Back to Journals » Infection and Drug Resistance » Volume 18
Molecular Identification of Aspergillus Species, Antifungal Susceptibility, and Phenotypic Identification of Azole-Resistant Mutations in Cyp51A Gene Isolated from Xinjiang
Authors Yusufu A, Aizezi Z, Nuermaimaiti X, Liu Y, Wang X
Received 23 October 2024
Accepted for publication 20 March 2025
Published 2 April 2025 Volume 2025:18 Pages 1699—1711
DOI https://doi.org/10.2147/IDR.S496489
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sandip Patil
Aikedai Yusufu, Zubaidanmu Aizezi, Xiyidan Nuermaimaiti, Yiting Liu, Xiaodong Wang
Department of Dermatology, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, People’s Republic of China
Correspondence: Xiaodong Wang, Department of Dermatology, The First Affiliated Hospital of Xinjiang Medical University, No. 393, Xinyi Road, Urumqi, Xinjiang, People’s Republic of China, Email [email protected]
Purpose: This study aimed to determine the clinical distribution characteristics, in vitro antifungal susceptibility, and cyp51A mutation types of clinically isolated Aspergillus species in Xinjiang.
Methods: In this study, a total of 111 Aspergillus species were identified by sequencing the internal transcribed spacer (ITS) and β-tubulin (BenA) genes for molecular identification, performed antifungal susceptibility testing on these isolates using Sensititre YeastOne, selected azole-resistant isolates based on the antifungal susceptibility results and amplified the cyp51A gene for identification of the azole resistance mutation phenotype in the selected isolates.
Results: The most common Aspergillus species was A. fumigatus (40.54%), followed by A. niger (18.02%), A. tubingensis (16.22%), A. terreus (13.51%), A. flavus (6.31%), A. welwitschiae (2.70%), A. fumigatiaffinis (1.80%), and A. lentulus (0.90%). The antifungal susceptibility test results showed that A. fumigatus, A. niger, A. tubingensis, A. flavus and A. terreus were completely sensitive to itraconazole, with sensitivity rates of posaconazole and voriconazole were 99.10% and 88.29%, respectively. The sensitivity rate to amphotericin B was the lowest (62.16%). The MIC values of amphotericin B and voriconazole for the two cryptic Aspergillus species, A. lentulus and A. fumigatiaffinis with high (> 1mg/L). The azole non-susceptible or non-wild type rate was (15/111, 13.51%). Eleven azole-resistant Aspergillus species had cyp51A mutations, while four strains did not have any cyp51A mutations.
Conclusion: In this study, the pathogenic Aspergillus species isolated from clinical cases in Xinjiang were diverse. Common pathogenic species showed the best in vitro antifungal activity against itraconazole, posaconazole, and echinocandins, whereas the MIC distribution of amphotericin B was significantly higher. Resistant strains may be mediated by point mutations in cyp51A, and phenotypic mutations are diverse. This information is of great significance for guiding the early diagnosis and antifungal therapy for aspergillosis.
Keywords: invasive aspergillosis, Aspergillus fumigatus, azole-resistance, antifungal susceptibility, cyp51A gene
Introduction
Invasive fungal infections can cause a range of serious human diseases, but little is known about increasingly serious invasive fungal diseases. Among them, invasive aspergillosis (IA) is a systemic fungal infection that endangers patients’ lives, mainly affecting patients with impaired immune function, with a mortality rate of up to 60%, and a mortality rate of around 20–30% when treated with voriconazole and isavuconazole as the first-line treatment.1 Pathogenic Aspergillus species are the main cause of infections and deaths. Approximately 30 species of Aspergillus, with common ones including A. fumigatus, A. flavus, A. niger and A. terreus.2 Among them, A. fumigatus has been reported to cause IA in human. Additionally, the warming climate and changes in the ecological niche of environmental fungi have led to the evolution and emergence of new Aspergillus pathogens, especially the emergence and spread of drug-resistant strains, which has exacerbated the difficulty in diagnosing and treating invasive aspergillosis.
Azole antifungal drugs are recommended as the first-line treatment and preventive drugs for Aspergillus infection.3 However, increasing reports of Aspergillus resistance to azoles have emerged in recent years.4 In recent years, it has also been reported that a number of A. fumigatus isolates from specimens of patients with novel coronavirus pneumonia-associated pulmonary Aspergillus have been reported to be resistant to multiple azole antifungal agents.5,6 The failure rate of azole-resistant aspergillosis treatment is very high, with a mortality rate of up to 88%, which is 3–4 times that of patients with azole-sensitive aspergillosis.7 Due to the limited types of antifungal drugs, some invasive aspergillosis, especially those affecting the central nervous system, will greatly increase the difficulty of clinical treatment if they are drug-resistant strains.
The mechanism of action of azoles is to inhibit ergosterol 14α-demethylase (cyp51), blocking the ergosterol biosynthesis pathway, leading to a reduction in ergosterol synthesis, and exerting antifungal effects. The cyp51A gene mutation is the most common drug resistance mechanism of A. fumigatus,8–10 the mutation types include point mutations, such as M220, G54, G138 and other hotspot amino acid substitutions, which reduce the affinity with azole antifungal drugs, these types of mutations are considered therapeutic drug-inducing mutations. The other type is tandem repeat non-synonymous mutations in the promoter region of cyp51A, such as TR34/L98H and TR46/Y121F/T289A, which are drug resistance mutations caused by exposure to environmental azole fungicides, and result in cross-resistance to multiple azole classes.11
Nevertheless, studies have reported that over 40% of A. fumigatus may be related to non-cyp51A mutations that lead to azole resistance among A. fumigatus isolates.12 One of the issues is the mutations in the 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase-encoding gene (hmg1) associated with triazole-resistance A. fumigatus isolates. Interestingly, a new study investigated the combination of hapE and hmg1 mutations as potential contributors to azole resistance.13,14 In addition to these mutations, other candidate genes such as crd1B, MDR1, MDR2, and erg6 have also been extensively studied and are believed to play a role in non-cyp51A azole resistance. The growing number of resistant strains and the continuous emergence of new resistance mechanisms have aroused great concern worldwide.
Despite the increasing threat of invasive fungal infections caused by known or novel Aspergillus pathogens, there is still no research on the epidemiological characteristics of Aspergillus infections, in vitro antifungal drug susceptibility, or drug resistance in Xinjiang. Therefore, the present study was conducted to investigate the epidemiological distribution characteristics, drug sensitivity, and azole-resistant mutant phenotype of Aspergillus strains in Xinjiang to provide useful information for the early diagnosis and treatment of invasive fungal infections.
Materials and Methods
Sample Collection and Strain Culture
A total of 111 Aspergillus clinical strains were collected from 2011 to 2023 from the fungal specimen library of the Dermatology Laboratory of the First Affiliated Hospital of Xinjiang Medical University. After resuscitating the frozen preserved strains at −80°C in a cryogenic refrigerator back to room temperature, a small amount of fungal liquid was collected with a sterile inoculation ring and inoculated onto Potato Dextrose Agar (PDA). The inoculated plates were incubated at 28°C for 3–5d. The colonies were observed for their macroscopic characteristics, including colony color, texture, and growth rate. For microscopic examination, a small portion of the colony was transferred to a glass slide and stained with lactophenol cotton blue. The slide was then observed under a light microscope at 400× magnification to examine the morphological features of the conidiophores, conidia, and vesicle structures.
Aspergillus Isolates and Molecular Identification
DNA was extracted from isolates grown on PDA plates using the Ezup DNA Isolation Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. The clinical isolates were molecularly identified by PCR and sequencing of the ITS and BenA genes using the primer pair ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), and the primer pair BT2a (5′-GGTAACCAAATCGGTGCTGCTTTC-3′) and BT2b (5′-ACCCCTCAGTGTAGTGACCCTTGGC-3′). The thermal cycling profiles for ITS and BenA amplification were as follows: 5 min at 95°C, followed by 30 cycles of 94°C for 30s, 57°C for 30s, and 72°C for 90s, with a final extension step at 72°C for 10 min. The PCR reaction system is as follows: 1 µL of 10 mm dNTPs, 2.5 µL of 10X PCR buffer, 1 µL of 10 µM of each primer, 1 U Taq DNA polymerase, 1 µL genomic DNA, and DNase-free water, up to a final reaction volume of 25 µL. The PCR products were analyzed by electrophoresis on a 1.5% agarose gel, the resultant PCR amplicons were purified and sequenced, and the obtained sequences were compared to reference sequences in GenBank.
Antifungal Susceptibility Testing
Sensititre YeastOne YO10 (Thermo Fisher) was used for antifungal susceptibility testing with the seven antifungal agents, according to the manufacturer’s instructions. The seven antifungal agents used in this study were amphotericin B (AMB), itraconazole (ITZ), voriconazole (VOR), posaconazole (POS), caspofungin (CAS), anidulafungin (AND), and micafungin (MCF). The antifungal agents were at a final concentration of 0.12–8 μg/mL for amphotericin B, 0.008–8 μg/mL for caspofungin, micafungin, voriconazole, and posaconazole, 0.015–16 μg/mL for itraconazole, and 0.015–8 μg/mL for anidulafungin. The conidial inoculum suspension was prepared at a turbidity of 0.5 McFarland units for the assay. With 48h of culturing (except for echinocandins, which were cultured for 24h), the antifungal drug sensitivity profiles of these inocula were determined according to the manufacturer’s instructions. All isolates were tested according to Clinical and Laboratory Standards Institute (CLSI) M38-A3. Quality control was ensured using the strain recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST): Candida Krusei ATCC6258. The clinical and epidemiological breakpoint values of different Aspergillus species for azole antifungal drugs are shown in Table 1.
Sequencing of cyp51A Gene
Azole-resistant strains were screened based on the results of the in vitro susceptibility testing. Genomic DNA of 15 strains of azole-resistant Aspergillus was extracted using a Fungus Genomic DNA Extraction Kit following the manufacturer’s instructions. The cyp51A sequences of non-WT Aspergillus isolates were amplified using previously described PCR primers (Table 2). The cyp51A thermal cycling profile for amplification was as follows: 95°C for 5 min, followed by 30 cycles of 94°C for 30s, 63°C for 30s, and 72°C for 30s, with a final step of 72°C for 10 min. DNA sequences were compared with the cyp51A sequences of the reference strains of A. fumigatus (GenBank accession AF338659), A. flavus (GenBank accession NRRL3357), A. niger (GenBank accession ATCC 1015), A. tubingensis (GenBank accession F13880), and A. terreus (GenBank accession NIH 2624) to detect point mutations associated with azole resistance.
 |
Table 1 The Clinical and Epidemiological Breakpoints of Different Aspergillus Species to Commonly Used Antifungal Drugs |
 |
Table 2 PCR Primers Used to Amplify the cyp51A Gene |
Statistical Analysis
Statistical analyses and graphing were performed using SPSS 22 and Origin 2018 software. Count data were presented as frequencies or constituent ratios. A P value <0.05 was considered statistically significant for all statistical analyses.
Results
Strain Clinical Information
Aspergillus spp. were isolated from 111 patients with invasive fungal infections between 2021 and 2023. Of the 111 isolates examined, 54 were obtained from male patients, and the remaining 57 were obtained from female patients. The patients ranged in age from 20 to 95 (57.87±17.47) years, with a mean age of 59 years. Regarding clinical origin types, 55.86% of the Aspergillus isolates (62/111) were recovered from ear canal secretions, 40.54% (45/111 isolates) from sputum, 1.80% (2/111 isolates) from bronchoalveolar lavage fluid, and 1.80% (2/111 isolates) from other body fluid secretions. The detailed clinical departments of each patient are presented in Figure 1.
 |
Figure 1 Distribution of Aspergillus isolates in the clinical department. |
Strain Identification and Department Distribution
Based on molecular identification, the most common species was A. fumigatus (n=45), followed by A. niger (n=20), A. tubingensis (n=18), A. terreus (n=15), A. flavus (n=7), A. welwitschiae (n=3), A. fumigatiaffinis (n=2), and A. lentulus (n=1) (Figure 2). In the otolaryngology department, A. tubingensis accounted for 30%, A. niger for 21.67%, A. fumigatus and A. terreus for 20%. In the respiratory department, A. fumigatus accounted for 60.60% and A. niger accounted for 18.18%. A. fumigatus accounted for 87.5% of patients in the department of intensive care medicine (Figure 3).
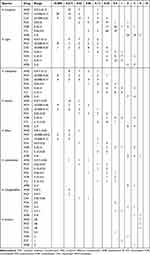 |
Table 3 MICs/MECs Range of Various Antifungal Agents Against Different Aspergillus Species |
 |
Figure 2 Molecular identification of Aspergillus species. |
 |
Figure 3 The source distribution of Aspergillus species specimens. The y axis shows the number of isolates. |
Antifungal Susceptibility Testing
The antifungal resistance of the 111 Aspergillus isolates against seven active antifungals are shown in Table 3. For echinocandins, all Aspergillus isolates revealed lower MECs and Modal MECs ≤0.25 μg/mL to the AND, CAS, and MCF. For AMB, AMB had significantly higher MICs for Aspergillus species than other antifungal drugs. According to EUCAST, all 111 strains of Aspergillus species had MICs ≥2.00 μg/mL to AMB. A. fumigatus and A. niger were resistant to AMB at 2 μg/mL, with 46.67% (21/45) A. fumigatus resistant to AMB and 45.00% (9/20) A. niger being resistant to AMB. A. flavus and A. terreus were resistant to AMB at 4 μg/mL, 14.29% (1/7) of A. flavus isolates were resistant to AMB, and 0% (0/20) of A. terreus isolates were resistant to AMB. For azoles, the most clinical Aspergillus species had MICs ≤0.50 μg/mL for POS (98.20%, n=109) and ITZ (91.90%, n=102). The prevalence of VOR resistance was 6.67% in A. fumigatus, 28.57% in A. flavus, 16.67% in A. tubingensis, 10.00% in A. niger, and 20.00% in A. terreus, which are summarized in Figure 4. In addition, the drug susceptibility characteristics of the six cryptic Aspergillus species found in this study showed that the three strains of A. welwitschiae were sensitive to seven antifungal agents. A. lentulus and A. fumigatiaffinisare resistant to VOR, and the MIC for AMB is 2–8 μg/mL. Of the 111 isolates, 15 that showed azole-resistance to the tested antifungal drugs (3 A. fumigatus, 2 A. flavus, 3 A. tubingensis, 3 A. terreus, 2 A. niger, 1 A. lentulus and 1 A. fumigatiaffinis) were identified as azole-resistant.
 |
Table 4 Characterisation of cyp51A Amino Acid Substitutions in Fifteen Clinical Aspergillus Species Isolates With Triazole MICs Above the ECV |
Cyp51A Mutation Phenotypes
Table 4 shows cyp51A mutations found in azole-resistant Aspergillus strains. Three A. fumigatus strains were azole resistant, had no tandem repeat in its cyp51A promoter region, but one of them (XJ-04) harbored the (F46Y, M172V, and D255E) mutation, while one strain (XJ-14) carried a G138 substitution, and another one (XJ-05) had no mutation in the cyp51A gene. Of the 3 non-WT A. tubingensis isolates, one (XJ-08) harbored the (G202, I335, and F345) mutation, two (XJ-01 and XJ-09) harbored the (G202 and F345) mutation. Two A. niger strains, one (XJ-11) had the T57A mutation, one (XJ-12) carried the (F29V and T57A) mutation in the cyp51A gene. In A. terreus, F200L were found only in (XJ-10) isolates, two (XJ-03, XJ-06) azole-resistant strains did not have mutations in the cyp51A gene. Likewise, two A. flavus isolates (XJ-02 and XJ-07) showed the amino acid substitution (P55, K130, F182, Y247F, and P388). Among the cryptic species, only A. lentulus showed the F46Y/N248T mutation combination.
Discussion
In this study, we report the findings from the survey of 111 Aspergillus species isolated from patients diagnosed with aspergillosis at the First Affiliated Hospital of Xinjiang Medical University. We characterized the species distribution and in vitro susceptibility to seven antifungal agents. For the azole-resistant Aspergillus species, we further determined the azole non-susceptible or non-wild type mechanisms by focusing on mutations in cyp51A. Our results provide a regional perspective on the management of patients with aspergillosis and pathogen characterization in China.
A total of 111 strains of Aspergillus species were collected and mainly isolated from ear secretion specimens, followed by sputum. The main sources of departments were otolaryngology department, respiratory department, and intensive medicine department. Sputum having A. fumigatus isolated in high proportion from specimens. The current study’s finding is similar to the previously reported 20-year retrospective study from China, indicating the endemic nature of A. fumigatus as a respiratory tract pathogen in the region.19 The A. niger and A. tubingensis were isolated in high proportions from ear secretion specimens. The association between A. niger, A. tubingensis and ear infection has been well documented in previous studies, which reported that almost more than half of otomycosis cases are caused by A. niger and A. tubingensis.20 Data reported by Li from China21 showed that A. tubingensis and A. niger were the main pathogenic fungi causing otomycosis in China, and there was no difference in their proportion. This may explain why the predominance of A. tubingensis in otomycosis in western China is likely to be due to dry, dusty, and windy environments.
In the present study, A. fumigatus was the most frequently isolated species, accounting for 40.54% of the isolates collected, similar to the rates observed in several epidemiological studies in other countries.22,24 In our study, A. niger was the most common non-fumigatus Aspergillus species, which is in accordance with previous studies in Switzerland and Korea.25,26 However, A. flavus was reported as the second most common species in the United States, Iran, and Brazil.27–29 A. terreus and A. niger were the third and fourth most common isolates, respectively, indicating geographical variations in the prevalence of different species. For example, this could be due to the arid climate in Iran, which favors the growth of thermo-tolerant fungi such as A. flavus. This would lead to a higher prevalence of A. flavus in these regions, which is supported by the aspergillosis surveys conducted in these countries.
Regarding the antifungal susceptibility testing, echinocandins showed good activity against the Aspergillus isolates in this collection, while anidulafungin and micafungin can effectively inhibit the growth of more than 98% of strains when MEC is ≤0.12μg/mL, and CAS can effectively inhibit the growth of all strains when MEC is ≤0.25μg/mL. AND and MCF appeared to be more potent than CAS, which is consistent with the results of several previous studies.30,31 Owing to the limited use of echinocandins in clinical settings in China, all three echinocandins presented ideal potency against Aspergillus in vitro, and no resistant isolates were detected. Amphotericin B is widely used in hospitals as a salvage and last-line drug to treat severe and urgent cases of triazole-resistant Aspergillus infections.32,33 In our study, the AMB MICs of all 111 isolates were ≥2 µg/mL. Although little is known about the worldwide susceptibility of Aspergillus species to AMB, up to now, the identical incidence of high AMB MIC has been described in Korea and Canada.26,34 Nonetheless, the reasons behind the emergence of high AMB resistance rates in these two geographic populations are still unknown. Resistance to AMB may involve two mechanisms. One is a mutation in the synthesis pathway that leads to a decrease in ergosterol concentration in the cell membrane and the other is an increase in the production of catalase, which can protect cells from oxidative stress caused by the drug. Resistance to AMB is usually due to a reduction in ergosterol or a change in the ergosterol biosynthesis pathway that results in a decrease in the target lipid on the plasma membrane, thereby reducing the ability to bind AMB,35 spore germination upon UV irradiation,36 and mutations in genes encoding sphingolipid FEN1 and SUR4.37 In our study, 37.84% of the Aspergillus species were resistant to AMB. Therefore, the physicians in the region under study should carefully consider the prescription of AMB for Aspergillus infection treatment, and further exploration should be conducted to investigate the reasons for the widespread emergence of AMB resistance in this area. ITZ was the most efficient antifungal agent against this set of isolates (100.00% of the isolates were susceptible to ITZ), followed by POS (99.10%) and VOR (88.29%). The local rates of resistance to azoles in Aspergillus species was 13.51%, with 6.67% in A. fumigatus. In agreement with our findings, the susceptibility profiles of 227 clinical Aspergillus species collected from the northern Portugal also showed that most of the isolates were susceptible to ITZ (95.8%), VOR (97.4%), and POS (84.7%).38 Moreover, in Anhui, China,39 a reduced susceptibility to VOR was observed in 4.11% of the Aspergillus isolates, whereas none of them were resistant to POS and ITZ. This rate of resistance has been attributed to long-term azole therapy in patients with chronic aspergillosis in addition to cross-resistance to agricultural triazoles.
We also identified three other cryptic species that contributed 5.41% of all isolated Aspergillus species. It has been reported that in Iran, A. welwitschiae (former name A. awamori) in section Nigri serves as the pathogen of otomycosis,40,41 and the species has also been isolated from human nails, causing onychomycosis.41 A. lentulus and A. fumigatiaffinis are cryptic species of A. fumigatus complex. They have been observed to exhibit high MICs for all triazoles in several studies and are considered to have intrinsic azole resistance. In this study, the A. lentulus isolates had MICs of 0.5 µg/mL, 2 µg/mL, and 1 µg/mL to POS, VOR, and ITR, respectively. Two A. fumigatiaffinis isolates were VOR resistant. Although these data were suggestive of intrinsic resistance to azoles in these species, it should not be overlooked that susceptibility varied among isolates. Notably, POS appears to retain remarkable antifungal activity against these cryptic species. This suggests the necessity of a surveillance program on azole resistance in non-fumigatus Aspergillus species as well as their genetic mechanisms.
Drug resistance mechanisms, such as mutations in cyp51A, increase in cyp51A expression, upregulation of efflux pumps, and other mechanisms, have been reported in A. fumigatus.33 However, the emergence of non-synonymous hotspot mutations in the cyp51 gene represents the main molecular mechanism associated with azole resistance. However, little information is available regarding other pathogenic Aspergillus species. Here, we attempted to understand the relationship between drug resistance and cyp51A mutations in Aspergillus species. In our study, we found amino acid substitutions (F46Y, M172V, D255E, and G138) in the cyp51A protein of resistant A. fumigatus. The resistance mutations (G138) tend to arise during prolonged treatment of chronic aspergillosis with azole drugs, which has been exemplified by several cases.42,43 Other mutation-harboring A. fumigatus strains found in our study, including amino acid substitution (F46Y, M172V, and D255E) are scarcely related to azole resistance.44,45 In addition, in one azole-resistant A. fumigatus isolate, none of the mutations were found, indicating that the elevated VRC MICs found for these strains could be due to other mechanisms. Three non-synonymous mutations (P55, K130, F182, Y247F, and P388) were found to be associated with drug resistance in the cyp51A sequence of resistant strains of A. flavus.18 Regarding our results in the A. niger, sequence analysis of cyp51A revealed no general correlation between amino acid changes and azole resistance, since most of the mutations were present in both WT and non-WT strains. This is the case for F29V and T57A substitutions in A. niger, which were previously reported by others as well.17,46 However, the G202, I335, and F345 amino acid substitutions might have a role in the triazole resistance of A. tubingensis, since the number of isolates tested in this study is limited, confirmatory studies should be performed in order to support our data. To our knowledge, the mutations F200L found in these isolates have not yet been reported. However, it remains unclear whether they are the cause of azole resistance in these strains, and further studies are required to clarify their exact role. Regarding A. terreus, only one resistant isolate of A. terreus showed the amino acid substitution F200L. To the best of our knowledge, this is the first report of this point mutation in the cyp51A gene of A. terreus, and no mutations were found in the cyp51A gene of the two other azole-resistant A. terreus isolates.
A recently conducted multicentre study showed that azole-resistant A. fumigatus was found in 1.3% of environmental and 3.3% of clinical isolates. TR34/L98H mutations in the cyp51A gene were detected in 47.4% of the azole-resistant A. fumigatus isolates. However, in 52.6% of phenotypically resistant isolates, no mutations were detected within the cyp51A gene.47 Moreover, the prevalence of azole-resistant A. fumigatus in a Danish national surveillance study was 6.1% (66/1083) at the patient level, and TR34/L98H was the most common alteration, but non-cyp51A-mediated resistance accounted for 19.7% (13/66).23 In our study, 26.7% (4/15) Aspergillus strains did not find mutation in the cyp51A gene. To fill a gap in our understanding of the mechanism for azole resistance in the non-cyp51A strains, we highly recommend further and more extensive monitoring of the soil exposure to fungicides in agricultural and hospital areas, to determine trends in the rate of azole-resistant Aspergillus species and to investigate the other mechanisms of resistance such as overexpression of efflux pumps, gain-of-function mutations in transcription factors, mutations in regulatory and sterol biosynthesis elements, and mutations within the HMG-CoA reductase-encoding gene (hmg1), which encodes HMG-CoA reductase in Aspergillus species.
Conclusion
In conclusion, this study provides a comprehensive analysis of the clinical distribution, antifungal susceptibility, and resistance mechanisms of Aspergillus species in Xinjiang, China. The findings highlight the predominance of A. fumigatus as the most common pathogenic species, alongside a diverse range of other Aspergillus species, including cryptic species with intrinsic resistance to azoles. The high prevalence of amphotericin B resistance observed in this region underscores the need for cautious use of this antifungal agent in clinical practice. Additionally, the identification of cyp51A mutations in azole-resistant strains suggests that these genetic alterations may play a significant role in mediating resistance, although further research is needed to fully elucidate the underlying mechanisms. Here, we emphasize the need for continuous surveillance of fungal infections in the hospital environment, which aids in overcoming the knowledge gap regarding the global fungal burden of infections and antifungal resistance, thus supporting public health interventions.
Ethics Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical approval for the study was obtained from the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University. All patients consented to being involved in this study (Approval number: K202405-15).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was financed by the Xinjiang Uygur Autonomous Region “Tianshan Elite Medical and Health High-level Talent Training Program Young Backbone Talents” (No. TSYC202301B029), the Xinjiang Nature Science Foundation of China (No. 2021D01E30).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Verweij PE, Chowdhary A, Melchers WJG, et al. Azole Resistance in Aspergillus fumigatus: can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin Infect Dis. 2016;62(3):362–368. doi:10.1093/cid/civ885
2. Lockhart SR, Frade JP, Etienne KA, et al. Azole Resistance in Aspergillus fumigatus Isolates from the ARTEMIS Global Surveillance Study Is Primarily Due to the TR/L98H Mutation in the cyp51A Gene. Antimicrob Agents Chemother. 2011;55(9):4465–4468. doi:10.1128/AAC.00185-11
3. Patterson TF, Thompson GR, Denning DW, et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;63(4):e1–e60. doi:10.1093/cid/ciw326
4. Lestrade PPA, Meis JF, Melchers WJG, et al. Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin Microbiol Infect. 2019;25(7):799–806. doi:10.1016/j.cmi.2018.11.027
5. Meijer EFJ, Dofferhoff ASM, Hoiting O, et al. Azole-Resistant COVID-19-Associated Pulmonary Aspergillosis in an Immunocompetent Host: a Case Report. J Fungi. 2020;6(2):79. doi:10.3390/jof6020079
6. Alastruey-Izquierdo A, Mellado E, Peláez T, et al. Population-Based Survey of Filamentous Fungi and Antifungal Resistance in Spain (FILPOP Study). Antimicrob Agents Chemother. 2013;57(9):4604. doi:10.1128/AAC.01287-13
7. van der Linden JWM, Arendrup MC, Warris A, et al. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg Infect Dis. 2015;21(6):1041–1044. doi:10.3201/eid2106.140717
8. Verweij PE, Howard SJ, Melchers WJG, et al. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat. 2009;12(6):141–147. doi:10.1016/j.drup.2009.09.002
9. Parker JE, Warrilow AGS, Price CL, et al. Resistance to antifungals that target CYP51. J Chem Biol. 2014;7(4):143–161. doi:10.1007/s12154-014-0121-1
10. Shishodia SK, Tiwari S, Shankar J. Resistance mechanism and proteins in Aspergillus species against antifungal agents. Mycology. 2019;10(3):151–165. doi:10.1080/21501203.2019.1574927
11. Buil JB, Hare RK, Zwaan BJ, et al. The fading boundaries between patient and environmental routes of triazole resistance selection in Aspergillus fumigatus. PLoS Pathog. 2019;15(8):e1007858. doi:10.1371/journal.ppat.1007858
12. Hagiwara D, Arai T, Takahashi H, et al. Non-cyp51A Azole-Resistant Aspergillus fumigatus Isolates with Mutation in HMG-CoA Reductase. Emerg Infect Dis. 2018;24(10):1889–1897. doi:10.3201/eid2410.180730
13. Hortschansky P, Misslinger M, Mörl J, et al. Structural basis of HapE(P88L)-linked antifungal triazole resistance in Aspergillus fumigatus. Life Sci Alliance. 2020;3(7):e202000729. doi:10.26508/lsa.202000729
14. Souza A, Ge W, Wiederhold NP, et al. hapE and hmg1 mutations are drivers of cyp51A-independent pan-triazole resistance in an aspergillus fumigatus clinical isolate. Microbiology Spectrum. 2023;11(3):e0518822. doi:10.1128/spectrum.05188-22
15. Bader O, Weig M, Reichard U, et al. cyp51A-Based Mechanisms of Aspergillus fumigatus Azole Drug Resistance Present in Clinical Samples from Germany. Antimicrob Agents Chemother. 2013;57(8):3513. doi:10.1128/AAC.00167-13
16. Arendrup MC, Jensen RH, Grif K, et al. In Vivo Emergence of Aspergillus terreus with Reduced Azole Susceptibility and a Cyp51a M217I Alteration. J Infect Dis. 2012;206(6):981–985. doi:10.1093/infdis/jis442
17. Hashimoto A, Hagiwara D, Watanabe A, et al. Drug Sensitivity and Resistance Mechanism in Aspergillus Section Nigri Strains from Japan. Antimicrob Agents Chemother. 2017;61(8). doi:10.1128/AAC.02583-16
18. Lucio J, Gonzalez-Jimenez I, Rivero-Menendez O, et al. Point Mutations in the 14-α Sterol Demethylase Cyp51A or Cyp51C Could Contribute to Azole Resistance in Aspergillus flavus. Genes. 2020;11(10):1217. doi:10.3390/genes11101217
19. Yang X, Chen W, Liang T, et al. A 20-Year Antifungal Susceptibility Surveillance (From 1999 to 2019) for Aspergillus spp. and Proposed Epidemiological Cutoff Values for Aspergillus fumigatus and Aspergillus flavus: a Study in a Tertiary Hospital in China. Front Microbiol. 2021;12:680884. doi:10.3389/fmicb.2021.680884
20. Nandyal CB, Choudhari AS, Sajjan NB. A Cross sectional study for Clinico mycological Profile of Otomycosis in North Karnataka. Int J Med Health Sci. 2015;4(1):64–69. doi:10.1128/mBio.02563-18
21. Li Y, Wan Z, Liu W, et al. Identification and susceptibility of Aspergillus section nigri in China: prevalence of species and paradoxical growth in response to echinocandins. J Clin Microbiol. 2015;53(2):702–705. doi:10.1128/JCM.03233-14
22. Krishnan S, Manavathu EK, Chandrasekar PH. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses. 2009;52(3):206–222. doi:10.1111/j.1439-0507.2008.01642.x
23. Risum M, Hare RK, Gertsen JB, et al. Azole resistance in Aspergillus fumigatus. The first 2-year’s data from the Danish National Surveillance Study, 2018-2020. Mycoses. 2022;65(4):419–428. doi:10.1111/myc.13426
24. Bilal H, Zhang D, Shafiq M, et al. Epidemiology and antifungal susceptibilities of clinically isolated Aspergillus species in South China. Epidemiol Infect. 2023;151. doi:10.1017/S095026882300167X
25. Ragozzino S, Goldenberger D, Wright PR, et al. Distribution of Aspergillus Species and Prevalence of Azole Resistance in Respiratory Samples From Swiss Tertiary Care Hospitals. Open Forum Infect Dis. 2022;9(2):ofab638. doi:10.1093/ofid/ofab638
26. Heo MS, Shin JH, Choi MJ, et al. Molecular Identification and Amphotericin B Susceptibility Testing of Clinical Isolates of Aspergillus From 11 hospitals in Korea. Ann Lab Med. 2015;35(6):602. doi:10.3343/alm.2015.35.6.602
27. Negri CE, Gonçalves SS, Xafranski H, et al. Cryptic and Rare Aspergillus Species in Brazil: prevalence in Clinical Samples and In Vitro Susceptibility to Triazoles. J Clin Microbiol. 2014;52(10):3633. doi:10.1128/JCM.01582-14
28. Xu X, Naseri A, Houbraken J, et al. Identification and in vitro antifungal susceptibility of causative agents of onychomycosis due to Aspergillus species in Mashhad, Iran. Sci Rep. 2021;11:1. doi:10.1038/s41598-021-86038-z
29. Balajee SA, Kano R, Baddley JW, et al. Molecular Identification of Aspergillus Species Collected for the Transplant-Associated Infection Surveillance Network. J Clin Microbiol. 2009;47(10):3138. doi:10.1128/JCM.01070-09
30. Schmitt HJ, Bernard EM, Andrade J, et al. MIC and fungicidal activity of terbinafine against clinical isolates of Aspergillus spp. Antimicrob Agents Chemother. 1988;32(5):780. doi:10.1128/aac.32.5.780
31. Pfaller MA, Boyken L, Hollis RJ, et al. In vitro Susceptibility of Clinical Isolates of Aspergillus spp. to Anidulafungin, Caspofungin, and Micafungin: a Head-to-Head Comparison Using the CLSI M38-A2 Broth Microdilution Method. J Clin Microbiol. 2009;47(10):3323. doi:10.1128/JCM.01155-09
32. Zavrel M, Esquivel BD, White TC. The Ins and Outs of Azole Antifungal Drug Resistance: molecular Mechanisms of Transport. In: Gotte M, Berghuis A, Matlashewski G, Wainberg M, Sheppard D, editors. Handbook of Antimicrobial Resistance. New York: Springer; 2014:1–27. doi:10.1007/978-1-4939-0667-3_29-1
33. Zakaria A, Osman M, Dabboussi F, et al. Recent trends in the epidemiology, diagnosis, treatment, and mechanisms of resistance in clinical Aspergillus species: a general review with a special focus on the Middle Eastern and North African region. J Infect Public Health. 2020;13(1):1–10. doi:10.1016/j.jiph.2019.08.007
34. Ashu EE, Korfanty GA, Samarasinghe H, et al. Widespread amphotericin B-resistant strains of Aspergillus fumigatus in Hamilton, Canada. Infect Drug Resist. 2018;11:1549–1555. doi:10.2147/IDR.S170952
35. Ellis D. Amphotericin B: spectrum and resistance. J Antimicrob Chemother. 2002;49(1):7–10. doi:10.1093/jac/49.suppl_1.7
36. Manavathu EK, Alangaden GJ, Chandrasekar PH. In-vitro isolation and antifungal susceptibility of amphotericin B-resistant mutants of Aspergillus fumigatus. J Antimicrob Chemother. 1998;41(6):615–619. doi:10.1093/jac/41.6.615
37. Sharma S, Alfatah M, Bari VK, Rawal Y, Paul S, Ganesan K. Sphingolipid biosynthetic pathway genes FEN1 and SUR4 modulate amphotericin B resistance. Antimicrob Agents Chemother. 2014;58(4):2409–2414. doi:10.1128/AAC.02130-13
38. Pinto E, Monteiro C, Maia M, et al. Aspergillus Species and Antifungals Susceptibility in Clinical Setting in the North of Portugal: cryptic Species and Emerging Azoles Resistance in A. fumigatus. Front Microbiol. 2018;9:1656. doi:10.3389/fmicb.2018.01656
39. Wang Y, Zhang L, Zhou L, et al. Epidemiology, Drug Susceptibility, and Clinical Risk Factors in Patients With Invasive Aspergillosis. Front Public Health. 2022;10:1. doi:10.3389/fpubh.2022.835092
40. Szigeti G, Sedaghati E, Mahmoudabadi AZ, et al. Species assignment and antifungal susceptibilities of black aspergilli recovered from otomycosis cases in Iran. Mycoses. 2012;55(4):333–338. doi:10.1111/j.1439-0507.2011.02103.x
41. Tsang CC, Hui TWS, Lee KC, et al. Genetic diversity of Aspergillus species isolated from onychomycosis and Aspergillus hongkongensis sp. nov. with implications to antifungal susceptibility testing. Diagnostic Microbiol Infect Dis. 2016;84(2):125–134. doi:10.1016/j.diagmicrobio.2015.10.027
42. Kang Y, Li Q, Yao Y, et al. Epidemiology and Azole Resistance of Clinical Isolates of Aspergillus fumigatus from a Large Tertiary Hospital in Ningxia, China. Infect Drug Resist. 2024;17:427–439. doi:10.2147/IDR.S440363
43. Bellete B, Raberin H, Morel J, et al. Acquired resistance to voriconazole and itraconazole in a patient with pulmonary aspergilloma. Med Mycol. 2010;48(1):197–200. doi:10.3109/13693780902717018
44. Howard SJ, Arendrup MC. Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol. 2011;49(1):S90–95. doi:10.3109/13693786.2010.508469
45. Escribano P, Rodríguez-Sánchez B, Díaz-García J, et al. Azole resistance survey on clinical Aspergillus fumigatus isolates in Spain. Clin Microbiol Infect. 2021;27(8):1170.e1–1170.e7. doi:10.1016/j.cmi.2020.09.042
46. Iatta R, Nuccio F, Immediato D, et al. Species Distribution and In Vitro Azole Susceptibility of Aspergillus Section Nigri Isolates from Clinical and Environmental Settings. J Clin Microbiol. 2016;54(9):2365–2372. doi:10.1128/JCM.01075-16
47. Ener B, Ergin Ç, Gülmez D, et al. Frequency of azole resistance in clinical and environmental strains of Aspergillus fumigatus in Turkey: a multicentre study. J Antimicrob Chemother. 2022;77(7):1894–1898. doi:10.1093/jac/dkac125
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Epidemiology of Clinically Significant Aspergillus Species from a Large Tertiary Hospital in Shanghai, China, for the Period of Two Years
Zhang Y, Wang S, Zhou C, Zhang Y, Pan J, Pan B, Wang B, Hu B, Guo W
Infection and Drug Resistance 2023, 16:4645-4657
Published Date: 17 July 2023


