Back to Journals » Infection and Drug Resistance » Volume 18
Possible Association of Carbapenemase Production with Susceptibility Pattern and Resistance Genes Among Carbapenemase Producing Enterobacterales from Blood Isolates or Bacteremic Patients
Authors Shakhatreh MAK, Al-Ghawrie HM, Meqdam MM, Shakhatreh Z, Khair IY, Tabnjh AK , Jacob JH, Khabour MO, Alzoubi KH
Received 15 February 2025
Accepted for publication 6 May 2025
Published 16 May 2025 Volume 2025:18 Pages 2569—2579
DOI https://doi.org/10.2147/IDR.S519561
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Muhamad Ali K Shakhatreh,1 Hadeel M Al-Ghawrie,1 Mamdoh M Meqdam,1 Zaid Shakhatreh,2 Iliya Y Khair,3 Abedelmalek Kalefh Tabnjh,4 Jacob H Jacob,5 Mohammed O Khabour,2 Karem H Alzoubi6,7
1Department of Medical Laboratory Sciences, Faculty of Applied Medical Sciences, Jordan University of Science and Technology, Irbid, Jordan; 2Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan; 3Department of Biotechnology and Genetic Engineering, Faculty of Science and Art, Jordan University of Science and Technology, Irbid, Jordan; 4Department of Applied Dental Sciences, Faculty of Applied Medical Sciences, Jordan University of Science and Technology, Irbid, Jordan; 5Department of Biological Sciences, Faculty of Science, Al Al-Bayt University, Al-Mafraq, Jordan; 6Department of Pharmacy Practice and Pharmacotherapeutics, University of Sharjah, Sharjah, United Arab Emirates; 7Department of Clinical Pharmacy, Faculty of Pharmacy, Jordan University of Science and Technology, Irbid, 22110, Jordan
Correspondence: Muhamad Ali K Shakhatreh, Department of Medical Laboratory Sciences, Faculty of Applied Medical Sciences, Jordan University of Science and Technology, P.O. Box 3030, Irbid, 22110, Jordan, Tel + 962-2-7201000, Fax + 962-2-7201071, Email [email protected]
Background: Bacteremia caused by Enterobacteriaceae strains is associated with increased mortality rates due to antibiotic resistance, including carbapenems. The current study investigated antimicrobial susceptibility, carbapenemase production, and the presence of resistance genes in Enterobacteriaceae isolated from blood cultures.
Methods: Eighty pure Enterobacteriaceae isolates were collected from positive blood cultures from four Jordanian hospitals. Antimicrobial susceptibility was investigated using the Kirby–Bauer method. Chromogenic culture media was used for the Hodge test, and the carbapenemase production was determined using the Carba NP test. The PCR technique was used to identify genes that confer resistance.
Results: Most isolates were positive for Klebsiella pneumoniae (55%), followed by Escherichia coli (37.5%) and Enterobacter cloacae (5%). The highest rates of resistance were observed against ampicillin (90%), cefazolin (76.7%), cefotaxime (70%), and ceftriaxone (65%). The lowest rate of resistance was observed against imipenem (13.7%). The frequencies of carbapenemase production, as determined by chromogenic culture media, the modified Hodge test, and the Carba NP Test, were 18.75%, 21.25%, and 10%, respectively. The identified carbapenemase resistance genes were bla-KPC (10%), bla-NDM (15%), bla-VIM (5%), and bla-OXA-48 (6.25%). A significant association (P < 0.05) was found between multidrug resistance and carbapenemase production.
Conclusion: A low percentage of carbapenem-resistant Enterobacteriaceae was observed among Jordanian patients with bacteremia. A significant association was observed between carbapenemase production and multi-drug resistance. The results can be used in the management of bacteremic patients in Jordan.
Keywords: Enterobacteriaceae, carbapenemase, bacteremia, antibiotic resistance, gene, Jordan
Introduction
Bacteremia is a severe medical condition defined by the presence of bacteria in the bloodstream.1,2 It is diagnosed with positive blood culture and is associated with systematic symptoms such as fever, chills, nausea, headache, low blood pressure, and high white blood cell counts.3,4 The condition is associated with high morbidity and mortality rates worldwide and is known to be a commonly acquired infection in healthcare settings.5,6
Bacteremia can be classified according to the site of infection into primary, which originates from the cardiovascular system, or secondary, which occurs due to the transmission of bacteria from another site of the body into the blood.7,8 It can also be classified as nosocomial (hospital-acquired) or community-acquired, with differences in infectious organisms and antibiotic susceptibility.9 Bacteremia can indicate the presence of an actual systemic infection, such as sepsis, which often stems from sources such as the urinary tract or lungs.10
Bacteremia in the United States is reported to be shared based on data published by the National Healthcare Safety Network.11 In Jordan, a study of healthcare-associated bloodstream infections (BSIs) indicates the overall incidence of health-care-associated BSIs is 8.1 per 1000 admissions, while the mortality rate caused by healthcare-associated BSIs is 5.8 per 1000 admissions.12
Gram-negative bacteria, including the Enterobacteriaceae, can lead to bacteremia.13 Members of Enterobacteriaceae are rod-shaped, Gram-negative bacteria, and some species in this family are part of the intestinal flora.14 Enterobacteriaceae encompasses common human pathogens, including Salmonella, Escherichia coli, Yersinia pestis, Klebsiella, Proteus, Enterobacter, and Shigella, which cause various infections such as cystitis, pyelonephritis, septicemia, pneumonia, peritonitis, meningitis, and device-associated infections.15,16 Some strains of Enterobacteriaceae have become resistant to antibiotics, including penicillin, cephalosporins, and carbapenems, considered the last line of antibiotics against resistant organisms.17,18 Mobile genes can explain this in plasmids that can spread through bacterial strains.19
Antimicrobial resistance (AMR) is one of the most pressing global health challenges, with multidrug-resistant organisms (MDROs) significantly contributing to morbidity and mortality worldwide. The World Health Organization (WHO) has identified AMR as a critical threat, with an estimated 4.95 million deaths linked to bacterial AMR in 2019.20 Bloodstream infections (BSIs) caused by MDROs, particularly carbapenem-resistant Enterobacteriaceae (CRE), are associated with high mortality rates due to limited treatment options, prolonged hospital stays, and increased risk of complications such as septic shock and organ failure.21,22
Carbapenems-resistant Enterobacteriaceae (CRE) are high-priority antibiotic-resistant pathogens identified by health reports as a major health threat.20 One study demonstrated that critically ill patients who developed bloodstream infections from multidrug-resistant organisms exhibited a significantly elevated risk of mortality compared to their non-infected counterparts.23
The prevalence of CRE poses a significant clinical challenge in Jordan and other regions with limited resources. Despite heightened awareness, critical scientific gaps remain; these include an incomplete understanding of the epidemiology and mechanisms underlying carbapenem resistance in diverse geographical settings.
This study aims to address these gaps by investigating antimicrobial susceptibility patterns, carbapenemase production, and resistance genes in Enterobacteriaceae isolated from blood cultures in Jordan. By providing insights into the prevalence and molecular characteristics of CRE, this research seeks to inform national prevention efforts and contribute to global strategies for combating AMR.
Therefore, emerging resistance of Enterobacteriaceae represents a significant challenge that requires immediate attention, and research studies are needed to inform national prevention efforts. In the current study, antimicrobial susceptibility, carbapenemase production, and resistance genes were investigated in Enterobacteriaceae isolated from blood cultures in Jordan.
Materials and Methods
Sample Collection, Identification, and Preservation
Eighty isolates of pure, non-repeated clinical Enterobacteriaceae from positive blood cultures were collected from patients in four Jordanian hospitals: King Abdullah University Hospital (KAUH), Prince Rahma Teaching Hospital, Zarqa New Governmental Hospital, and Prince Hashim bin Al-Hussein Military Hospital during the period from October 2019 to February 2020. The Research Ethics Committee (REC) of Jordan University of Science and Technology approved the study procedures. All samples were identified in the diagnostic microbiology laboratories of included hospitals using microbiological techniques/automated systems (VITEK@2, bioMérieux), and further confirmation was done in our research laboratory using the Microgen GNA-ID (Microgen Bioproducts Ltd, UK) system as the VITEK@2 identification system is not absolutely accurate.21 The data obtained by the Microgen GNA-ID microwell strip were designed to generate a 4-digit octal code for Enterobacteriaceae, which was used to interpret the result using the Microgen Identification System Software. Escherichia coli ATCC 25922 and Acinetobacter baumannii ATCC 19606 were used as experimental quality control. The Microgen GNA-ID system differentiates Enterobacteriaceae through simultaneous biochemical reactions, identifying microorganisms by color change after 18–24 hours at 35 ± 2°C. It uses biochemical tests based on statistical probabilities for numeric identification and compares profiles to a reference database. For sample preparation, the collected samples were sub-cultured on blood agar at 37°C for 24 hours. Fresh colonies were inoculated into nutrient broth and incubated for 18 hours. The broth was mixed with 50% sterile glycerol (1:1 ratio) and stored at −80°C for future testing.
Antimicrobial Susceptibility Testing
The Kirby-Bauer disc diffusion method on Mueller-Hinton Agar was used to examine the susceptibility of isolates to fifteen antimicrobial agents selected according to CLSI guidelines (M100, 27th ed. January 2018).15 Isolated colonies were grown for 18–24 hours on agar plates; then a 0.5 McFarland standard turbidity inoculum was then mixed with the saline. A sterile cotton swab was used to streak the inoculum across the agar surface. After drying for 5 minutes, the antibiotic discs were applied with a minimum distance of 24 mm between them. The plates were then placed under aerobic incubation at 37°C for 16–18 hours. Following the incubation period, the inhibition zone (mm) around each disc, if present, was measured in millimeters using a transparent ruler. Escherichia coli ATCC 25922 was used for quality control purposes as the control strain in susceptibility testing.
Detection of Carbapenemase Production
Carbapenemase production was assessed using three methods: chromogenic media (CHROMagar, Paris, France), the Modified Hodge Test (MHT), and the Carba NP test, to cross-validate findings.22–24 The media were prepared according to the manufacturer’s instructions under aseptic conditions; K. pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC 700603 were used for quality control according to the manufacturer’s instructions. Then, all samples were streaked onto two different chromogenic agars (CHROMagarm SuperCARBA, CHROMagar KPC) to cross-validate the results and to minimize false negatives. The plates were then incubated in aerobic conditions at 35–37°C for 18–24 hours; the results were interpreted as described by the manufacturer.
The Modified Hodge Test (MHT) was also performed to detect carbapenemase-producing Enterobacteriaceae according to CLSI guidelines (2018). A 0.5 McFarland standard suspension of E. coli ATCC® 25922 was diluted at a 1:10 ratio and streaked on Mueller–Hinton agar, with a carbapenem susceptibility disc (Ertapenem or Meropenem) placed at the center. Colonies of the test organism were streaked from the disc’s edge to the plate’s edge, ranging between 20 and 25 mm in length, then incubated at 35°C ± 2°C for 16–20 hours. After incubation, a cloverleaf indentation at the intersection of the test organism and E. coli indicated a positive result, while no growth indicated a negative result. Positive and negative controls were K. pneumoniae ATCC® BAA-1705 and K. pneumoniae ATCC® BAA-1706, respectively.
In the current study, the RAPIDEC CARBA NP® (bioMérieux, France) test was used to measure carbapenemase activity by growing isolates on Mueller–Hinton agar for 18–24 hours and as described by the manufacturer.25 The results were visually assessed by comparing the colors of the test and control wells at 30 minutes and finalized within 2 hours.
Detection of Resistance Genes
Bacterial DNA was extracted from all samples using a Zymogen DNA extraction kit (Zymo Research crop. Irvine, USA) according to the protocol provided by the manufacturer. Polymerase Chain Reaction (PCR) was used to investigate the presence of resistance genes using primers targeting bla-VIM, bla-IMP, bla-KPC, bla-OXA-48, and bla-NDM. Primers were synthesized by Integrated DNA Technologies, and sequences, annealing temperature, and product size were as previously described.26 A liquid stock (100 µM) of each primer was prepared from the lyophilized primers using nuclease-free water. 10 µM working stock solution was prepared for each primer for PCR use. Stock solutions were saved at −20°C until use.
PCR was performed on a BIO-RAD T100 Thermal Cycler. For amplification, 3μL of template DNA (50 ng/μL) was mixed with 2.5μL of forward and reverse primers (10μM each), 12.5μL of master mix (i-Taq DNA Polymerase), and 4.5 μL of nuclease-free water, resulting in a final volume of 25μL. The cycling conditions included an initial denaturation at 94°C for 5 minutes, followed by 40 cycles: denaturation at 94°C for 45 seconds, annealing (52°C for bla-VIM, bla-NDM, bla-OXA-48; 45°C for bla-IMP; and 62°C for bla-KPC) for 1 minute, and elongation at 72°C for 1 minute. A final extension at 72°C for 10 minutes completed the process.26
After amplification, 5 µL of each PCR product was analyzed using agarose gel electrophoresis (1% agarose in Tris-Borate EDTA buffer) stained with 5 µL of Red Safe dye per 100 mL of gel (iNtron, RedSafe™ Nucleic Acid Staining Solution (20,000x)). To ensure the detection of amplicons, 100 base-pair DNA ladder (iNtron, Sizer ™ −100 DNA Marker Solution) was used for comparison. Genotyping and molecular procedures were performed at the Princess Haya Biotechnology Center.
Statistical Analysis
Statistical analysis of data was done using The Statistical Package for Social Science (SPSS) software (IBM, USA). Data were analyzed by the Pearson Chi-Square test. P value < 0.05 was considered statistically significant.
Results
Study Isolates
A total of 80 clinically pure Enterobacteriaceae isolates were collected from positive blood cultures. Most isolates were positive for Klebsiella pneumoniae (55%) followed by Escherichia coli (37.5%) and Enterobacter cloacae (5%). One isolate for each Citrobacter freundii and one isolate of Pantoea agglomerans were identified.
Antimicrobial Susceptibility Profile
Antimicrobial susceptibility testing revealed high rates of resistance among isolates to ampicillin (95%), cefazolin (81.2%), cefotaxime (70%), and ceftriaxone (67.5%). In contrast, the highest susceptibility was observed with imipenem (86.3%), meropenem (82.5%), and ertapenem (82.5%). The study also showed a high prevalence of multidrug-resistant (MDR) strains, represented by the resistant nature of 68.75% of isolates in three or more categories, conserving a consistent minimum of one drug per category. Carbapenem-resistant Enterobacteriaceae (CRE) were identified if they were resistant to ertapenem, meropenem, or imipenem with resistance rates of 13.7%, 17.5%, and 17.5% were found, respectively. Additionally, Klebsiella pneumoniae (80%) and Escherichia coli (20%) were mostly common among isolates. Table 1 summarizes the antimicrobial susceptibility profile.
 |
Table 1 Antimicrobial Susceptibility of the Collected Isolates |
Detection of Carbapenemase Production
Carbapenemase production was detected using chromogenic media (CHROMagar mSuperCARBA and CHROMagar KPC). A total of 18.75% of all isolates were Carbapenem-resistant Enterobacteriaceae (CRE), while (81.25%) were non-CRE (Table 2). Modified Hodge Test (MHT) was also performed to detect CRE (Table 2). Based on MHT results, 21.25% of all isolates were identified as CRE. Finally, RAPIDEC CARBA NP® test (bioMérieux, France) was performed to detect Carbapenemase activity. Carba NP test detects only (10%); this might be due to less sensitivity of Carba NP test (Table 2).
 |
Table 2 Carbapenemase Production Detection Test Results as Positive and Negative |
The Association Between Carbapenemase Production and Antimicrobial Susceptibility
The association between Carbapenemase production and Antimicrobial Susceptibility results is shown in Supplementary Table 1. According to the previous tests, several significant associations (P<0.05) were observed between antibiotics and Carbapenemase production. Carbapenemase production was strongly associated (P<0.05) with resistance to all carbapenem antibiotics (Ertapenem, Imipenem, and Meropenem). Carbapenemase production was also significantly associated (P<0.05) with resistance to Cefepime, Ceftriaxone, Cefoxitin, Ciprofloxacin, and Levofloxacin.
Table 3 shows the association between multi-drug-resistant strains, Carbapenemase-resistant strains, and Carbapenemase production. Nearly all the results show a significant association (P<0.05) between the Carbapenemase production with multi-drug resistant and CRE strains.
 |
Table 3 Association Between Multi-Drug Resistance Strains, Carbapenemase-Resistant Strains, and Carbapenemase Production |
All Carbapenem Resistant Enterobacteriaceae strains were strongly associated (P<0.05) with Carbapenemase production, which means that Carbapenemase production leads to carbapenem resistance in nearly all isolates in this study. Furthermore, there was a strong concordance between multi-drug resistance strain and Carbapenemase production detected by Chromogenic media and the Modified Hodge tests (Table 3).
Detection of Resistance Genes
PCR was applied to all bacterial isolates to investigate the presence of resistance genes (bla-KPC, bla-NDM, bla-IMP, bla-VIM, and bla-OXA-48). Based on the PCR assays, 23 (28.75%) isolates were positive for one or more Carbapenemase genes. Of the 23 positive isolates for Carbapenemase genes, 6 (26%) were positive for more than one gene. Most of the carbapenemase-encoding Enterobacteriaceae isolates were Klebsiella pneumoniae (74%), followed by E. coli (26%). Overall, the NDM gene was the most predominant Carbapenemase gene detected in 12 isolates (15%), followed by KPC gene 8 (10%), OXA-48 5 (6.25%), VIM 4 (5%), and all isolates were negative to IMP-types. Table 4 represents the distribution of Carbapenemase genes among all study isolates. Figure 1 shows gels for the detection of resistance genes.
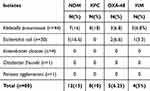 |
Table 4 The Distribution of Carbapenemase Genes Among All Study Isolates |
Supplementary Table 2 shows a significant association between Carbapenemase resistance genes and Antimicrobial Susceptibility results. Bla-KPC, bla-NDM and bla-OXA-48 were significantly associated (P<0.05) with resistance to all carbapenems (Ertapenem, Imipenem and Meropenem). There was no significant association between bla-VIM and carbapenems resistance. bla-NDM showed the most significant association (P<0.05) with resistance to other antibiotic classes – other than carbapenems-.
The association between carbapenemase resistance genes and carbapenemase production is shown in Table 5. Carbapenemase production was significantly associated (P<0.05) with the presence of Carbapenemase resistant genes, except bla-VIM which showed no significant association with Carbapenemase production.
 |
Table 5 Association Between Carbapenemase Resistance Genes and Carbapenemase Production |
Discussion
Carbapenemase Resistant Enterobacteriaceae (CRE) are among the broad spectrum of Multi-Drug Resistant (MDR) Enterobacteriaceae that represents a threat to human and public health, according to the World Health Organization (WHO).27 The current study explores the potential antimicrobial susceptibility, carbapenemase production, and the presence of resistance genes of Enterobacteriaceae isolated from blood cultures of bacteremic patients, which were investigated.
Among the collected pure isolates of Enterobacteriaceae-positive blood cultures, 55% of all isolates were Klebsiella pneumoniae, 37.5% were Escherichia coli, and 5% were Enterobacter cloacae. In a recent study on Carbapenem-resistant Enterobacteriaceae from patients with bacteremia in South Africa (2020), it was found that Klebsiella pneumoniae was the predominant species (78%) isolated from positive blood culture infected with Enterobacteriaceae.28 Our findings are also consistent with those of the Egyptian study (2018), which found that Escherichia coli constituted 30.7% of all Enterobacteriaceae species isolated from various clinical samples, second to Klebsiella pneumoniae, which formed 38.6% of all samples.29 Additionally, in a study from the United States (2017), it was shown that 90% of bloodstream infections were caused by K. pneumoniae.30 Furthermore, a Jordanian study (2010) reported that Klebsiella species and Escherichia coli are the most common gram-negative microorganisms that cause bacteremia among children.31
Enterobacteriaceae strains become resistant to the last line antibiotics, including penicillin, cephalosporins, and carbapenems.32 In the current study, the highest rates of resistance were observed against ampicillin (90%), followed by cefazolin (76.7%), cefotaxime (70%) and ceftriaxone (65%). At the same time, the highest susceptibility rates were observed for imipenem (86.3%), followed by meropenem (82.5%) and ertapenem (81.3%). Similar trend was found in a recent study about bacteremia among Jordanian children.31 Much lower susceptibility rates to the carbapenem group (40%) have been observed in Enterobacteriaceae strains isolated from blood culture in South Africa.28
High or intermediate Enterobacteriaceae resistance among all isolates was considered CRE. The resistance rate to imipenem was 13.7%, to meropenem was 17.5%, and to ertapenem was 17.5%. Comparable findings have been reported in Egypt.29 In the disc diffusion method, 18.75% of all Enterobacteriaceae species studied were classified as CRE. Comparable findings have been reported that the resistance rate against carbapenem in nosocomial-infected patients with gram-negative bacilli was 27.17%.33 On the contrary, two Egyptian studies found that the rates of CRE were 45% and 47%, simultaneously,34,35 demonstrating a higher carbapenem resistance rate compared to this current study. A lower rate of carbapenem resistance was found among Enterobacteriaceae in previous studies from Jordan, Turkey, the United States, Lebanon, and Malaysia.34,36–41
In this study, Klebsiella pneumoniae was the most common CRE species (64%), followed by Escherichia coli (20%). These findings align with a previous study from Jordan, where K. pneumoniae was also the most prevalent CRE strain (82.1%).36 Similarly, a study in Egypt found K. pneumoniae (51.4%) and E. coli (28.6%) to be the dominant CRE species.29
Carbapenemase production was significantly associated (P<0.05) with resistance to all carbapenem antibiotics (Ertapenem, Imipenem, and Meropenem). Carbapenemase production was also significantly associated (P<0.05) with cefepime, ceftriaxone, cefoxitin, ciprofloxacin, and levofloxacin resistance. All CRE strains were significantly associated with Carbapenemase production, which means that Carbapenemase production led to carbapenem resistance in nearly all isolates in this study. Also, there was a significant association between Multi-Drug Resistance strain (MDR) and carbapenemase production that was detected by Chromogenic media and the Modified Hodge test but not by the Carba NP test. The observed differences observed among the different detection methods could likely stem from variations in the sensitivity and specificity of each technique.
Based on the PCR assays in the current study, 28.75% of all isolates were positive for one or more of the carbapenemase genes. Of the 23 isolates positive for carbapenemase genes, 26% were positive for more than one gene. Overall, the NDM gene was the most predominant carbapenemase gene detected in 15% of isolates, followed by KPC gene (10%), OXA-48 (6.25%), VIM (5%), and all isolates were negative to IMP gene. KPC, NDM and OXA-48 were associated with resistance to all carbapenems. There was no association between VIM and carbapenem resistance. This could be due to the low prevalence of this gene in the collected sample (5%), expressivity of the gene, and utilization of alternative resistance mechanisms that do not require the expression of VIM gene. NDM showed the most significant association with resistance to other antibiotic classes – other than carbapenems-. Similar patterns, with OXA-48 and NDM as predominant genes, have been reported in nearby regions like the Arabian Gulf, Morocco, and Palestine.42–44
In the current study, the predominant gene was NDM; this was in contrast with a study performed in the United States that showed KPC detected in more than 90% of CRE isolates from blood cultures.45 In a study from South African, the most common Carbapenemase genes reported were OXA-48 and NDM among CRE isolated from blood culture.28 In a study from Egypt, the predominant genes were KPC and VIM.29 Contrary to our findings, some studies have reported KPC, IMP, and VIM genes lacking in Klebsiella pneumoniae isolates.46,47 The variation in the prevalence of Carbapenemase genes highlights the importance of examining the spectrum of these genes in different settings/countries.
CRE remains a significant clinical challenge in Jordanian hospitals and may be spreading. Our findings provide a valuable baseline for ongoing CRE surveillance and infection control efforts across different hospital wards. In summary, this study found a relatively low prevalence of CRE compared to regional and global levels. However, the findings serve as an early warning, highlighting the need for enhanced laboratory capabilities to detect, identify, and characterize CRE more effectively.
Conclusions
The isolated Enterobacteriaceae strains demonstrated the highest resistance against Ampicillin, followed by Cefazolin, Cefotaxime, and Ceftriaxone, and the highest susceptibility rates to Imipenem, followed by Meropenem and Ertapenem. The rate of resistance strains against carbapenems was 18.75%. The prevalence of carbapenemase production according to Chromogenic Culture Media, Modified Hodge Test, and Carba NP Test was 18.75%, 21.25%, and 10%, respectively. Carba NP test showed less sensitivity in carbapenemase detection compared with other carbapenemase production detection methods. Carbapenemase production was significantly associated with resistance to all of carbapenems antibiotics. Carbapenemase production was also significantly associated with resistance to Cefepime, Ceftriaxone, Cefoxitin, Ciprofloxacin, and Levofloxacin. All Carbapenem-Resistant Enterobacteriaceae strains were significantly associated with carbapenemase production. There was a significant association between multi-drug resistance strain and carbapenemase production detected by Chromogenic media and the Modified Hodge test. The prevalence of carbapenemase resistance genes bla-KPC, bla-NDM, bla-IMP, bla-VIM, and bla-OXA-48 was 10%, 15%, 0%, 5% and 6.25%, respectively. Bla-KPC, bla-NDM and bla-OXA-48 were significantly associated with resistance to all carbapenems. bla-NDM showed the most significant association with resistance to other antibiotic classes other than carbapenems. Carbapenemase production was significantly associated with the presence of carbapenemase-resistant genes. Bla-VIM showed no significant association with carbapenem resistance or carbapenemase production.
One of the limitations of this study lies in its relatively small sample size of 80 isolates, which may not comprehensively represent the broader population of bacteremic patients in Jordan. Additionally, the study was conducted in only four hospitals, potentially limiting the generalizability of the findings to other regions or healthcare settings. Furthermore, the reliance on specific diagnostic methods, such as the Carba NP test, showed lower sensitivity compared to other techniques like the Modified Hodge Test and chromogenic media. These limitations should be considered when interpreting the results and designing future studies.
Data Sharing Statement
Data will be available upon reasonable request via e-mailing the corresponding author.
Ethics Approval and Consent to Participate
The institutional review board of Jordan University of Science and Technology, Irbid, Jordan, approved the study (approval number: 393/2019). All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
Consent for Publication
Written informed consent was obtained from the study participants.
Funding
This study was funded by the Deanship of Research, Jordan University of Science and Technology, Irbid, Jordan (grant number 393/2019).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hassoun-Kheir N, Guedes M, Ngo Nsoga MT, et al. A systematic review on the excess health risk of antibiotic-resistant bloodstream infections for six key pathogens in Europe. Clin Microbiol Infect. 2024;30(Suppl 1):S14–s25. doi:10.1016/j.cmi.2023.09.001
2. Namikawa H, Imoto W, Yamada K, et al. Predictors of mortality from extended-spectrum beta-lactamase-producing Enterobacteriaceae bacteremia. Emerg Microbes Infect. 2023;12(1):2217951. doi:10.1080/22221751.2023.2217951
3. Rasmussen M, Gilje P, Fagman E, Berge A. Bacteraemia with gram-positive bacteria-when and how do I need to look for endocarditis? Clin Microbiol Infect. 2024;30(3):306–311. doi:10.1016/j.cmi.2023.08.027
4. Rogers R, Rice LB. State-of-the-art review: persistent enterococcal bacteremia. Clin Infect Dis. 2024;78(1):e1–e11. doi:10.1093/cid/ciad612
5. Bonten M, Johnson JR, van den Biggelaar AHJ, et al. Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin Infect Dis. 2021;72(7):1211–1219. doi:10.1093/cid/ciaa210
6. Kłos M, Wójkowska-Mach J. Hospital-acquired Enterobacteriaceae bloodstream infections in children. Dev Period Med. 2019;23(2):131–136. doi:10.34763/devperiodmed.20192302.131136
7. Timsit JF, Ruppé E, Barbier F, Tabah A, Bassetti M. Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med. 2020;46(2):266–284. doi:10.1007/s00134-020-05950-6
8. Kwiecińska-Piróg J, Skowron K, Gospodarek-Komkowska E. Primary and secondary bacteremia caused by proteus spp.: epidemiology, strains susceptibility and biofilm formation. Pol J Microbiol. 2018;67(4):471–478. doi:10.21307/pjm-2018-055
9. Andersen CT, Langendorf C, Garba S, et al. Risk of community- and hospital-acquired bacteremia and profile of antibiotic resistance in children hospitalized with severe acute malnutrition in Niger. Int J Infect Dis. 2022;119:163–171. doi:10.1016/j.ijid.2022.03.047
10. Niederman MS, Baron RM, Bouadma L, et al. Initial antimicrobial management of sepsis. Crit Care. 2021;25(1):307. doi:10.1186/s13054-021-03736-w
11. Prestel C, Fike L, Patel P, et al. A review of pediatric central line-associated bloodstream infections reported to the national healthcare safety network: United States, 2016-2022. J Pediatric Infect Dis Soc. 2023;12(9):519–521. doi:10.1093/jpids/piad066
12. Al-Rawajfah OM, Cheema J, Hewitt JB, Hweidi IM, Musallam E. Laboratory-confirmed, health care-associated bloodstream infections in Jordan: a matched cost and length of stay study. Am J Infect Control. 2013;41(7):607–611. doi:10.1016/j.ajic.2012.08.014
13. Soneda K, Uda K, Araki K, et al. Clinical characteristics and treatment of IMP-type carbapenemase-producing Enterobacteriaceae bacteremia: case series and literature review. J Infect Chemother. 2023;29(1):26–32. doi:10.1016/j.jiac.2022.09.003
14. Flokas ME, Karanika S, Alevizakos M, Mylonakis E. Prevalence of ESBL-producing Enterobacteriaceae in pediatric bloodstream infections: a systematic review and meta-analysis. PLoS One. 2017;12(1):e0171216. doi:10.1371/journal.pone.0171216
15. Sutton SP, Reinert JP. Evaluation of the efficacy of piperacillin/tazobactam in extended-spectrum beta-lactamase-producing Enterobacteriaceae urinary tract infections: a systematic review of the literature. Ann Pharmacother. 2024;10600280241291604. doi:10.1177/10600280241291604
16. Shao Y, Jia W, Li G. Related factors of bloodstream infections associated with urinary tract infections and pathogenetic characteristics analysis. Eur J Med Res. 2024;29(1):566. doi:10.1186/s40001-024-02152-4
17. Chiotos K, Hayes M, Gerber JS, Tamma PD. Treatment of carbapenem-resistant Enterobacteriaceae infections in children. J Pediatric Infect Dis Soc. 2020;9(1):56–66. doi:10.1093/jpids/piz085
18. Pascale R, Giannella M, Bartoletti M, Viale P, Pea F. Use of meropenem in treating carbapenem-resistant Enterobacteriaceae infections. Expert Rev Anti Infect Ther. 2019;17(10):819–827. doi:10.1080/14787210.2019.1673731
19. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–s36. doi:10.1093/infdis/jiw282
20. Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nat News. 2017;543(7643):15. doi:10.1038/nature.2017.21550
21. Nimer NA, Al-Saa’da RJ, Abuelaish O. Accuracy of the VITEK® 2 system for a rapid and direct identification and susceptibility testing of gram-negative rods and gram-positive cocci in blood samples. East Mediterr Health J. 2016;22(3):193–200. doi:10.26719/2016.22.3.193
22. Amladi AU, Sudarsanam TD, Kandasamy S, Kekre N, Veeraraghavan B, Sahni RD. Evaluation of CHROMagar™ TMmSuperCARBA™ as a phenotypic test for detection of carbapenemase producing organisms. J Clin Diagn Res. 2019;13(9).
23. Panagea T, Galani I, Souli M, Adamou P, Antoniadou A, Giamarellou H. Evaluation of CHROMagar™ KPC for the detection of carbapenemase-producing Enterobacteriaceae in rectal surveillance cultures. Int J Antimicrob Agents. 2011;37(2):124–128. doi:10.1016/j.ijantimicag.2010.10.010
24. Genc O, Aksu E. Chromogenic culture media or rapid immunochromatographic test: which is better for detecting Klebsiella pneumoniae that produce OXA-48 and can they be used in blood and urine specimens. J Microbiol Methods. 2018;148:169–173. doi:10.1016/j.mimet.2018.04.014
25. Kabir MH, Meunier D, Hopkins KL, Giske CG, Woodford N. A two-centre evaluation of RAPIDEC® CARBA NP for carbapenemase detection in Enterobacteriaceae, pseudomonas aeruginosa and Acinetobacter spp. J Antimicrob Chemother. 2016;71(5):1213–1216. doi:10.1093/jac/dkv468
26. Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. Biomed Res Int. 2014;2014:1–6. doi:10.1155/2014/303104
27. Suay-García B, Pérez-Gracia MT. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics. 2019;8(3). doi:10.3390/antibiotics8030122
28. Perovic O, Ismail H, Quan V, et al. Carbapenem-resistant Enterobacteriaceae in patients with bacteraemia at tertiary hospitals in South Africa, 2015 to 2018. Eur J Clin Microbiol Infect Dis. 2020;1–8.
29. Mohamed T, Yousef LM, Darweesh EI, Khalil AH, EL-zahraa MM. Detection and characterization of carbapenem resistant Enterobacteriaceae in Sohag University Hospitals. Egyptian Journal of Medical Microbiology. 2018;27(4):61–69. doi:10.21608/ejmm.2018.285747
30. Satlin MJ, Chen L, Patel G, et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother. 2017;61(4):e02349–16. doi:10.1128/AAC.02349-16
31. Mohammad A. Bacteremia among Jordanian children at princess Rahmah hospital: pathogens and antimicrobial susceptibility patterns. Iranian J Microbiol. 2010;2(1):22.
32. Elshamy AA, Aboshanab KM. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future Sci OA. 2020;6(3):FSO438. doi:10.2144/fsoa-2019-0098
33. El-Rehewy MS, Saboor EA, Afifi NA, Ibrahim MA, Qayed SS. Detection and characterization of nosocomial carbapenem-resistant gram negative bacilli from assuit university hospitals. Egypt J Med Microbiol. 2016;38(79):1–9.
34. Elraghy NA, Zahran WA, Makled AF, et al. Multidrug-resistant Enterobacteriaceae nosocomial uropathogens at Menoufia University Hospitals: phenotypic characterization and detection of resistance genes using real-time PCR. Menoufia Med J. 2016;29(4):855.
35. El-Kazzaz SS, Abou El-khier NT. AmpC and metallo beta-lactamases producing Gram negative bacteria in patients with hematological malignancy. African J Microbiol Res. 2015;9(18):1247–1254. doi:10.5897/AJMR2015.7435
36. Aqel AA, Findlay J, Al-Maayteh M, et al. Characterization of carbapenemase-producing Enterobacteriaceae from patients in Amman, Jordan. Microb Drug Resist. 2018;24(8):1121–1127. doi:10.1089/mdr.2017.0238
37. Aqel AA, Giakkoupi P, Alzoubi H, Masalha I, Ellington MJ, Vatopoulos A. Detection of OXA-48-like and NDM carbapenemases producing Klebsiella pneumoniae in Jordan: a pilot study. J Infection Public Health. 2017;10(2):150–155. doi:10.1016/j.jiph.2016.02.002
38. Baran I, Aksu N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clinic Microbiol Antimicrob. 2016;15(1):20. doi:10.1186/s12941-016-0136-2
39. Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries R. Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J Clin Microbiol. 2014;52(11):4003–4009. doi:10.1128/JCM.01397-14
40. Hammoudi D, Moubareck CA, Aires J, et al. Countrywide spread of OXA-48 carbapenemase in Lebanon: surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Inter J Infect Dis. 2014;29:139–144. doi:10.1016/j.ijid.2014.07.017
41. Hamzan NI, Yean CY, Rahman RA, Hasan H, Rahman ZA. Detection of bla IMP4 and bla NDM1 harboring Klebsiella pneumoniae isolates in a university hospital in Malaysia. Emerg Health Threats J. 2015;8(1):26011. doi:10.3402/ehtj.v8.26011
42. Boyd SE, Livermore DM, Hooper DC, Hope WW. Metallo-β-lactamases: structure, function, epidemiology, treatment options, and the development pipeline. Antimicrob Agents Chemother. 2020;64(10). doi:10.1128/AAC.00397-20
43. El Wartiti MA, Bahmani F-Z, Elouennass M, Benouda A. Prevalence of carbapenemase-producing Enterobacteriaceae in a university Hospital in Rabat, Morocco: a 19-months prospective study. Int Arabic J Antimicrobial Agents. 2012;2(3).
44. Liddawi R, Siryani I, Ghneim R, et al. Emergence of Klebsiella pneumoniae carbapenemase (blaKPC-2) in members of the Enterobacteriaceae family in Palestine. Int Arabic J Antimicrobial Agents. 2012;2(2).
45. Tamma PD, Goodman KE, Harris AD, et al. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clinl Infect Dis. 2017;64(3):257–264. doi:10.1093/cid/ciw741
46. Sahin K, Tekin A, Ozdas S, et al. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann Clinic Microbiol Antimicrob. 2015;14(1):1–6. doi:10.1186/s12941-015-0105-1
47. Iraz M, Özad Düzgün A, Sandallı C, et al. Distribution of β-lactamase genes among carbapenem-resistant Klebsiella pneumoniae strains isolated from patients in Turkey. Ann Lab Med. 2015;35(6):595–601. doi:10.3343/alm.2015.35.6.595
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Multi-Drug Resistance Profile, Prevalence of Extended-Spectrum Beta-Lactamase and Carbapenemase-Producing Gram Negative Bacilli Among Admitted Patients After Surgery with Suspected of Surgical Site Nosocomial Infection North East Ethiopia
Tilahun M
Infection and Drug Resistance 2022, 15:3949-3965
Published Date: 26 July 2022
Metagenomic Analysis of Urban Wastewater Treatment Plant Effluents in Tokyo
Sekizuka T, Itokawa K, Tanaka R, Hashino M, Yatsu K, Kuroda M
Infection and Drug Resistance 2022, 15:4763-4777
Published Date: 23 August 2022
Antibiotic Resistance, Molecular Characteristics and Risk Factors of Carbapenem-Resistant Klebsiella pneumoniae in Clinical Isolates
Zhu J, Chen Y, Yang X
Infection and Drug Resistance 2022, 15:6671-6680
Published Date: 15 November 2022


