Back to Journals » Journal of Inflammation Research » Volume 18
Molecular Mechanisms of Immunoinflammatory Infiltration and Ferroptosis in Arthritis Revealed by a Combination of Bioinformatics and Single-Cell Analysis
Authors Song C, Song W, Liu Y, Zhou D, Cai W, Mei Y, Liu F , Jiang F, Chen F, Liu Z
Received 18 November 2024
Accepted for publication 12 February 2025
Published 18 February 2025 Volume 2025:18 Pages 2409—2432
DOI https://doi.org/10.2147/JIR.S503618
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Kattepura Krishnappa Dharmappa
Chao Song,1,2 Weijun Song,1,3 Yong Liu,1 Daqian Zhou,1 Weiye Cai,1 Yongliang Mei,1 Fei Liu,1,2 Feng Jiang,1 Feng Chen,1,2 Zongchao Liu1,4
1Department of Orthopedics and Traumatology, The Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University, Luzhou, Sichuan Province, People’s Republic of China; 2Department of Orthopedics, RuiKang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning, People’s Republic of China; 3Department of Orthopedics, Affiliated Sport Hospital of CDSU, Chengdu, Sichuan Province, People’s Republic of China; 4Luzhou Longmatan District People’s Hospital, Luzhou, Sichuan Province, People’s Republic of China
Correspondence: Zongchao Liu; Feng Chen, The Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University, No. 182 Chunhui Road, Longmatan District, Luzhou City, Sichuan Province, People’s Republic of China, Email [email protected]; [email protected]
Background: Osteoarthritis (OA) is a widespread chronic inflammatory disease in orthopedics, and its molecular mechanisms are still poorly understood.
Objective: The purpose of this work was to detect the immunological infiltration of OA and the manner of cell death utilizing bioinformatics and single-cell analysis in order to provide guidelines for clinical therapy and medicine.
Methods: Ferroptosis -associated genes were sourced from the ferroptosis Database, single-cell and bioinformatic expression profiles were chosen from the Gene Expression Comprehensive Database, and OA gene information was taken from GeneCards. To ascertain the categorization status of OA cells, single-cell analysis was conducted. Protein-protein interaction networks were established by SRING analysis, and functional enrichment was examined in the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases. The important proteins of immune-ferroptosis death in OA were elucidated through co-analysis. Last but not least, network pharmacology and molecular docking support the mechanism by which resveratrol controls Ferroptosis in OA.
Results: The development of OA was found to be tightly related to chondrocytes and immune cells, particularly T and macrophage cells, according to single-cell analysis profile. In patients with OA, immune infiltration also revealed a notable infiltration of T cells, B cells, NK cells, monocytes, and macrophages. The hub genes were shown to be enriched in immunological responses, chemokine-mediated signaling pathways, and inflammatory responses, according to enrichment analysis. The main signaling pathways included autophagy, ferroptosis, the HIF-1 signaling pathway, the PI3K-Akt signaling pathway, and the FoxO signaling pathway. Ferroptosis is a significant cell death mechanism that contributes to the advancement of osteoarthritis. Ferroptosis in chondrocytes is lessened by resveratrol regulation of GPX4, TFRC, and SLC7A11.
Conclusion: Various immune cell infiltrates, especially T cells and macrophages, play an important role in the progression of OA, and resveratrol ameliorates OA by modulating chondrocyte ferroptosis.
Keywords: osteoarthritis, immune infiltration, macrophages, T cells, ferroptosis, resveratrol
Introduction
Degeneration and persistent inflammation of joint cartilage-related structures are the primary pathological manifestations of osteoarthritis (OA), a common chronic degenerative disease in orthopedics that is a major cause of disability in middle-aged and older adults.1,2 Inflammation, immunity, and chondrocyte death—which includes apoptosis, senescence, autophagy, Ferroptosis, and pyroptosis—have all been revealed to be intimately associated with the molecular process of OA.3–5 Chondrocyte death, a significant biological component of cartilage, is a significant contributor to the onset and progression of OA. Studying Ferroptosis and associated pathways in OA is very essential because Ferroptosis, a recently identified mode of programmed cell death, is a key molecular mechanism that contributes to the development of OA. Iron is a necessary trace element that can take part in a number of significant metabolic processes. Thus, preserving the organism’s iron homeostasis is essential for regular cellular functions.6 According to studies, patients with OA have much more iron in their synovial fluid than people without the disease, and the amount of damage to their knee cartilage is positively connected with their serum ferritin levels.7 According to other research, OA progression is accelerated by chondrocyte iron mortality, and iron-overloaded mice show increased chondrocyte destruction. According to these findings, the body’s aberrant iron buildup may cause apoptosis and cartilage degradation, which would accelerate the development and course of OA.8 There are not many studies on immune cells and iron-death, but a lot of research has just started to explore the intricate functions of iron-death-related pathways in various facets of immune function. On the one hand, immune response may be hampered by Ferroptosis in immune cells. Conversely, Ferroptosis in non-immune cells triggers the production of inflammatory mediators and immune-associated complexes, which in turn notify immune cells of alterations in the internal environment.9 ACSL4 and CD36 have been shown to enhance Ferroptosis in CD8 T cells, whereas SLC7A11, GPX4, or AIFM2 Ferroptosis critical proteins have been shown to suppress Ferroptosis. However, IFNG from CD8 T cells activated STAT3-dependent down-regulation of the XC system, which in turn made Ferroptosis proteins (SLC3A2 and SLC7A11) more expressed.10,11 In addition to T cells, macrophages are also impacted by Ferroptosis.Lipid peroxidation in macrophages is inhibited by activation of the SLC7A110-GSH-GPX4 axis or an increase in iron store proteins brought on by ferritin autophagy mediated by nuclear receptor coactivator 4. On the other hand, exogenous iron ions or erythropoietic effects can cause macrophages to undergo iron-dependent Ferroptosis.12 Dysregulated immune responses are linked to excessive or deficient Ferroptosis, which is linked to an increasing variety of physiological and pathological processes. In addition to clarifying the relationship between immunity and cell death, an understanding of the immunological characterisation and activity of Ferroptosis may help design novel treatment strategies for immunopathological conditions. Thus, we will investigate the connection between immunity and iron mortality during OA in more detail.
Bulk tissue RNA-seq-based bioinformatics techniques, which frequently distinguish between cell types using marker genes and cell shape, are gaining traction. Research in immunology, cancer, and genetics in particular will be greatly impacted by the more precise and objective cell grouping made possible by single-cell sequencing technology. By analyzing each cell’s transcriptome, single-cell sequencing can shed light on the endogenous and external processes that influence immunotherapy response and resistance, as well as the main mechanisms behind disease progression. Thus, scRNA-seq and bulk RNA-seq together can offer fresh insights in biological study.13,14 Resveratrol is a naturally occurring polyphenol that is extremely physiologically active. It can be found in many different foods, including peanuts, tiger nuts, and grapes. According to recent pharmacological research, resveratrol possesses hepatoprotective, antibacterial, anti-inflammatory, anti-aging, anticancer, antioxidant, and estrogen-like properties.15 The other method involving resveratrol is oxidative stress, which neutralizes ROS and boosts the activity of certain antioxidant enzymes to function as a direct antioxidant. Since ROS is a major contributor to Ferroptosis, we proposed that resveratrol may help OA by taking part in both the immune response and the Ferroptosis process.
The immune cells and important markers in the development of OA were systematically elucidated in the current study through the creative use of bioinformatic analysis in conjunction with single-cell analysis. Additionally, by enriching genes linked to Ferroptosis, we were able to clarify the associated immune-Ferroptosis and the possible molecular mechanisms of OA. Finally, in order to provide direction for the clinical therapy of OA, we employed network pharmacology and experimental validation to clarify the molecular mechanism of immune-iron mortality in OA treated with resveratrol.
Materials and Methods
Single-Cell Analysis
GSE254840 sample data was obtained from the Gene Expression Omnibus (GEO) network, this sample data was made public on June 21, 2024.16 We processed the data using the Sangshin Bean Sprout Analysis Tool, which calculates the mitochondrial content and the content of rRNA using the Percentage Feature Set function in the Seurat package of the R language. It then performs a correlation analysis to examine the relationship between the mitochondrial content and the ncount (UMI) and the nFeature (number of genes). Based on the QC data, cell filtration criteria were established. nFeature < 4000 < 100. The percentage of mitochondrial content was fixed at 10%. When the tsne/UMAP plot of each sample’s cell distribution following cellular filtering is combined with it, cells scattered in the tissue outliers must be handled using the de-batch effect; harmony de-batch should be used this time.17
We employed 2000 highly variable genes for Principal Component Analysis (PCA) dimensionality reduction. The “FindNeighbors” and “FindClusters” (resolution = 0.05) functions, along with Seurat’s Stochastic Neighbor Embedding (t-SNE) technique, were then used to choose the best clusters for display. The top 5 (Top5) expressed genes were employed as a reference in this case, and findmarker was used to select subgroups of marker genes; the higher the ranking, the higher the expression. To elucidate the cell types, we annotate the cells of various subpopulations using the Top5 marker genes, the “SingleR” program, the Human Protein Atlas, and a mix of literature search techniques.18–20 In addition, cell subpopulation abundance calculations were performed using the R package to clarify the proportion of different cell types in different samples.
Bioinformatic Analysis Based on Transcriptome Sequencing
Differential Gene Analysis
The NCBI Gene Expression Public Database provided the bioinformatic information about OA, choosing the GSE55235 dataset, this data was made public on February 21, 2014.21 The GPL96 was used to sequence the GSE55235 data, and Sangerbox 3.0’s Gene Name Conversion tool was used to convert the microarray matrix data into gene name matrix expression data. All public database programs involving human data were approved by the Ethics Committee of the Affiliated Hospital of Traditional Chinese Medicine of Southwest Medical University (Ethics No. V3.0 20250110). After that, the “limma” tool was chosen for differential gene analysis in order to determine which genes were different between samples with osteoarthritis and samples from normal controls. The 1.5-fold and P.value < 0.05 thresholds for differentially expressed genes were established, with positive values signifying the increase of differentially expressed genes (DEGs). Likewise, down-regulation of differentially expressed genes is indicated by a negative number, a P.value < 0.05, and a 1.5-fold multiplicity of differences. Heat maps and volcanoes to display the effects of differently expressed genes.22
Weighted Gene Co-Expression Network Analysis
Weighted Gene Co-expression Network Analysis (WGCNA) is a systems biology methodology used to characterize patterns of gene association between different samples, which can be used to identify highly synergistically varying sets of genes, and to identify candidate biomarker genes or therapeutic targets based on gene-set end-connectivity and gene-set-phenotype associations.23 Using the “WGCNA” package in Sangerbox 3.0 (in R software) to assess the associations between modules and phenotypes, we created gene co-expression networks (MEs) to investigate the co-expression relationship between genes as well as the relationship between genes and phenotypes. Key modules for additional examination were determined by looking at the modules with the greatest absolute values of correlation coefficients. The correlation coefficient between a gene’s expression value and a module’s ME, which shows the relationship between the gene and the module, is called module affiliation (MM). The relationship between a gene’s expression value and a phenotype is represented by the correlation coefficient known as gene significance (GS), which measures the relationship between the two. The hub gene was retrieved with the following thresholds set: 0.8 for the MM, 0.1 for the GS, and 0.1 for the weighting.24,25
Disease Dataset Acquisition and Biomarker Identification
GeneCards (https://www.genecards.org/), a disease database, was searched for illness target proteins using the keyword “arthritis” in order to investigate the targets and consequences of arthritic diseases.26 Genes associated with ferroptosis were obtained from the literature and the FerrDb database (http://www.zhounan.org/ferrdb/current/).27 Differentially expressed genes, modular gene data, and GeneCards data were taken to intersect with ferroptosis-related genes, respectively, and these genes were identified as genes related to the process of arthritis ferroptosis- Osteoarthritis ferroptosis gene set (Osteoarthritis- Ferroptosis gene data OFGD). In this study, PPI network analyses were performed using the string database (http://string-db.org/) on OFGDs obtained for the species restricted to “Homo sapiens” with a confidence value > 0.4. The PPI networks were constructed using Cytoscape software (version 3.9.1).28 In addition, the CytoHubba algorithm, a plug-in for Cytoscape software, was utilized to construct a gene regulatory network for the arthritis-ferroptosis mechanism.29
Enrichment Analysis
To analyze the relevant signaling pathways and mechanisms of action in arthritis, OFGD was subjected to GO and KEGG and Reactome enrichment analysis.30 Import the gene set into Ouyi BioCloud, select Enrichment Analysis in the tool center, and limit the species to “H.sapiens”. Enter the Gene Symbol of the gene set in the common parameters and submit, the results will be displayed in different graphs.
Recognition of Disease Immune Infiltrating Cells
An essential tool for forecasting the progression of an illness and how a patient will react to therapy is the analysis of immune cell infiltration.A preliminary analysis of the immune response during an illness can be conducted by estimating the number of immune cells in a sample using RNA sequencing data, and the CIBERSORT algorithm can use linear support vector regression to deconvolve gene expression profiles.31 We computed the immune cell types of patients with distinct immune patterns in the GSE55235 dataset using the CIBERSORT algorithm in the Sangerbox 3.0 program. We then used stacked graphs to display the immune cell composition of patients with varied immune patterns. Lastly, correlation heatmaps were utilized to illustrate the degree of association between less immune cells, and difference box line plots were employed to display the statistical disparities between various immune cells.
6 Combined analysis of transcriptome intersection genes and single-cell marker genes The intersection genes of OAferroptosis obtained based on general transcriptome sequencing were intersected with Marker genes screened by single-cell analysis to obtain the co-analyzed intersection genes, which further validated the expression and molecular mechanisms of the related genes in different cells. Similarly, the obtained set of intersecting genes was subjected to PPI network analysis using the string database (http://string-db.org/), and the co-analyzed intersecting genes were screened using the CytoHubba algorithm to take the first few core genes. To shed light on the expression of linked genes in various cells, R language packages were used to create UMAP maps of particular gene expression. The screened pivotal genes were queried in the Human Protein Expression Profiling Database (THPA) (https://www.proteinatlas.org/) to obtain the expression of the relevant proteins in the cartilage-associated cells and immune cells, to further validate our specific gene expression results. The database summarizes and aggregates the expression of common human proteins in different tissues, different cell lines, and different immune cells.32,33 Furthermore, the dataset’s dimensionality is decreased by the analytical method known as Uniform Mobility Approximation and Projection (UMAP).34 The Subcellular Localization UMAP was generated using a large number of confocal microscopy images that show patterns of subcellular localization of human proteins. In THPA, this tool provides a new method to visualize and explore high-dimensional protein localization data that constitute the subcellular fraction. To further elucidate the localization and potential mechanisms of action of hub genes in cells, we used subcellular 3D localization and confocal microscopy images to search for subcellular localization of hub genes.
Mechanistic Analysis of Resveratrol for OA
Resveratrol Target Screening and Mechanism Research
The chemical composition and chemical formula of resveratrol were collected from TCMSP (http://tcmspw.com/tcmsp.php) database, and the targets corresponding to the active ingredients were queried. The targets of the obtained compounds were intersected with the OFGD targets, and the “resveratrol-target protein-signaling pathway-arthritis” visualization linkage was constructed using Cytoscape 3.9.1 software. Based on the visualized targets, KEGG enrichment analysis was performed to clarify the molecular mechanism of resveratrol in the treatment of OA.
Molecular Docking and Validation
- Ligand processing: obtain the 3D structure of the proposed docking target in mol2 format from the PubChem database, open the small ligand molecule with AutodockTools 1.5.6, hydrogenate it, charge it, detect the ligand root, search for and define rotatable bonds, and then save it as a pdbqt file.
- Receptor processing: download the core 3D structure of the target protein from the RCSB protein database (www.rcsb.org/) as a docking protein. Add all hydrogen atoms open in AutodockTools 1.5.6, calculate the Gasteiger charge, bind nonpolar hydrogen, define it as a receptor, and save it as a pdbqt file (30).
- Docking parameter settings: determine the coordinates and box size for docking of the Vina molecule, set the parameter exhaustivity to 15, and take the default values for the other parameters.
- Operation and output. Autodock vina 1.1.2 was used for semi-flexible docking, and the best affinity conformation was selected as the final docked conformation.
In vitro Experimental Validation
Experimental Materials
Human chondrocytes were purchased from pricella biological reagent company, Item No.: CP-H107, the cells were derived from human cartilage tissues, prepared by collagenase-neutral protease combined digestion, with a purity of more than 90%. Resveratrol was purchased from Aladdin Company, China. Human articular chondrocyte complete medium was purchased from pricella biological reagent company, item number: CM-H096. GPX4, TFRC, SLC7A11 antibody and GAPDH antibody were purchased from Baxter (Shanghai, China). The supplier of the CCK-8 kit was Beijing Solepol Science and Technology Co.
Resveratrol Solution Preparation
A 100 mmol/L resveratrol stock solution was prepared by dissolving 10 mg of resveratrol solution in 438 μL of DMSO solution, packed and stored in a refrigerator at −20°C. The solution was diluted to the following concentrations using human articular chondrocyte complete medium: 0.01, 0.1, 1, 10 and 100 μmol/L.
Cell Culture
The Human Chondrocyte Culture System was cultured at 37°C in a 5% CO2 incubator, and all experiments were performed when the cells had grown to more than 80%.
CCK-8 Method to Detect the Effect of Drugs on Cell Proliferation Function
To create a cell suspension, chondrocytes were cultivated to 80% fusion and then digested using a digest containing 0.25% trypsin. The cells were cultivated at 37°C in an incubator with 5% CO2 after being injected on 96-well plates with 3000 cells per well. Following a 24-hour period of cell attachment, 200 μL of resveratrol-containing media (containing 0.01, 0.1, 1, 10, and 100 μmol/L) was introduced, and the cells were incubated for another 24- and 48-hour period. After adding 10 μL of CCCK-8 solution to each well, the incubator was left to incubate for two hours. After that, the cells were kept in an incubator with 5% CO 2 for two hours at 37°C. With the aid of a multifunctional enzyme marker, the OD value at 450 nm was measured.
Mitochondrial Membrane Potential Staining
Normal chondrocytes were used as the control group, and the model group was constructed by intervening chondrocytes with 10 ng/mLIL-1β for 24 hours to make OA cell model according to literature search and pre-test. With CCK-8 results, the optimal time and concentration of resveratrol intervention was selected as the treatment group. Take three groups of cells, aspirate the culture fluid, wash the cells twice with PBS, take 1× Incubation Buffer and preheat to 37°C, aspirate 500ul 1× Incubation Buffer, add 1ul JC-1, vortex mix to form JC-1 working solution, take 500ul JC-1 working solution to cover the cells evenly, incubate at 37°C, 5% CO2 The cells were incubated in an incubator at 37°C with 5% CO2 for 15–20 min, washed twice with 1× Incubation Buffer, digested and centrifuged, resuspended with 1× Incubation Buffer, and detected by flow cytometry (BD, USA).
qRT-PCR Validation
As above, RAN was extracted using control, OA model, and OA treatment groups for qRT-PCR analysis. qRT-PCR procedure: total RNA was extracted from the three groups using the Total RNA Extraction Kit. iScriptcDNA Synthesis Kit was then used to reverse-transcribe all the RNA into cDNA and calculate the relative mRNA levels. gAPDH was normalized using the 2 ΔΔCt technique for normalization and analyzed using a Bio-RadCFX96 device for real-time fluorescence quantitative polymerase chain reaction (qPCR) analysis.
Statistical Analysis
Statistical analyses and plots were performed using GraphPad Prism 9.0 software. For data from three or more independent experiments, data were presented as histograms of mean ± standard error (SEM) values. A t-test was used to compare the two groups if the samples fell under a normal distribution, and a non-parametric test was used if they did not. One-way analysis of variance (ANOVA) was used to compare the samples of different groups.Tukey’s method test was used to compare any two groups with *P<0.05 indicating statistically significant difference and **P<0.01 indicating statistically significant difference.
Results
Data Quality Control and Cell Clustering
Single cell sample data were initially processed, filtered, de-batched and analyzed for cell clustering (Supplementary Figures 1–4). Subclusters were categorized using the singleR package, and eight cell clusters were analyzed. The marker gene axis distribution map shows the characteristic genes of different subpopulations (Figure 1A), and through literature query and TOP5 gene match screening, we defined annotations for 8 cell populations (Figure 1B). After screening, we obtained the key marker genes for subgroup 0 as CCL5, GZMA, HOPX, CD3E, CTSW, for subgroup 1 as CSF1R, SAT1, CXCL1, C1QB, C4BPA, C1QC, and for subgroup 2 as APOE, PLTP, LGMN, DRB.1, CD68, SMAP2, key marker genes for subgroup 3 are TIMP1, S100A11, FABP5, FTL, LDHA, NME1, key marker genes for subgroup 4 are CPVL, IRF8, CST3, GPIHBP1, DQA, key marker genes for subgroup 5 are FN1, CLU, CCDC80, DCN, CST6, BGN, key marker subgroup 6 genes were SNX10, SOD2, ILT11B, IFIT2, CCRL2, and key marker genes for subgroup 7 were MS4A1, CD79A, TCF4, S100A10, VIM, ITM2B, BANK1 (Supplement Table S1). In the end, we annotated cell subpopulations 0–7, which are T_cell, DC, monocyte:CD14+, macrophage, monocyte, T_cell:CD8+, and B_cell (Figure 1C). The subpopulation abundance ratios demonstrated that macrophages, dendritic cells, monocytes, and T-cell:CD8+ constituted the majority of the cellular infiltration in the OA samples (Figure 1D).
Results of Differential Gene Analysis
To create an integrated dataset, OA-related data were extracted from GEO data (Supplement Table S2). 3234 DEGs out of 12823 genes were found using differential analysis between the OA and normal groups; of them, 1717 had up-regulated gene expression and 1517 had down-regulated gene expression (Figure 2A and B, Supplement Table S3).
 |
Figure 2 Differential gene analysis: (A) Differential volcano map of GSE55235 samples, (B) Differential heat map of GSE55235 samples. |
Results of WGCNA
In order to create the WGCNA, 12,823 genes were taken out of the combined matrix dataset after the aberrant samples were eliminated and the genes were screened. The average connection value was 1.23 and the scale independence reached 0.86 when the soft threshold power was adjusted to 14 (Figure 3A and B). A total of 20 functional modules were clustered when the cutting height was set to 0.25 and the minimum module size to 30, according to the gene clustering tree (Figure 3C). The individual modules were then subjected to module eigenvector clustering analysis, which demonstrated that the distance between multiple associated modules was large and that the module genes were of research significance (Figure 3D).Then, the modules were correlated with clinical characteristics, black module was positively correlated with osteoarthritis (correlation coefficient=0.90, p=7.5e-8), and brown module was negatively correlated with osteoarthritis (correlation coefficient=−0.82, p=1.1e-5) (Figure 3E).The scatter plot of GS and MM correlation also suggested that the black module was positively associated with osteoarthritis (p=1.7e-154, r=0.75) (Figure 3F). Finally, we extracted 731 module-associated genes. (Supplement Table S4).
Results of Biomarker Identification
In GeneCards, 5215 pertinent illness targets were found after searching disease databases (Supplement Table S5). Ferroptosis-related genes totaling 565 were retrieved from the FerrDb database and the literature (Supplement Table S6). After using intersections with ferroptosis, modular, and differential genes as the GeneCards disease targets of action, a final set of 20 important intersecting genes was generated (Supplement Table S7, Figure 4A and B). Fifteen hub genes were identified by PPI and Cytoscape screening (Figure 4C).
Enrichment Analysis Results
We enriched a set of 20 osteoarthritis ferroptosis genes as a means of exploring the pathogenesis of OA disease. GO enrichment analysis showed that the hub gene was enriched to a total of 646 GO entries, including 464 biological process (BP) entries, 77 cellular component (CC) entries, and 105 molecular_function (MF) entries (Supplement Table S8). Among them, the top 10 enrichment results in BP,CC,MF are shown in chord diagrams and bubble plots (Figure 5A and B), and the results show that the BP analysis during OA is mainly involved in negative regulation of transcription by RNA polymerase II, lipoxygenase pathway, regulation of cellular response to oxidative stress, negative regulation of sprouting angiogenesis, locomotor rhythm, positive regulation of gene expression, viral entry into host cell, long-chain fatty acid biosynthetic process, response to amino acid. CC mainly in melanosome, apical plasma membrane, basolateral plasma membrane, cell surface chromatin, endolysosome lumen, perinuclear region of cytoplasm myelin sheath adaxonal region, MF major influence protein homodimerization activity, protein kinase binding, DNA-binding transcription factor activity, transcription cis-regulatory region binding, collagen binding, double-stranded RNA binding, virus receptor activity. KEGG pathway analysis showed that the OA process involved the expression of 143 signaling pathways (Supplement Table S9), and filtering showed that Cellular Processes mainly included Cellular senescence, Ferroptosis. Environmental Information Processing mainly involved HIF-1 signaling pathway, PI3K-Akt signaling pathway, FoxO signaling pathway (Figure 5C and D). Among them, ferroptosis plays an important role in OA, and querying the KEGG signaling pathway further labeled the articular genes for the ferroptosis process (Figure 6).
 |
Figure 6 Ferroptosis signaling pathway map and joint target protein labeling. |
Analysis of Immune Infiltration Results
We obtained the immune infiltration scores of potential therapeutic genes by CIBERSORT algorithm, and the results of the immune infiltration scores are shown in a stacked plot (Supplement Table S10). The immune infiltration stacked plot showed that the immune cells involved in potential therapeutic genes were mainly CD4_naïve, CD8_naive, Cytotoxic, Exhausted, Tr1, nTreg, iTreg, Th1, Th2, Th17, Tfh, DC, Bcell, Monocyte, Macrophage, NK and other 24 immune cells (Figure 7A). Figure 7B shows the statistical significance of different immune cell’s between OA group and normal group. The results showed that Cytotoxic, Th17, Tfh, Macrophage, and NK were statistically significant (* P<0.01). Figure 7C shows the results of Reactome enrichment analysis of 20 arthritis ferroptosis hub genes, which also showed a close correlation with the immune response.
Results of Combined Transcriptome and Single-Cell Analysis
In order to identify the intersection genes of OA with the marker genes of single cells, transcriptome sequencing was used. Six co analyzed intersection genes—CTSB, KLF2, BRD2, GPX4, TFRC, and SLC7A11—were identified. Through UMAP mapping of the specific gene expression, the distribution of the six important genes in various cellular subpopulations was determined. CTSB, KLF2, BRD2, GPX4, TFRC, and SLC7A11 were shown to be considerably distributed in various subpopulations, according to the fusion of the six pivotal genes map analysis (Figure 8A).
Subsequent examination of individual gene UMAP maps based on these six hub genes revealed that GPX4 was extensively distributed in immune cells and chondrocytes, primarily in T_cell, T_cell:CD8+, with significant distribution; TFRC was primarily distributed in Macrophages; SLC7A11 was distributed with negligible expression in T_cell, Macrophages and expressed in small amounts; CTSB was primarily expressed in Macrophages, Monocytes, T_cell:CD8+, and expressed in significant amounts; BRD2 was not significantly distributed and expressed in small amounts in T_cell and Macrophage; KLF2 was primarily expressed in Macrophage, Monocyte, T_cell:CD8+, and expressed in significant amounts and there was significance in the expression. Using the Human Protein Expression Profile database, we gathered single-cell expression profiles of the six hub genes in skeletal tissues and associated cells to confirm the cellular distribution of the six hub genes in OA. According to the data, T cells and macrophages exhibited the highest levels of expression for the six hub genes (Figure 8B).
Furthermore, 3D and confocal localization of cells revealed that GPX4 was primarily localized in the mitochondria and nucleoplasm of cells; TFRC was localized in vesicles, Golgi, and nucleosomes; SLC7A11 was primarily localized in the vesicles of cells; CTSB was localized in vesicles, Golgi, and nucleoli; KLF2 was localized in the nucleoplasm of human fibroblasts; BRD2 localized to nucleoplasm, cytoplasmic lysosomes, and nuclear speckles; and 3D localization revealed that it was more localized to nuclear speckles (Figure 9A–E). Therefore, we believe that CTSB, KLF2, BRD2, GPX4, TFRC, and SLC7A11 targets are important in the disease prediction of OA, and their molecular mechanisms are closely related to immune cells, especially macrophages. In addition, single-cell subpopulation clustering and bioinformatic immune infiltration analysis suggested that the development of OA was closely related to T cells and macrophages.
Mechanisms of Resveratrol in the Treatment of OA
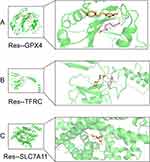
Numerous studies have found that resveratrol can repair chondrocytes to ameliorate arthritis, so we further analyzed its potential ferroptosis-related mechanisms through network pharmacology. Through TCMSP we obtained 106 targets of action for resveratrol, and took the intersection with the OFGD dataset again to obtain 8 therapeutic targets (Figure 10A). In addition, we queried again the target mechanism associated with resveratrol in the Ingredient Target Prediction Database (Figure 10B). The data showed that 40% of the targets of action of resveratrol were related to oxidoreductase. Based on the analysis of KEGG enrichment of 8 therapeutic targets, we found that resveratrol could improve OA by participating in ferroptosis (Supplement Tables S11, 12 and Sheet 1, Figure 10C). Comparing single-cell and coanalyzed intersecting genes by raw letter analysis, we found that GPX4, TFRC, and SLC7A11 are key targets for therapy. Therefore, molecular docking of resveratrol with GPX4, TFRC, and SLC7A11 was performed. The small molecule ligand can spontaneously bind to the protein receptor when the binding energy is <0 kJ mol. If the binding energy is <-5.0 kJ mol or lower, it indicates that both have better binding ability. By docking simulation, both of them have a binding energy <-5 kJ mol, which means they both bind well. This molecular docking result suggests that resveratrol regulates ferroptosis through GPX4, TFRC, and SLC7A11 for the treatment of OA (Figure 11A–C).
By CCK-8 assay, we analyzed that the optimal time and dose of resveratrol intervention in chondrocytes was 10 μmol/L for 24 hours (Figure 12B). Mitochondrial membrane potential staining suggested that the mitochondrial membrane was damaged after modeling of chondrocytes, whereas the mitochondrial membrane potential was restored after intervention with resveratrol compared with the model group, the more normal the mitochondrial structure, the more prominent the red glow. Standard resveratrol could lessen the structural damage of mitochondria, as seen by the considerable reduction of red light in the model group with damaged mitochondria and the partial restoration of red fluorescence in the treatment group (Figure 12A). Ferroptosis signaling pathway related markers GPX4, TFRC, and SLC7A11 were highly expressed in the OA model group (*P<0.05) as detected by qRT-PCR, and the related RNA expression could be inhibited by resveratrol intervention (Figure 12C). The above experimental results preliminarily demonstrated that Ferroptosis-related RNAs were expressed in inflammation-induced chondrocytes, and resveratrol could reverse this process. And the process of Ferroptosis was accompanied by impaired mitochondrial function, which was consistent with the results of colocalization.
Discussion
OA is the primary cause of disability among middle-aged and older adults globally. It is attributed to a multifaceted interaction between genetic predisposition and environmental exposures, encompassing various pathological processes such as oxidative stress, cell death, inflammation, and mediators. Additionally, autoimmunity and ferroptosis have been reported recently.35 Since ferroptosis and OA have been linked in numerous studies, there may be renewed hope for understanding the pathophysiology of arthropathies and identifying potential treatment targets.36 Furthermore, immunological problems and inflammation are the primary etiology of OA, and synovitis and increasing cartilage and subchondral bone disintegration are the primary pathologic alterations.36 Numerous cells have a role in the development of osteoarthritis (OA); immune cells mostly consist of monocytes, macrophages, T-cells, B-cells, and neutrophils, whereas pathogenic process cells primarily consist of chondrocytes, fibroblast-like synoviocytes, and synoviocyte macrophages.37,38 Ferroptosis in T cells and macrophages has been linked in some studies to OA; additionally, there is evidence that ferroptosis may be related to chondrocyte destruction.39,40 However, the relationship between immune cells and chondrocytes as well as ferroptosis has been less well studied, and therefore it is of great importance to study the crosstalk between these three.
The current single-cell analysis demonstrated the immunoinflammatory characteristics of OA by elucidating the presence of immune cells and important pathogenic cells in OA. Eight resident cell populations were found to be T_cell, B_cell, DC, monocyte:CD14+, macrophage, monocyte, T_cell:CD8+, and chondrocytes.As osteoarthritis (OA) advances, innate immune cells release cytokines, chemokines, and matrix metalloproteinases, which cause cartilage and bone to break down. In rheumatoid arthritis, several cytokines have a direct effect on monocytes/macrophages and a pathological role in disease progression.41 IL-1β is a potent pro-inflammatory cytokine that plays a role in bone erosion and cartilage degradation. In a streptococcal cell wall-induced arthritis model, IL-1 mice showed reduced late cell infiltration and cartilage damage, while joint swelling was unaffected.42 It was discovered that IL-1β stimulates macrophages and causes them to produce prostaglandins, reactive oxygen intermediates, and cytokines.43 TNFα is regarded as the primary inflammatory cytokine in OA and plays a key role in the development and upkeep of synovitis, which is generated and activated by macrophages, resulting in an autocrine inflammatory effect.44 The immunopathogenesis of osteoarthritis (OA) remains poorly understood; aside from the recognized involvement of B cells, dysregulated CD4+ T helper cell homeostasis plays a role in the formation and evolution of OA. It has been noted that there is an imbalance between Th17 cells and regulatory T cells, which could result in chronic inflammation.45 An increasing amount of data points to the potential involvement of peripheral helper T cells and circulating follicular T cells in the pathophysiology of OA and its response to treatment.46 In conclusion, whether monocytes/macrophages, T-cells or B-cells, of immune cells play an important role in the development of OA. Furthermore, cell death with apoptotic morphological and molecular features has been detected in OA cartilage, suggesting that chondrocyte death/survival plays a key role in the pathogenesis of OA. Research have demonstrated a strong correlation between immune cells and joint disorders including cartilage degradation. Immune cells like macrophages may produce substances that influence cartilage regeneration, either pro- or anti-inflammatory. But pro-inflammatory substances can also cause chondrocyte death and hasten the deterioration of cartilage.47 Thus, we think that one key molecular mechanism affecting how OA progresses is immune cells’ control on chondrocytes.
We were able to determine the functions of CTSB, KLF2, BRD2, GPX4, TFRC, and SLC7A11 markers in OA by combining single-cell and bioinformatic research. Furthermore, three-dimensional and confocal localization revealed that SLC7A11 was mostly found in cell vesicles, TFRC was found in vesicles, the Golgi apparatus, and nucleosomes, and GPX4 was primarily found in the mitochondria of cells. GPX4 has been reported to be a key regulator of ferroptosis and protective against ferritin damage in cells under various conditions. In a study, reduced levels of GPX4 were found in degenerating chondrocytes, suggesting a potential association between GPX4 expression and OA development.48 In addition, it has been found that GPX4 has a dual function in OA, regulating ferroptosis or oxidative stress and modulating ECM degradation through the MAPK/NFκB signaling pathway.6 The condition of ferroptosis had an impact on both the structure and function of the mitochondria; however, the inhibitors were able to restore the former. Additionally, the inhibition of either transferrin receptor 1 (TFRC) or hypoxia-inducible factor 1α (HIF-1α) prevented the loss of mitochondrial membrane potential caused by IL-1β, which is in line with the findings of GPX4 localization to mitochondria. While TFRC inhibition did not decrease IL-1β-induced HIF-1α expression in chondrocytes, it did downregulate TFRC expression when HIF-1α was inhibited. In addition, upregulation of TFRC promoted elevated 2+ of Fe into chondrocytes, inducing Fenton reaction and lipid peroxidation, which in turn caused ferroptosis, chondrocyte dysfunction.49 An essential part of the amino acid countertransporter protein system xc-, solute carrier family 7 member 11 (SLC7A11) primarily regulates intercellular glutathione (GSH) levels. By suppressing SLC7A11, glutathione activity and intracellular GSH depletion are inhibited, which raises lipid peroxidation and ferroptosis.Numerous investigations have demonstrated that a range of miRNAs can control the activity of SLC7A11.50 Icariin upregulated the expression of GPX4, SLC7A11, and SLC3A2L, and activated the System Xc-/GPX4 axis to decrease ferroptosis in synoviocytes in an erastin-induced cellular model of OA. As a result, we have discovered that GPX4, TFRC, SLC7A11, and other essential ferroptosis proteins are involved in OA.
Even while non-steroidal anti-inflammatory medicines (NSAIDs), corticosteroids, and disease-modifying anti-rheumatic drugs (DMARD) are currently used to treat osteoarthritis (OA), they have a number of negative side effects. In this context, natural compounds possessing strong anti-inflammatory and antioxidant qualities could be a valuable asset for the creation of potential treatments for osteoarthritis. Resveratrol has anti-inflammatory, anticancer, and ROS-scavenging properties.51 Previous studies have shown that resveratrol has beneficial effects on the development and progression of rheumatoid arthritis. Resveratrol can reduce the infiltration of T cells and macrophages into synovial tissue by inhibiting the expression of MCP-1. Numerous studies have shown that one of the main causes of OA and cartilage degeneration is the overexpression of TNF-α, the interaction between T and B cells, and the interaction between macrophages and synovial-like fibroblasts. In addition, TNF-β expressed by natural killer (NK) cells, T cells and B cells triggers further inflammatory cascades. Therefore, resveratrol supplementation by targeting TNF-β may be a potential treatment for RA.52,53 In addition, it has been found that resveratrol can inhibit ferroptosis by activating the SIRT3/FoxO3a pathway, increasing the expression of SOD2 and catalase, decreasing the production of ROS and LPO, compensating the GSH/GPX4 pathway and.54 Another study found that resveratrol may alleviate diabetic periodontitis-induced cellular ferroptosis by regulating SLC7A11/GPX4.55 However, there are fewer studies on whether resveratrol can modulate GPX4, TFRC, and SLC7A11 to improve the ferroptosis of OA. Finally, our research showed that OA is caused by the infiltration of different immune cells, primarily T and macrophage cells, which cause chondrocyte ferroptosis. Natural resveratrol binds to GPX4, TFRC, and SLC7A11 on chondrocyte mitochondria or vesicles. It then mediates mitochondrial autophagy to reduce ROS and inflammation, which in turn prevents chondrocyte ferroptosis and lessens osteoarthritis. There are some shortcomings in this study, one is that the molecular association mechanism of various immune cells with chondrocyte ferroptosis needs to be further discovered and verified, and the other is that whether resveratrol is involved in the regulation of immune cells in arthritis needs to be further discovered and verified. We will further address the above issues in future studies.
Conclusion
In conclusion, we discovered that chondrocyte ferroptosis and different immune cell infiltration, among which T-cell and monocyte/macrophage infiltration are significant, are strongly associated with the development of OA. Resveratrol may alleviate OA by controlling the essential proteins that suppress chondrocyte ferroptosis—GPX4, TFRC, and SLC7A11—that are found in mitochondria and vesicles.
Data Sharing Statement
Publicly available datasets (GSE55235, GSE254840) were analyzed in this study. The dataset was obtained from the GEO (http://www.ncbi.nlm.nih.gov/geo) database. All data are in the manuscript and/or supporting information files. Figure 6 is from the KEGG database and Figure 9 is from the Human Protein Expression Database, which has been reasonably cited in the relevant literature.
Ethics Approval
All public databases involving human data have undergone review and approval by the Ethics Committee of the The Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University.
Acknowledgments
We thank the BioBean (Sheng-Xin-Dou-Ya-Cai) team for providing the user-friendly bioinformatics platform (http://www.sxdyc.com/), which has significantly streamlined and accelerated our research process.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The present study was supported in part by research grants from the Program for Special project of Traditional Chinese Medicine scientific research of Sichuan Science and Traditional Chinese Medicine Administration (nos.2020LC0228), Luzhou’s major scientific and technology research and development project (nos.2022-SYF-42), Joint Innovation Special of the Sichuan Provincial Science and Technology Plan (nos.2022YFS0609/2022YFS0609-B3). Southwest Medical University (SWMU) School-level Scientific Research Program (nos. 2024ZKZ009). There was no additional external funding received for this study. All the funding’s funder was Zongchao Liu, the funder had role in study conceptualization, methodology, supervision, funding acquisition.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Jiang Y. Osteoarthritis year in review 2021: biology. Osteoarthritis Cartilage. 2022;30:207–215. doi:10.1016/j.joca.2021.11.009
2. Abramoff B, Caldera FE. Osteoarthritis: pathology, diagnosis, and treatment options. Med Clin North Am. 2020;104:293–311. doi:10.1016/j.mcna.2019.10.007
3. Liu S, Pan Y, Li T, et al. The role of regulated programmed cell death in osteoarthritis: from pathogenesis to therapy. Int J mol Sci. 2023;25:24. doi:10.3390/ijms25010024
4. Riegger J, Schoppa A, Ruths L, Haffner-Luntzer M, Ignatius A. Oxidative stress as a key modulator of cell fate decision in osteoarthritis and osteoporosis: a narrative review. Cell Mol Biol Lett. 2023;28:76. doi:10.1186/s11658-023-00489-y
5. Rim YA, Nam Y, Ju JH. The Role of Chondrocyte Hypertrophy and Senescence in Osteoarthritis Initiation and Progression. Int J mol Sci. 2020;22:21. doi:10.3390/ijms22010021
6. Miao Y, Chen Y, Xue F, et al. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine. 2022;76:103847. doi:10.1016/j.ebiom.2022.103847
7. Yang J, Hu S, Bian Y, et al. Targeting cell death: pyroptosis, ferroptosis, apoptosis and necroptosis in osteoarthritis. Front Cell Dev Biol. 2021;9:789948. doi:10.3389/fcell.2021.789948
8. Yao X, Sun K, Yu S, et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Translat. 2021;27:33–43. doi:10.1016/j.jot.2020.09.006
9. Chen X, Kang R, Kroemer G, Tang D. Ferroptosis in infection, inflammation, and immunity. J Exp Med. 2021;218.
10. Garg SK, Yan Z, Vitvitsky V, Banerjee R. Differential dependence on cysteine from transsulfuration versus transport during T cell activation. Antioxid Redox Signal. 2011;15:39–47. doi:10.1089/ars.2010.3496
11. Drijvers JM, Gillis JE, Muijlwijk T, et al. Pharmacologic screening identifies metabolic vulnerabilities of CD8(+) T cells. Cancer Immunol Res. 2021;9:184–199. doi:10.1158/2326-6066.CIR-20-0384
12. Klöditz K, Fadeel B. Three cell deaths and a funeral: macrophage clearance of cells undergoing distinct modes of cell death. Cell Death Discov. 2019;5:65. doi:10.1038/s41420-019-0146-x
13. Mikami Y, Grubb BR, Rogers TD, et al. Chronic airway epithelial hypoxia exacerbates injury in muco-obstructive lung disease through mucus hyperconcentration. Sci Transl Med. 2023;15:eabo7728. doi:10.1126/scitranslmed.abo7728
14. Gémes N, Balog J, Neuperger P, et al. Single-cell immunophenotyping revealed the association of CD4+ central and CD4+ effector memory T cells linking exacerbating chronic obstructive pulmonary disease and NSCLC. Front Immunol. 2023;14:1297577. doi:10.3389/fimmu.2023.1297577
15. Li YR, Li S, Lin CC. Effect of resveratrol and pterostilbene on aging and longevity. Biofactors. 2018;44:69–82. doi:10.1002/biof.1400
16. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi:10.1093/nar/gks1193
17. Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257–272. doi:10.1038/s41576-019-0093-7
18. Massimino M, Martorana F, Stella S, et al. Single-Cell Analysis in the Omics Era: technologies and Applications in Cancer. Genes. 2023;15:14. doi:10.3390/genes15010014
19. Zucha D, Kubista M, Valihrach L. Tutorial: guidelines for Single-Cell RT-qPCR. Cells. 2021;11:10. doi:10.3390/cells11010010
20. Lee TL. Single cell genomics. Int J Biochem Cell Biol. 2019;116:105596. doi:10.1016/j.biocel.2019.105596
21. Hu X, Ni S, Zhao K, Qian J, Duan Y. Bioinformatics-Led Discovery of Osteoarthritis Biomarkers and Inflammatory Infiltrates. Front Immunol. 2022;13:871008. doi:10.3389/fimmu.2022.871008
22. Xia L, Gong N. Identification and verification of ferroptosis-related genes in the synovial tissue of osteoarthritis using bioinformatics analysis. Front Mol Biosci. 2022;9:992044. doi:10.3389/fmolb.2022.992044
23. Zeng J, Lai C, Luo J, Li L. Functional investigation and two-sample Mendelian randomization study of neuropathic pain hub genes obtained by WGCNA analysis. Front Neurosci. 2023;17:1134330. doi:10.3389/fnins.2023.1134330
24. Tian Z, He W, Tang J, et al. Identification of Important Modules and Biomarkers in Breast Cancer Based on WGCNA. Onco Targets Ther. 2020;13:6805–6817. doi:10.2147/OTT.S258439
25. Quan Q, Xiong X, Wu S, Yu M. Identification of Immune-Related Key Genes in Ovarian Cancer Based on WGCNA. Front Genet. 2021;12:760225. doi:10.3389/fgene.2021.760225
26. Mo L, Ma C, Wang Z, et al. Integrated Bioinformatic Analysis of the Shared Molecular Mechanisms Between Osteoporosis and Atherosclerosis. Front Endocrinol. 2022;13:950030. doi:10.3389/fendo.2022.950030
27. Zhou N, Yuan X, Du Q, et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2023;51:D571–d582. doi:10.1093/nar/gkac935
28. Zhao C, Sahni S. String correction using the Damerau-Levenshtein distance. BMC Bioinf. 2019;20:277. doi:10.1186/s12859-019-2819-0
29. Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network Analysis and Visualization of Proteomics Data. J Proteome Res. 2019;18:623–632. doi:10.1021/acs.jproteome.8b00702
30. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–d592. doi:10.1093/nar/gkac963
31. Craven KE, Gökmen-Polar Y, Badve SS. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci Rep. 2021;11:4691. doi:10.1038/s41598-021-83913-7
32. Uhlén M, Hallström BM, Lindskog C, Mardinoglu A, Pontén F, Nielsen J. Transcriptomics resources of human tissues and organs. mol Syst Biol. 2016;12:862. doi:10.15252/msb.20155865
33. Chen W, Yang Q, Hu L, et al. Shared diagnostic genes and potential mechanism between PCOS and recurrent implantation failure revealed by integrated transcriptomic analysis and machine learning. Front Immunol. 2023;14:1175384. doi:10.3389/fimmu.2023.1175384
34. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018. doi:10.1038/nbt.4314
35. Jiang W, Chen H, Lin Y, et al. Mechanical stress abnormalities promote chondrocyte senescence - The pathogenesis of knee osteoarthritis. Biomed Pharmacother. 2023;167:115552. doi:10.1016/j.biopha.2023.115552
36. Chang S, Tang M, Zhang B, Xiang D, Li F. Ferroptosis in inflammatory arthritis: a promising future. Front Immunol. 2022;13:955069. doi:10.3389/fimmu.2022.955069
37. Zhao H, Tang C, Wang M, Zhao H, Zhu Y. Ferroptosis as an emerging target in rheumatoid arthritis. Front Immunol. 2023;14:1260839. doi:10.3389/fimmu.2023.1260839
38. He X, Zhang J, Gong M, et al. Identification of potential ferroptosis-associated biomarkers in rheumatoid arthritis. Front Immunol. 2023;14:1197275. doi:10.3389/fimmu.2023.1197275
39. Lai B, Wu CH, Wu CY, Luo SF, Lai JH. Ferroptosis and Autoimmune Diseases. Front Immunol. 2022;13:916664. doi:10.3389/fimmu.2022.916664
40. Zhang Y, Huang X, Qi B, et al. Ferroptosis and musculoskeletal diseases: “Iron Maiden” cell death may be a promising therapeutic target. Front Immunol. 2022;13:972753. doi:10.3389/fimmu.2022.972753
41. Drexler SK, Kong PL, Wales J, Foxwell BM. Cell signalling in macrophages, the principal innate immune effector cells of rheumatoid arthritis. Arthritis Res Ther. 2008;10:216. doi:10.1186/ar2481
42. van den Berg WB, Joosten LA, Kollias G, van De Loo FA. Role of tumour necrosis factor alpha in experimental arthritis: separate activity of interleukin 1beta in chronicity and cartilage destruction. Ann Rheum Dis. 1999;58(Suppl 1):I40–48. doi:10.1136/ard.58.2008.i40
43. Zwerina J, Redlich K, Polzer K, et al. TNF-induced structural joint damage is mediated by IL-1. Proc Natl Acad Sci U S A. 2007;104:11742–11747. doi:10.1073/pnas.0610812104
44. Wijngaarden S, van de Winkel JG, Bijlsma JW, Lafeber FP, van Roon JA. Treatment of rheumatoid arthritis patients with anti-TNF-alpha monoclonal antibody is accompanied by down-regulation of the activating Fcgamma receptor I on monocytes. Clin Exp Rheumatol. 2008;26:89–95.
45. Povoleri GAM, Durham LE, Gray EH, et al. Psoriatic and rheumatoid arthritis joints differ in the composition of CD8+ tissue-resident memory T cell subsets. Cell Rep. 2023;42:112514. doi:10.1016/j.celrep.2023.112514
46. Lucas C, Perdriger A, Amé P. Definition of B cell helper T cells in rheumatoid arthritis and their behavior during treatment. Semin Arthritis Rheum. 2020;50:867–872. doi:10.1016/j.semarthrit.2020.06.021
47. Yan M, Sun Z, Wang J, et al. Single-cell RNA sequencing reveals distinct chondrocyte states in femoral cartilage under weight-bearing load in rheumatoid arthritis. Front Immunol. 2023;14:1247355. doi:10.3389/fimmu.2023.1247355
48. Wang S, Li W, Zhang P, et al. Mechanical overloading induces GPX4-regulated chondrocyte ferroptosis in osteoarthritis via Piezo1 channel facilitated calcium influx. J Adv Res. 2022;41:63–75. doi:10.1016/j.jare.2022.01.004
49. Chen BY, Pathak JL, Lin HY, et al. Inflammation triggers chondrocyte ferroptosis in TMJOA via HIF-1α/TFRC. J Dent Res. 2024;103:712–722. doi:10.1177/00220345241242389
50. Wang D, Fang Y, Lin L, et al. Upregulating miR-181b promotes ferroptosis in osteoarthritic chondrocytes by inhibiting SLC7A11. BMC Musculoskelet Disord. 2023;24:862. doi:10.1186/s12891-023-07003-7
51. Zhang J, Song X, Cao W, et al. Autophagy and mitochondrial dysfunction in adjuvant-arthritis rats treatment with resveratrol. Sci Rep. 2016;6:32928. doi:10.1038/srep32928
52. Deng Q, Peng Z, Liu N, Peng J. Bioinformatics analysis of resveratrol-induced autophagy in rheumatoid arthritis. Asian J Surg. 2024.
53. Karimi A, Azar PS, Kadkhodayi M, et al. A comprehensive insight into effects of resveratrol on molecular mechanism in rheumatoid arthritis: a literature systematic review. Int J Rheum Dis. 2022;25:827–843. doi:10.1111/1756-185X.14356
54. Wang X, Shen T, Lian J, et al. Resveratrol reduces ROS-induced ferroptosis by activating SIRT3 and compensating the GSH/GPX4 pathway. Mol Med. 2023;29:137. doi:10.1186/s10020-023-00730-6
55. Li Y, Huang Z, Pan S, et al. Resveratrol alleviates diabetic periodontitis-induced alveolar osteocyte ferroptosis possibly via regulation of SLC7A11/GPX4. Nutrients. 2023;16:15. doi:10.3390/nu16010015
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Characterization of Ferroptosis-Related Molecular Subtypes with Immune Infiltrations in Neuropathic Pain
Bi YH, Wang J, Guo ZJ, Jia KN
Journal of Pain Research 2022, 15:3327-3348
Published Date: 22 October 2022
Identification and Validation of Ferroptosis-Related LncRNAs Signature as a Novel Prognostic Model for Chronic Lymphocytic Leukemia
Xu Z, Pan B, Li Y, Xia Y, Liang J, Kong Y, Zhang X, Tang J, Wang L, Li J, Xu W, Wu J
International Journal of General Medicine 2023, 16:1541-1553
Published Date: 26 April 2023
Identification of Ferroptosis-Related Biomarkers for Diagnosis and Molecular Classification of Staphylococcus aureus-Induced Osteomyelitis
Shi X, Tang L, Ni H, Li M, Wu Y, Xu Y
Journal of Inflammation Research 2023, 16:1805-1823
Published Date: 26 April 2023
Identification and Analysis of Neutrophil Extracellular Trap-Related Genes in Osteoarthritis by Bioinformatics and Experimental Verification
Luan T, Yang X, Kuang G, Wang T, He J, Liu Z, Gong X, Wan J, Li K
Journal of Inflammation Research 2023, 16:3837-3852
Published Date: 31 August 2023
Bioinformatics and Integrative Experimental Method to Identifying and Validating Co-Expressed Ferroptosis-Related Genes in OA Articular Cartilage and Synovium
Ma J, Yu P, Ma S, Li J, Wang Z, Hu K, Su X, Zhang B, Cheng S, Wang S
Journal of Inflammation Research 2024, 17:957-980
Published Date: 12 February 2024