Back to Journals » Cancer Management and Research » Volume 17
Prediction of Ki-67 Expression in HIV-Associated Lung Adenocarcinoma Patients Using Multiple Machine Learning Models Based on CT Imaging Radiomics
Authors Song C, Chen J, Zhao C, Song S, Yang T, Huang A, Liu R, Pan Y, Xu C, Chen C, Zhu Q
Received 19 November 2024
Accepted for publication 17 April 2025
Published 25 April 2025 Volume 2025:17 Pages 881—892
DOI https://doi.org/10.2147/CMAR.S505390
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Chang Song,1,* Jingsong Chen,2,* Chunyan Zhao,1,* Shulin Song,3,* Tong Yang,4 Aichun Huang,1 Renhao Liu,1 Yanxi Pan,3 Chaoyan Xu,1 Canling Chen,1 Qingdong Zhu1
1Tuberculosis Department, Nanning Fourth People’s Hospital, Nanning, Guangxi, 530023, People’s Republic of China; 2Gastroenterology Department, Hepu County People’s Hospital, Beihai, Guangxi, 536100, People’s Republic of China; 3Radiology Department, Nanning Fourth People’s Hospital, Nanning, Guangxi, 530023, People’s Republic of China; 4Rehabilitation Department, Hepu County People’s Hospital, Beihai, Guangxi, 536100, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qingdong Zhu, Tuberculosis Department, Nanning Fourth People’s Hospital, Nanning, Guangxi, 530023, People’s Republic of China, Email [email protected]
Purpose: The incidence of lung adenocarcinoma (LUAD) in HIV-infected individuals is significantly increased. However, invasive procedures for Ki-67 assessment may increase the risk of complications. Therefore, developing a non-invasive and accurate method for Ki-67 prediction holds significant clinical importance. This study aims to explore the feasibility and value of a radiomics model based on preoperative CT images in predicting Ki-67 expression levels in HIV-associated LUAD.
Patients and Methods: A total of 237 patients with HIV-associated LUAD were included. Of these, 102 were classified into the high Ki-67 expression group, and 135 into the low Ki-67 expression group. The patients were randomly divided into a training group (n=189) and a validation group (n=48) in a 4:1 ratio. Feature selection was based on intra-class correlation coefficient (ICC), Spearman correlation coefficient, and Least Absolute Shrinkage and Selection Operator (LASSO) regression, yielding 16 optimal radiomic features for building a logistic regression model. Model performance was evaluated by sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), F1 score, and the area under the receiver operating characteristic curve (AUC).
Results: 1834 CT image features were extracted, with 16 retained for further analysis. The Support Vector Machine (SVM) model demonstrated the most balanced and optimal performance among the seven developed models. It achieved robust sensitivity (training set: 0.89; testing set: 0.86), specificity (training set: 0.92; testing set: 0.89), PPV (training set: 0.89; testing set: 0.86), NPV (training set: 0.92; testing set: 0.89), F1 score (training set: 0.89; testing set: 0.86), and AUC (training set: 0.975; testing set: 0.905), indicating excellent predictive accuracy.
Conclusion: This study first demonstrates that a preoperative CT-based radiomics model can non-invasively predict Ki-67 expression levels in HIV-associated LUAD patients. This finding not only provides a precise assessment tool for the HIV-infected population to avoid the risks of invasive examinations but also paves new interdisciplinary research avenues for exploring tumor heterogeneity under immunodeficiency conditions.
Keywords: HIV, lung adenocarcinoma, Ki-67, radiomics, machine learning, SVM
Introduction
Despite the use of antiretroviral therapy (ART), HIV-related pulmonary diseases remain a leading cause of morbidity and mortality among HIV/AIDS patients.1 Due to immune system deficiencies, individuals with HIV/AIDS face a substantially higher risk of various malignancies.2 Among these, lung adenocarcinoma, one of the most common types of non-small cell lung cancer (NSCLC), is especially prevalent. Research shows that cancer incidence and mortality rates are significantly higher in HIV/AIDS patients compared to those without HIV. This elevated risk is driven by chronic immune suppression, direct carcinogenic effects of the virus, and factors related to ART, resulting in distinct differences in tumor biology and survival outcomes compared to non-HIV-infected individuals.3,4 The Ki-67 protein, a key marker for assessing cell proliferation and tumor heterogeneity,5 increases rapidly from the G1 phase to mitosis, making it an important indicator of tumor malignancy, disease-free survival, progression-free survival, and overall survival.6 Ki-67, as a nuclear protein marker, has become an important indicator for evaluating tumor proliferation activity and prognosis by quantifying the proportion of tumor cells in the active phase of the cell cycle. Previous studies have shown that as the gold - standard marker of cell proliferation activity, the expression level of Ki-67 is associated with the invasiveness of LUAD, treatment resistance, and differences in patient survival.7–9 Moreover, the Ki-67 proliferation index is an independent risk factor for recurrence in patients with early-stage LUAD after segmentectomy.10 However, in the context of HIV infection, the biological behavior and clinical management of LUAD face unique challenges. HIV - infected patients often have comorbid opportunistic infections and multidrug resistance issues. As a result, patients with high Ki-67 LUAD may not tolerate intensive chemotherapy regimens. There is an urgent need to explore individualized treatment strategies that balance efficacy and safety. Therefore, predicting Ki-67 levels in patients with HIV-associated lung adenocarcinoma is crucial for prognosis and treatment planning.
In recent years, the rapid advancement of imaging technology and computational science has positioned radiomics as a valuable tool in oncology research. Radiomics leverages the wealth of information embedded in medical images related to tumor biology, behavior, and pathophysiology, which is often undetectable through conventional visual assessments.11 By reducing diagnostic subjectivity, radiomics enhances accuracy, enabling earlier tumor detection and facilitating timely treatment. This approach also supports personalized medicine by identifying specific tumor characteristics, allowing for tailored treatment plans.12 Moreover, radiomics predicts tumor responses to therapies, helping clinicians adjust treatment strategies to improve patient outcomes. Using radiomics minimizes unnecessary invasive procedures, reducing patient discomfort and risk, while improving research efficiency and accelerating the development of new diagnostic and therapeutic methods. By fostering interdisciplinary collaboration across medical imaging, computer science, bioinformatics, and clinical practice, radiomics advances medical imaging technology. Its data-driven model provides evidence-based support for clinical decision-making, contributing to more informed and effective healthcare strategies.
For patients with HIV-associated lung adenocarcinoma, radiomics offers a promising method for identifying and quantifying imaging features linked to Ki-67 expression. Currently, a number of studies have demonstrated significant associations between radiomic features and Ki-67 expression. Early diagnosis is crucial for prognosis assessment.9 For instance, Fu et al successfully developed a predictive model for Ki-67 expression in lung adenocarcinoma by integrating CT radiomics with clinical characteristics.13 Moreover, a recent meta-analysis further highlighted that the use of CT-based radiomics for predicting Ki-67 expression in lung cancer shows promising diagnostic performance. The pooled sensitivity, specificity, and area under the curve (AUC) in the training cohort were 0.78, 0.81, and 0.85, respectively. In the validation cohort, these values were 0.78, 0.70, and 0.8114. However, it is noteworthy that existing studies have primarily focused on immunocompetent populations, leaving a significant gap in research targeting HIV-positive individuals. The present study utilizes CT imaging radiomics features and various machine learning models to predict Ki-67 expression levels to better assess tumor proliferative activity and develop more precise treatment strategies. In addition, we aim to expand the application of radiomics and machine learning in cancer research, offering methodological insights for future studies.
Patients and Methods
Study Subjects and Data Collection
This study used a retrospective analysis to systematically gather detailed clinical data from 237 hIV-positive patients with concurrent lung adenocarcinoma treated at Nanning Fourth People’s Hospital between October 2019 and August 2024. The inclusion criteria were: (1) confirmed HIV-positive status; (2) histopathological diagnosis of lung adenocarcinoma through biopsy or surgical resection; (3) no prior treatment with radiation therapy, chemotherapy, or surgery before diagnosis; (4) complete clinical records. Exclusion criteria included: (1) substandard image quality, such as artifacts or other issues in CT images; (2) patients who did not undergo CT examination; (3) Patients who have undergone cancer treatment, such as radiation, chemotherapy, or surgery (Figure 1). Ethical approval was obtained from the Ethics Committee of the Fourth People’s Hospital of Nanning (Approval No: [2022]64). This retrospective study did not contain any identifiable human images and was an anonymous data collection that waived patients’ written informed consent. The study was conducted in compliance with the Declaration of Helsinki, and all patient data were anonymized and handled with strict confidentiality to protect patient privacy. Immunohistochemical staining was conducted using a mouse anti-human Ki-67 monoclonal antibody. Tumor slices with the highest hotspots were selected, and at least five high-power fields were randomly chosen from each stained slice to ensure that the Ki-67 expression level accurately represented the entire tumor. The percentage of Ki-67-positive cells relative to the total number of cells was quantified. Patients were classified based on the Ki-67 labeling index, with low Ki-67 expression defined as ≤ 40% positive staining and high Ki-67 expression defined as > 40% positive staining, in line with previous studies and the expression levels observed in this study.13–16 Two pathologists reviewed all pathological results, and any discrepancies were resolved through discussion with a third pathologist.
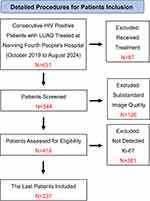 |
Figure 1 Patient screening process. |
Imaging Data Acquisition
Imaging data for this study were acquired using high-precision GE NEW Revolution ES 128-slice spiral CT and GE LightSpeed 64-slice spiral CT scanners. The CT equipment was first prepared to ensure optimal working conditions. Patients were instructed to lie supine on the scanning table and remain still, avoiding movement or speech to minimize motion artifacts. A scan thickness of 5 mm was set for routine scanning without the use of contrast agents. Contrast-enhanced scanning was performed following intravenous administration of non-ionic iodine contrast agent [Iohexol 300mg(I)/mL, Beijing, China] via the median cubital vein using a high-pressure injector, with an injection flow rate of 2.5–3.0 mL/s and a dosage of 1.5 mL/kg. Following the scan, images were reconstructed to produce high-resolution CT images. These images were analyzed to identify structural features associated with lung adenocarcinoma. The imaging data were then securely stored and transmitted to the hospital’s imaging management system or the patient’s electronic medical record, with imaging reports and professional diagnostic recommendations provided as clinically needed.
Image Processing, ROI Definition, and Feature Analysis
DICOM format CT images were exported from the imaging workstation and standardized using ITK-SNAP software. The slice thickness was adjusted to 1×1×1 mm, and the window width and level were set to 40 and 400, respectively, to optimize image display. Two experienced specialists (Q.D.Z. and S.L.S.) then collaboratively delineated and defined the tumor boundaries, manually outlining all layers of the regions of interest without knowledge of the Ki-67 index pathology results. After combining ROIs from all layers, a three-dimensional volume of interest (VOI) was created (Figure 2). Radiomics feature analysis was performed on the VOI using the PyRadiomics open-source software. In the feature selection pipeline, we established a rigorous multi-level quality control system. Firstly, the Intraclass Correlation Coefficient (ICC) was employed to evaluate feature reproducibility, retaining only features with ICC values ≥0.8 between two independent readers, thereby ensuring the stability of feature extraction. Subsequently, Pearson correlation coefficient matrices were utilized for multicollinearity detection, with a threshold of 0.9 to eliminate highly linearly correlated features, reducing the risk of model overfitting. Building upon this foundation, the Minimum Redundancy Maximum Relevance (mRMR) algorithm was introduced for feature optimization. This algorithm maximizes the correlation between features and the target variable while minimizing redundancy among features, compressing the feature space to 32 most representative features. Finally, the L1-regularized Least Absolute Shrinkage and Selection Operator (LASSO) regression model was adopted for feature selection. The optimal penalty parameter λ was determined through 10-fold cross-validation, achieving sparse constraints on regression coefficients and automatically screening out the most predictive feature subset. This systematic feature selection pipeline, ranging from feature stability assessment to final model optimization, ensures the predictive performance and generalization capability of the constructed model. The extracted features were categorized into three main types: geometric features describing the tumor’s three-dimensional shape, intensity features representing the statistical distribution of voxel intensities within the tumor, and texture features reflecting the spatial distribution patterns and complexity of intensities.
 |
Figure 2 Image segmentation process. Notes: (A) Case 1; (B) Case 2. |
Data Analysis Methods
Data analysis was performed using SPSS 23.0 software. Quantitative data with a normal distribution were expressed as mean ± standard deviation, and group differences were assessed using independent sample t-tests. For categorical data, differences between groups were evaluated using Pearson’s chi-square test. In the training set, dimensionality reduction of numerous radiomics features was carried out using the Least Absolute Shrinkage and Selection Operator (LASSO) algorithm with 10-fold cross-validation. Moreover, all radiological features were subjected to Mann–Whitney U-tests, with features having a significance level of less than 0.05 being selected. The Spearman rank correlation coefficient was used to assess correlations among highly repeatable features. Features with correlation coefficients greater than 0.9 were reduced to one to avoid multicollinearity. To enhance the descriptive capability of the feature set, a greedy recursive elimination strategy was employed to sequentially remove the most redundant features, thereby retaining the most representative ones.
Results
Clinical Baseline Characteristics
This study included 237 patients, with 135 having low Ki-67 expression and 102 having high Ki-67 expression. The patients were randomly divided into training and testing sets in a 4:1 ratio. Baseline information and clinical symptoms were collected for all patients, including sex, age, BMI, heart disease, diabetes, fever, smoking, and alcohol consumption, as detailed in Table 1. Laboratory test results, including immunological markers and routine blood tests, were also recorded, as shown in Table 2.
 |
Table 1 Patient Characteristics |
 |
Table 2 Laboratory Examination of Patients |
Radiomic Feature Selection
Based on an Intraclass Correlation Coefficient greater than 0.8, we retained 1,834 manually extracted features across seven categories with good consistency (Figure 3A), with the detailed information provided in Table 3. These features were obtained using the internal feature extraction program implemented in PyRadiomics (http://pyradiomics.readthedocs.io). Through 10-fold cross-validation and LASSO regression for dimensionality reduction and feature selection, 16 optimal radiomic features were identified (Figure 3B and C). The coefficient values for these final 16 non-zero features are displayed in Figure 3D.
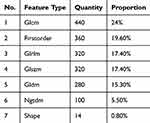 |
Table 3 Number and Proportion of Feature Types |
The Rad score is as follows:
label = 0.43037974683543606
–0.063736 * exponential_glszm_SmallAreaLowGrayLevelEmphasis
+0.116259 * lbp_3D_k_gldm_SmallDependenceLowGrayLevelEmphasis
+0.134215 * lbp_3D_m1_firstorder_90Percentile
-0.049984 * lbp_3D_m1_glrlm_LongRunLowGrayLevelEmphasis
+0.002011 * lbp_3D_m2_firstorder_Range
-0.043006 * lbp_3D_m2_firstorder_Skewness
+0.062097 * log_sigma_2_0_mm_3D_ngtdm_Busyness
-0.089358 * original_firstorder_Kurtosis
+0.042893 * original_gldm_DependenceVariance
-0.096638 * squareroot_glszm_LargeAreaLowGrayLevelEmphasis
+0.074894 * wavelet_HHH_firstorder_Kurtosis
+0.022393 * wavelet_HHH_firstorder_Maximum
-0.022679 * wavelet_HLH_ngtdm_Complexity
+0.027256 * wavelet_LLH_firstorder_Maximum
-0.024895 * wavelet_LLH_glcm_Imc2
-0.033751 * wavelet_LLL_glszm_LargeAreaHighGrayLevelEmphasis.
Development and Evaluation of Predictive Models Using Different Machine Learning Algorithms
Herein, six predictive models were constructed using the following radiomic features: SVM, K-Nearest Neighbors (KNN), RandomForest, Extremely Randomized Trees (Extra Trees), Light Gradient Boosting Machine (LightGBM), and Multi-Layer Perceptron (MLP). The performance of these models was evaluated based on sensitivity, specificity, PPV, NPV, F1 score, and AUC (Figure 4 and Table 4).
 |
Table 4 Comparison of Prediction Performance for Ki-67 Among Various Models |
 |
Figure 4 ROC curves for predicting Ki-67 using various models. (A). Training Set; (B). Testing Set. |
Evaluation of the Predictive Performance of the Optimal SVM Model
Figure 5A and B present the diagnostic contingency tables for the SVM model applied to the training and testing sets. In the training set, the model accurately identified Ki-67 expression status in 171 patients, with only 18 misclassification cases. In the testing set, the model correctly identified the Ki-67 expression status in 42 patients, with 6 cases of misclassification. Decision curve analysis demonstrates the clinical utility of the radiomic SVM model (Figure 5C and D). Figure 5E and F illustrate the construction of the radiomic score, which is derived from a linear combination of radiomic features multiplied by their corresponding coefficients.
Discussion
The pivotal role of Ki-67 in tumor growth and proliferation, along with its diagnostic significance, makes it an essential tool in oncology research and clinical practice. An in-depth analysis of the Ki-67 index enhances our understanding of tumor biology, providing valuable insights for patient treatment and prognosis.17 Radiomics, an emerging technology, shows considerable promise for both prospective and clinical applications. Its primary advantage is the non-invasive extraction of numerous quantitative features from medical images (such as CT, MRI, PET), which reveals the microstructure and heterogeneity of tumors or lesions. This capability provides critical information for disease diagnosis, prognosis assessment, and treatment decisions.18,19 Radiomics surpasses traditional imaging diagnostics that depend on subjective visual assessments by uncovering hidden features in imaging data, thereby supplementing conventional biomarker testing and excelling in tumor heterogeneity assessment and personalized medicine.20 Radiomics involves extracting a large volume of features from imaging data and converting them into actionable insights. This process leverages advanced artificial intelligence techniques, including machine learning, deep learning, and convolutional neural networks, to enhance predictive accuracy.21 The radiomic analysis of medical images comprises several critical steps: image preprocessing, segmentation, feature extraction, feature selection, and classification. Image preprocessing is essential for ensuring the validity and reproducibility of radiomic features and typically involves normalization and resampling. Feature extraction can be performed in 2D or 3D using either internal or commercial software, with the number of features depending on the texture and higher-order characteristics of the images. Feature selection focuses on eliminating redundant features to reduce dimensionality and prevent issues such as multicollinearity and overfitting.22 Radiomic features encompass dimensions such as texture, shape, and intensity, which capture internal heterogeneity and microchanges within tumors, offering more comprehensive tumor information than single biomarkers. By transforming medical imaging from a macro diagnostic tool into a method for detailed micro-level analysis of tumor biology, radiomics provides objective evidence that supports the development of personalized treatment strategies. This approach enables clinicians to choose the most appropriate treatment plan based on specific radiomic features, ultimately improving treatment outcomes.
Although the potential of radiomics in predicting Ki-67 expression in lung adenocarcinoma has been demonstrated in individuals with normal immune function,23–25 there remains a notable research gap for patients with acquired immunodeficiency. To date, no evidence supports the effectiveness or feasibility of radiomic models specifically for this patient group, underscoring the need for further development and validation of predictive tools tailored to these patients. Herein, we aimed to address this gap by constructing and evaluating several machine learning models to predict Ki-67 expression levels based on radiomic features.
The results of this study underscore the effectiveness of radiomic features in predicting Ki-67 expression levels. We developed and assessed several machine learning models, including SVM, KNN, Random Forest, Extra Trees, LightGBM, and MLP. Each model’s performance was evaluated using sensitivity, specificity, PPV, NPV, F1 score, and AUC. Among these models, SVM demonstrated superior performance across all metrics. In the training set, the SVM model accurately identified 153 out of 171 patients, while in the testing set, it correctly identified 36 out of 42 patients. The SVM model’s robust performance can be attributed to its ability to handle high-dimensional data by using kernel functions to map low-dimensional data into higher-dimensional space, which enables the model to capture complex patterns and nonlinear relationships. Moreover, SVM’s approach to maximizing the margin between classes enhances its robustness and stability, making it less sensitive to noisy data and overfitting. This characteristic is particularly advantageous in radiomics, where feature dimensions are high and there may be redundancy among features. Furthermore, SVM’s suitability for small sample sizes allows it to perform well even with limited patient data by using support vectors to define classification boundaries, avoiding over-reliance on large-scale training data. Overall, the SVM model’s unique algorithmic attributes make it a valuable tool for predicting Ki-67 expression levels in HIV-associated lung adenocarcinoma patients.26 Finally, the decision-making process of the SVM model is more transparent due to its reliance on support vectors, which helps clinicians interpret model predictions. In contrast, KNN, Random Forest, Extra Trees, LightGBM, and MLP models showed slightly lower performance. We further evaluated the clinical utility of the SVM model through decision curve analysis, which highlighted its significant practical value as a non-invasive method for predicting Ki-67 expression levels. By combining radiomic features with their corresponding coefficients to create a radiomic score, we provided stronger evidence for the clinical application of radiomics technology. A meta-analysis combining 10 retrospective studies showed that imaging omics demonstrated encouraging diagnostic performance in predicting Ki-67 expression in lung cancer. The combined sensitivity, specificity, and AUC in the validation cohort were 0.78, 0.70, and 0.81, respectively. In this study, these values were 0.86, 0.89 and 0.905, respectively. These data indicate that, compared with previous studies, the imaging omics model used in this study shows equally superior diagnostic efficacy in predicting Ki-67 expression in HIV-infected LUAD patients.27 This radiomic score offers a quantifiable measure of tumor proliferation activity, delivering intuitive and reliable references for clinicians. This quantitative approach not only complements traditional pathological methods but also paves the way for advancements in precision medicine and personalized treatment.
This study also faces certain limitations, including a small sample size and reliance on single-center data, which may affect the model’s generalizability. Future research could address these limitations by validating the model’s accuracy and robustness using multi-center, large-scale clinical datasets. We implemented a random sampling strategy to ensure the balance of confounding factors, although some degree of heterogeneity remains inevitable To address the intrinsic heterogeneity of lung adenocarcinoma and the variability in Ki-67 expression across different tumor subtypes, future studies will proportionally represent major molecular subtypes (EGFR, ALK, KRAS mutations), ensuring biological diversity within the cohort while maintaining statistical power. To elucidate the interaction between radiomic features and molecular determinants, a multivariable mixed-effects model will be established, explicitly incorporating tumor purity estimates, treatment history parameters, and molecular subtype classifications. The integration of digital pathology quantification will further enable precise characterization of tumor microenvironmental factors, particularly stromal content, through computational histopathological analysis of three key texture features.
Conclusion
This study utilized CT radiomics to assess the application value of various machine learning models in predicting Ki-67 index expression in patients with HIV-related pulmonary adenocarcinoma. By extracting a large number of quantitative features from CT images, radiomics reveals biological information embedded in imaging data, enabling non-invasive assessment of tumor molecular characteristics. Among the models evaluated, the SVM effectively captures the complex relationships between radiomic features and the Ki-67 index, demonstrating superior predictive accuracy. This approach serves as a non-invasive tool that supports the evaluation of the Ki-67 index in HIV-related lung adenocarcinoma patients, reducing reliance on tissue biopsies and offering a refined strategy for personalized clinical treatment. Ultimately, it aids in formulating more precise treatment plans and improving patient prognosis and quality of life.
Data Sharing Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Statement
Ethical approval was obtained from the Ethics Committee of the Fourth People’s Hospital of Nanning (Approval No: [2022]64). This retrospective study did not contain any identifiable human images and was an anonymous data collection that waived patients’ written informed consent. The study was conducted in compliance with the Declaration of Helsinki, and all patient data were anonymized and handled with strict confidentiality to protect patient privacy.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research project funded by the Guangxi Zhuang Autonomous Region Health Commission (Z-A20231211); Nanning Science Research and Technology Development Program project (20233069).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cribbs SK, Crothers K, Morris A. Pathogenesis of HIV-related lung disease: immunity, infection, and inflammation. Physiol Rev. 2020;100(2):603–632. Physiological reviews. doi:10.1152/physrev.00039.2018
2. Sigel K, Makinson A, Thaler J. Lung cancer in persons with HIV. Current Opinion in HIV and AIDS. 2017;12(1):31–38. doi:10.1097/coh.0000000000000326
3. Hysell K, Yusuf R, Barakat L, et al. Decreased overall survival in HIV-associated non-small-cell lung cancer. Clinical Lung Cancer. 2021;22(4):e498–e505. doi:10.1016/j.cllc.2020.11.006
4. Rengan R, Mitra N, Liao K, Armstrong K, Vachani A. Effect of HIV on survival in patients with non-small-cell lung cancer in the era of highly active antiretroviral therapy: a population-based study. The Lancet Oncology. 2012;13(12):1203–1209. doi:10.1016/s1470-2045(12)70466-7
5. Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. Journal of Immunology. 1984;133(4):1710–1715. doi:10.4049/jimmunol.133.4.1710
6. Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review). Molecular Medicine Reports. Mar. 2015;11(3):1566–1572. doi:10.3892/mmr.2014.2914
7. Wang J, Dong L, Zheng Z, et al. Effects of different KRAS mutants and Ki67 expression on diagnosis and prognosis in lung adenocarcinoma. Scientific Reports. 2024;14(1):4085. doi:10.1038/s41598-023-48307-x
8. Li Z, Li F, Pan C, et al. Tumor cell proliferation (Ki-67) expression and its prognostic significance in histological subtypes of lung adenocarcinoma. Lung cancer. 2021;154:69–75. doi:10.1016/j.lungcan.2021.02.009
9. Yang Y, Shao X, Li Z, et al. Prognostic heterogeneity of Ki67 in non-small cell lung cancer: a comprehensive reappraisal on immunohistochemistry and transcriptional data. Journal of Cellular and Molecular Medicine. 2024;28(14):e18521. doi:10.1111/jcmm.18521
10. Liu Z, Feng H, Ma S, et al. Clinicopathological characteristics of peripheral clinical stage IA lung adenocarcinoma with high Ki-67 expression. Trans Cancer Res. 2021;10(1):152–161. Translational cancer research. doi:10.21037/tcr-20-2608
11. Russo L, Charles-Davies D, Bottazzi S, Sala E, Boldrini L. Radiomics for clinical decision support in radiation oncology. Clinical Oncology (Royal College of Radiologists. 2024;36(8):e269–e281. doi:10.1016/j.clon.2024.03.003
12. Horvat N, Papanikolaou N, Koh DM. Radiomics beyond the hype: a critical evaluation toward oncologic clinical use. Radiology Artificial Intelligence. 2024;6(4):e230437. doi:10.1148/ryai.230437
13. Fu Q, Liu SL, Hao DP, et al. CT radiomics model for predicting the Ki-67 Index of lung cancer: an exploratory study. Frontiers in Oncology. 2021;11:743490. doi:10.3389/fonc.2021.743490
14. Yao W, Liao Y, Li X, et al. Noninvasive method for predicting the expression of Ki67 and prognosis in non-small-cell lung cancer patients: radiomics. Journal of Healthcare Engineering. 2022;2022:7761589. doi:10.1155/2022/7761589
15. Liu F, Li Q, Xiang Z, et al. CT radiomics model for predicting the Ki-67 proliferation index of pure-solid non-small cell lung cancer: a multicenter study. Frontiers in Oncology. 2023;13:1175010. doi:10.3389/fonc.2023.1175010
16. Ahn HK, Jung M, Ha SY, et al. Clinical significance of Ki-67 and p53 expression in curatively resected non-small cell lung cancer. Tumour Biology: the Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(6):5735–5740. doi:10.1007/s13277-014-1760-0
17. Qin L. Application value of Ki67 and serum CA125 in the deep myometrial invasion of endometrial adenocarcinoma. BMC Cancer. 2023;23(1):240. doi:10.1186/s12885-023-10711-x
18. Zhang W, Liu J, Jin W, et al. Radiomics from dual-energy CT-derived iodine maps predict lymph node metastasis in head and neck squamous cell carcinoma. La Radiologia medica. 2024;129(2):252–267. doi:10.1007/s11547-023-01750-2
19. Yu H, Yang Z, Wei Y, et al. Computed tomography-based radiomics improves non-invasive diagnosis of Pneumocystis jirovecii pneumonia in non-HIV patients: a retrospective study. BMC Pulmonary Medicine. 2024;24(1):11. doi:10.1186/s12890-023-02827-4
20. Maino C, Vernuccio F, Cannella R, et al. Radiomics and liver: where we are and where we are headed? European Journal of Radiology. 2024;171:111297. doi:10.1016/j.ejrad.2024.111297
21. Wang XM, Zhang XJ. Role of radiomics in staging liver fibrosis: a meta-analysis. BMC Med Imaging. 2024;24(1):87. BMC medical imaging. doi:10.1186/s12880-024-01272-x
22. Park HJ, Park B, Lee SS. Radiomics and deep learning: hepatic applications. Korean Journal of Radiology. Apr. 2020;21(4):387–401. doi:10.3348/kjr.2019.0752
23. Huang Z, Lyu M, Ai Z, Chen Y, Liang Y, Xiang Z. Pre-operative prediction of Ki-67 expression in various histological subtypes of lung adenocarcinoma based on CT radiomic features. Frontiers in Surgery. 2021;8:736737. doi:10.3389/fsurg.2021.736737
24. Yan J, Xue X, Gao C, et al. Predicting the Ki-67 proliferation index in pulmonary adenocarcinoma patients presenting with subsolid nodules: construction of a nomogram based on CT images. Quantitative Imaging in Medicine and Surgery. 2022;12(1):642–652. doi:10.21037/qims-20-1385
25. Zhu M, Yang Z, Zhao W, et al. Predicting Ki-67 labeling index level in early-stage lung adenocarcinomas manifesting as ground-glass opacity nodules using intra-nodular and peri-nodular radiomic features. Cancer Medicine. 2022;11(21):3982–3992. doi:10.1002/cam4.4719
26. Zhou S. Sparse SVM for sufficient data reduction. IEEE Trans. Pattern Anal. Mach. Intell. 2022;44(9):5560–5571. doi:10.1109/tpami.2021.3075339
27. Luo X, Zheng R, Zhang J, et al. CT-based radiomics for predicting Ki-67 expression in lung cancer: a systematic review and meta-analysis. Frontiers in Oncology. 2024;14:1329801. doi:10.3389/fonc.2024.1329801
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



