Back to Journals » Journal of Pain Research » Volume 18
Pain Intensity in Patients with Opioid Use Disorder on Extended-Release Naltrexone or Opioid Agonists; The Role of COMT rs4680 and OPRM1 rs1799971: An Exploratory Study
Authors Juya F , Sannes AC , Solli KK, Weimand B, Gjerstad J, Tanum L, Mordal J
Received 15 October 2024
Accepted for publication 9 February 2025
Published 21 February 2025 Volume 2025:18 Pages 827—836
DOI https://doi.org/10.2147/JPR.S500984
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Timothy Atkinson
Farid Juya,1 Ann Christin Sannes,2,3 Kristin Klemmetsby Solli,1,2,4 Bente Weimand,2,4 Johannes Gjerstad,2,5 Lars Tanum,2,3 Jon Mordal1
1Division of Mental Health and Addiction, Vestfold Hospital Trust, Tønsberg, Norway; 2Mental Health Services, Akershus University Hospital, Lørenskog, Norway; 3Faculty of Health Sciences, Oslo Metropolitan University, Oslo, Norway; 4Centre for Mental Health and Substance Abuse, University of South-Eastern Norway, Drammen, Norway; 5School of Health Sciences, Kristiania University College, Oslo, Norway
Correspondence: Farid Juya, Division of Mental Health and Addiction, Vestfold Hospital Trust, Kirkeveien 275, Nøtterøy, 3140, Norway, Tel +4791167061, Email [email protected]
Purpose: To examine whether reported pain intensity over time is related to the single nucleotide polymorphisms of the catechol-O-methyltransferase (COMT rs4680) and mu-opioid receptor (OPRM1 rs1799971) in patients with opioid use disorder (OUD) choosing treatment with extended-release naltrexone (XR-NTX) or opioid agonist treatment (OAT).
Patients and Methods: This exploratory study was part of a 24-week, open-label clinical prospective trial of patients with OUD who chose intramuscular XR-NTX, and patients receiving OAT. Men and women aged 18 to 65 years with OUD per the Diagnostic and Statistical Manual of Mental Disorders, fifth edition were included. Pain intensity was measured at baseline and at 24-week follow-up using the Numerical Pain Rating Scale-11 and genotyping was performed by TaqMan technology. Data were analyzed with ordinal logistic regression.
Results: Of 317 participants included at baseline, 210 samples were obtained and analyzed. In the OAT group, there was a negative significant association between pain intensity and having the Val/Val allele of COMT rs4680 (wild-type = most common type) and the rare allele G of OPRM1 rs1799971 at 24-week follow-up. No such effects were seen in the XR-NTX group.
Conclusion: The wild-type allele Val/Val of COMT rs4680 and the rare allele G of OPRM1 rs1799971 may have a possible protective effect regarding pain intensity in patients with OUD receiving OAT. Given relatively low sample size, particularly low number of female participants in the XR-NTX group and other possible confounders, our findings should be interpreted with caution.
Keywords: pain, genetics, opioid receptor, agonist, antagonist, naltrexone
Introduction
Patients with opioid use disorder (OUD) have a higher prevalence of chronic pain and pain intensity than the populace.1–3 Also, continued and prolonged use of opioids for persistent pain has been linked to higher risk of harmful use, dependence and overdose.4,5 The reasons remain unclear, but long-term opioid use may result in reduced pain tolerance, increased pain sensitivity and self-medication.6–8 It is, however, not known whether the increased pain intensity in patients with OUD is mainly caused by opioid-induced hyperalgesia or a genetic predisposition. Due to the wide range of inter-individual pain variations, further knowledge regarding the role of genetic factors in patients with OUD may improve the understanding of pain and potentially treatment outcomes.
The single nucleotide polymorphisms (SNPs) of the catechol-O-methyltransferase (COMT rs4680) and mu-opioid receptor (OPRM1 rs1799971) are proposed to be associated with the risk of chronic pain, self-medication, and opioid addiction.9–12 However, to our knowledge, the impact of SNP effects on pain intensity has not yet been studied in patients with OUD who receive opioid antagonists such as extended-release naltrexone (XR-NTX), or opioid agonist treatment (OAT) with methadone or buprenorphine.
The enzyme COMT is expressed in several tissues, including the brain. COMT catalyzes the metabolism of catecholamines such as dopamine, adrenaline, and noradrenaline, which affect mood, cognition, and stress responses.13,14 Notably, all of these functions may be related to pain mechanisms.12 One SNP that may affect pain is the SNP rs4680 G > A in the gene that encodes COMT.15 COMT rs4680 G > A leads to substitution of the amino acid valine (Val) with methionine (Met), where the Met alleles display three to four times reduced enzyme activity.16 Earlier studies suggest that COMT rs4680 may be associated with chronic postsurgical pain as well as sensitivity to experimental pain.15,17 The wild-type allele Val (the most common) may be associated with better long-term recovery from back pain and a decreased probability of pain in patients during social stress.9 As a result, there is reason to suspect that COMT rs4680 could affect pain intensity.
The opioid receptor mu 1 (MOR1), which is encoded by the genetic locus OPRM1, has a significant affinity for β-endorphin and encephalin as well as many exogenous opioids.18 The MOR1 function is also essential for the analgesic effects of opioids.19 The SNP rs1799971 A > G, located in exon 1 of the OPRM1 gene, induces a change from the amino acid asparagine to aspartic acid at the 40th amino acid residue (Asn40Asp).20,21 Previous data show that the minor allele G (Asp) in men may be associated with better long-term recovery of back pain.22 Moreover, the same rare allele is linked to the need for higher doses of opioids for cancer pain, acute post-surgical pain, as well as subacute pain management.23–25 Additionally, medications for treatment of OUD act differently on mu-opioid receptors; naltrexone being an antagonist, methadone and buprenorphine agonists (full and partial, respectively). Given the importance of mu-opioid receptors for the analgesic effects of opioids, one might expect the SNP OPRM1 rs1799971 to affect pain intensity.
For patients with OUD, OAT with methadone or buprenorphine is recommended by the World Health Organization.26 However, antagonist treatment with a monthly intramuscular injection of XR-NTX has in the past decade gained recognition as a viable treatment alternative for OUD.27 Patients with OUD who were treated with XR-NTX reported no increase in pain intensity, thus emphasizing the possible positive potential of this treatment.28
Given the important role of the SNPs COMT rs4680 and OPRM1 rs1799971 in pain and OUD, this study aimed to explore the possible influence of these SNPs on pain intensity among patients with OUD who chose treatment with XR-NTX as compared to OAT.
Materials and Methods
Design
This exploratory study was part of a larger 24-week, open-label clinical prospective study of monthly intramuscular injections of XR-NTX with an optional 28-week follow-up (NaltRec study), which is described in detail by Weimand et al.29 In this study, participants were patients with OUD who chose intramuscular XR-NTX and patients receiving OAT (methadone or buprenorphine).
Ethical Approval and Informed Consent
Ethical approval for the NaltRec trial, including the present study, was granted by the Norwegian Regional Ethical Committees for Medical and Health Research Ethics committee South East Norway (# 2018/132), by the Personal Data Protection Representative for each of the sites, and by the Norwegian Medicine Agency. The trial is registered at ClinicalTrials.gov (#NCT03647774), the European Union Clinical Trials Register (#2017-004706-18), and complies with the Declaration of Helsinki. All participants gave written and informed consent for their participation. The study treatment was provided free of charge, and participants received no payment or economic compensation for their participation, except for reimbursement of travel expenses if public transportation was used.
Participants and Setting
Participants were recruited from addiction clinics in five urban hospitals in Norway (both in- and outpatient). Eligible participants were men and women aged 18 to 65 years with OUD according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition criteria. Exclusion criteria were severe alcohol use disorder or serious somatic (eg, liver failure) or psychiatric illness (eg, psychosis), or the need for intensive medical treatment that would clearly interfere with study participation. Women of childbearing potential were required to confirm they were not pregnant or lactating and agree to use effective birth control if receiving study medication. The MINI International Neuropsychiatric Interview was used to screen for psychiatric disorders, and a medical doctor examined participants for serious somatic diseases.30 Prior to inclusion, 64% of participants in XR-NTX and all in the OAT group were in the Norwegian OAT program.31 Inclusion lasted from September 2018 to September 2020.
Genotyping
During baseline assessments, study participants were asked to provide a saliva sample for genotyping in accordance with instructions from the manufacturer (OrageneRNA sample collection kits, DNA Genotek Inc., Kanata, Ontario, Canada). Genotyping was conducted by a predesigned TaqMan SNP assay (Applied Biosystems, Foster City, CA, USA). Approximately 10 ng of genomic DNA were amplified in 5 µL of a reaction mixture in a 384-well plate that contained 1x TaqMan genotyping master mix (Applied Biosystems) and 1x assay mix; the latter contained the respective primers and probes. The probes were labeled with the reporter dye FAM or VIC to distinguish between the two alleles. In accordance with the procedure in previous studies an ABI 79000HT sequence detection system was used.32,33 Negative controls were included in every run. Approximately 10% of the samples were genotyped again, and the concordance rate was 100%.
Study Interventions
Screened eligible participants seeking treatment with XR-NTX were referred to an inpatient unit for medically managed withdrawal and induction with XR-NTX. Following induction, participants receiving XR-NTX attended study visits every 4 weeks, during which they received 380 mg XR-NTX intramuscularly (Vivitrol®) and completed the study interviews. The OAT group attended study visits at baseline and at week 24, and were otherwise followed up in accordance with the Norwegian OAT program. During study inclusion, all participants had to be enrolled in the Norwegian OAT program. This precaution ensured that participants had immediate access to individual counselling and pharmacological treatment (methadone or buprenorphine) if they withdrew from XR-NTX treatment. The individual counselling is a part of the Norwegian OAT program, which did not include specific psychosocial interventions for pain management.
Measurements
Data on demographic and clinical characteristics were collected at baseline by trained researchers using the European Addiction Severity Index.34 Pain intensity was measured with the Numerical Pain Rating Scale-11 (NPRS-11), at baseline and at 24-week follow-up. The NPRS-11 is a validated numeric rating scale from 0 to 10, where 0 is anchored with “no pain” and 10 with “worst pain imaginable”, and has also been used in a previous study by our study group.1,35 The NPRS-11 was adopted to following instruction: Rate your current pain intensity from 0 (no pain) to 10 (worst pain imaginable).
Statistical Analyses
Descriptive statistics were used to summarize the study sample. Chi-squared tests were conducted to assess whether the genotype distributions conformed to Hardy–Weinberg equilibrium. The outcome, pain intensity as measured by the NPRS-11 is considered an ordinal value. Hence an ordinal logistic regression adjusted for age, gender and pain at baseline was conducted to assess the association between genotype and treatment with XR-NTX or OAT on pain intensity at 24-week follow-up. The results were presented as odds ratios (OR) with corresponding 95% confidence intervals (CI).
Analyses of the interaction effect of genotype versus XR-NTX group on pain intensity were conducted for both the COMT rs4680 and OPRM1 rs1799971 SNPs. Due to the low number of OPRM1 rs1799971 GG carriers (n=1) a decision was made to combine the GG and AG (n= 34) carriers, and to assess AA versus G carriers. This model may lead to an increased statistical certainty and has commonly been used earlier.36–38 The analyses included data only from participants (XT-NTX or OAT) who adhered to their medication and completed assessments at the 24-week follow-up. Power analysis was not performed due to the exploratory nature of the trial, and being part of a larger 52-week study.29 The analyses were performed using STATA/SE 16.0 (StataCorp, College Station, TX, USA). A p-value < 0.05 was deemed statistically significant.
Results
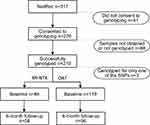
Of 317 patients included at baseline, 276 consented to genotyping (Figure 1). For 66 participants, saliva samples were either not obtained due to retraction of consent, participants being out of reach, hyposalivation or saliva samples were too contaminated for use (mostly tobacco snuff). Thus, a total of 210 samples were successfully genotyped for the COMT rs4680 and 209 samples for the OPRM1 rs1799971. In the OAT group, 21% of the participants used methadone and 79% used buprenorphine. Participants in the XR-NT (n = 88) were compared with those in the OAT group (n = 119) regarding baseline demographic and clinical characteristics (Table 1). Compared to the OAT group, the XR-NTX group was on average six years younger, had fewer female participants, fewer positive lifetime hepatitis, fewer poor dental health (p = 0.001, 0.047, 0.002, and 0.005 respectively), and reported lower mean pain intensity at baseline (2.6 vs 3.5, p = 0.014). Almost half of the participants (47.6%) rated their pain intensity ≤2, thereby indicating an overall low mean pain intensity. These participants aside, the average baseline pain intensity was 5.4 in the XR-NTX group and 5.1 in the OAT group.
 |
Table 1 Baseline Demographic and Clinical Characteristics Based on Groups (n=210)* |
Genotype distributions for the COMT rs4680 and OPRM1 rs1799971 were not significantly different between participants in XR-NTX and OAT group, neither were the genotype distributions versus methadone and buprenorphine among participants in the OAT group (p = 0.958, 0.171, 0.684, and 0.458 respectively) (Table 2). Both COMT rs4680 and OPRM1 rs1799971 genotype distributions were consistent with Hardy–Weinberg equilibrium (p = 0.948 and 0.980, respectively).
 |
Table 2 COMT rs4680, OPRM1 rs179991 Allele Frequency and Allele Frequency versus OAT Medication (n=210) * |
The COMT rs4680 Val/Val carriers showed a negative association with pain intensity compared to Met/Met carriers (OR: 0.302, 95% CI: 0.109 to 0.863) at the 24-week follow-up. In addition, a negative significant association when comparing the XR-NTX group to the OAT group was seen (OR: 0.248, 95% CI: 0.075 to 0.814) at the 24-week follow-up. However, no significant interaction effect (COMT rs4680 versus XR-NTX group) was observed (Table 3).
 |
Table 3 Ordinal Logistic Regression of Pain Intensity at 24-week Follow-Up Split on Genotype Conducted Separately for COMT and OPRM1 (N = 210) * |
The same analysis was conducted to investigate OPRM1 rs1799971. G carriers had a negative significant association with pain intensity compared to AA carriers (OR: 0.305, 95% CI: 0.107 to 0.869) at the 24-week follow-up. However, no statistically significant findings were seen for between group differences (XR-NTX versus OAT group) nor for the interaction effects (OPRM1 rs1799971 versus XR-NTX group) (Table 3).
Since differences were seen between the groups, two post hoc analyses, stratified by group, were conducted to further investigate the effects of the COMT rs4680 and OPRM1 rs1799971 (Table 4). No statistically significant findings were seen in the XR-NTX group. However, a statistically significant association between both SNPs and pain intensity was seen in the OAT group. The COMT rs4680 Val/Val carriers (OR: 0.282, 95% CI: 0.095 to 0.832) and OPRM1 rs1799971 G carriers (OR: 0.245, 95% CI: 0.081 to 0.736) showed a negative significant association with pain intensity at the 24-week follow-up (Table 4).
 |
Table 4 COMT and OPRM1 and Ordinal Logistic Regression of Pain Intensity at 24-week Follow-Up, Stratified by Group (n = 210) * |
Discussion
To our knowledge, this study is the first to explore the effect of the COMT rs4680 and OPRM1 rs1799971 SNPs on pain intensity in patients with OUD who were treated with XR-NTX or OAT. The COMT Val/Val (wild-type) and OPRM1 G carriers showed a negative association with pain intensity at 24-week follow-up. The possible protective effect of the COMT Val/Val and the OPRM1 G was, however, only seen in the OAT group. Additionally, the XR-NTX group showed a negative association with pain intensity compared to the OAT group.
Our data support earlier findings in the populace, where COMT rs4680 Val carriers experienced less stress-induced pain than subjects with Met/Met carriers, and OPRM1 rs1799971 G carriers reported less persistent back pain than subjects with OPRM1 rs1799971 AA.9,22,25,39 Also, our data pointed to a possible interaction between the COMT rs4680 and the OPRM1 rs1799971 genotype and OAT versus XR-NTX treatment. This suggest that COMT rs4680 Val and OPRM1 rs1799971 G carriers with OUD being treated with opioid agonists may have better long-term pain outcomes. Stratification of the data regarding treatment clearly demonstrated that blocking the opioid receptors with XR-NTX attenuates the effect of genetic factors COMT rs4680 Val/Val and OPRM1 rs1799971 G. Thus, our data indicates that the effect of these genotypes may be dependent on the type of opioid treatment—the pharmacological activation or blocking of the opioid systems.
Several earlier studies have reported the need for higher doses of opioids in the OPRM1 rs1799971 GG carriers in the populace.23–25 Surprisingly, our finding in patients with OUD being treated with opioid agonists, show that OPRM1 rs1799971 G (GA pooled with GG due to low number of GG carriers) are associated with reduced pain intensity. The reasons remain unclear and are beyond the scope of this exploratory study. Presumably, increasing levels of pain intensity may amplify genetic differences (baseline pain intensity in the XR-NTX versus OAT group; 2.6 vs 3.5). Perhaps the molecular mechanisms underlying the effects of the OPRM1 rs1799971 on pain intensity measures in patients with OUD treated with opioid agonists, differ from those in healthy individuals.
This exploratory study was open-label and participants’ expectancies about the effects of the treatments (agonist or antagonist) may have influenced the findings. Also, the study was limited by its relatively low sample size, particularly low number of female participants in the XR-NTX group and missing data for a number of participants at the 24-week follow-up. Sub-group analyses (eg methadone versus buprenorphine) were not performed due to lack of statistical power, as genetic effects are often small, and detecting these requires larger sample sizes. The analyses were adjusted only for age, gender, and pain at baseline. However, a recent paper from our study group reported that the XR-NTX and the OAT groups were similar with regard to other possible factors influencing pain (eg depression, anxiety, number of previous hospitalizations for mental or physical health problems, marital status and years of education).31
Given the small number of female participants in the XR-NTX group, the generalizability of our findings may be limited, and recruitment of more female participants in future studies is needed to confirm our findings. We also lacked relevant information of pain duration, location and functional interference. Moreover, the overall reported pain intensity in both XR-NTX and OAT group is considered to be mild. This might limit the clinical significance of our findings. However, it may also highlight their clinical relevance. The perceived notion is that XR-NTX is less suitable for OUD patients with pain comorbidity due to the medication’s opioid receptor blockade. A recent study reported that pain management concerns are among the common barriers for treatment with XR-NTX.40 Therefore, overall low reported pain intensity is particularly expected in patients with OUD opting for treatment with XR-NTX.
Future Improvements
We recommend that future studies include larger number of participants. In genetic research, as in other fields, larger sample sizes tend to yield more robust and clinically meaningful conclusions. In this study, several saliva samples used for genotyping were contaminated, primarily due to the use of tobacco snuff (a common form of tobacco in Norway). While saliva samples are less invasive, the DNA yields from blood samples are higher and have lower levels of DNA contamination.41,42
In this exploratory study, the association between genotype, treatment with XR-NTX or OAT, and pain intensity at 24-week follow-up was assessed with ordinal logistic regression adjusted for age, gender and pain at baseline. For future studies with larger samples, particularly those involving multiple repeated measures, other statistical methods, such as mixed model ANOVA may be considered. We also recommend the addition of other factors influencing pain (eg sex, income and years of education, access to treatment, physical health and body mass index, pain diagnosis, depression, anxiety, and trauma) as covariates in the analyses. Additionally, because of the different analgesic properties of OAT medications, conducting subgroup analyses in the OAT group by medication type (methadone or buprenorphine) and dose is recommended. Moreover, given that pain is a multidimensional biopsychosocial entity, we recommend including multidisciplinary pain variables such as pain duration, location, and functional interference.
Conclusion
This exploratory study showed that COMT rs4680 Val/Val (wild-type) and OPRM1 rs1799971 G may have a potential protective effect regarding pain intensity in patients with OUD in OAT. Though, this effect may be hidden or absent in individuals choosing treatment with opioid antagonists such as XR-NTX. Given relatively low sample size, particularly low number of female participants in the XR-NTX group and other possible confounders, the impact of our findings on clinical care is limited but can be a potential contribution to future research.
Data Sharing Statement
The ethical approval for this study does not open up for sharing data with any third party not mentioned in the protocol. However, anonymous data can be made available based on an agreement with the National Coordinating Investigator (Lars Tanum) and after having notified the Regional Board of Research Ethics for South-East Norway.
Acknowledgments
The manufacturer, Alkermes, Inc., provided the study medication extended-release naltrexone injection at no cost and in accordance with an investigator-initiated trial agreement. No editorial control or access to study data were given to the manufacturer or administrative bodies. We wish to thank all of the participants enrolled in the study, the participating study sites, and the staff members.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by unrestricted grants from the Research Council of Norway (grant 269864) and the South-Eastern Norway Regional Health Authority (grant 2019105).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Latif ZH, Skjaervø I, Solli KK, Tanum L. Chronic pain among patients with an opioid use disorder. Am J Addict. 2021;30(4):366–375. doi:10.1111/ajad.13153
2. Breivik H, Collett B, Ventafridda V, Cohen R, Gallacher D. Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur J Pain. 2006;10(4):287–333. doi:10.1016/j.ejpain.2005.06.009
3. Dahlhamer J, Lucas J, Zelaya C, et al. Prevalence of chronic pain and high-impact chronic pain among adults - United States, 2016. MMWR Morb Mortal Wkly Rep. 2018;67(36):1001–1006. doi:10.15585/mmwr.mm6736a2
4. Coloma-Carmona A, Carballo J-L, Rodríguez-Marín J, Pérez-Carbonell A, Alonso-Garrido C. Medical and psychological predictors of prescription opioids dependence during chronic pain treatment. Eur Rev Appl Psychol. 2018;68(6):199–204. doi:10.1016/j.erap.2018.10.002
5. Chou R, Deyo R, Devine B, et al. The effectiveness and risks of long-term opioid treatment of chronic pain. Evid Rep Technol Assess. 2014;(218):1–219. doi:10.23970/ahrqepcerta218
6. Fishbain DA, Cole B, Lewis JE, Gao J, Rosomoff RS. Do opioids induce hyperalgesia in humans? An evidence-based structured review. Pain Med. 2009;10(5):829–839. doi:10.1111/j.1526-4637.2009.00653.x
7. Voon P, Hayashi K, Milloy MJ, et al. Pain among high-risk patients on methadone maintenance treatment. J Pain. 2015;16(9):887–894. doi:10.1016/j.jpain.2015.06.003
8. Koller G, Schwarzer A, Halfter K, Soyka M. Pain management in opioid maintenance treatment. Expert Opin Pharmacother. 2019;20(16):1993. doi:10.1080/14656566.2019.1652270
9. Jacobsen LM, Schistad EI, Storesund A, et al. The COMT rs4680 Met allele contributes to long-lasting low back pain, sciatica and disability after lumbar disc herniation. Eur J Pain. 2012;16(7):1064–1069. doi:10.1002/j.1532-2149.2011.00102.x
10. Zhang X, Liang Y, Zhang N, et al. The relevance of the OPRM1 118A>G genetic variant for opioid requirement in pain treatment: a meta-analysis. Pain Physician. 2019;22(4):331–340.
11. Li T, Liu X, Zhu ZH, et al. Association analysis of polymorphisms in the μ opioid gene and heroin abuse in Chinese subjects. Addict Biol. 2000;5(2):181–186. doi:10.1080/13556210050003775
12. Belfer I, Segall S. COMT genetic variants and pain. Drugs Today. 2011;47(6):457–467. doi:10.1358/dot.2011.47.6.1611895
13. Guldberg HC, Marsden CA. Catechol-O-methyl transferase: pharmacological aspects and physiological role. Pharmacol Rev. 1975;27(2):135–206. doi:10.1016/S0031-6997(25)06681-5
14. Männistö PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51(4):593–628.
15. Zubieta JK, Heitzeg MM, Smith YR, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003;299(5610):1240–1243. doi:10.1126/science.1078546
16. Lotta T, Vidgren J, Tilgmann C, et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34(13):4202–4210. doi:10.1021/bi00013a008
17. Frangakis SG, MacEachern M, Akbar TA, et al. Association of genetic variants with postsurgical pain: a systematic review and meta-analyses. Anesthesiology. 2023;139(6):827–839. doi:10.1097/aln.0000000000004677
18. Zhang H, Luo X, Kranzler HR, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15(6):807–819. doi:10.1093/hmg/ddl024
19. Uhl GR, Sora I, Wang Z. The mu opiate receptor as a candidate gene for pain: polymorphisms, variations in expression, nociception, and opiate responses. Proc Natl Acad Sci U S A. 1999;96(14):7752–7755. doi:10.1073/pnas.96.14.7752
20. Schwantes-An TH, Zhang J, Chen LS, et al. Association of the OPRM1 variant rs1799971 (A118G) with non-specific liability to substance dependence in a collaborative de novo meta-analysis of European-ancestry cohorts. Behav Genet. 2016;46(2):151–169. doi:10.1007/s10519-015-9737-3
21. Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–9613. doi:10.1073/pnas.95.16.9608
22. Bjorland S, Røe C, Moen A, Schistad E, Mahmood A, Gjerstad J. Genetic predictors of recovery in low back and lumbar radicular pain. Pain. 2017;158(8):1456–1460. doi:10.1097/j.pain.0000000000000934
23. Yu Z, Wen L, Shen X, Zhang H. Effects of the OPRM1 A118G polymorphism (rs1799971) on opioid analgesia in cancer pain: a systematic review and meta-analysis. Clin J Pain. 2019;35(1):77–86. doi:10.1097/ajp.0000000000000636
24. Chou WY, Wang CH, Liu PH, Liu CC, Tseng CC, Jawan B. Human opioid receptor A118G polymorphism affects intravenous patient-controlled analgesia morphine consumption after total abdominal hysterectomy. Anesthesiology. 2006;105(2):334–337. doi:10.1097/00000542-200608000-00016
25. Olsen MB, Jacobsen LM, Schistad EI, et al. Pain intensity the first year after lumbar disc herniation is associated with the A118G polymorphism in the opioid receptor mu 1 gene: evidence of a sex and genotype interaction. J Neurosci. 2012;32(29):9831–9834. doi:10.1523/jneurosci.1742-12.2012
26. WHO Guidelines Approved by the Guidelines Review Committee. Guidelines for the Psychosocially Assisted Pharmacological Treatment of Opioid Dependence. World Health Organization. World Health Organization Copyright ©. 2009
27. Krupitsky E, Nunes EV, Ling W, Illeperuma A, Gastfriend DR, Silverman BL. Injectable extended-release naltrexone for opioid dependence: a double-blind, placebo-controlled, multicentre randomised trial. Lancet. 2011;377(9776):1506–1513. doi:10.1016/s0140-6736(11)60358-9
28. Latif ZE, Solli KK, Opheim A, et al. No increased pain among opioid-dependent individuals treated with extended-release naltrexone or buprenorphine-naloxone: a 3-month randomized study and 9-month open-treatment follow-up study. Am J Addict. 2019;28(2):77–85. doi:10.1111/ajad.12859
29. Weimand BM, Solli KK, Reichelt WH, Tanum L. Enablers and hindrances for longer-term abstinence in opioid dependent individuals receiving treatment with extended-release naltrexone: a Norwegian longitudinal recovery trial (NaltRec study). Contemp Clin Trials Commun. 2021;21:100728. doi:10.1016/j.conctc.2021.100728
30. Sheehan DV, Lecrubier Y, Sheehan KH, et al. The mini-international neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59 Suppl 20:22–33; quiz34–57.
31. Weimand B, Karlsson AT, KKS J-KV, Mordal J, Wergeland Digranes L, Tanum L. Characteristics of opioid-dependent patients choosing antagonist treatment with extended-release naltrexone compared with patients in opioid maintenance treatment in Norway. Heroin Addict Relat Clin Probl. 2023;25(N4):5–14.
32. Jacobsen DP, Nielsen MB, Einarsen S, Gjerstad J. Negative social acts and pain: evidence of a workplace bullying and 5-HTT genotype interaction. Scand J Work Environ Health. 2018;44(3):283–290. doi:10.5271/sjweh.3704
33. Rajalingam D, Jacobsen DP, Nielsen MB, Einarsen SV, Gjerstad J. Exposure to workplace bullying, distress, and insomnia: the moderating role of the miR-146a genotype. Front Psychol. 2019;10:1204. doi:10.3389/fpsyg.2019.01204
34. McLellan AT, Luborsky L, Woody GE, O’Brien CP. An improved diagnostic evaluation instrument for substance abuse patients. The addiction severity index. J Nerv Ment Dis. 1980;168(1):26–33. doi:10.1097/00005053-198001000-00006
35. Hjermstad MJ, Fayers PM, Haugen DF, et al. Studies comparing numerical rating scales, verbal rating scales, and visual analogue scales for assessment of pain intensity in adults: a systematic literature review. J Pain Symptom Manage. 2011;41(6):1073–1093. doi:10.1016/j.jpainsymman.2010.08.016
36. Peciña M, Love T, Stohler CS, Goldman D, Zubieta JK. Effects of the Mu opioid receptor polymorphism (OPRM1 A118G) on pain regulation, placebo effects and associated personality trait measures. Neuropsychopharmacology. 2015;40(4):957–965. doi:10.1038/npp.2014.272
37. Dunn KE, Huhn AS, Finan PH, et al. Polymorphisms in the A118G SNP of the OPRM1 gene produce different experiences of opioids: a human laboratory phenotype–genotype assessment. Addict Biol. 2024;29(1):e13355. doi:10.1111/adb.13355
38. Saad Z, Hibar D, Fedgchin M, et al. Effects of Mu-opiate receptor gene polymorphism rs1799971 (A118G) on the antidepressant and dissociation responses in esketamine nasal spray clinical trials. Int J Neuropsychopharmacol. 2020;23(9):549–558. doi:10.1093/ijnp/pyaa030
39. Christensen JO, Nielsen MB, Sannes AC, Gjerstad J. Leadership style, headache, and neck pain: the moderating role of the catechol-O-methyltransferase (COMT. Genotype J Occup Environ Med. 2021;63(2):151–158. doi:10.1097/jom.0000000000002103
40. Gauthier P, Greco P, Meyers-Ohki S, Desai A, Rotrosen J. Patients’ perspectives on initiating treatment with extended-release naltrexone (XR-NTX). J Subst Abuse Treat. 2021;122:108183. doi:10.1016/j.jsat.2020.108183
41. Abraham JE, Maranian MJ, Spiteri I, et al. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics. 2012;5:19. doi:10.1186/1755-8794-5-19
42. Yao RA, Akinrinade O, Chaix M, Mital S. Quality of whole genome sequencing from blood versus saliva derived DNA in cardiac patients. BMC Med Genomics. 2020;13(1):11. doi:10.1186/s12920-020-0664-7
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Opioid Receptor Mu 1 Gene (OPRM1) A118G Polymorphism and Emotional Modulation of Pain
Trimble EA, Kell PA, Avella MA, France CR, Rhudy JL
Journal of Pain Research 2024, 17:489-500
Published Date: 1 February 2024