Back to Journals » Infection and Drug Resistance » Volume 18
Nontuberculous Mycobacterial Pneumonia Caused by Mycobacterium immunogenum: A Case Report and Literature Review
Authors Huang S, Shangguan Y, Guo W, Ji Z, Jin X, Zhao R, Zheng L, Wang Y, Jiang L, Xu K
Received 16 December 2024
Accepted for publication 28 March 2025
Published 13 April 2025 Volume 2025:18 Pages 1859—1865
DOI https://doi.org/10.2147/IDR.S512539
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shujuan Huang,1,* Yanwan Shangguan,2,* Wanru Guo,1 Zhongkang Ji,1 Xiuyuan Jin,1 Ruihong Zhao,1 Lin Zheng,1 Yuping Wang,1 Liangxiu Jiang,1 Kaijin Xu1
1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, People’s Republic of China; 2Infection Control Department, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Kaijin Xu, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University, Hangzhou, People’s Republic of China, Tel +86-0571-87236440, Email [email protected]
Background: Mycobacterium immunogenum is a rare nontuberculous mycobacterium belonging to the Mycobacterium chelonae-abscessus group. Most cases reported in the past decade have been extrapulmonary infections, and reports of nontuberculous mycobacterial pulmonary disease (NTM-PD) caused by M. immunogenum are rare. Herein, we report a case of NTM-PD caused by M. immunogenum.
Case Presentation: An 81-year-old man with a history of chronic kidney disease and cardiovascular disease was hospitalised in 2021 owing to pneumonia. Two sputum cultures tested positive for Mycobacterium. The mycobacterium biochip test identified M. abscessus and Mycobacterium gilvum, and whole-genome sequencing confirmed the identity as M. immunogenum. Antimicrobial drug susceptibility testing showed that the isolate was resistant to imipenem, moxifloxacin, and doxycycline; intermediately sensitive to tobramycin and linezolid; and sensitive to amikacin, cefoxitin, and clarithromycin. The patient was treated with cefoperazone sodium and sulbactam sodium (2 g twice daily) and switched to meropenem (0.5 g every 6 hours) for anti-infection but died due to acute respiratory failure and severe pneumonia before targeted treatment for NTM-PD could be initiated.
Conclusion: NTM-PD is frequently diagnosed at an advanced stage, primarily because its clinical presentation is often atypical and definitive laboratory tests are not readily available. Therefore, greater attention should be paid to the diagnosis and treatment of NTM-PD. Nontuberculous mycobacterial infections, especially clinically rare infections, need to be diagnosed without delay and their antibiotic susceptibility needs to be determined.
Keywords: nontuberculous mycobacterial pulmonary disease, whole-genome sequencing, antimicrobial susceptibility testing, treatment
Introduction
Nontuberculous mycobacteria (NTM), characterized by slow growth and intrinsic antibiotic resistance, are ubiquitous environmental organisms.1 Notably, the prevalence of pulmonary nontuberculous mycobacteria (PNTM) infections has shown a marked upward trend in recent decades. Epidemiological data from the United States reveal a substantial increase in PNTM incidence rates, escalating from 20 to 47 cases per 100,000 population between 1997 and 2007.2 Patients with NTM-PD typically present with a constellation of respiratory and systemic symptoms, including chronic cough, sputum production, hemoptysis, dyspnea, fever, night sweats, weight loss, and fatigue, which may occur in various combinations.3 NTM are categorised as rapidly growing mycobacteria (RGM) or slowly growing mycobacteria.4 RGM form bacterial colonies within 7 days and include the Mycobacterium fortuitum group, Mycobacterium chelonae-abscessus complex, Mycobacterium smegmatis, Mycobacterium mucogenicum, Mycobacterium mageritense, Mycobacterium wolinskyi, and pigmented RGM.4 Mycobacterium immunogenum was first identified in 2000 in metalworking fluids and is part of the M. chelonae-abscessus group.5 Its biological characteristics and high-performance liquid chromatography features are similar to those of M. abscessus and M. chelonae; 16S rRNA sequencing indicates that M. immunogenum differs from M. abscessus and M. chelonae by only 8 and 10 base pairs, respectively.5 The first case report of human M. immunogenum infection was in 2001; since then, most reported cases of M. immunogenum infection have been extrapulmonary. Infection is often associated with surgical procedures,6 trauma,7 or catheter sites.8 Herein, we report on the case of an 81-year-old man with severe pneumonia caused by M. immunogenum.
Case Presentation
Case Report
An 81-year-old man was hospitalised for a cough in August 2021. The patient had chronic kidney disease and had been undergoing regular haemodialysis three times a week for 6 years. In 2019, he had undergone “left arm graft vascular fistula surgery” and pacemaker implantation surgery for coronary heart disease. His blood pressure is 140/67 mmHg, and the body temperature is 37°C. The chest computed tomography revealed multiple nodular high-density opacities in both lungs, indicative of pneumonia, chronic bronchitis, and emphysema. Three smear test samples for acid-fast bacilli were negative, and general bacterial or fungal cultures of sputum and blood showed no bacterial growth. Moxifloxacin (0.4 g daily) administration did not lead to an improvement; therefore, the antibiotic regimen was changed to cefoperazone sodium and sulbactam sodium (1 g twice daily). The patient’s nutritional status was poor; therefore, intravenous immunoglobulin was administered to improve his immune function. His clinical condition improved, and he was discharged from hospital after 4 days. He did not return for a follow-up. Two days after his discharge, two different sputum cultures tested positive for mycobacteria, and subsequent whole-genome sequencing identified the isolate as M. immunogenum. Therefore, he was diagnosed with NTM-PD.
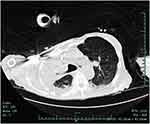
In February 2022, he was re-hospitalised because of chest tightness and shortness of breath. Computed tomography revealed multiple patchy high-density opacities in both lungs, with some areas of consolidation and decreased volume of the right lung (Figure 1). The patient was initially treated with cefoperazone sodium and sulbactam sodium (2 g twice daily) and subsequently switched to meropenem (0.5 g every 6 hours), but died on Day 10 of admission due to acute respiratory failure and severe pneumonia before targeted treatment for NTM-PD could be initiated.
Sputum Testing Methods
The patient’s sputum specimen was tested in September 2021 at the Tuberculosis Laboratory of the First Affiliated Hospital, Zhejiang University School of Medicine.
Smear
The sputum sample was spread evenly on the front side of a glass slide to form an oval sputum film measuring 10 mm × 20 mm and was fixed by heating. After acid-fast staining, the cells were observed under a microscope (Olympus, Tokyo, Japan) using a 100× oil immersion lens.
Culture
First, 2% NALC-NaOH specimen pretreatment solution (1–2× volume of the specimen) was added to the sputum sample; the mixture was transferred to a 50 mL centrifuge tube and vortexed to mix evenly. The mixture was left to stand at room temperature for 15 to 20 minutes,9 then phosphate-buffered saline was added and mixed well. Finally, 0.8 mL of the specimen was added to the Mycobacteria Growth Indicator Tube (MGIT) culture tube and measured using a MIGT 960 Automated Mycobacterial Detection System (BD Biosciences, Franklin Lakes, NJ, USA).
Mycobacterium Biochip Test
A 20-µL aliquot of the suspension was pipetted from the MGIT liquid culture tube, and nucleic acids were extracted, followed by polymerase chain reaction amplification. Load the amplified product onto the chip for hybridization. The chip was then washed and scanned (Capital Bio, Beijing, China).
Whole-Genome Sequencing
A single colony was detected on blood agar medium and Middlebrook 7H10 agar medium on Days 4 and 7, respectively. Monoclonal clones were used for the enrichment analysis. Logarithmic bacterial cultures were collected, and DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA). Sequencing was performed using an Illumina NovaSeq PE150 platform (Novogene Bioinformatics Technology Co., Beijing, China). Bacterial species were identified using KmerFinder 3.2 (Center for Genomic Epidemiology, Technical University of Denmark).
M. immunogenum Susceptibility Testing
After obtaining definitive results, the tuberculosis laboratory performed susceptibility testing for M. immunogenum by broth microdilution following the Clinical and Laboratory Standards Institute M24s-2nd ed. 202310 (Table 1).
 |
Table 1 Susceptibility Testing for Mycobacterium immunogenum |
Sputum Results
Three sputum samples were obtained for acid-fast bacillus smears; however, no acid-fast bacilli were observed. Two sputum samples were positive for growth and clustered microscopically. The mycobacterium biochip test identified the isolates as M. abscessus and M. gilvum. Whole-genome sequencing revealed that the microorganism was M. immunogenum. In the drug susceptibility tests, the isolate was resistant to imipenem, moxifloxacin, and doxycycline; had intermediate susceptibility to tobramycin and linezolid; and was sensitive to amikacin, cefoxitin, and clarithromycin. Drug susceptibility tests for rifampicin, rifabutin, azithromycin, streptomycin, tigecycline, and omadacycline were conducted as references.
Discussion
Determining the clinical significance of an infection without species identification is challenging. Because NTM can be isolated from environmental or contaminated clinical specimens, it is generally necessary to have more than one culture-positive specimen for diagnostic purposes.11 This patient tested positive in two cultures, which, combined with the patient’s symptoms and pulmonary imaging findings, confirmed that he had NTM-PD.
Antibiotic susceptibility differs among various species, directly affecting treatment decisions; thus, identifying mycobacterial isolates at the species level rather than the group level is strongly recommended.11 The M. chelonae-abscessus group includes three species: M. chelonae, M. abscessus, and M. immunogenum.12 In this case, the Gene Chip method did not identify M. immunogenum; therefore, the isolate was sent to a reference laboratory and underwent whole-genome sequencing. To confirm whether the patient had a mixed infection, the isolated monoclonal colonies were retested using the Gene Chip method, which again identified only M. abscessus and M. gilvum, demonstrating the limitations of the Gene Chip method. For mycobacteria that are not among the 17 species identifiable by this method, the results can read as “uninterpretable” or can yield results misidentifying the species. Therefore, closer attention must be paid to assessing the reliability of the experimental results, and whole-genome sequencing is recommended if the result of the Gene Chip method is in doubt.
Although M. immunogenum is extremely similar to M. chelonae and M. abscessus, most isolates have unique resistance patterns to cefoxitin and tobramycin.5 In this case, the drug susceptibility tests showed that the isolate had intermediate sensitivity to tobramycin and was sensitive to cefoxitin. The Clinical and Laboratory Standards Institute Standard (CLSI) has updated the susceptibility testing standard for RGM. However, the clinical significance of this cut-off has not yet been confirmed for M. immunogenum, and data validating the susceptibility of many NTM species to antibiotics are lacking.11 We conducted drug susceptibility tests for drugs for which the CLSI M24s guidelines do not provide cut-off values, including rifampicin, rifabutin, azithromycin, streptomycin, tigecycline, and omadacycline. These results may inform the choice of antimicrobial treatment for future cases of M. immunogenum-induced NTM-PD. Importantly, these results demonstrate that drug susceptibility testing is crucial for informing the choice of treatment.
We also searched the PubMed database for English-language reports of M. immunogenum infection published from 2014 to 2024.6–8,13–21 We identified 11 articles, reporting 12 cases, summarised in Table 2. The patients originated in Japan, the United States, Spain, China, and Australia and included four male and seven female patients (and one patient with unspecified sex) aged 7 to 81 years. Infection was detected in the lungs in only one patient (9%); the other infection sites included the skin or soft tissue (n = 7, 64%), catheter insertion site, faeces, pleural and abdominal fluid, and a brain abscess (one case each, 9%). Thus, most M. immunogenum infections occur outside the lungs, and some individuals contract the infection after surgery or following trauma.
 |
Table 2 Reported Mycobacterium immunogenum Infections from 2014 to 2024 |
The most recent discovery of M. immunogenum was in a Japanese patient with tenosynovitis.13 Her biopsy culture was positive, and M. immunogenum was identified using mass spectrometry (MALDI-TOF-MS). The result was further confirmed by 16S rRNA sequencing. The patient improved after undergoing surgery and receiving a 6-month course of amikacin treatment, with no recurrence observed for over a year. This article recommends sequencing, and the effective treatment with amikacin is consistent with the experimental conclusions of this study, which may provide insights for clinical treatment.
Owing to the lack of typical clinical symptoms and guidelines and the lack of laboratory identification methods for rare NTM, delayed diagnosis of NTM-PD is common.3 In this case, the rarity of M. immunogenum and the testing limitations led to repeated laboratory testing and a delayed diagnosis. The second hospitalisation was further complicated by the patient’s advanced age, severe clinical condition, and multiple underlying diseases, requiring clinicians to prioritise more serious issues. The patient died before targeted treatment for NTM-PD could be initiated.
Several factors should be considered when choosing treatment. This patient had multiple treatment considerations, such as advanced age, low body mass index, and comorbidities. Moreover, his sputum culture was positive, and he had lobar lung involvement, indicating a need for targeted NTM-PD treatment.22 Despite the patient’s advanced age and other underlying conditions, pneumonia was the primary cause of death.
Conclusion
In clinical practice, the results of bacterial species identification should be considered and should be confirmed through whole-genome sequencing, if the identity is in doubt. M. immunogenum is pathogenic, and the clinical significance of M. immunogenum-induced infections should be recognised in the context of NTM-PD. In managing complex cases, the multi-disciplinary team (MDT) involving clinicians, microbiologists, and radiologists is essential to ensure timely and accurate diagnosis and treatment. The treatment of NTM-PD should be guided by individualized antibiotic regimens based on susceptibility testing to avoid ineffective therapies and reduce the risk of treatment failure. Moreover, microbiology laboratories should be able to rapidly detect NTM species, particularly those encountered infrequently in clinical practice, and should diligently perform antimicrobial susceptibility testing to ensure appropriate antimicrobial selection.
Ethical Approval
The patient’s family provided written informed consent for publication of his case details and the accompanying images, and the publication of this report was approved by the Ethical Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Key Research and Development Program (2021YFC2301800) and the National Natural Science Foundation (82100640).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Primm TP, Lucero CA, Falkinham JO. Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004;17(1):98–106. doi:10.1128/CMR.17.1.98-106.2004
2. Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185(8):881–886. doi:10.1164/rccm.201111-2016OC
3. Kumar K, Loebinger MR. Nontuberculous mycobacterial pulmonary disease: clinical epidemiologic features, risk factors, and diagnosis: the nontuberculous mycobacterial series. Chest. 2022;161(3):637–646. doi:10.1016/j.chest.2021.10.003
4. Brown-Elliott BA, Philley JV. Rapidly growing mycobacteria. Microbiol Spectr. 2017;5(1). doi:10.1128/microbiolspec.TNMI7-0027-2016
5. Wilson RW, Steingrube VA, Böttger EC, et al. Mycobacterium immunogenum sp. nov. a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int J Syst Evol Microbiol. 2001;51(5):1751–1764. doi:10.1099/00207713-51-5-1751
6. Kong Y, Chen H, Xiong J, Hao Z. Infection with Mycobacterium immunogenum after an injection lipolysis procedure. Br J Dermatol. 2021;185(3):e68. doi:10.1111/bjd.20392
7. Mitchell CB, Isenstein A, Burkhart CN, Groben P, Morrell DS. Infection with Mycobacterium immunogenum following a tattoo. J Am Acad Dermatol. 2011;64(5):e70–e71. doi:10.1016/j.jaad.2009.12.037
8. Shenoy A, El-Nahal W, Walker M, et al. Management of a Mycobacterium immunogenum infection of a peritoneal dialysis catheter site. Infection. 2018;46(6):875–880. doi:10.1007/s15010-018-1199-0
9. Siddiqui S, Rusch-Gerdes S. MGIT Procedure Manual for BACTEC MGIT 960 TB System. Geneva, Switzerland: Foundation for Innovative Diagnostics; 2006.
10. CLSI. Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia Spp. and Other Aerobic Actinomycetes.
11. Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. doi:10.1164/rccm.200604-571ST
12. Brown-Elliott BA, Wallace RJ. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev. 2002;15(4):716–746. doi:10.1128/CMR.15.4.716-746.2002
13. Okinaka T, Fujimura K, Hamasaki Y, Hasegawa Y, Matono T. Significance of early diagnosis and surgical management in treating Mycobacterium immunogenum-related pyogenic extensor tenosynovitis: a case report. BMC Infect Dis. 2024;24(1):395. doi:10.1186/s12879-024-09249-5
14. Jia F, Nick SE, Davidson RM, Baker AW. Complete genome sequence of Mycobacterium immunogenum strain C14-1.MIM isolated from a hospitalized lung transplant recipient. Microbiol Resour Announc. 2023;12(12):e0061823. doi:10.1128/MRA.00618-23
15. Greninger AL, Langelier C, Cunningham G, et al. Two rapidly growing mycobacterial species isolated from a brain abscess: first whole-genome sequences of Mycobacterium immunogenum and Mycobacterium llatzerense. J Clin Microbiol. 2015;53(7):2374–2377. doi:10.1128/JCM.00402-15
16. Iroh Tam PY, Kline S, Ward G, Ferrieri P. Non-tuberculous mycobacterial infection in hospitalized children: a case series. Epidemiol Infect. 2015;143(15):3173–3181. doi:10.1017/S0950268815000333
17. Garcia-Zamora E, Sanz-Robles H, Elosua-Gonzalez M, Rodriguez-Vasquez X, Lopez-Estebaranz JL. Cutaneous infection due to Mycobacterium immunogenum: an European case report and review of the literature. Dermatol Online J. 2017;23(10). doi:10.5070/D32310036992
18. McNeil EP, Goldfarb N, Hannon GR, Miller DD, Farah RS. Mycobacterium immunogenum folliculitis on the lower extremities of a healthy young adult. Clin Exp Dermatol. 2019;44(3):328–330. doi:10.1111/ced.13703
19. Aryee JNA, Akinleye SD, Freilich AM, Deal DN. Mycobacterium immunogenum flexor tenosynovitis: a case report. J Wrist Surg. 2021;10(3):241–244. doi:10.1055/s-0040-1715803
20. Yeon J, Chan RC, Fallah H. Infection with Mycobacterium immunogenum following botulinum toxin injection. Australas J Dermatol. 2021;62(1):79–80. doi:10.1111/ajd.13406
21. Damronglerd P, Higgins E, Fida M, et al. Characteristics and management of periprosthetic joint infections caused by rapidly growing mycobacteria: a retrospective study and a review of the literature. J Bone Jt Infect. 2024;9(1):99–106. doi:10.5194/jbji-9-99-2024
22. Cowman S, van Ingen J, Griffith DE, Loebinger MR. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J. 2019;54(1):1900250. doi:10.1183/13993003.00250-2019
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.