Back to Journals » International Journal of Nanomedicine » Volume 20
Recent Advances in HPV Detection: From Traditional Methods to Nanotechnology and the Application of Quantum Dots
Authors He Z , Zheng X, Liu R, Zhao K, Mao D , Zhang L, Wan R , Zhang H, Wang X
Received 6 March 2025
Accepted for publication 2 May 2025
Published 21 May 2025 Volume 2025:20 Pages 6333—6356
DOI https://doi.org/10.2147/IJN.S524518
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kamakhya Misra
Zhenglin He,1 Xuepeng Zheng,1 Ruiqi Liu,1 Kai Zhao,2,3 Dezhi Mao,4 Lingkai Zhang,5 Runshan Wan,6 Hongyang Zhang,1 Xue Wang1,7
1China-Japan Union Hospital of Jilin University, Jilin University, Changchun, 130033, People’s Republic of China; 2School of Engineering & Applied Science, Yale University, New Haven, CT, 06520, USA; 3School of Medicine, Yale University, New Haven, CT, 06520, USA; 4Clinical Stem Cell Research Center, Peking University Third Hospital, Beijing, 100191, People’s Republic of China; 5State Key Laboratory of Female Fertility Promotion, Center for Reproductive Medicine, Department of Obstetrics and Gynecology, Peking University Third Hospital, Beijing, 100191, People’s Republic of China; 6College of Chemistry, Jilin University, Changchun, 130012, People’s Republic of China; 7Department of Clinical Nutrition and Toxicology, China-Japan Union Hospital of Jilin University, Changchun, 130033, People’s Republic of China
Correspondence: Xue Wang, Department of Clinical Nutrition and Toxicology, China-Japan Union Hospital of Jilin University, Changchun, 130033, People’s Republic of China, Tel/Fax +86 0431 84995322, Email [email protected]
Abstract: Cervical cancer, a significant public health concern, demands precise and expeditious detection methods to curb the spread of human papillomavirus (HPV). The early detection of cervical cancer remains a critical challenge in developing reliable and efficient screening tools to meet the demand for controlling cervical cancer. Traditional detection techniques are often cumbersome, costly, and inadequate for on-site HPV testing. Nanotechnology, with its unique electrical, chemical, and optical properties, has emerged as a pivotal component in the development of biosensors for rapid and reliable HPV detection. This article provides a comprehensive review of the advancements in cervical cancer detection, encompassing traditional methods, emerging protocols, and novel quantum dots (QDs)-based approaches for detection. The review examines the application of various nanomaterials in electrochemical and photoelectrochemical biosensors for the diagnosis of cervical cancer, with these innovations offering a significant improvement over conventional approach. Furthermore, we detail the synthesis methods of QDs and their properties, illustrate the substantial enhancement in sensor performance achieved through their applications, and elucidate the improvements and challenges associated with these new protocols while highlighting the potential application prospects of novel QDs technology in HPV detection.
Keywords: cervical cancer, human papillomavirus detection, nanotechnology, quantum dots, biosensors, early diagnosis
Introduction
Cervical cancer ranks as the fourth most prevalent malignancy among women worldwide, constituting a significant menace to women’s health.1 Despite being preventable through vaccination, cervical cancer screening remains indispensable due to limitations in vaccine coverage and immunogenicity.2,3 The World Health Organization (WHO) recommends nucleic acid detection of human papillomavirus (HPV) as the preferred method for cervical cancer screening,4 which has drawn scholars’ attention to advancements in cervical cancer detection technology.5 Apart from high-income countries exhibiting screening rates of 84%, women aged 30–49 years residing in upper-middle- (48%), lower-middle- (9%), and low-income countries (11%)6 may leave generations of girls and women at risk, though global strategies are paralleled with the goal of a 70% screening coverage by 2030.4 Given the vast population size, there is an urgent imperative to explore innovative methods for promoting cervical cancer screening.7
Over the past two decades, traditional diagnostic modality has evolved from a cytology adjunct to a primary co-testing methodology with cytology, establishing an optimized cost-benefit paradigm for cervical cancer screening.8–10 Persistent infection with high-risk HPV genotypes, particularly HPV16 and HPV18, represents the leading etiological factor in cervical carcinogenesis and precancerous lesions development.11,12 Molecular detection of these oncogenic HPV types constitutes a critical component of evidence-based strategies for early cervical cancer detection and precision management.13,14 The integration of nanotechnology into clinical diagnostics has achieved transformative breakthroughs. In the battle against cervical cancer, timely detection is crucial for effective treatment outcomes. Nanotechnology, with its exceptional sensitivity, precision, and capability for multiplex measurements, has emerged as a potent tool to enhance the early detection of cervical cancer.15,16
Quantum dots (QDs), known as semiconductor nanocrystals, are composed of nanoparticles that typically range in size from 1 nm to 100 nm, closely aligning with the exciton Bohr radius.17 Unlike traditional enzyme-linked immunosorbent assays and organic fluorescent dyes, QDs offer a broader and more effective excitation spectrum,18 along with a narrow and symmetrical emission spectrum. They also exhibit high fluorescence quantum efficiency,19 excellent photochemical stability,20 flexible surface chemistry, and the ability to be biologically modified. Among various traditional and novel HPV detection technologies, QDs stand out as a remarkable fluorescent marker due to their superior specificity, heightened sensitivity, and enhanced fluorescence stability.21
Overall, this review provides a thorough examination of recent advancements in HPV detection, covering both conventional strategies and cutting-edge approaches that incorporate various nanotechnologies, particularly the integration of QDs. It examines the advantages and disadvantages of traditional methods and highlights the exciting potential of QD technology for HPV detection.
Traditional Methods for HPV DNA Detection
The HPV genome is intricately organized into three principal functional regions: the early transcription region (E region), the late transcription region (L region), and the long control region (LCR). The E1 and E2 genes within the E region are instrumental in viral replication and transcription, respectively.22,23 Overexpression of E6 and E7 can immortalize cells by interfering with host cell cycle, inhibiting cell apoptosis, causing gene instability and affecting telomerase activity, which leads to the occurrence of cervical cancer.24,25 The L1 and L2 genes encode structural proteins of the viral capsid, pivotal for HPV infectivity.25–27 In the context of persistent HPV infection, integration of the viral genome into the host’s genomic DNA may lead to the disruption and loss of regions such as L1, E1, and E2, while the E6 and E7 regions are often preserved.25,28 This observation suggests that detection methods focusing on the E6 and E7 regions could offer a more specific assessment of HPV DNA, regardless of its integrated state.29 Current HPV molecular detection technologies in clinical practice are developed into nucleic acid hybridization signal amplification, nucleic acid amplification technologies, and other developed biological technologies.30
Nucleic Acid Hybridization Signal Amplification Technology
Nucleic acid hybridization signal amplification technology represents a direct method for identifying HPV nucleic acid sequences in clinical samples, circumventing the need for PCR amplification. This approach minimizes the risk of contamination, although it falls short in discerning specific HPV genotypes. The domain encompasses three principal techniques: hybrid capture technology, enzyme digestion signal amplification technology, and branched DNA signal amplification technology.
Hybrid Capture Technology
Hybrid capture technology’s detection principle is based on hybridization of the target HPV genomic DNA with the corresponding RNA probe, which is captured by the corresponding antibody on a microplate. Finally, HPV detection is carried out by enzyme-linked secondary antibody and catalytic substrate chemiluminescence.31–33 A pioneering clinical trial of cervical cancer screening published in The New England Journal of Medicine demonstrated that the second-generation hybrid capture (HC2) product has significantly better clinical detection sensitivity in detecting grade 2 or 3 cervical intraepithelial neoplasia (CIN2 or CIN3) than the traditional cytological detection (94.6% vs 55.4%), but its specificity is slightly lower than that of conventional cytological detection (94.1% vs 96.8%).34 In addition, studies have shown that HC2 undergoes certain cross-reactivity with certain non-targeted and non-carcinogenic HPV genotypes.35 After optimizing probes, this series of products can effectively reduce potential cross-reactivity between HPV genotypes. There is no need for a professional PCR laboratory to use hybrid capture technology for testing work, which has the potential to penetrate into grassroots medical units and remote areas and has great application prospects.
Enzyme Digestion Signal Amplification Technology
Enzyme digestion signal amplification technology’s detection principle is to detect specific target nucleic acid sequences through two synchronous isothermal reaction signals and amplification technology. Based on this technology, Cervista™ HPV HR was developed and approved by the US FDA in 2009.36 A comparative study between Cervista and HC2 for detecting high-risk HPV in cervical lesions showed that the consistency rates of positive and negative percentages were 90.8% and 64.5%, respectively. Cervista and HC2 methods have the same sensitivity of 90% in detecting cervical lesions of CIN2, with specificity of 47% and 43%, respectively.37,38 Enzyme digestion signal amplification technology excels in its good specificity, providing the capability to semi-quantitatively assess mRNA transcription levels. Despite these strengths, the method encounters limitations, notably its inability to discriminate between specific HPV subtypes and the complex procedural steps involved, which can be a deterrent in clinical settings seeking streamlined operations.
Branched DNA Signal Amplification Technology
The representative product of branched DNA signal amplification technology for HPV detection is QuantiVirus™ HPV E6/E7 mRNA test reagent developed by DiaCarta. This reagent adopts the principle of nucleic acid sandwich hybridization technology and can directly detect HPV mRNA in liquid-based samples.39,40 Studies have shown that the positive rate of E6/E7 mRNA detection is notably higher than that of HC2 detection (75.3% vs 62.3%). HPV mRNA detection and HC2 detection demonstrate equivalent sensitivity to high-grade cervical intraepithelial neoplasia (CIN2+), reaching a rate of 82.4% (14/17). However, the specificity of HPV mRNA detection for CIN2+ is significantly lower than that of HC2 detection.39 Leveraging the precision of branched-chain DNA signal amplification technology, Kodia Biotechnology has crafted a qualitative detection kit targeting 14 high-risk HPV mRNA species with commendable specificity and simplified pretreatment process. Nonetheless, the methodology faces a trade-off, as the detection protocol involves several incubation cycles that may be perceived as cumbersome and time-intensive.40
Nucleic Acid Amplification Technology
HPV nucleic acid amplification technology mainly includes PCR amplification or constant temperature amplification, which first enriches a portion of the DNA sequence in the HPV genome, and then detects it by fluorescent probes, reverse dot hybridization, gene chips, flow cytometry fluorescence hybridization, and other methods.
Real-Time Fluorescence PCR Technology
Real-time fluorescence PCR technology involves the incorporation of fluorescent markers into the PCR reaction system, allowing for the real-time monitoring of DNA amplification through the accumulation of fluorescence signals. This method enables the simultaneous amplification and detection within a closed tube system, culminating in the quantitative analysis of the target sample using standard curves. It offers high sensitivity and specificity for the detection of specific HPV types and provides several operational advantages, including full automation, expedited detection times, high throughput, and the capability to measure viral load. Moreover, it significantly minimizes the risk of contamination from amplification products.41–43 However, the technology faces constraints due to the finite number of fluorescence channels on real-time PCR instruments, which limits the typing detection scheme within a single tube. To overcome this, multiple HPV types must be tested in grouped sets across multiple reactions, compensating for the instrument’s limitations.
Reverse Dot Hybridization Technology
Reverse line blot (RLB) method employs biotin-labeled PCR amplification products that hybridize with probes fixed on nitrocellulose or nylon membranes, and generates detection signals through chemical colorimetry.44,45 This technique offers broad coverage for the identification of HPV genotypes and is characterized by a stable hybridization process, coupled with a robust adaptability to diverse sample types. However, RLB technology encounters some challenges, including a heightened susceptibility to contamination from PCR amplification products, a multi-step procedural requirement, and the potential for subjective bias in the interpretation of colorimetric results, which could introduce human error into the diagnostic process.
Gene Chip Technology
Gene chip technology employs universal primers to amplify the HPV E1 gene, followed by denaturation of the amplified product. Simultaneously, a multitude of probe molecules are affixed onto solid-phase supports, generating a two-dimensional DNA probe array designed to hybridize specifically with the labeled amplification products. HPV genotyping is subsequently accomplished through enzyme-labeled colorimetry or fluorescence labeling methods.46–49 This technology offers the capability to simultaneously identify multiple HPV types and concurrent infections, meeting the requirements of high-throughput, rapid analysis, and minimal sample volume. It is tailored to clinical needs with its sensitivity and specificity aligning well with HPV typing, featuring stable fluorescence signal interpretation and clear, actionable results. Nevertheless, gene chip technology faces certain limitations. The cost of DNA chips is relatively high, and the necessity for a gene chip scanner for analysis elevates the overall detection expense. Moreover, the HPV viral load usually cannot be determined using this method. A comprehensive summary of the specific products of molecular assay for HPV detection is provided in Table 1.50–75
 |
Table 1 Specific Products of Molecular Assay for HPV Detection |
Other Developed Biological Technologies
RNAscope is an innovative method for RNA in situ hybridization that combines the benefits of traditional RNA in situ hybridization techniques and fluorescence in situ hybridization (FISH) technology to visualize the transcription of individual RNA molecules. Unlike PCR technology, RNAscope can confirm whether the HPV DNA content exceeds a specific threshold, allowing for a more straightforward and intuitive screening of HPV infections.76 This technique addresses several challenges, including the inability of PCR to provide location data, non-specific binding observed in immunohistochemistry, the limitation of protein detection only, and the restricted availability of antibodies.
With the progress of molecular biology, the importance of isothermal amplification methods—those that do not depend on temperature cycling instruments—has increasingly been recognized by researchers. Recombinase polymerase amplification (RPA) technology is particularly significant due to its straightforward and rapid nature, yielding results within approximately 30 minutes. This method does not necessitate sophisticated laboratory environments or equipment; rather, it requires only a simple heating device,77 which enables individuals without extensive professional training to conduct the tests. The sensitivity of RPA can reach levels of 100 to 101 copies per reaction.78 Nonetheless, as the entire RPA process occurs at a constant temperature, there may be instances of non-specific amplification.
The field of nucleic acid detection is currently focused on three primary research areas: CRISPR/Cas9, CRISPR/Cas12, and CRISPR/Cas13. Within the CRISPR/Cas12a system, two essential components are the guide RNA (gRNA) and the Cas12a proteins.79 Recently, a novel CRISPR-Cas12a-mediated colorimetric detection platform was developed for MPXV and HPV DNA sensing by applying probe DNA to reprogram the catalytic properties of MoS2 QDs, featuring subpicomolar detection limits, high specificity/sensitivity and applicability in human sera biosamples, with its colorimetric results analysable via a smartphone platform.80 The CRISPR/Cas system employs various signal transmission methods, with the most classic being real-time fluorescence detection, lateral flow chromatography strips, and hydrogels. Although a portable constant temperature fluorescence detector can measure the fluorescence intensity during the reaction, the viral load in the original sample was low. Transferring it to the CRISPR/Cas12a system for detection can significantly enhance sensitivity; however, this also raises the risk of aerosol contamination.
Nanotechnology in the Diagnosis of Cervical Cancer
In the late 1990s, nanotechnologies advanced clinical diagnosis, notably with the use of gold nanoparticles (AuNPs). The nanogold-mediated catalyzed reporter deposition (CARD) technique has demonstrated superior diagnostic performance compared to conventional polymerase chain reaction (PCR), exhibiting 90% detection sensitivity for HPV-positive cervical cancer specimens versus only 20% for PCR.81 This breakthrough catalyzed the application of nanotechnology in virus detection.
Ultrahigh Sensitivity with Fluorescent Nanoparticles
The surface of the nanoparticle is modified with antibodies or ligands with biomarkers related to cervical cancer, which can be targeted by biomarkers to achieve visualization or electrochemical signal detection. In a groundbreaking study, Palantavia et al developed an ultrabright fluorescent mesoporous silica nanoparticle for the early detection of cervical cancer. The nanoparticle, loaded with Rhodamine 6G at a concentration significantly higher than its aqueous solubility, was labeled by folic acid for the specific targeting of cervical cancer cells, which overexpress folate (FA) receptors. The research revealed that the fluorescence intensity from pre-cancerous cervical cells was substantially greater than that from normal cells. This novel approach also enabled significantly better sensitivity (95–97% vs 30–80%) and maintained specificity (94–95%) compared with current clinical tests, indicating a promising method for the enhanced diagnosis of early-stage cervical cancer.82,83 Recently, a colorimetric nanosensor based on AuNPs for detecting high-risk HPV 16 and 18 was developed, showing high specificity (77.8%–87.3%) and excellent negative predictive value (>96%) in clinical evaluation of 173 patients.84 The innovative use of more nanoparticles with various biomarker-specific targeting offered a significant advancement in the field of cervical cancer diagnostics, with the potential to revolutionize early detection methods and improve patient outcomes.
Cutting-Edge Nanotechnology in Biosensors
The unique properties of nanotechnology of biosensors platform emerged as a highly promising tool, offering rapid and precise diagnostics for cervical cancer. A recent study by Wang et al introduced a novel electrochemiluminescence (ECL) sensor for HPV16 DNA detection, utilizing Eu3+ doped polydopamine nanoparticles (PDA:Eu NPs). To enhance sensor sensitivity, a hydrogel reductive copper (I) particle catalyst was integrated, inducing more hydroxyl radicals for signal strengthening. By employing a T7 exonuclease-mediated cycling process, the sensor demonstrated a linear response to target DNA concentrations from 1 nmol/L to 100 nmol/L, with a limit of detection (LOD) of only 0.6 nmol/L.85 Another highly sensitive ECL biosensor, named “signal-off” for HPV16 detection, was developed. This biosensor utilized a novel PCN-224/nano-zinc oxide composite enhanced by polyacrylamide to improve water solubility and stability. Signal amplification is achieved through exonuclease-based cycling cleavage and hybridization chain reaction (Figure 1A).86 A novel study presented a highly sensitive ECL biosensing strategy for HPV16 DNA detection, utilizing CRISPR/Cas12a to regulate Pdots-DNA binding and enhance ECL emission through the local surface plasmon resonance effect of AuNPs, achieving a detection limit of 3.2 fM.87 Recently, Wang et al (2025) developed a modularized electrochemical sensing strategy that integrates CRISPR/Cas12 with nanoporous materials, enabling ultrasensitive nucleic acid detection. This strategy achieved a detection limit of 0.41 fM for HPV16 and demonstrated universality in detecting HPV18 (Figure 1B).88 In a similar vein, Yue et al (2025) created a label-free electrochemical platform that combines CRISPR/Cas14a with DNA walkers and magnetic self-assembly. This innovative approach allows for the ultrasensitive detection of HPV16 E7, with a detection limit of 67.17 fg/mL and recovery rates of 98.46%–115.78% in serum samples (Figure 1C).89 Additionally, Wang et al presented a label- and modification-free Cas12a-based ECL biosensor, which can detect HPV16 DNA at an ultralow concentration of 0.63 pM in approximately 60 minutes, showcasing great potential for point-of-care diagnostics.90 These developments highlight the power of combining advanced technologies like CRISPR systems with nanomaterials to enhance detection capabilities.
 |
Figure 1 (A) Schematic diagram for the ECL biosensing platform based on PCN-224/ZnO nanocomposites coupled with cyclic amplification and chain reaction for HPV-16 assay. Reproduced from Wu D, Dong W, Yin T, et al. PCN-224/Nano-Zinc oxide nanocomposite-based electrochemiluminescence biosensor for Hpv-16 detection by multiple cycling amplification and hybridization chain reaction. Sensors and Actuat B Chem. 2022;372:132659. © 2022 Elsevier B.V. All rights reserved.86 (B) Illustration of the modularized electrochemical sensing strategy based on CRISPR/Cas-mediated controllable MB release/enrichment system for ultrasensitive determination of HPV-16, including target recognition module (a), signal amplification module (b), and signal transduction module (c).Reproduced from Wang H, Niu Y, Liu H, et al. A modularized universal strategy by integrating CRISPR/Cas with nanoporous materials for ultrasensitive determination of nucleic acids. Chem Eng J. 2025;506:160065. © 2025 Elsevier B.V. All rights are reserved, including those for text and data mining, AI training, and similar technologies.88 (C) Schematic diagram of an electrochemical nanobiosensor with CRISPR/Cas14a system integrated with bipedal DD walker to detect HPV16 E7 serum samples. Reproduced from Yue Y, Liu M, Ma M, et al. CRISPR/Cas14a integrated with DNA walker based on magnetic self-assembly for human papillomavirus type 16 oncoprotein E7 ultrasensitive detection. Biosens Bioelectron. 2025;272:117135. © 2025 Elsevier B.V. All rights are reserved, including those for text and data mining, AI training, and similar technologies.89 (D) Schematic illustration of the fabrication procedures of Fe3O4@Au@PEI@HPP NPs and the strategy of HPV genotype detection in buffer or in 100% serum based on Fe3O4@Au@PEI@HPP NPs. Reproduced from Chen L, Liu M, Tang Y, et al. Preparation and Properties of a Low Fouling Magnetic Nanoparticle and Its Application to the HPV Genotypes Assay in Whole Serum. ACS Appl Mater Interfaces. 2019;11(20):18637–18644. Copyright © 2019 American Chemical Society.91 |
Based on electrochemical genosensor array for the simultaneous and sensitive detection of high-risk HPV DNA sequences,92–94 the optimization of novel nanocomposite materials provided a promising platform, such as a 3-aminopropyltriethoxysilane (APTES) modified gold electrode coupled with a super sandwich structure,95 a graphitic nano-onion/molybdenum disulfide (MoS2) nanosheet composite,96 a novel nano-composite of Perylene Tetra carboxylic acid functionalized copper nanoparticles and reduced graphene oxide (Cu-PTCA/rGO),97 and rGO and DNA nano-biohybrid-coated carbon screen-printed flexible electrodes (CSPEs).98
Breakthroughs in Biochip Technologies
In recent advances, a biochip allows hundreds of samples to be analyzed simultaneously with high efficiency, thereby making it possible for detecting multiple HPV subtypes.99 A DNA microarray system utilizing a bipolar integrated circuit photodiode array (PDA) chip, through a gold nanoparticle-mediated silver enhancement technique, improved the detection limit from 1.2 nM to 30 pM and expanded the range of detectable DNA concentrations by adjusting the silver development time.100 Chen et al’s study presented an innovative nanoparticle probe, featuring gold magnetic particles modified with polyethyleneimine and hyperbranched polyether polyol, which exhibited low fouling and high stability in complex biological systems, and have been successfully applied for the sensitive and selective fluorescence detection of high-risk HPV genotypes 18 and 16 in buffer and whole serum, demonstrating strong linearity and low detection limits (Figure 1D).91
QDs for HPV DNA Detection
Types of QDs
QDs, as intricate assemblies of atoms and molecules, can be crafted from a single semiconductor material or a combination of multiple semiconductors.101 Predominantly, QDs are composed of atoms from IIB-VI, III–V, or IV–VI groups of the periodic table that have 1~10 nm size dimensions, with CdY (Y represents S, Se, Te) being a common choice, alongside composite and multilayer structures.102–104 QDs mainly encompasses four primary types: mononuclear, core-shell, doped, and alloyed. Mononuclear QDs, characterized by a high density of surface defects, typically exhibit low quantum yield and inferior luminescence stability. In contrast, the latter three categories of QDs have been engineered to address and ameliorate these deficiencies. QDs transcend conventional notions of “dots”, as they consist of hundreds to thousands of atoms, yet confine their internal electron motion to a very limited spatial scale.101 This unique confinement endows QDs with distinctive physical and chemical properties, most notably their exceptional optical characteristics. These attributes have catapulted QDs to the forefront of applications in in vitro diagnostics and live cell imaging, underscoring their broad potential and value in the biomedical sciences.105–107
Preparation of QDs
The preparation of QDs has undergone significant refinement over the years, converging on two predominant methodologies: physical and chemical approaches, with the latter frequently taking precedence in contemporary practice. Within the chemical domain, two principal strategies have emerged for the fabrication of QDs: one is to synthesize them in organic systems using colloidal chemistry, and the other is to synthesize them in aqueous solutions.
Metal Organic Compound Synthesis Method
The metal-organic compound synthesis method refers to the method of preparing QDs based on the high-temperature cracking reaction between organic compounds and inorganic metal compounds or organic metal compounds in the presence of ligands. This approach is a significant breakthrough in the early field of QD research. In a seminal work in 1993, Bawendi et al108 pioneered the synthesis of highly luminescent CdSe QDs, employing dimethyl cadmium (Cd(CH3)2), trioctylphosphine selenide (TOPSe), and trioctylphosphine oxide (TOPO) as precursors within a coordinated solvent system. The insolubility of CdSe nanoparticles in methanol allows for the acquisition of nanoparticles with favorable quantum yield through a simple centrifugation process post-methanol addition. Nonetheless, this technique is encumbered by several significant limitations, including intricate procedural steps, the challenging control of reaction conditions, and the use of highly toxic and flammable precursors. These constraints have impeded the broader adoption of this method in the synthesis of CdSe QDs, suggesting that researchers need to make improvements in synthesis methods and reaction reagents to synthesize QDs with higher quality.
TOPO is the most commonly used solvent for synthesizing colloidal nanocrystals. Common methods include preparing CdSe nanoparticles by mixing Cd(CH3)2 and TOPSe through nucleation, followed by maturation, annealing, and selective deposition to ultimately separate high-quality CdSe QDs. The particle size can be controlled by changing the temperature.109 Furthermore, Zhang et al110 developed a new method for producing hyperbranched Co2P nanocrystals with uniform size, shape, and symmetry using TOPO as a solvent and phosphorus source. The morphology of the nanocrystals can be controlled from a layered structure to a hexagonal symmetric structure by changing the concentration of surfactants. The synthesis of colloidal small-size CdS QDs is typically fraught with challenges, including low particle yields and the concurrent formation of byproducts such as precursor compounds (PCs) associated with magic-size clusters (MSC). However, Li et al111 have illuminated a novel pathway in the field. Their work demonstrates that the introduction of TOPO can effectively fragmentize the PCs, thereby facilitating the nucleation and growth of small-size QDs at room temperature. This innovation presents a groundbreaking method for the production of small-size QDs, eliminating the issue of PC coexistence and significantly enhancing particle yield (Figure 2A).
 |
Figure 2 (A) Schematic drawing for our comprehension on the use of TOPO to fragmentize the PC that has formed in a prenuclation stage sample to facilitate the nucleation and growth of colloidal small-size CdS QDs with enhanced particle yield and without the coexistence of the PC and/or MSCs. Reproduced from Li L, Zhang J, Zhang M, et al. Fragmentation of Magic-Size Cluster Precursor Compounds into Ultrasmall CdS Quantum Dots with Enhanced Particle Yield at Low Temperatures. Angew Chem Int Ed Engl. 2020;59(29):12013–12021. © 2020 Wiley‐VCH Verlag GmbH & Co. KGaA, Weinheim.111 (B) Schematic diagram of the synthesis and multifunctional applications of N-SiQDs. Reproduced from Wang YF, Pan MM, Song YL, et al. Beyond the fluorescence labelling of novel nitrogen-doped silicon quantum dots: the reducing agent and stabilizer for preparing hybrid nanoparticles and antibacterial applications. J Mater Chem B. 2022;10(36):7003–7013. © 2022 Royal Society of Chemistry.112 (C) This schematic represents the key features of QDs that are critical for HPV detection, emphasizing their size-adjustable luminescence, tunable emission for wavelength specificity, and robust photostability. It also illustrates the integration of QDs with various materials to form composite structures, enhancing their application in biosensing through magnetic separation, carbon and graphene interfaces, and metal-organic frameworks. |
Aqueous Inorganic Synthesis Method
QDs synthesized via organic methods are often limited in their solubility, being soluble primarily in specific non-polar or weakly polar organic solvents. This limitation hinders their direct application in aqueous environments, which are prevalent in many analytical and biological systems. To surmount this, the surface of QDs must be modified with appropriate ligands, enabling their transfer into the aqueous phase for further analysis and application. In this context, the exploration of direct aqueous-phase synthesis of QDs is of paramount importance, promising to expand the versatility and applicability of QDs in scientific research and clinical diagnostics.
The direct synthesis of QDs in the aqueous phase offers several unparalleled advantages, including operational simplicity and cost-effectiveness compared to the intricate organic phase synthesis methods. It is characterized by high repeatability, minimal environmental impact, controllable surface charge and properties, excellent biocompatibility, suitability for mass production, and the ease of introducing functional groups. Consequently, this approach has emerged as a prominent research topic.113,114 Pioneering work by Sondi et al115 successfully synthesized CdSe QDs at room temperature by rapidly mixing aqueous solutions of either sodium selenide or selenourea with those of cadmium chloride in the presence of aminoglycan as stabilizing agent. While the luminescence efficiency of the resulting QDs is relatively low, and the preparation time for red fluorescent QDs is extended, the method’s unparalleled advantages for biological applications make the refinement of this aqueous phase synthesis technique highly significant. Currently, the direct synthesis of water-soluble QDs in aqueous phase predominantly employs water-soluble thiol reagents, such as acetic acid (TGA) and propionic acid (MPA), as stabilizing agents. These thiol compounds are capable of coordinating and binding with the metal cadmium on the QD surface, effectively repairing surface defects and enhancing the stability of the QDs. Concurrently, the functional groups present in thiol reagents, including NH2, COOH, and OH, act as functional modification groups, which significantly improve the water solubility of the QDs.116 In recent years, methods for preparing water-soluble QDs using other types of reagents as stabilizers have emerged. Wang et al117 successfully prepared CdTe:Zn2+ QDs with strong fluorescence performance, low biological toxicity, and good biocompatibility by using glutathione as a stabilizer and incorporating Zn2+ ions through an aqueous inorganic synthesis method. Besides, Yang et al118 successfully prepared CdTexSe1-x core-shell QDs modified with mercaptopropionic acid and L-cysteine using a high-temperature hydrothermal synthesis method. High-quality CdxZn1-xSe and CdxZn1-xSe/ZnS core/shell QDs were prepared by using a one-step hydrothermal method, while the fluorescence properties and stability were significantly enhanced after capping with a ZnS layer. The prepared products have low cytotoxicity and can also detect Hg2+ ions with high sensitivity and selectivity.119 These studies113,118–120 also indicate that the core/shell/shell QD structure with doping in the shell layer is a versatile method for synthesizing and ameliorating doped QDs.
Notably, hydrothermal synthesis has been extensively utilized for fabricating multifarious QDs, catering to a multitude of applications. For instance, Paul et al121 introduced a one-pot hydrothermal synthesis of gelatin quantum dots (GeQDs) with high photoluminescence quantum yield and remarkable stability, which were successfully applied for cell imaging across various clinical cell types, demonstrating their potential as non-toxic biomarkers for stable and long-term fluorescent imaging. Regarding carbon dots (CDs), Xian et al’s study successfully synthesized red fluorescent CDs with high quantum yield and monochromaticity using a hydrothermal method, revealing a logarithmic correlation between their aggregation-induced emission wavelength and concentration.122 Furthermore, Gao et al123 ulteriorly proposed nitrogen-doped CDs as a versatile ratiometric fluorescence probe for the visual detection of hypochlorite and thiosulfate, which offers high sensitivity, selectivity, and biocompatibility, paving the way for the development of efficient fluorescent probes for visual detection and biomedical applications. Intriguingly, sulfur quantum dots (SQDs) have emerged as promising candidates, distinguished by their low toxicity and exceptional luminescent capabilities. Most recent research by Shen et al synthesized red-light-emitting SQDs with high fluorescence efficiency and stability via a one-step hydrothermal process, demonstrating their potential as luminophores for fluorescence and ECL analysis, as well as for bio-labeling and imaging applications, by leveraging the use of an etching agent to tune emission and resorcinol to enhance ECL intensity.124 Wang et al125 presented a novel H2O2-assisted top-down synthesis of SQDs with high photoluminescence quantum yield and color tunability, enabling the creation of down-conversion white light emitting diodes with excellent color rendering, and highlighting the potential of these eco-friendly, water-soluble SQDs as luminescent materials derived from abundant precursors. Silicon quantum dots (SiQDs) have thoroughly proven their utility, underscored by their fluorescent properties. Wang et al’s research presented the one-pot hydrothermal synthesis of nitrogen-doped silicon quantum dots (N-SiQDs) with multifaceted applications, including bacterial imaging due to their biocompatibility and fluorescence, the synthesis of N-SiQDs-stabilized gold nanoparticles with enhanced catalytic performance, and intrinsic antibacterial activity against both Gram-positive and Gram-negative bacteria, thereby expanding the horizons for SiQDs in nanocomposite and biomedicine applications (Figure 2B).112
Application of QDs in HPV DNA Detection
Conventional virus detection methods often suffer from low sensitivity and specificity, long time consumption, and high cost. Rapid, efficient, selective, and sensitive virus detection remains problematic. As shown in Figure 2C, QD has a series of remarkable characteristics, making it more suitable for detecting HPV.126–130 A summary of the QDs-based developed biosensors for HPV detection is provided in Table 2.
 |
Table 2 A Summary of the QDs-Based Developed Biosensors for HPV Detection |
The main application of QDs in HPV DNA detection is presented in various forms, such as fluorescence sensors, electrochemical sensors and the emerging carbon and silicon quantum dot detection photoelectric analysis, fluorescence analysis, electroanalysis and colorimetric analysis, etc.134,135,137–139 The function of quantum dots mainly depends on its structural regulation, including the surface of quantum dots, doping and alloying of quantum dots, and composite structure of quantum dots (Figure 2C). Innovative applications of QDs are also combined with traditional methods in situ hybridization and PCR synergies. The application of QDs-based detection technology in the diagnosis and monitoring of cervical cancer has demonstrated its versatility and efficiency, positioning it as a pivotal tool for future screening and diagnostic procedures.
Size Adjustment of QDs and HPV Detection
The luminescence mechanism of quantum dots reveals that their fluorescence properties are intrinsically linked to the size of the energy gap. Specifically, the width of the energy gap between the valence band and the conduction band is directly influenced by the size of the QDs. An innovative photoelectrochemical (PEC) biosensor, engineered with precise quantum size control to harness quantum confinement effects, was developed for the detection of HPV16. This was achieved through the CRISPR-Cas12a (Cpf1)-mediated disassembly of a Z-scheme heterojunction, leveraging the biosensor’s unique response to the targeted viral DNA. Li et al perfectly exploited the sensor’s ability to combine biomolecular recognition with photon-to-electron conversion capabilities and designed a PEC biosensor that can detect HPV16 by controlling the quantum size, thus improving the PEC reaction performance (Figure 3C).140
 |
Figure 3 (A) Schematic illustration of the synthesis of POM@CdS QD composites. (B) Schematic illustration of the PEC sensor for detecting HPV 16 DNA. Reproduced from Cheng Y, Sun C, Chang Y, et al. Photoelectrochemical biosensor based on SiW12@CdS quantum dots for the highly sensitive detection of HPV 16 DNA. Front Bioeng Biotechnol. 2023;11:1193052. Copyright © 2023 Cheng, Sun, Chang, Wu, Zhang, Liu, Ge, Li, Li, Sun and Zang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY).141 (C) Schematic illustration of CRISPR-Cas12a-based PEC biodetection toward HPV-16 by disassembly of Z-Scheme heterojunction among TiO2, Au NPs, and CdS QDs. Reproduced from Li Y, Zeng R, Wang W, et al. Size-Controlled Engineering Photoelectrochemical Biosensor for Human Papillomavirus-16 Based on CRISPR-Cas12a-Induced Disassembly of Z-Scheme Heterojunctions. ACS Sens. 2022;7(5):1593–1601. Copyright © 2022 American Chemical Society.140 |
Structure Adjustment of QDs and HPV Detection
Surface Modification of QDs
Through the use of the photoelectric analysis technique, AuNPs’ surface can accumulate metallic silver. The target DNA will ultimately be quantified into the corresponding electrical signal after a sequence of steps.142 The Au NP-modified Keggin-type polyoxometalate (SiW12)-grafted CdS quantum dots (SiW12@CdS QDs) demonstrated exceptional selectivity and sensitivity in target DNA detection, attributed to its outstanding PEC response (Figure 3A and B).141 Sun et al’s work presented a novel photoelectrochemical biosensor array (PEBA) platform for HPV genotyping that utilizes TiO2@AuNPs and CdS QDs-labeled DNA probes. The presence of HPV targets triggers a conformational change in the probes, altering the PEC signal and enabling sensitive detection with a low limit of 0.1 copies/μL across a linear range for nine HPV types (Figure 4).134 Researchers have crafted an ultrasensitive nanobiosensor for the detection of HPV18 by leveraging the synthesis of water-soluble CdTe QDs. These dots were functionalized with amino-modified oligonucleotides to form QDs-DNA conjugates that, upon interaction with target DNA and a Cy5-labeled oligonucleotide, assemble into sandwich hybrids. This assembly facilitates a precise and sensitive fluorescence resonance energy transfer (FRET)-based detection assay, demonstrating a linear detection range from 1.0 to 50.0 nM and an impressively low limit of 0.2 nM.132
 |
Figure 4 (A) Schematic illustration of the PEBA setup for HPV genotyping and a comparison between the proposed PEBA and conventional photoelectrochemical testing system. (B) Schematic depiction of the fabricated PEBA for detecting HPV-related genes. (C) Schematics of the PEBA assembly process. (D) Photocurrent responses of the PEBA toward different concentrations of synthetic HPV16 oligonucleotide standard with from 0 fM to 1 nM. Insert: the calibration curve of ΔI/I0 versus HPV16 target concentration. (E) Photocurrent responses of the PEBA to different concentrations of HPV DNA subtype plasmids (0, 0.6, 3, 6, 60, 300, 600 copies/μL). (F) The calibration curves of photocurrent responses versus the HPV DNA concentrations from 0.6 to 600 copies/μL. Error bars represented the standard deviations of three independent experiments. Reproduced from Sun Y, Liu J, Peng X, et al. A novel photoelectrochemical array platform for ultrasensitive multiplex detection and subtype identification of HPV genes. Biosens Bioelectron. 2023;224:115059. © 2023 Published by Elsevier B.V.134 |
Doping and Alloying of QDs
Doped QDs in which foreign atoms or ions are introduced into the lattice structure with new optical and electrical properties, are commonly used to improve their luminescence efficiency, adjust the wavelength of light emission, or enhance their stability. Nie et al133 developed an ECL sensor that capitalizes on the synergistic enhancement strategy of Zn-doped MoS2 QDs and reductive Cu(I) particles. This biosensor achieved sensitive detection of HPV16 DNA over a range from 0.1 nmol/L to 200 nmol/L, with a LOD as low as 0.03 nmol/L. Notably, the ECL signal, captured by a smartphone, can be transformed into high-resolution images through software processing. This innovation presents a significant potential for point-of-care HPV16 DNA testing in the future (Figure 5).
 |
Figure 5 (A) The ECL performance. (B) ECL response of the biosensor with (a) 0.1 nM, (b) 1 nM, (c) 10 nM, (d) 20 nM, (e) 50 nM, (f) 100 nM and (g) 200 nM HPV 16 DNA. (C) Schematic illustration of reductive Cu(I) particles catalyzed Zn-doped MoS2 QD-based ECL biosensor. (D) Visualized ECL images processed by the self-developed software with (a) 0.1 nM, (b) 10 nM, (c) 50 nM, (d) 100 nM and (e) 200 nM HPV 16 DNA. Reproduced from Nie Y, Zhang X, Zhang Q, et al. A novel high efficient electrochemiluminescence sensor based on reductive Cu(I) particles catalyzed Zn-doped MoS2 QDs for HPV 16 DNA determination. Biosens Bioelectron. 2020;160:112217. © 2020 Elsevier B.V. All rights reserved.133 |
Composite Structure of QDs and HPV Detection
QDs can be further expanded by combining with magnetic materials, carbon and graphene, and metal-organic frameworks (MOFs), presenting improved performance, or new functionalities in probe or biosensor platform for HPV detection (Figure 2C).
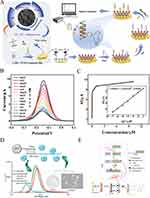 |
Figure 6 (A) Schematic representation of the proposed electrochemical DNA biosensor. (B) DPV responses of the proposed biosensor with various TD concentrations for HPV-16 detection. (C) the correlation between DPV peak current ΔI and TD concentrations, obtained by an inserted calibration with the logarithm of the TD concentrations, error bars = SD (n = 3). Reproduced from Yu J, Dong C, Yang Y, et al. Electrochemical DNA biosensor for HPV-16 detection based on novel carbon quantum dots/APTES composite nanofilm. Microchem J. 2024;204:110949. © 2024 Elsevier B.V. All rights are reserved, including those for text and data mining, AI training, and similar technologies.143 (D) Optical biosensing system utilizing MGPs and CdTe/ZnSe QDs coupled with nucleic acid probes. Reproduced from Jimenez Jimenez AM, Moulick A, Bhowmick S, et al. One-step detection of human papilloma viral infection using quantum dot-nucleotide interaction specificity. Talanta. 2019;205:120111. © 2019 Elsevier B.V. All rights reserved.144 (E) Schematic representation for principle of sensitive DNA detection based on GQDs ECL coupled with cycling amplification technique. Reproduced from Jie G, Zhou Q, Jie G. Graphene quantum dots-based electrochemiluminescence detection of DNA using multiple cycling amplification strategy. Talanta. 2019;194:658–663. © 2018 Elsevier B.V. All rights reserved.145 |
Combining with Magnetic Materials
A novel hybridization assay for the detection of HPV16 infections has been developed, leveraging the combined advantages of QDs and superparamagnetic NPs. This assay offers a rapid and straightforward method that is significantly faster and more convenient than the conventional type-specific PCR method.136 Furthermore, researchers have developed a magnetic glass particle (MGP) with a DNA probe attached to its surface to hybridize with the target DNA. Eventually, the MGP probe DNA hybrid combines with CdTe/ZnSe core/shell QDs to detect HPV infection using quantum dot-nucleotide specific interactions (Figure 6D).144 These studies have successfully substantiated the clinical efficacy of these innovative assays. The marked reduction in detection time, coupled with the simplicity of the process, endows the approach with immense potential for clinical applications, particularly in the realm of large-scale epidemiological screening. Notably, aside from detecting HPV16, QDs-based biosensors can also help identify different types of HPV. A novel biobarcoded analytical method was developed to detect multiple DNA sequences simultaneously based on the CdSe/ZnS QDs and magnetic microparticle, offering enhanced sensitivity, rapid preparation, and simplified analysis compared to traditional biobarcode assays.146
Combining with Carbon and Graphene
Carbon quantum dots (CQDs) and graphene quantum dots (GQDs) are nanomaterials with excellent biocompatibility and non-toxicity, featuring high charge transfer capabilities and useful for analytical detection and quantification in electrochemical applications.131,147 Utilizing a novel CQDs/(3-aminopropyl)triethoxysilane (APTES) composite nanofilm to enhance sensitivity, researchers have developed a highly sensitive electrochemical biosensor for the detection of HPV16, with remarkable linear range and LOD of 100 fM-100 nM and 0.73 fM, respectively (Figure 6A–C).143 Furthermore, Léony S Oliveira et al148 introduced an innovative electrochemical genosensor for the detection of HPV oncogenes E6 and E7. The sensor utilized a nanostructure based on cysteine and GQDs, which not only provided a rich environment for functional groups and surface area but also enhanced electrochemical properties. The sensor demonstrated high sensitivity and selectivity, with a detection limit of 26 fM and a quantification limit of 79.6 fM. Researchers have developed a highly sensitive ECL biosensor for DNA detection by harnessing the ECL properties of GQDs and a multiple cycling amplification technique, achieving good selectivity and high sensitivity for HPV16 DNA and showcasing potential for point-of-care screening (Figure 6E).145
Combining with Metal-Organic Frameworks (MOFs)
Metal-organic frameworks (MOFs) exhibit great potential in applications as fluorescent sensors and luminescent probes.149,150 Functionalizing MOFs with QDs that possess ECL activity results in a synergistic combination that leverages the high accumulation and catalytic capabilities of MOFs with the luminescent properties of QDs, thereby significantly boosting the ECL emission.151,152 Yang et al’s study demonstrated a marked improvement in ECL efficiency by integrating CdTe QDs into isoreticular metal organic framework-3 (IRMOF-3), through both internal encapsulation and external surface decoration.152 In 2024, researchers have successfully developed an innovative “off-on” fluorescent sensor, leveraging the properties of water-soluble dual-color emitting CdTe QDs conjugated with ssDNA probes. The sensor utilizes porphyrin derivatives (NiTPP) to quench the fluorescence, enabling a highly sensitive response. In vitro detection of HPV16 and HPV18 demonstrated excellent linearity within the ranges of 0.1–40 nM and 0.05–40 nM, respectively, with detection limits reaching as low as 62 pM and 40 pM. This method has been successfully developed and validated using spiked urine and cervical swab samples, showcasing its capability for the quantitative and sensitive detection of HPV16 and HPV18 concurrently (Figure 7).153
 |
Figure 7 (A) Schematic outlining the method for analyzing HPV16 and HPV18. (B) Fluorescence spectra of QDs-ssDNA reacting with NiTPP solutions of different concentrations (Ex=300 nm, EmgQDs=567 nm, EmrQDs=678 nm). (C) Fluorescence spectra of QDs-ssDNA reacting with different concentrations of HPV16 and HPV18 (Inset: fluorescence changes under ultraviolet lamp). (D) Linear relationship between gQDs-gDNA and HPV16. (E) Linear relationship between rQDs-rDNA and HPV18. reproduced from Jiang X, Yin C, Wu M, et al. Fluorescent switch based on QDs modified DNA probe and NiTPP for simultaneous dual color sensitive sensing of HPV16 and HPV18. Sensors and Actuat B Chem. 2024;403:135128. © 2023 Elsevier B.V. All rights reserved.153 |
Conclusions and Perspectives
Currently, nanotechnology has brought revolutionary changes to various fields. Among them, QDs have emerged as a transformative tool in the detection of HPV. Their unique optical properties, such as broad absorption spectra, size-tunable emission, and exceptional photostability, have made them a powerful tool for improving the accuracy and sensitivity of HPV detection regarding the limitations of traditional methods. The development of biosensors based on QDs has opened up new possibilities for the diagnosis of cervical cancer based on photoelectric analysis, fluorescence-based detection, and electrochemical genosensors. These biosensors can detect multiple genotypes of HPV simultaneously, providing more comprehensive information for screening. In addition, the combination of QDs with other nanomaterials, such as AuNPs, heavy-metal-free carbon and graphene, has further enhanced the detection efficiency and reduced the complexity of the detection process. The transition from laboratory-based testing to point-of-care testing is gaining momentum, with the development of portable devices and simplified protocols.
However, the trajectory of HPV detection technology is poised for further innovation and refinement. The pursuit of point-of-care testing solutions will continue, with a focus on miniaturization, user-friendliness, and cost-effectiveness. Long-term use could lead to the release of toxic ions, damaging cells and organs. To address this, researchers are exploring the development of biocompatible and non-toxic QDs.154 Protocols for QD green synthesis and surface modification are being optimized. For instance, coating QDs with protective layers can reduce ion release. Also, the development of heavy-metal-free QDs is a promising direction. These approaches can ensure the reproducibility and reliability of QDs-based diagnostic tools for broader application. Green synthesis methods, such as using plant extracts or microorganisms, can produce QDs with lower toxicity. Surface modification can introduce functional groups to enhance stability and reduce immunogenicity, minimizing potential adverse reactions.155,156 For instance, PEGylation of QDs has been shown to significantly reduce their cytotoxicity and improve their biocompatibility.157 Adverse effects, such as inflammatory responses or oxidative stress, should be closely monitored. Long-term animal experiments and clinical trials are needed to assess potential toxicity. Studies have indicated that QDs with ZnS shell and PEG coating are more beneficial to cell proliferation compared to naked QDs, demonstrating the effectiveness of surface modification in enhancing QDs’ biocompatibility and reducing toxicity.154,157
Looking ahead, several key development directions should be prioritized to further advance the field: (1) Enhanced novel materials and designs for QDs: The development of biocompatible and non-toxic QDs is a priority. Protocols for QD green synthesis and surface modification are being optimized to ensure the reproducibility and reliability of QDs-based diagnostic tools for the broader application. (2) Improvement in detection efficiency: By coupling various high-performance materials, the detection efficiency of biosensors will see continuous improvement. This will address the current limitations of insufficient LODs, poor stability, and meet the urgent need for efficient on-site detection and diagnosis. (3) Advances in multifunctional sensing: The combination of magnetic separation, microfluidic systems, and dielectrophoresis technologies with existing sensors is expected to yield highly sensitive sensors with multiple detection functions. (4) Suitability for promotion: The successful translation of QD technology from the laboratory to clinical settings will necessitate the ability to process a diverse range of specimens, including blood, urine, paraffin-embedded tissue, frozen tissue, and cytological samples. Additionally, it is essential to conduct further metabolism studies to ensure the safety of these technologies in humans. (5) Convergence with advanced technologies: The integration with mobile health technologies, artificial intelligence, and machine learning algorithms will enable promising real-time monitoring and data analysis, potentially transforming the way HPV and other diseases are detected and managed.
Significantly, to achieve these improvements, the role of collaborative efforts in driving research forward cannot be overstated. International partnerships, interdisciplinary research, and knowledge sharing will be vital in overcoming the remaining challenges and in translating these innovative technologies into routine clinical practice in the forthcoming years.
Data Sharing Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Finance Department of Jilin Province (Grant No. 2023SCZ65) and the Development and Reform Commission of Jilin Province (Grant No. 2024C017-10).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi:10.3322/caac.21763
2. Falcaro M, Castañon A, Ndlela B, et al. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: a register-based observational study. Lancet. 2021;398(10316):2084–2092. doi:10.1016/S0140-6736(21)02178-4
3. Kamamoto S, Murayama A, Hamaki T, et al. HPV vaccination and cervical cancer screening. Lancet. 2022;399(10339):1939–1940. doi:10.1016/S0140-6736(22)00106-4
4. Zarocostas J. Renewed calls to scale-up cervical cancer screening. Lancet. 2024;403(10429):797. doi:10.1016/S0140-6736(24)00408-2
5. Xu M, Cao C, Wu P, et al. Advances in cervical cancer: current insights and future directions. Cancer Commun. 2025;45(2):77–109. doi:10.1002/cac2.12629
6. Bruni L, Serrano B, Roura E, et al. Cervical cancer screening programmes and age-specific coverage estimates for 202 countries and territories worldwide: a review and synthetic analysis. Lancet Glob Health. 2022;10(8):e1115–e1127. [published correction appears in Lancet Glob Health. 2023 Jul;11(7):e1011. doi: 10.1016/S2214-109X(23)00240-1]. doi:10.1016/S2214-109X(22)00241-8.
7. Ma Y, Di J, Bi H, et al. Comparison of the detection rate of cervical lesion with TruScreen, LBC test and HPV test: a Real-world study based on population screening of cervical cancer in rural areas of China. PLoS One. 2020;15(7):e0233986. doi:10.1371/journal.pone.0233986
8. Lew J-B, Simms KT, Smith MA, et al. Primary HPV testing versus cytology-based cervical screening in women in Australia vaccinated for HPV and unvaccinated: effectiveness and economic assessment for the National Cervical Screening Program. Lancet Public Health. 2017;2(2):e96–e107. doi:10.1016/S2468-2667(17)30007-5
9. Fontham ETH, Wolf AMD, Church TR, et al. Cervical cancer screening for individuals at average risk: 2020 guideline update from the American Cancer Society. CA Cancer J Clin. 2020;70(5):321–346. doi:10.3322/caac.21628
10. Demarco M, Hyun N, Carter-Pokras O, et al. A study of type-specific HPV natural history and implications for contemporary cervical cancer screening programs. EClinicalMedicine. 2020;22:100293. doi:10.1016/j.eclinm.2020.100293
11. Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
12. Molina MA, Steenbergen RDM, Pumpe A, et al. HPV integration and cervical cancer: a failed evolutionary viral trait. Trends mol Med. 2024;30(9):890–902. doi:10.1016/j.molmed.2024.05.009
13. Pinkowish MD. Human papillomavirus genotype distributions inform screening and vaccination policy. CA Cancer J Clin. 2009;59(5):280–281. doi:10.3322/caac.20035
14. de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11(11):1048–1056. doi:10.1016/S1470-2045(10)70230-8
15. Zhang Y, Li M, Gao X, et al. Nanotechnology in cancer diagnosis: progress, challenges and opportunities. J Hematol Oncol. 2019;12(1):137. doi:10.1186/s13045-019-0833-3
16. Song X, Li X, Tan Z, et al. Recent status and trends of nanotechnology in cervical cancer: a systematic review and bibliometric analysis. Front Oncol. 2024;14:1327851. doi:10.3389/fonc.2024.1327851
17. Rosenthal SJ. Bar-coding biomolecules with fluorescent nanocrystals. Nat Biotechnol. 2001;19(7):621–622. doi:10.1038/90213
18. Larson DR, Zipfel WR, Williams RM, et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300(5624):1434–1436. doi:10.1126/science.1083780
19. Dirks RW, Tanke HJ. Advances in fluorescent tracking of nucleic acids in living cells. Biotechniques. 2006;40(4):489–496. doi:10.2144/000112121
20. Wu X, Liu H, Liu J, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21(1):41–46. doi:10.1038/nbt764
21. Zhang H, Yee D, Wang C. Quantum dots for cancer diagnosis and therapy: biological and clinical perspectives. Nanomedicine. 2008;3(1):83–91. doi:10.2217/17435889.3.1.83
22. Van Doorslaer K, McBride AA. Molecular archeological evidence in support of the repeated loss of a papillomavirus gene. Sci Rep. 2016;6:33028. doi:10.1038/srep33028
23. Gelbard MK, Munger K. Human papillomaviruses: knowns, mysteries, and unchartered territories. J Med Virol. 2023;95(10):e29191. doi:10.1002/jmv.29191
24. Doorbar J, Egawa N, Griffin H, et al. Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25(Suppl Suppl 1):2–23. doi:10.1002/rmv.1822
25. Sanclemente G, Gill DK. Human papillomavirus molecular biology and pathogenesis. J Eur Acad Dermatol Venereol. 2002;16(3):231–240. doi:10.1046/j.1473-2165.2002.00419.x
26. Ribeiro AL, Caodaglio AS, Sichero L. Regulation of HPV transcription. Clinics. 2018;73(suppl 1):e486s. doi:10.6061/clinics/2018/e486s
27. Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol mol Biol Rev. 2004;68(2):362–372. doi:10.1128/MMBR.68.2.362-372.2004
28. Molijn A, Kleter B, Quint W, et al. Molecular diagnosis of human papillomavirus (HPV) infections. J Clin Virol. 2005;32(1):S43–S51. doi:10.1016/j.jcv.2004.12.004
29. Volkova LV, Pashov AI, Omelchuk NN. Cervical Carcinoma: oncobiology and Biomarkers. Int J mol Sci. 2021;22(22):12571. doi:10.3390/ijms222212571
30. Kundrod KA, Barra M, Wilkinson A, et al. An integrated isothermal nucleic acid amplification test to detect HPV16 and HPV18 DNA in resource-limited settings. Sci Transl Med. 2023;15(701):eabn4768. doi:10.1126/scitranslmed.abn4768
31. Bhatla N, Singla S, Awasthi D. Human papillomavirus deoxyribonucleic acid testing in developed countries. Best Pract Res Clin Obstet Gynaecol. 2012;26(2):209–220. doi:10.1016/j.bpobgyn.2011.11.003
32. Vince A, Kutela N, Iscic-Bes J, et al. Clinical utility of molecular detection of human papillomavirus in cervical samples by hybrid capture technology. J Clin Virol. 2002;25(3):S109–S112. doi:10.1016/s1386-6532(02)00184-1
33. Iftner T, Neis KJ, Castanon A, et al. Longitudinal Clinical Performance of the RNA-Based Aptima Human Papillomavirus (AHPV) Assay in Comparison to the DNA-Based Hybrid Capture 2 hPV Test in Two Consecutive Screening Rounds with a 6-Year Interval in Germany. J Clin Microbiol. 2019;57(1):e01177–18. doi:10.1128/JCM.01177-18
34. Mayrand MH, Duarte-Franco E, Rodrigues I, et al. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med. 2007;357(16):1579–1588. doi:10.1056/NEJMoa071430
35. Castle PE, Solomon D, Wheeler CM, et al. Human papillomavirus genotype specificity of hybrid capture 2. J Clin Microbiol. 2008;46(8):2595–2604. doi:10.1128/JCM.00824-08
36. Day SP, Hudson A, Mast A, et al. Analytical performance of the Investigational Use Only Cervista HPV HR test as determined by a multi-center study. J Clin Virol. 2009;45(1):S63–S72. doi:10.1016/S1386-6532(09)70010-1
37. Tao K, Yang J, Yang H, et al. Comparative study of the cervista and hybrid capture 2 methods in detecting high-risk human papillomavirus in cervical lesions. Diagn Cytopathol. 2014;42(3):213–217. doi:10.1002/dc.23025
38. Boehmer G, Wang L, Iftner A, et al. A population-based observational study comparing Cervista and Hybrid Capture 2 methods: improved relative specificity of the Cervista assay by increasing its cut-off. BMC Infect Dis. 2014;14:674. doi:10.1186/s12879-014-0674-1
39. Shen Y, Gong J, He Y, et al. Quantivirus® HPV E6/E7 RNA 3.0 assay (bDNA) is as sensitive, but less specific than Hybrid Capture 2 test. J Virol Methods. 2013;187(2):288–293. doi:10.1016/j.jviromet.2012.11.024
40. Liu TY, Xie R, Luo L, et al. Diagnostic validity of human papillomavirus E6/E7 mRNA test in cervical cytological samples. J Virol Methods. 2014;196:120–125. doi:10.1016/j.jviromet.2013.10.032
41. Mackay J, Landt O. Real-time PCR fluorescent chemistries. Methods mol Biol. 2007;353:237–261. doi:10.1385/1-59745-229-7:237
42. Nie L, Qin H, Li S, et al. The establishment of a multiplex fluorescent polymerase chain reaction coupled with capillary electrophoresis analysis technology enables the simultaneous detection of 16 genotypes of human papillomavirus. J Clin Lab Anal. 2023;2023:e24996. doi:10.1002/jcla.24996
43. Santos FLSG, Invenção MCV, Araújo ED, et al. Comparative analysis of different PCR-based strategies for HPV detection and genotyping from cervical samples. J Med Virol. 2021;93(11):6347–6354. doi:10.1002/jmv.27118
44. Greenfield EA. Hybridoma Screening by Antigen Capture: reverse Dot Blot. Cold Spring Harb Protoc. 2022;2022(1):
45. Kong F, Gilbert GL. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)--a practical epidemiological and diagnostic tool. Nat Protoc. 2006;1(6):2668–2680. doi:10.1038/nprot.2006.404
46. Dalstein V, Merlin S, Bali C, et al. Analytical evaluation of the PapilloCheck test, a new commercial DNA chip for detection and genotyping of human papillomavirus. J Virol Methods. 2009;156(1–2):77–83. doi:10.1016/j.jviromet.2008.11.002
47. Vieira L, Almeida A. The cytology and DNA detection by the PapilloCheck(®) test in the diagnosis of human papillomavirus infection. Eur J Microbiol Immunol. 2013;3(1):61–67. doi:10.1556/EuJMI.3.2013.1.9
48. Bryant D, Rai N, Rowlands G, et al. Human papillomavirus type distribution in vulval intraepithelial neoplasia determined using PapilloCheck DNA Microarray. J Med Virol. 2011;83(8):1358–1361. doi:10.1002/jmv.22107
49. Crosbie EJ, Bailey A, Sargent A, et al. The PapilloCheck Assay for Detection of High-Grade Cervical Intraepithelial Neoplasia. J Clin Microbiol. 2015;53(11):3553–3559. doi:10.1128/JCM.01578-15
50. Rabaan AA, Taylor DR, Dawamneh MF, et al. Comparison of Xpert® HPV and Hybrid Capture® 2 DNA Test™ for detection of high-risk HPV infection in cervical atypical squamous cells of undetermined significance. J Infect Public Health. 2017;10(2):219–223. doi:10.1016/j.jiph.2016.04.017
51. Latsuzbaia A, Vanden Broeck D, Van Keer S, et al. Validation of BD Onclarity HPV Assay on Vaginal Self-Samples versus Cervical Samples Using the VALHUDES Protocol. Cancer Epidemiol Biomarkers Prev. 2022;31(12):2177–2184. doi:10.1158/1055-9965.EPI-22-0757
52. Martinelli M, Giubbi C, Sechi I, et al. Evaluation of BD Onclarity™ HPV Assay on Self-Collected Vaginal and First-Void Urine Samples as Compared to Clinician-Collected Cervical Samples: a Pilot Study. Diagnostics. 2022;12(12):3075. doi:10.3390/diagnostics12123075
53. He Y, Chen W, Guo Z, et al. The performance of Cobas HPV test for cervical cancer screening in Chinese female migrant workers. Epidemiol Infect. 2021;149(e196). doi:10.1017/S0950268821001904
54. Effah K, Tekpor E, Klutsey GB, et al. Antenatal and postnatal cervical precancer screening to increase coverage: experience from Battor, Ghana. Ecancermedicalscience. 2023;17:1616. doi:10.3332/ecancer.2023.1616
55. Xue P, Gao LL, Yin J, et al. A direct comparison of four high-risk human papillomavirus tests versus the cobas test: detecting CIN2+ in low-resource settings. J Med Virol. 2019;91(7):1342–1350. doi:10.1002/jmv.25451
56. Wang M, Ji X, Dou X, et al. Consistency evaluation of Liferiver, Yaneng, Darui, and the Cobas 4800 test for high-risk human papillomavirus screening. J Clin Lab Anal. 2020;34(12):e23536. doi:10.1002/jcla.23536
57. Sun Z, Zhang R, Liu Z, et al. Development of a fluorescence-based multiplex genotyping method for simultaneous determination of human papillomavirus infections and viral loads. BMC Cancer. 2015;15:860. doi:10.1186/s12885-015-1874-9
58. Xu L, Oštrbenk A, Poljak M, et al. Assessment of the Roche Linear Array HPV Genotyping Test within the VALGENT framework. J Clin Virol. 2018;98:37–42. doi:10.1016/j.jcv.2017.12.001
59. Donà MG, Ronchetti L, Giuliani M, et al. Performance of the linear array HPV genotyping test on paired cytological and formalin-fixed, paraffin-embedded cervical samples. J mol Diagn. 2013;15(3):373–379. doi:10.1016/j.jmoldx.2013.01.002
60. Xu L, Padalko E, Oštrbenk A, et al. Clinical Evaluation of INNO-LiPA HPV Genotyping EXTRA II Assay Using the VALGENT Framework. Int J mol Sci. 2018;19(9):2704. doi:10.3390/ijms19092704
61. Ngou J, Gilham C, Omar T, et al. Comparison of analytical and clinical performances of the digene HC2 hPV DNA assay and the INNO-LiPA HPV genotyping assay for detecting high-risk HPV infection and cervical neoplasia among HIV-positive African women. J Acquir Immune Defic Syndr. 2015;68(2):162–168. doi:10.1097/QAI.0000000000000428
62. Yin J, Peng S, Li X, et al. Head-to-head comparison of genotyping of human papillomavirus by GP5+/6+-PCR-based reverse dot blot hybridization assay and SPF10-PCR-based line probe assay. J Med Virol. 2023;95(2):e28435. doi:10.1002/jmv.28435
63. Low HC, Silver MI, Brown BJ, et al. Comparison of Hybribio GenoArray and Roche human papillomavirus (HPV) linear array for HPV genotyping in anal swab samples. J Clin Microbiol. 2015;53(2):550–556. doi:10.1128/JCM.02274-14
64. Didelot MN, Boulle N, Damay A, et al. Comparison of the PapilloCheck® assay with the digene HC2 hPV DNA assay for the detection of 13 high-risk human papillomaviruses in cervical and anal scrapes. J Med Virol. 2011;83(8):1377–1382. doi:10.1002/jmv.22148
65. Li Y, Fu Y, Cheng B, et al. A Comparative Study on the Accuracy and Efficacy Between Dalton and CINtec® PLUS p16/Ki-67 Dual Stain in Triaging HPV-Positive Women. Front Oncol. 2022;11:815213. doi:10.3389/fonc.2021.815213
66. Bottari F, Iacobone AD, Radice D, et al. Anyplex II HPV test in detection and follow-up after surgical treatment of CIN2+ lesions. J Med Virol. 2021;93(11):6340–6346. doi:10.1002/jmv.26862
67. Liao Y, Li Q. Profile of MeltPro® HPV test for human papillomavirus genotyping and cervical precancer screening. Expert Rev mol Diagn. 2019;19(10):857–862. doi:10.1080/14737159.2019.1662299
68. Zeng Z, Yang H, Li Z, et al. Prevalence and Genotype Distribution of HPV Infection in China: analysis of 51,345 hPV Genotyping Results from China’s Largest CAP Certified Laboratory. J Cancer. 2016;7(9):1037–1043. doi:10.7150/jca.14971
69. Wei B, Mei P, Huang S, et al. Evaluation of the SureX HPV genotyping test for the detection of high-risk HPV in cervical cancer screening. Virol J. 2020;17(1):171. doi:10.1186/s12985-020-01417-8
70. Derbie A, Mekonnen D, Woldeamanuel Y, et al. HPV E6/E7 mRNA test for the detection of high grade cervical intraepithelial neoplasia (CIN2+): a systematic review. Infect Agent Cancer. 2020;15:9. doi:10.1186/s13027-020-0278-x
71. Jaworek H, Koudelakova V, Drabek J, et al. A Head-to-Head Analytical Comparison of Cobas 4800 hPV, PapilloCheck HPV Screening, and LMNX Genotyping Kit HPV GP for Detection of Human Papillomavirus DNA in Cervical and Cervicovaginal Swabs. J mol Diagn. 2018;20(6):849–858. doi:10.1016/j.jmoldx.2018.07.004
72. Heard I, Cuschieri K, Geraets DT, et al. Clinical and analytical performance of the PapilloCheck HPV-Screening assay using the VALGENT framework. J Clin Virol. 2016;81:6–11. doi:10.1016/j.jcv.2016.05.004
73. Alameda F, Garrote L, Mojal S, et al. Cervista HPV HR test for cervical cancer screening: a comparative study in the Catalonian population. Arch Pathol Lab Med. 2015;139(2):241–244. doi:10.5858/arpa.2014-0012-OA
74. Haedicke J, Iftner T. A review of the clinical performance of the Aptima HPV assay. J Clin Virol. 2016;76(1):S40–S48. doi:10.1016/j.jcv.2015.10.027
75. Arbyn M, Roelens J, Cuschieri K, et al. The APTIMA HPV assay versus the Hybrid Capture 2 test in triage of women with ASC-US or LSIL cervical cytology: a meta-analysis of the diagnostic accuracy. Int, J, Cancer. 2013;132(1):101–108. doi:10.1002/ijc.27636
76. Chen R, Zhang R, Zhang M, et al. CIN grades possessing different HPV RNA location patterns and RNAscope is helpful tool for distinguishing squamous intraepithelial lesions in difficult cervical cases. Diagn Pathol. 2023;18(1):23. doi:10.1186/s13000-023-01308-w
77. Wang Y, Tang Y, Chen Y, et al. Ultrasensitive one-pot detection of monkeypox virus with RPA and CRISPR in a sucrose-aided multiphase aqueous system. Microbiol Spectr. 2024;12(1):e0226723. doi:10.1128/spectrum.02267-23
78. Moore MD, Jaykus LA. Development of a Recombinase Polymerase Amplification Assay for Detection of Epidemic Human Noroviruses. Sci Rep. 2017;7:40244. doi:10.1038/srep40244
79. Solanki D, Murjani K, Singh V. CRISPR-Cas based genome editing for eradication of human viruses. Prog mol Biol Transl Sci. 2024;208:43–58. doi:10.1016/bs.pmbts.2024.07.012
80. Tao Y, Wang H, Ju E, et al. CRISPR-Cas12a-regulated DNA adsorption on MoS2 quantum dots: enhanced enzyme mimics for sensitive colorimetric detection of human monkeypox virus and human papillomavirus DNA. Talanta. 2025;283:127153. doi:10.1016/j.talanta.2024.127153
81. Zehbe I, Hacker GW, Su H, et al. Sensitive in situ hybridization with catalyzed reporter deposition, streptavidin-Nanogold, and silver acetate autometallography: detection of single-copy human papillomavirus. Am J Pathol. 1997;150(5):1553–1561.
82. Palantavida S, Guz NV, Woodworth CD, et al. Ultrabright fluorescent mesoporous silica nanoparticles for prescreening of cervical cancer. Nanomedicine. 2013;9(8):1255–1262. doi:10.1016/j.nano.2013.04.011
83. Palantavida S, Guz NV, Sokolov I. Functionalized ultrabright fluorescent mesoporous silica nanoparticles. Part Part Syst Charact. 2013;30:804–811. doi:10.1002/ppsc.201300143
84. Navarro Chica CE, Alfonso Tobón LL, López Abella JJ, et al. Nanoparticle-based colorimetric assays for early and rapid screening of the oncogenic HPV variants 16 and 18. Clin Chim Acta. 2025;568:120144. doi:10.1016/j.cca.2025.120144
85. Wang L, Nie Y, Zhang X, et al. A novel Eu3+ doped polydopamine nano particles/reductive copper particle hydrogel-based ECL sensor for HPV 16 DNA detection. Microchem J. 2022;181:107818. doi:10.1016/j.microc.2022.107818
86. Wu D, Dong W, Yin T, et al. PCN-224/Nano-Zinc oxide nanocomposite-based electrochemiluminescence biosensor for Hpv-16 detection by multiple cycling amplification and hybridization chain reaction. Sensors and Actuat B Chem. 2022;372:132659. doi:10.1016/j.snb.2022.132659
87. Li L, Yu S, Wu J, et al. Regulation of Target-activated CRISPR/Cas12a on Surface Binding of Polymer Dots for Sensitive Electrochemiluminescence DNA Analysis. Anal Chem. 2023;95(18):7396–7402. doi:10.1021/acs.analchem.3c01521
88. Wang H, Niu Y, Liu H, et al. A modularized universal strategy by integrating CRISPR/Cas with nanoporous materials for ultrasensitive determination of nucleic acids. Chem Eng J. 2025;506:160065. doi:10.1016/j.cej.2025.160065
89. Yue Y, Liu M, Ma M, et al. CRISPR/Cas14a integrated with DNA walker based on magnetic self-assembly for human papillomavirus type 16 oncoprotein E7 ultrasensitive detection. Biosens Bioelectron. 2025;272:117135. doi:10.1016/j.bios.2025.117135
90. Wang H, Hang X, Wang H, et al. Label/immobilization-free Cas12a-based electrochemiluminescence biosensor for sensitive DNA detection. Talanta. 2024;275:126114. doi:10.1016/j.talanta.2024.126114
91. Chen L, Liu M, Tang Y, et al. Preparation and Properties of a Low Fouling Magnetic Nanoparticle and Its Application to the HPV Genotypes Assay in Whole Serum. ACS Appl Mater Interfaces. 2019;11(20):18637–18644. doi:10.1021/acsami.9b04147
92. Gulliksen A, Solli LA, Drese KS, et al. Parallel nanoliter detection of cancer markers using polymer microchips. Lab Chip. 2005;5(4):416–420. doi:10.1039/b415525d
93. Wang W, Pang DW, Tang HW. Sensitive multiplexed DNA detection using silica nanoparticles as the target capturing platform. Talanta. 2014;128:263–267. doi:10.1016/j.talanta.2014.05.011
94. Civit L, Fragoso A, Hölters S, et al. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal Chim Acta. 2012;715:93–98. doi:10.1016/j.aca.2011.12.009
95. Yang Y, Qing Y, Hao X, et al. APTES-Modified Remote Self-Assembled DNA-Based Electrochemical Biosensor for Human Papillomavirus DNA Detection. Biosensors. 2022;12(7):449. doi:10.3390/bios12070449
96. Kim Y, Kang E. A graphitic nano-onion/molybdenum disulfide nanosheet composite as a platform for HPV-associated cancer-detecting DNA biosensors. J Nanobiotechnology. 2023;21(1):187. doi:10.1186/s12951-023-01948-6
97. Reema R, Sonam S, Souradeep R, et al. Design and development of an electroanalytical genosensor based on Cu-PTCA/rGO nanocomposites for the detection of cervical cancer. Mater Chem Phys. 2023;295:127050. doi:10.1016/j.matchemphys.2022.127050
98. Rawat R, Roy S, Goswami T, et al. An Electroanalytical Flexible Biosensor Based on Reduced Graphene Oxide-DNA Hybrids for the Early Detection of Human Papillomavirus-16. Diagnostics. 2022;12(9):2087. doi:10.3390/diagnostics12092087
99. Chen J, Gu W, Yang L, et al. Nanotechnology in the management of cervical cancer. Rev Med Virol. 2015;25(1):72–83. doi:10.1002/rmv.1825
100. Baek TJ, Park PY, Han KN, et al. Development of a photodiode array biochip using a bipolar semiconductor and its application to detection of human papilloma virus. Anal Bioanal Chem. 2008;390(5):1373–1378. doi:10.1007/s00216-007-1814-x
101. Fölsch S, Martínez-Blanco J, Yang J, et al. Quantum dots with single-atom precision. Nat Nanotechnol. 2014;9(7):505–508. doi:10.1038/nnano.2014.129
102. Liu H, Wang X, Wang H, et al. Synthesis and biomedical applications of graphitic carbon nitride quantum dots. J Mater Chem B. 2019;7(36):5432–5448. doi:10.1039/c9tb01410a
103. Ma F, Li CC, Zhang CY. Development of quantum dot-based biosensors: principles and applications. J Mater Chem B. 2018;6(39):6173–6190. doi:10.1039/c8tb01869c
104. Sheikhzadeh E, Beni V, Zourob M. Nanomaterial application in bio/sensors for the detection of infectious diseases. Talanta. 2021;230:122026. doi:10.1016/j.talanta.2020.122026
105. Zhao JY, Chen G, Gu YP, et al. Ultrasmall Magnetically Engineered Ag2Se Quantum Dots for Instant Efficient Labeling and Whole-Body High-Resolution Multimodal Real-Time Tracking of Cell-Derived Microvesicles. J Am Chem Soc. 2016;138(6):1893–1903. doi:10.1021/jacs.5b10340
106. Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi:10.1126/science.1104274
107. Jing L, Ding K, Kershaw SV, et al. Magnetically engineered semiconductor quantum dots as multimodal imaging probes. Adv Mater. 2014;26(37):6367–6386. doi:10.1002/adma.201402296
108. Murray C, Norris DJ, Bawendi MG. Synthesis and characterization of nearly monodisperse CdE (E= sulfur, selenium, tellurium) semiconductor nanocrystallites. J Am Chem Soc. 1993;115(19):8706–8715. doi:10.1021/ja00072a025
109. Kopping JT, Patten TE. Identification of acidic phosphorus-containing ligands involved in the surface chemistry of CdSe nanoparticles prepared in tri-N-octylphosphine oxide solvents. J Am Chem Soc. 2008;130(17):5689–5698. doi:10.1021/ja077414d
110. Zhang H, Ha DH, Hovden R, et al. Controlled synthesis of uniform cobalt phosphide hyperbranched nanocrystals using tri-n-octylphosphine oxide as a phosphorus source. Nano Lett. 2011;11(1):188–197. doi:10.1021/nl103400a
111. Li L, Zhang J, Zhang M, et al. Fragmentation of Magic-Size Cluster Precursor Compounds into Ultrasmall CdS Quantum Dots with Enhanced Particle Yield at Low Temperatures. Angew Chem Int Ed Engl. 2020;59(29):12013–12021. doi:10.1002/anie.202001608
112. Wang YF, Pan MM, Song YL, et al. Beyond the fluorescence labelling of novel nitrogen-doped silicon quantum dots: the reducing agent and stabilizer for preparing hybrid nanoparticles and antibacterial applications. J Mater Chem B. 2022;10(36):7003–7013. doi:10.1039/d2tb01304e
113. Nishimura H, Enomoto K, Pu YJ, et al. Hydrothermal synthesis of water-soluble Mn- and Cu-doped CdSe quantum dots with multi-shell structures and their photoluminescence properties. RSC Adv. 2022;12(10):6255–6264. doi:10.1039/d1ra08491g
114. Nammahachak N, Aup-Ngoen KK, Asanithi P, et al. Hydrothermal synthesis of carbon quantum dots with size tunability via heterogeneous nucleation. RSC Adv. 2022;12(49):31729–31733. doi:10.1039/d2ra05989d
115. Sondi I, Siiman O, Matijević E. Synthesis of CdSe nanoparticles in the presence of aminodextran as stabilizing and capping agent. J Colloid Interface Sci. 2004;275(2):503–507. doi:10.1016/j.jcis.2004.02.005
116. Abd Rahman S, Ariffin N, Yusof NA, et al. Thiolate-Capped CdSe/ZnS Core-Shell Quantum Dots for the Sensitive Detection of Glucose. Sensors. 2017;17(7):1537. doi:10.3390/s17071537
117. Wang Q, Fang T, Liu P, et al. Direct synthesis of high-quality water-soluble CdTe:Zn2+ quantum dots. Inorg Chem. 2012;51(17):9208–9213. doi:10.1021/ic300473u
118. Yang F, Yang P, Cao Y. Hydrothermal synthesis of high-quality thiol-stabilized CdTe(x)Se(1-x) alloyed quantum dots. J Fluoresc. 2013;23(6):1247–1254. doi:10.1007/s10895-013-1256-0
119. Lai L, Sheng SY, Mei P, et al. Hydrothermal synthesis for high-quality glutathione-capped Cdx Zn1 - x Se and Cdx Zn1 - x Se/ZnS alloyed quantum dots and its application in Hg(II) sensing. Luminescence. 2017;32(2):231–239. doi:10.1002/bio.3174
120. Makkar M, Viswanatha R. Frontier challenges in doping quantum dots: synthesis and characterization. RSC Adv. 2018;8(39):22103–22112. doi:10.1039/c8ra03530j
121. Paul S, Banerjee SL, Khamrai M, et al. Hydrothermal synthesis of gelatin quantum dots for high-performance biological imaging applications. J Photochem Photobiol B. 2020;212:112014. doi:10.1016/j.jphotobiol.2020.112014
122. Xian Y, Li K. Hydrothermal Synthesis of High-Yield Red Fluorescent Carbon Dots with Ultra-Narrow Emission by Controlled O/N Elements. Adv Mater. 2022;34(21):e2201031. doi:10.1002/adma.202201031
123. Gao Y, Liu Y, Zhang H, et al. One-pot synthesis of efficient multifunctional nitrogen-doped carbon dots with efficient yellow fluorescence emission for detection of hypochlorite and thiosulfate. J Mater Chem B. 2022;10(43):8910–8917. doi:10.1039/d2tb01695h
124. Shen L, Tang J, Li M, et al. Facile synthesis of sulfur quantum dots with red light emission: implications for electrochemiluminescence analysis application. Spectrochim, Acta a mol, Biomol, Spectrosc. 2024;2024:124878. doi:10.1016/j.saa.2024.124878
125. Wang H, Wang Z, Xiong Y, et al. Hydrogen Peroxide Assisted Synthesis of Highly Luminescent Sulfur Quantum Dots. Angew Chem Int Ed Engl. 2019;58(21):7040–7044. doi:10.1002/anie.201902344
126. Song M, Yang M, Hao J. Pathogenic Virus Detection by Optical Nanobiosensors. Cell Rep Phys Sci. 2021;2(1):100288. doi:10.1016/j.xcrp.2020.100288
127. Mukherjee A, Shim Y, Myong Song J. Quantum dot as probe for disease diagnosis and monitoring. Biotechnol J. 2016;11(1):31–42. doi:10.1002/biot.201500219
128. Chan WC, Maxwell DJ, Gao X, et al. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13(1):40–46. doi:10.1016/s0958-1669(02)00282-3
129. Smith AM, Duan H, Mohs AM, et al. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv Drug Deliv Rev. 2008;60(11):1226–1240. doi:10.1016/j.addr.2008.03.015
130. Nabil M, Megahed F. Quantum Dot Nanomaterials: preparation, Characterization, Advanced Bio-Imaging and Therapeutic Application. J Fluoresc. 2023;34:2467–2484. doi:10.1007/s10895-023-03472-0
131. Chung S, Revia RA, Zhang M. Graphene Quantum Dots and Their Applications in Bioimaging, Biosensing, and Therapy. Adv Mater. 2021;33(22):e1904362. doi:10.1002/adma.201904362
132. Shamsipur M, Nasirian V, Mansouri K, et al. A highly sensitive quantum dots-DNA nanobiosensor based on fluorescence resonance energy transfer for rapid detection of nanomolar amounts of human papillomavirus 18. J Pharm Biomed Anal. 2017;136:140–147. doi:10.1016/j.jpba.2017.01.002
133. Nie Y, Zhang X, Zhang Q, et al. A novel high efficient electrochemiluminescence sensor based on reductive Cu(I) particles catalyzed Zn-doped MoS2 QDs for HPV 16 DNA determination. Biosens Bioelectron. 2020;160:112217. doi:10.1016/j.bios.2020.112217
134. Sun Y, Liu J, Peng X, et al. A novel photoelectrochemical array platform for ultrasensitive multiplex detection and subtype identification of HPV genes. Biosens Bioelectron. 2023;224:115059. doi:10.1016/j.bios.2023.115059
135. Zhang H, Liu L, Li CW, et al. Multienzyme-nanoparticles amplification for sensitive virus genotyping in microfluidic microbeads array using Au nanoparticle probes and quantum dots as labels. Biosens Bioelectron. 2011;29(1):89–96. doi:10.1016/j.bios.2011.07.074
136. Yu-Hong W, Rui C, Ding L. A quantum dots and superparamagnetic nanoparticle-based method for the detection of HPV DNA. Nanoscale Res Lett. 2011;6(1):461. doi:10.1186/1556-276X-6-461
137. Palomino-Vizcaino G, Valencia Reséndiz DG, Benítez-Hess ML, et al. Effect of HPV16 L1 virus-like particles on the aggregation of non-functionalized gold nanoparticles. Biosens Bioelectron. 2018;100:176–183. doi:10.1016/j.bios.2017.09.001
138. Gao Y, Fan X, Zhang X, et al. HCR/DNAzyme-triggered cascaded feedback cycle amplification for self-powered dual-photoelectrode detection of femtomolar HPV16. Biosens Bioelectron. 2023;237:115483. doi:10.1016/j.bios.2023.115483
139. Maria Jimenez Jimenez A, Moulick A, Richtera L, et al. Dual-color quantum dots-based simultaneous detection of HPV-HIV co-infection. Sens Actuat B-Chem. 2018;258:295–303. doi:10.1016/j.snb.2017.11.074
140. Li Y, Zeng R, Wang W, et al. Size-Controlled Engineering Photoelectrochemical Biosensor for Human Papillomavirus-16 Based on CRISPR-Cas12a-Induced Disassembly of Z-Scheme Heterojunctions. ACS Sens. 2022;7(5):1593–1601. doi:10.1021/acssensors.2c00691
141. Cheng Y, Sun C, Chang Y, et al. Photoelectrochemical biosensor based on SiW12@CdS quantum dots for the highly sensitive detection of HPV 16 DNA. Front Bioeng Biotechnol. 2023;11:1193052. doi:10.3389/fbioe.2023.1193052
142. Pan L, Li B, Chen J, et al. Nanotechnology-Based Weapons to Combat Human Papillomavirus Infection Associated Diseases. Front Chem. 2021;9:798727. doi:10.3389/fchem.2021.798727
143. Yu J, Dong C, Yang Y, et al. Electrochemical DNA biosensor for HPV-16 detection based on novel carbon quantum dots/APTES composite nanofilm. Microchem J. 2024;204:110949. doi:10.1016/j.microc.2024.110949
144. Jimenez Jimenez AM, Moulick A, Bhowmick S, et al. One-step detection of human papilloma viral infection using quantum dot-nucleotide interaction specificity. Talanta. 2019;205:120111. [published correction appears in Talanta. 2022 May 15;242:123172. doi: 10.1016/j.talanta.2021.123172]. doi:10.1016/j.talanta.2019.07.006
145. Jie G, Zhou Q, Jie G. Graphene quantum dots-based electrochemiluminescence detection of DNA using multiple cycling amplification strategy. Talanta. 2019;194:658–663. doi:10.1016/j.talanta.2018.10.098
146. Xiang DS, Zeng GP, He ZK. Magnetic microparticle-based multiplexed DNA detection with biobarcoded quantum dot probes. Biosens Bioelectron. 2011;26(11):4405–4410. doi:10.1016/j.bios.2011.04.051
147. Barrientos K, Arango JP, Moncada MS, et al. Carbon dot-based biosensors for the detection of communicable and non -communicable diseases. Talanta. 2023;251:123791. doi:10.1016/j.talanta.2022.123791
148. Oliveira LS, Goel S, Amreen K, et al. Microfluid biosensor for detection of hpv in patient scraping samples: determining E6 and E7 oncogenes. Bioelectrochemistry. 2024;160:108795. doi:10.1016/j.bioelechem.2024.108795
149. Guo X, Zhou L, Liu X, et al. Fluorescence detection platform of metal-organic frameworks for biomarkers. Colloids Surf B Biointerfaces. 2023;229:113455. doi:10.1016/j.colsurfb.2023.113455
150. Luo D, Huang J, Jian Y, et al. Metal-organic frameworks (MOFs) as apt luminescent probes for the detection of biochemical analytes. J Mater Chem B. 2023;11(29):6802–6822. doi:10.1039/d3tb00505d
151. Zhou J, Li Y, Wang W, et al. Metal-organic frameworks-based sensitive electrochemiluminescence biosensing. Biosens Bioelectron. 2020;164:112332. doi:10.1016/j.bios.2020.112332
152. Yang X, Yu YQ, Peng LZ, et al. Strong Electrochemiluminescence from MOF Accelerator Enriched Quantum Dots for Enhanced Sensing of Trace cTnI. Anal Chem. 2018;90(6):3995–4002. doi:10.1021/acs.analchem.7b05137
153. Jiang X, Yin C, Wu M, et al. Fluorescent switch based on QDs modified DNA probe and NiTPP for simultaneous dual color sensitive sensing of HPV16 and HPV18. Sensors and Actuat B Chem. 2024;403:135128. doi:10.1016/j.snb.2023.135128
154. Lin X, Chen T. A Review of in vivo Toxicity of Quantum Dots in Animal Models. Int J Nanomed. 2023;18:8143–8168. doi:10.2147/IJN.S434842
155. Nikazar S, Sivasankarapillai VS, Rahdar A, et al. Revisiting the cytotoxicity of quantum dots: an in-depth overview. Biophys Rev. 2020;12(3):703–718. doi:10.1007/s12551-020-00653-0
156. Abdellatif AAH, Younis MA, Alsharidah M, et al. Biomedical Applications of Quantum Dots: overview, Challenges, and Clinical Potential. Int J Nanomed. 2022;17:1951–1970. doi:10.2147/IJN.S357980
157. Zhong L, Zhang L, Li Y, et al. Assessment of the Toxicity of Quantum Dots through Biliometric Analysis. Int J Environ Res Public Health. 2021;18(11):5768. doi:10.3390/ijerph18115768
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

