:10.1080/10408398.2015.1077192
27. Zhu Y, Huang WE, Yang QW. Clinical perspective of antimicrobial resistance in bacteria. Infect Drug Resist. 2022;15:735–746. doi:10.2147/IDR.S345574
28. Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139. doi:10.1016/j.mib.2018.04.004
29. World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021. Geneva: World Health Organization; 2021.
30. Chang F, Wang X, Huang X, et al. Analysis on bacterial distribution and change of drug resistance rate in ICUs across southwest China from 2018 to 2022. Infect Drug Resist. 2023;16:5685–5696. doi:10.2147/IDR.S421357
31. Gonzalez-Ferrer S, Peñaloza HF, Budnick JA, et al. Finding order in the chaos: outstanding questions in Klebsiella pneumoniae pathogenesis. Infect Immun. 2021;89(4):e00693–00720. doi:10.1128/IAI.00693-20
32. Chen J, Zhang H, Liao X. Hypervirulent Klebsiella pneumoniae. Infect Drug Resist. 2023;16:5243–5249. doi:10.2147/IDR.S418523
33. Kao CY, Zhang YZ, Bregente CJB, et al. A 24-year longitudinal study of Klebsiella pneumoniae isolated from patients with bacteraemia and urinary tract infections reveals the association between capsular serotypes, antibiotic resistance, and virulence gene distribution. Epidemiol Infect. 2023;151:e155. doi:10.1017/S0950268823001486
34. Jin M, Jia T, Liu X, et al. Clinical and genomic analysis of hypermucoviscous Klebsiella pneumoniae isolates: identification of new hypermucoviscosity associated genes. Front Cell Infect Microbiol. 2023;12:1063406. doi:10.3389/fcimb.2022.1063406
35. Chen Z, Liu M, Cui Y, et al. A novel PCR-based genotyping scheme for clinical Klebsiella pneumoniae. Future Microbiol. 2014;9(1):21–32. doi:10.2217/fmb.13.137
36. Huang LD, Yang MJ, Huang YY, et al. Molecular characterization of predominant serotypes, drug resistance, and virulence genes of Streptococcus pneumoniae isolates from East China. Front Microbiol. 2022;13:892364. doi:10.3389/fmicb.2022.892364
37. Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi:10.18388/abp.2015_1148
38. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing.
39. Wang G, Zhao G, Chao X, et al. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17):6278. doi:10.3390/ijerph17176278
40. Follador R, Heinz E, Wyres KL, et al. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom. 2016;2(8):e000073. doi:10.1099/mgen.0.000073
41. Ikeda M, Mizoguchi M, Oshida Y, et al. Clinical and microbiological characteristics and occurrence of Klebsiella pneumoniae infection in Japan. Int J Gen Med. 2018;11:293–299. doi:10.2147/IJGM.S166940
42. Wyres KL, Nguyen TNT, Lam MMC, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 2020;12(1):11. doi:10.1186/s13073-019-0706-y
43. Martin RM, Cao J, Brisse S, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere. 2016;1(5):e00261–00316. doi:10.1128/mSphere.00261-16
44. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi:10.1111/joim.13007
45. Matsuda N, Aung MS, Urushibara N, et al. Prevalence, clonal diversity, and antimicrobial resistance of hypervirulent Klebsiella pneumoniae and Klebsiella variicola clinical isolates in northern Japan. J Glob Antimicrob Resist. 2023;35:11–18. doi:10.1016/j.jgar.2023.08.009
46. Parrott AM, Shi J, Aaron J, et al. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin Microbiol Infect. 2021;27(4):583–589. doi:10.1016/j.cmi.2020.05.012
47. Yadav B, Mohanty S, Behera B. Occurrence and genomic characteristics of hypervirulent Klebsiella pneumoniae in a tertiary care hospital, eastern India. Infect Drug Resist. 2023;16:2191–2201. doi:10.2147/IDR.S405816
48. Kline KA, Dodson KW, Caparon MG, et al. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18(5):224–232. doi:10.1016/j.tim.2010.03.002
49. Chen Q, Wang M, Han M, et al. Molecular basis of Klebsiella pneumoniae colonization in host. Microb Pathog. 2023;177:106026. doi:10.1016/j.micpath.2023.106026
50. Kot B, Piechota M, Szweda P, et al. Virulence analysis and antibiotic resistance of Klebsiella pneumoniae isolates from hospitalised patients in Poland. Sci Rep. 2023;13(1):4448. doi:10.1038/s41598-023-31086-w
51. Ali S, Alam M, Hasan GM, et al. Potential therapeutic targets of Klebsiella pneumoniae: a multi-omics review perspective. Brief Funct Genomics. 2022;21(2):63–77. doi:10.1093/bfgp/elab038
52. Jacobs LMC, Consol P, Chen Y. Drug discovery in the field of β-lactams: an academic perspective. Antibiotics. 2024;13(1):59. doi:10.3390/antibiotics13010059
53. Lima LM, Silva BNMD, Barbosa G, et al. β-lactam antibiotics: an overview from a medicinal chemistry perspective. Eur J Med Chem. 2020;208:112829. doi:10.1016/j.ejmech.2020.112829
54. Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. doi:10.1093/cid/cit675
55. Rolain J M, Abat C, Jimeno M T, et al. Do we need new antibiotics? Clin Microbiol Infect. 2016;22(5):408–415. doi:10.1016/j.cmi.2016.03.012
56. Qiao LD, Chen S, Yang Y, et al. Characteristics of urinary tract infection pathogens and their in vitro susceptibility to antimicrobial agents in China: data from a multicenter study. BMJ Open. 2013;3(12):e004152. doi:10.1136/bmjopen-2013-004152
57. Naber KG, Wagenlehner FME. Novel Antibiotics in the treatment of urinary tract infections. Eur Urol Focus. 2019;5(1):10–12. doi:10.1016/j.euf.2018.11.012
58. Tang M, Kong X, Hao J, et al. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2020;11:581543. doi:10.3389/fmicb.2020.581543
59. Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi:10.3389/fcimb.2017.00483
60. Salawudeen A, Raji YE, Jibo GG, et al. Epidemiology of multidrug-resistant Klebsiella pneumoniae infection in clinical setting in South-Eastern Asia: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2023;12(1):142. doi:10.1186/s13756-023-01346-5
61. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
62. Lai YC, Lu MC, Hsueh PR. Hypervirulence and carbapenem resistance: two distinct evolutionary directions that led high-risk Klebsiella pneumoniae clones to epidemic success. Expert Rev Mol Diagn. 2019;19(9):825–837. doi:10.1080/14737159.2019.1649145
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Antibiotic Resistance, Molecular Characteristics and Risk Factors of Carbapenem-Resistant Klebsiella pneumoniae in Clinical Isolates
Zhu J, Chen Y, Yang X
Infection and Drug Resistance 2022, 15:6671-6680
Published Date: 15 November 2022





