Back to Journals » Drug Design, Development and Therapy » Volume 19
Serum Pharmacochemistry and Network Pharmacology Reveal Active Compounds and Mechanisms of the Huaxian Formula in Alleviating Radiation-Induced Pulmonary Fibrosis
Authors Gong C, Chen J, Zou P, Fang Z, Quan L, Wang J, Yin J, Lin B, Lang J , Chen M
Received 24 September 2024
Accepted for publication 22 January 2025
Published 27 January 2025 Volume 2025:19 Pages 627—644
DOI https://doi.org/10.2147/DDDT.S490844
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
CuiCui Gong,1– 3 Junyang Chen,2 Pingjin Zou,2 Zengyi Fang,2 Li Quan,1– 3 Jing Wang,2,4 Jie Yin,2,5 Bing Lin,1 Jinyi Lang,1– 5 Meihua Chen2,3
1School of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, 610032, People’s Republic of China; 2Department of Radiation Oncology, Radiation Oncology Key Laboratory of Sichuan Province, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, University of Electronic Science and Technology of China, Chengdu, 610041, People’s Republic of China; 3Institute of Integrated Traditional Chinese and Western Medicine Cancer Research, Chengdu University of Traditional Chinese Medicine, Chengdu, 610032, People’s Republic of China; 4Department of Oncology, School of Clinical Medicine, Southwest Medical University, Luzhou, Sichuan, 646000, People’s Republic of China; 5School of Medicine, University of Electronic Science and Technology of China, Chengdu, Sichuan, 611731, People’s Republic of China
Correspondence: Jinyi Lang, School of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, 610032, People’s Republic of China, Email [email protected] Meihua Chen, Department of Radiation Oncology, Radiation Oncology Key Laboratory of Sichuan Province, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, University of Electronic Science and Technology of China, Chengdu, 610041, People’s Republic of China, Email [email protected]
Purpose: Radiation-induced pulmonary fibrosis (RIPF) is a serious complication of radiotherapy that lacks effective treatment options. The Huaxian formula (HXF), a traditional Chinese herbal remedy, shows promise in alleviating RIPF; however, its active ingredients and underlying mechanisms remain poorly understood.
Methods: Through serum pharmacochemistry, network pharmacology, molecular docking, and experimental validation, we investigate the potential mechanisms of HXF in the prevention and treatment of radiation-induced pulmonary fibrosis (RIPF).
Results: Histological examination and non-invasive computed tomography (CT) scans in animal experiments revealed that HXF improved extracellular matrix collagen deposition in the lung tissue of irradiated mice and reduced fibrosis manifestations on CT images. Analysis of post-HXF administration serum samples identified 21 enriched compounds as potential active compounds, with 430 corresponding prospective targets. Overlapping these compounds with 991 RIPF-related genes yielded 127 genes primarily associated with the PI3K-Akt signaling pathway, EGFR tyrosine kinase inhibitor resistance, and the MAPK signaling pathway. Molecular docking indicated that key compounds in HXF serum, 5,7,8-trimethoxyflavone, and hyperoside, exhibited strong affinity with key targets. Finally, animal experiments confirmed that HXF significantly inhibited the expression of p-Akt and p-PI3K proteins in the lung tissue of irradiated mice.
Conclusion: Our research results indicate that HXF may exert its effects on the prevention and treatment of radiation-induced pulmonary fibrosis (RIPF) through multiple pathways and targets, with the PI3K-Akt signaling pathway likely playing the most crucial role in this process.
Keywords: radiation-induced pulmonary fibrosis, Huaxian formula, serum pharmacochemistry, network pharmacology, active compounds
Graphical Abstract:

Introduction
Radiotherapy is a cornerstone in the management of various cancers, but its collateral damage to healthy lung tissue can result in long-term complications such as radiation-induced pulmonary fibrosis (RIPF).1 RIPF is characterized by the progressive and irreversible scarring of lung tissue, leading to impaired lung function and reduced quality of life for affected patients. The underlying pathogenesis of RIPF involves a complex interplay of inflammatory responses, oxidative stress, fibroblast activation, and excessive extracellular matrix (ECM) deposition.2 Despite advancements in radiation therapy techniques, the incidence of RIPF remains concerning, especially after radiation therapy for thoracic malignancies.3 The lack of effective interventions to prevent or ameliorate RIPF underscores the urgent need for novel therapeutic strategies.
Recently, traditional Chinese medicine (TCM) has gained attention for its potential in the management of various diseases, including fibrotic disorders.4–6 Within the realm of TCM, the Huaxian formula (HXF) has emerged as a promising traditional Chinese herbal remedy for alleviating respiratory conditions, including RIPF. Previous studies have shown that HXF can alleviate RIPF by inhibiting the activation of fibroblasts and collagen production in mice after irradiation.7 The specific medicinal components of HXF, such as Huangqi (Astragalus), Danggui (Angelica), and Gacao (Licorice), have demonstrated significant efficacy in reducing RIPF occurrence.8–10 Wogonin, a key component of Huangqin (Scutellaria baicalensis) in HXF, protects against acute lung injury by modulating Akt phosphorylation and RhoA activation.11 Other individual herbs or active ingredients within HXF have shown effectiveness in inhibiting lung diseases,12,13 suggesting a theoretical pharmacological basis for RIPF prevention and treatment. The therapeutic impact of HXF is not merely the sum of its parts but results from novel components formed through ingredient interactions post-absorption. Consequently, the active compounds and mechanisms underlying HXF’s therapeutic effects in RIPF prevention and treatment remain incompletely understood.
To address this gap, we employ the combined approaches of serum pharmacochemistry and network pharmacology. Serum pharmacochemistry, utilizing liquid chromatography-tandem mass spectrometry (LC-MS), identifies active constituents of TCM. This technique is particularly valuable for multiple-herb TCM formulations, rapidly identifying potential active ingredients in serum after oral administration.14,15 Network pharmacology, an emerging discipline that combines systems biology theory and biological system network analysis,16 facilitates exploring the mechanism of TCM ingredients on diseases.17
In this study, we conducted animal experiments to assess the pharmacological effects of HXF on RIPF. Ultra-high-performance liquid chromatography-quadrupole-extractive-orbitrap mass spectrometry (UHPLC-Q-Extractive-Orbitrap-MS) detected HXF’s chemical constituents. Comparing these with serum components identified prototype blood-entry compounds. Network pharmacology screened corresponding targets, and molecular docking and visualization analyses confirmed interactions with key genes. This research bridges the knowledge gap, elucidating the active compounds and pharmacological mechanisms of HXF in RIPF. These findings have the potential to advance effective RIPF treatments, enhancing outcomes and quality of life for cancer patients undergoing radiotherapy.
Materials and Methods
Chemicals and Animals
All ingredients of HXF were sourced from Sichuan Neo-Green Pharmaceutical Technology Development Co., Ltd. (Chengdu, China). The prescription consisted of 15 herbal medicines (Table 1): Jinyinhua, Huangqin, Pipaye, Sangbaipi, Zhuru, Huangqi, Baihe, Maidong, Fabanxia, Chenpi, Fuling, Gancao, Danshen, Honghua, and Sanqi.
 |
Table 1 Composition of Huaxian Formula |
Studies have shown that after ionizing radiation, C57BL/6 male mice have a longer survival time after fibrosis compared to female mice, which facilitates experimental observation.18 Therefore, we used specific pathogen-free (SPF) C57BL/6 male mice (8-week-old, 20–25 g) obtained from Beijing HFK Bio-Technology Co., Ltd for animal studies. All animals were housed in a conventional animal housing environment, and every effort was made to reduce the number of animals used in the experiment and mitigate animal suffering. The studies adhered to the EU Directive 2010/63/EU for animal experiments. We used the ARRIVE guidelines 2.0.
Pharmacodynamic Assessment of HXF for the Treatment of RIPF
In vivo RIPF Models and Treatments
The mice were randomly assigned to four groups: control, HXF, ionizing radiation (IR), and IR+HXF, each consisting of 20 mice. The control and IR groups received sterile physiological saline via gavage, while the HXF and IR+HXF groups were administered HXF via gavage. Medication started one week prior to irradiation and was administered once daily. Within 2 hours after the seventh day of medication, the mice’s entire chest was locally irradiated with 17 Gy of X-ray, according to previous studies.19,20 The dose rate was set at 75 cGy/min, with the remaining body shielded using lead bricks. The X-RAD 320 irradiator (Precision X-Ray, CT) was used in this study. Daily medication was resumed on the second day after irradiation and continued for 4 weeks, resulting in a total medication duration of 5 weeks. After 4 months of irradiation, lung tissues from each group were collected. The collected lung tissues were subjected to hematoxylin-eosin (H&E) staining, Masson staining, and immunohistochemistry (IHC) staining for analysis. The extent of lung injury and fibrosis was assessed using the Szapiel score21 based on H&E staining and the Ashcroft score22 based on Masson’s staining.
Micro-Computed Tomography Scans of Mice
After irradiation in mice, we selected four time point-4, 8, 12, and 16-weeks and anesthetized the mice using 2% isoflurane. High-resolution lung scans were then performed using a Quantum GX Micro-CT (Rigaku Corporation). The total scan time was 4 minutes, with the X-ray tube voltage set at 80 kV and the X-ray tube current at 100 μ A. A 3D dataset was constructed using 360 projections over a 360-degree rotation, with an isotropic voxel size of 50 μ m, and saved in DICOM format. Hounsfield units (HU) provide a relative quantitative measurement of tissue absorption of X-rays in CT images. Clinically, HU values are used to distinguish between normal ventilation ([−900, −500] HU) and poor ventilation ([−500, −100] HU).23 Subsequently, the data were imported into the 3D Slicer software to calculate the HU values for the lungs of mice in different groups.
Staining and Immunohistochemistry
Lung tissues were subjected to fixation, sectioning, and staining with H&E and Masson’s trichrome stain. Subsequently, the stained sections were examined under an optical microscope. IHC staining was performed on the lung tissue sections to detect α-smooth muscle actin (α-SMA). For image analysis, the integrated optical density of each image was calculated using the professional imaging acquisition and analysis system, Image Pro Plus6.0.24,25
Sample Preparation for in vitro Chemical Identification
Preparation of Calibration Materials
Luming Biotech Co., Ltd. (Shanghai, China) established the LuMet-TCM database of reference substances.
Preparation of HXF
The HXF formula solution was thoroughly mixed with pure water in a 1:9 ratio. After ultrasonic treatment in an ice-water bath, the mixture was allowed to stand for 30 minutes. Following this standing period, the mixture was centrifuged to collect the supernatant, which was then incubated overnight at 4°C. The supernatant was subsequently diluted 1:1 with pure water and filtered through a 0.22 μ m membrane. Finally, the filtered solution was transferred to an LC-MS vial with an inner lining for analysis.
Preparation of Serum Samples to Identify Chemicals Absorbed in vivo
C57BL/6 male mice were randomly assigned to two groups: a control group (n = 15) and an HXF treatment group (n = 15). The HXF group underwent continuous gastric lavage with 31.85 g/kg of HXF for three consecutive days, while the control group received an equivalent volume of saline. Considering the impact of bioavailability on the detection of active components in the blood, and to minimize individual variation, we ensured an adequate sample size and incorporated multiple time points for optimized sample collection. Blood samples were obtained from the ocular orbit at 15 and 30 minutes, and 1, 2, and 4 hours following the most recent administration, as indicated in the relevant literature.26 After incubation at room temperature for 30 minutes, the samples were centrifuged at 4 °C and 4000 rpm for 20 minutes to obtain serum. Pooled serum samples were prepared by combining equal amounts of serum from different time points.
Samples were obtained by mixing 150 μ L of serum with 450 μ L of methanol-acetonitrile, vortex, and then subjecting the mixture to ultrasonic treatment in an ice-water bath for 10 minutes. After standing at −40 °C for 30 minutes, the mixture was centrifuged at 12,000 rpm and 4 °C for 10 minutes. The supernatant (500 μ L) was dried and reconstituted with 150 μ L of methanol-acetonitrile, vortexed, and sonicated for 3 minutes. After leaving at −40 °C overnight, centrifuge the samples again at 12,000 rpm and 4 °C for 10 min. Ultimately, 100 μ L of leftovers was moved to vials for LC-MS analysis.
UHPLC-Q-Exactive Orbitrap MS/MS Analysis
LC-MS Conditions
The analytical instrument used was an ACQUITY UPLC I-Class HF ultra-high-performance LC-MS system coupled with a QE high-resolution mass spectrometer. The mobile phase consisted of 0.1% formic acid in water and acetonitrile. The injection volume of the instrument was set to 5 μ L and the flow rate was 0.35 mL/min. The Q-Exactive Orbitrap MS, which was equipped with a heated electrospray ionization (HESI) source, was capable of operating in both negative and positive modes, scanning m/z of 100–1200.
Preprocessing and Statistical Analysis of MS Data
The identification of compounds in compounds relies on accurate mass measurements of compounds, fragmentation analysis by MS/MS, and examination of isotope distributions. This was done using the LuMet-TCM database provided by Luming Biotech Co., Ltd., located in Shanghai, China. The total assay score and secondary match score of the reference compound are selected as part of the original formulation. Substances with a fold change (FC) value of ≥ 10 between the treatment group (serum with administration) and the control group (blank serum) were noted.
Network Pharmacology Analysis
Discovery of the Targets of the Potential Active Compounds
The targets of the putative bioactive molecules were identified utilizing multiple databases: the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database27 (http://tcmspw.com/tcmsp.php), DrugBank (https://go.drugbank.com/), SwissTargetPrediction28 (http://www.swisstargetprediction.ch/about.php), and PharmMapper29 (https://www.lilab-ecust.cn/pharmmapper/).
Identification of RIPF-Related Genes
Using multiple databases to identify genes related to RIPF: GeneCards30 (https://www.genecards.org/), the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/), OMIM31 (http://www.omim.org/), and Disease Gene network (DisGeNET) (https://www.disgenet.org/). Duplicate values from these databases were removed, and the gene lists were merged to create a comprehensive set of RIPF-related genes.
Construction of Component-Target Network Graph
Active compound targets from TCMSP, DrugBank, SwissTargetPrediction, and PharmMapper databases were intersected with RIPF-related targets from GeneCards, NCBI, OMIM, and DisGeNET databases. The resulting intersected targets and their corresponding active chemical components were imported into Cytoscape 3.9.1 for network visualization.
Enrichment Analysis of the GO and KEGG Pathways
Gene Ontology (GO) analysis can be divided into three categories: biological processes (BP), molecular functions (MF), and cellular components (CC). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to characterize candidate targets. To study the biological function of targets at the intersection of drugs and diseases, GO and KEGG data were collected using the DAVID database32 (https://david.ncifcrf.gov/). The results were sorted by the number of participating genes and P values. Visualization of the results was done using the Bioinformatics (www.bioinformatics.com.cn).
Protein-Protein Interaction (PPI) Network Construction and Analysis
Potential targets for HXF treatment with RIPF were identified as overlapping targets and candidate genes. The intersection of RIPF-related genes and predicted HXF targets was determined, and a Venn diagram was created using OmicStudio33 (https://www.omicstudio.cn/) for visualization. Then import these overlapping targets into the STRING database34 (https://string-db.org/) to obtain information on the protein interaction network. The protein-protein interaction (PPI) data were visualized and a PPI network was created using Cytoscape 3.9.1 (https://www.cytoscape.org/). The critical topological parameter, degree centrality (DC), was used to screen for core composite targets within the PPI network.35
Molecular Docking
Molecular docking verification was conducted using AutoDockTools-1.5.6 and PyMOL software to reveal the interactions between active components and target proteins. First, the 2D structure files (SDF format) of the key active compounds were downloaded from PubChem, and then imported into Open Babel 2.4.1 for conversion into Mol2 format.36 The 3D structures of the targets were obtained from the Protein Data Bank (PDB) (http://www.rcsb.org/).
The Mol2 files of active components were opened with AutoDockTools-1.5.6,37 where water molecules were removed, hydrogen atoms and charges were added, and the components were defined as ligands and saved as pdbqt files. The pdb files containing the 3D structure of the protein were analyzed using AutoDockTools-1.5.6. The receptors were altered, assigned as receptors, and stored as pdbqt files. We used a semi-flexible docking method to dock the ligands and receptors, selecting the conformation with the lowest binding energy for output and analysis. Finally, PyMOL software38 was used to generate the docking interaction images.
Protein Extraction and Western Blot Analysis
The lung tissue was extracted, washed with pre-cooled PBS, and lysed using radio immunoprecipitation assay (RIPA) buffer containing protease inhibitors and phosphatase inhibitors (PC101, EpiZyme, Shanghai, China). The lysate was centrifuged at 12,000×g for 10 minutes at 4°C, and the supernatant protein was collected. SDS-PAGE was prepared using the Omni-Easy™ One-Step PAGE Gel Fast Preparation Kit (10%) (PG212, EpiZyme, Shanghai, China). Equal amounts of protein samples were loaded onto the 10% SDS-PAGE gel and then transferred to polyvinylidene fluoride membranes. The membranes were blocked with 5% BSA for 1 hour. Primary antibodies were incubated overnight at 4°C: AKT (1:1000, ET1609-51, HuaBio), p-AKT (1:5000, ab81283, Abcam), PI3K (1:1000, 67071-1-lg, ProteinTech), p-PI3K (1:1000, AF3241, Affinity), GAPDH (1:2000, AC002, Abclonal). The membranes were then incubated with secondary antibodies goat anti-rabbit IgG (1:5000, AC002, Abclonal) and goat anti-mouse IgG (1:5000, SA00001-1, ProteinTech) at room temperature for 1 hour. Visualization was performed using an ultra-sensitive ECL chemiluminescence kit. The band intensities were analyzed using Image J software.
Statistical Analysis
GraphPad Prism 9 was employed for statistical analysis of the data, which are presented as mean ± SEM. One-way analysis of variance (ANOVA) was used to compare the means among the groups. A probability value of P < 0.05 was considered statistically significant.
Results
HXF Attenuated RIPF in vivo
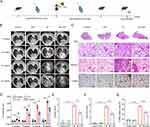
We established a mouse model of radiation-induced pulmonary fibrosis (RIPF) to assess the therapeutic effect of HXF. The experimental procedure is represented Figure 1A. In Figure 1B, the micro-CT images of mice from different groups at −4, 8, 12, and 16-weeks post-irradiation are shown. The longitudinal micro-CT scans of the lung structure reveal that the IR group exhibited significant progressive damage in the lungs following irradiation. By week 16, typical radiological features such as irregular septal thickening, patchy peripheral reticular abnormalities, and honeycombing were observed. Oral administration of HXF significantly improved the morphological signs of lung fibrosis in irradiated mice. On the other hand, the quantitative analysis of HU values was used to assess the extent of lung fibrosis. In the IR group, there was a significant increase in lung density at 8 weeks post-irradiation (P < 0.0001) (Figure 1D). The increase in lung density was significantly inhibited after the application of HXF (P < 0.0001), effectively reducing the CT manifestations of lung fibrosis. Histological examination of lung tissues stained with H&E at 16 weeks post-irradiation revealed that the lung structure in the HXF and control groups appeared normal, with clear alveoli and no signs of significant inflammatory cell infiltration. In contrast, the IR group exhibited evident structural damage, characterized by severe alveolar atrophy, thickened alveolar walls, and extensive infiltration of inflammatory cells. The IR+HXF group showed reduced lung tissue damage, decreased alveolar wall thickness, and reduced inflammatory cell infiltration compared to the IR group (Figure 1C). The Szapiel score indicates that, compared to the control group and the HXF-only group, the IR group exhibited significantly higher scores (P < 0.0001). However, the lung tissue inflammation in the IR+HXF group was significantly lower than that in the IR group (P < 0.0001) (Figure 1E).
Masson staining showed that lung tissues from the HXF and control groups exhibited normal structure without typical fibrosis. In contrast, the IR group displayed significant collagen deposition and severe fibrosis. The IR+HXF group exhibited reduced collagen deposition and alleviated fibrosis compared to the IR group. Semi-quantitative analysis using the Ashcroft score showed a significant increase in pulmonary fibrosis in the IR group, while HXF treatment attenuated radiation-induced pulmonary fibrosis (P < 0.0001) (Figure 1F).
Furthermore, IHC analysis demonstrated a notable increase in α-SMA expression in lung tissues following irradiation, which was effectively reduced by HXF therapy (Figure 1G). These findings indicate that HXF alleviates radiation-induced lung tissue damage by reducing tissue inflammation and suppressing collagen deposition.
In vitro Characterization of the Chemical Constituents in HXF
The composition of HXF was analyzed using LC-MS. Figure 2A and B presents the base peak chromatograms (BPC) of HXF in different modes of positive and negative ions. Initial identification of the compounds revealed the presence of terpenes, flavonoids, alkaloids, amino acids, and organic acids, and other components. A comprehensive comparison of accurate mass-to-charge ratios (m/z), secondary fragments, and isotopic distribution with the LuMet-TCM reference database led to the identification of 578 compounds in HXF (Supplementary Table 1). Mass spectrometry data for 5,7,8-trimethoxyflavone and hyperoside, along are presented in Figure 2C and D. The mass spectra of the remaining 19 common components can be found in Supplementary Figures 1–19.
Serum Chemical Profiling Following HXF Administration
To identify potential therapeutic compounds in HXF, we conducted chemical analysis on serum collected following HXF administration. To facilitate comprehensive analysis, serum samples from all time points were pooled to create a dosed serum sample. We used the UHPLC-Q Exactive Orbitrap-MS method to analyze and identify the absorbed HXF components in mice. Due to effects of absorption rate, distribution sites, and metabolic pathways of the components in the HXF formula within the mice, we analyzed data from the in vitro HXF components, blank serum, and serum containing HXF components. As a result, we identified 40 prototype ingredients and 29 metabolites in the blood (Supplementary Table 2). Among these prototype components, 21 common constituents were confirmed through comparison with standard reference substances, as shown in Table 2. While LC-MS technology enables accurate identification of blood components from HXF, there is still significant ongoing work required in sample processing, method optimization, and standardization. The detection of drug components in blood continues to be a key challenge and an important area of focus in the modernization of traditional Chinese medicine and drug analysis research.
 |
Table 2 Comparison of 21 Prototype Blood Components in HXF |
Analysis of the Target Inhibition of RIPF by HXF
A total of 991 target genes associated with RIPF were identified from the GeneCards, OMIM, NCBI, and DisGeNET databases. Additionally, the TCMSP, DrugBank, SwissTargetPrediction, and PharMapper databases provided 430 target genes related to the 21 blood components found in HXF. By intersecting the target genes associated with the HXF constituents and those associated with the disease, we identified 127 overlapping target genes (Figure 3A). Subsequently, we constructed a compound-overlapping target network using the 127 potential targets and their corresponding compounds. This network revealed that 5,7,8-trimethoxyflavone, hyperoside, helichrysetin, and isoformononetin target the highest number of proteins related to RIPF, with 50, 47, 40, and 39 targets, respectively (Figure 3B). The network consisted of 148 nodes and 537 edges.
GO, KEGG Enrichment Analysis and PPI Network Analysis
To gain insights into the molecular mechanisms of the 127 target molecules, we performed GO enrichment analysis and KEGG pathway analysis. This analysis revealed 662 BP, 74 CC, 154 MF, and 152 KEGG pathways associated with these targets (Supplementary Tables 3 and 4). We focused on the top 10 BP, CC, and MF terms related to the 127 targets (Figure 3C). Notably, the significantly upregulated BP terms included negative regulation of gene expression, adverse regulation of apoptotic processes, and positive regulation of the MAPK cascade. The CC terms comprised the nucleus, cytosol, and macromolecular complex. The highly enriched MF terms were enzyme binding, protein binding, and identical protein binding.
To further elucidate the pharmacological mechanism of HXF against RIPF, we conducted pathway analysis to identify potential pathways influenced by HXF. The 152 KEGG pathways can be divided into three categories: environmental information processing, organismal systems, and human disease, with predominant focus on human diseases (Supplementary Figure 20). After excluding irrelevant pathways and focusing on the top 20 enriched pathways, we pinpointed the PI3K-Akt signaling pathway, EGFR tyrosine kinase inhibitor resistance, and the MAPK signaling pathway as potential critical routes through which HXF impacts RIPF (Figure 3D). The identified molecular functions, biological processes, and pathways are closely associated with the development of RIPF, indicating that HXF exerts its inhibitory effects on RIPF through multiple targets and pathways. The P-value analysis of KEGG enrichment pathways indicates that a lower P-value signifies higher significance.39 Hence, we will concentrate on the PI3K-Akt pathway, given its lowest P-value and robust correlation. To explore the interactions among the 127 potential targets, we utilized the STRING platform for PPI network analysis. The resulting network was visualized using Cytoscape, where the size of each node represents its degree value, and the thickness of the lines reflects the strength of the interactions (Figure 3E). The degree value sorting can be found in Supplementary Table 5. The PPI network consisted of 120 nodes and 619 edges. Through topological analysis using CytoNCA, we identified 20 critical targets from this network (Figure 3F). Based on KEGG pathway analysis, AKT1, EGFR, and MAPK1 may be potential key targets for HXF’s action on RIPF.
Molecular Docking Analysis
To investigate the potential targets of HXF for treating RIPF, molecular docking analysis was performed to evaluate the binding energy between the active components and selected targets. AKT1, MAPK1, and EGFR were identified as probable targets. The results revealed that two compounds, specifically 5,7,8-trimethoxyflavone and hyperoside, exhibited binding interactions with multiple sites on AKT1 (PDBID: 7MYX), MAPK1 (PDBID: 7AUV), and EGFR (PDBID: 7OM5) proteins. The binding energies (kcal/mol) for these active compounds and significant targets are presented in Table 3. A lower binding energy value indicates a stronger affinity between the ligand molecules and receptor proteins, with a binding energy below −5 kcal/mol suggesting a strong binding affinity between them.40 The interactions between the components and targets with the six lowest free binding energy scores, along with their binding modes, were visualized using PyMOL (Figure 4). The binding energy scores were as follows: −5.25 kcal/mol (5,7,8-trimethoxyflavone-AKT1), −6.2 kcal/mol (5,7,8-trimethoxyflavone-MAPK1), −5.86 kcal/mol (hyperoside-AKT1), −5.02 kcal/mol (hyperoside-MAPK1), and −4.15 kcal/mol (hyperoside-EGFR). It can be observed that, apart from hyperoside with EGFR, all other compounds exhibit strong binding affinity with key targets and central genes. Notably, 5,7,8-trimethoxyflavone exhibited the highest affinity for EGFR, with a binding energy of −6.8 kcal/mol. These findings suggest that these two key constituents in HXF may exert anti-RIPF effects by targeting the PI3K-Akt, MAPK, and EGFR pathways.
 |
Table 3 The Docking Scores |
Experimental Validation of the HXF Action Pathway
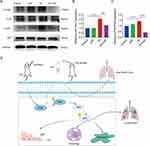
Through network pharmacology, we predicted that the PI3K-Akt signaling pathway is the most critical pathway through which HXF exerts its effects in the prevention and treatment of RIPF. Subsequently, Western blot experiments confirmed the expression of relevant proteins in the PI3K-Akt signaling pathway. As shown in Figure 5A–C, the expression levels of p-Akt and p-PI3K proteins were significantly higher in the IR group compared to the control group (p < 0.0001). After HXF intervention, the expression levels of p-Akt and p-PI3K proteins in the mice were significantly reduced compared to the IR group (p < 0.0001). These results indicate that HXF can modulate the PI3K-Akt signaling pathway. HXF may exert its anti-RIPF effect by inhibiting the activation of the PI3K-Akt signaling pathway during the progression of RIPF (Figure 5D).
Discussion
In recent years, traditional Chinese medicine natural products have emerged as promising candidates for managing RIPF. Yangyinqingfei decoction, for instance, shows potential in alleviating radiation-induced pulmonary fibrosis by targeting MMP-12 and TIMP-1.41 Similarly, rosmarinic acid exhibits inhibitory effects on RIPF by modulating NF-κB phosphorylation levels.42 Notably, the culinary spice Amomum subulatum has demonstrated anti-inflammatory and antioxidant properties, effectively impeding the progression of RILI.43 Research across traditional Chinese medicine formulas, natural product monomers, and dietary interventions supports their inhibitory effects on radiation-induced lung injury. Thus, exploring the active components and specific mechanisms of the traditional Chinese medicine formula HXF for RIPF prevention and treatment warrants investigation in this study.
In this study, the results of Micro-CT scans at different time points in animal experiments indicate that HXF effectively inhibits the increase in lung density and mitigates lung damage post-irradiation. Additionally, pathological experiments show that HXF significantly improves lung tissue inflammation, reduces the degree of fibrosis, inhibits the expression of α-SMA protein, and alleviates lung damage in mice after irradiation. Based on these findings, we further investigated the potential molecular mechanisms underlying the protective effects of HXF. Using LC-MS, we identified 21 common prototype components of HXF in mouse serum. By integrating RIPF-related genes, we identified 127 targets primarily involved in the MAPK, PI3K-Akt, and EGFR signaling pathways. Component-target network topology analysis revealed that 5,7,8-trimethoxyflavone and hyperoside are the main active compounds in HXF. Molecular docking results also confirmed the strong affinity of these active compounds with the central targets of the relevant pathways.
The MAPK signaling pathway involved in KEGG analysis can be activated by ROS generated by ionizing radiation, inducing EMT and inflammation, thereby promoting pulmonary fibrosis.44 In the progression of RIPF, the accumulation of epithelial-mesenchymal transition (EMT) is a direct contributor to lung tissue fibrosis.45 Quercetin can exert radioprotective effects by inhibiting the MAPK pathway. According to reports, there is a significant upregulation of EGFR expression in the lung epithelium of patients with various forms of pulmonary fibrosis, driving the development of pulmonary fibrosis.46 Studies have shown that EGFR inhibitors can alleviate RIPF by inhibiting the proliferation of lung fibroblasts and have been suggested as a recommended method to counteract and mitigate RIPF.47
Excluding cancer-related pathways, the most significantly enriched pathway in the KEGG enrichment analysis is the PI3K-Akt signaling pathway. The PI3K-Akt pathway is a critical pathway for promoting metabolism, cell proliferation, survival, growth, and angiogenesis, making it a promising target for interventions in various human diseases such as tumors, neurodegenerative diseases, pulmonary fibrosis, and diabetes.48 The activation of the PI3K-Akt signaling pathway plays a key role in EMT.49 AKT, as the most crucial downstream effector of PI3K, shows significantly elevated phosphorylation levels after irradiation. Myrtol can mitigate the progression of RIPF by reducing phosphorylated AKT expression, inhibiting vimentin and α-SMA levels, and decreasing collagen deposition.50 Additionally, TBK1, a potential therapeutic target for RIPF, can directly inhibit the AKT signaling pathway by targeting TBK1, thereby inhibiting EMT in alveolar type II epithelial cells.51 In mice and A549 cells, the deficiency of DEC1 protein can inhibit the PI3K-Akt signaling pathway, playing a key role in relieving EMT development and thereby ameliorating bleomycin-induced pulmonary fibrosis.52 Diseases characterized by excessive cell proliferation, such as pulmonary fibrosis, can be attenuated by autophagy.53 The activation of mTOR, a downstream target of the PI3K-AKT signaling pathway, can inhibit autophagy through the Atg1-Atg3 complex, leading to organ fibrosis and the development of pulmonary fibrosis.54 Additionally, aerobic glycolysis plays a crucial role in pulmonary fibrosis.55 Lipopolysaccharide mediates glycolysis in lung fibroblasts through the activation of the PI3K-Akt-mTOR/PFKFB3 pathway, promoting collagen synthesis and thus contributing to pulmonary fibrosis.56 Therefore, the overactivation of the PI3K-Akt signaling pathway can induce EMT, inhibit autophagy, and participate in glycolysis, leading to the accumulation of fibroblasts, deposition of collagen and extracellular matrix, and promotion of pulmonary fibrosis formation. In our in vivo experiments, we validated the PI3K-Akt signaling pathway, exhibiting the most robust correlation with RIPF. The findings demonstrated significant downregulation of p-Akt and p-PI3K protein expression levels in lung tissues of irradiated mice after HXF treatment, suggesting that this pathway may play a pivotal role in HXF’s inhibition of RIPF progression.
Both 5,7,8-trimethoxyflavone and hyperoside, flavonoids commonly found in natural plants, exhibit diverse pharmacological properties such as anti-inflammatory, antioxidant, anti-tumor, radioprotective, and anti-fibrotic effects.57 Inflammation is implicated in the pathogenesis of RIPF.58 5,7,8-Trimethoxyflavone demonstrates significant anti-inflammatory activity by NO production inhibition.59 Sinensetin, akin to 5,7,8-trimethoxyflavone, hinders pulmonary fibrosis by suppressing the Wnt/β-Catenin pathway, reducing myofibroblast and ECM production.60 Hyperoside showcases pharmacological activities countering pulmonary fibrosis development, including anti-inflammatory, EMT reversal, autophagy regulation, and antioxidant effects.61–64 The potential active compounds in HXF, 5,7,8-trimethoxyflavone and hypericin, identified in our study, necessitate further exploration of their combined or individual effects on RIPF inhibition. Our forthcoming research will delve into elucidating their specific mechanisms of action.
Building on previous studies exploring the progression of RIPF, it has been demonstrated that a 17 Gy irradiation dose induces the progressive loss of pulmonary epithelial cells, significant upregulation of EMT-related gene expression, and increased secretion of fibrotic factors such as TGF-β and Mmps—important indicators of post-irradiation pulmonary fibrosis.19,20 A 17 Gy dose is thus considered sufficient to cause irreversible fibrotic damage to the lungs. In this study, we irradiated the mice with a 17 Gy dose, and the results revealed typical RIPF lesions in the mice’s lungs four months post-irradiation. The results showed that the mice’s lungs exhibited typical RIPF lesions four months after irradiation. RIPF is characterized by its progressive and irreversible nature. Studies have shown that following total chest irradiation of 12.5 Gy, RIPF initiates around 12 weeks post-irradiation in mice and advances until mortality.65 A previous study noted a significant rise in collagen and α-SMA levels in mouse lung tissue four months post-irradiation.66 Similarly, at this time point, increases in alveolar density, septal thickness, collagen deposition, and Slug expression, a key EMT-associated transcription factor, were observed.67 Another study reported a notable increase in M2 macrophages and their secretion of fibrosis-promoting factors, including TGF-β1, IL-10, and Arginase-1 in mice four months post-irradiation.68 Collectively, these results support the successful establishment of the RIPF mouse model at four months post-irradiation. In our study, we monitored experimental mice for four months, during which the model group exhibited typical fibrotic manifestations including heightened lung density, alveolar atrophy, thickened alveolar walls, extensive collagen deposition, and a significant increase in α-SMA protein levels. Given the progressive nature of RIPF, we plan to extend the observation period to 6 months in future studies to more comprehensively assess the experimental effects.
In this study, we identified the absorbed components of HXF in the blood using LC-MS, explored its mechanism of action through network pharmacology and molecular docking, and validated these finding with animal experiments. However, this study has some limitations. Our findings suggest that 5,7,8-trimethoxyflavone and hyperoside hold promise as active compounds with potential RIPF-inhibiting properties, laying a foundational groundwork for the utilization of HXF in RIPF therapeutics. Nevertheless, our study currently lacks animal and cell experiments to substantiate the effects and mechanisms of these identified active compounds within HXF. Therefore, future investigations are essential to validate the anti-RIPF attributes of these compounds comprehensively. We will further investigate the downstream mechanisms involved in the relevant pathways, such as EMT, inflammation, autophagy, and glycolysis, to explore the specific mechanisms by which potential key active components in HXF inhibit RIPF. Additionally, we monitored the mice for 4 months in this study. Given the progressive nature of RIPF, we plan to extend the observation period to 5–6 months in future studies to more comprehensively assess the experimental effects. Through network pharmacology, we have identified potential pathways for RIPF prevention and treatment by HXF, based on targets available in public databases. Future research will adopt a more comprehensive approach, encompassing established RIPF-associated genes and exploring novel mechanistic targets. This broader perspective aims to reveal new associations and mechanisms, enhancing our understanding of RIPF pathogenesis and HXF’s therapeutic effects.
Conclusions
Through serum pharmacochemistry, network pharmacology, and molecular docking analyses, this study pinpointed the potential targets of HXF in RIPF inhibition-AKT1, MAPK1, and EGFR-along with the active compounds 5,7,8-trimethoxyflavone and hyperoside. Experimental validation indicates that HXF potentially hinders RIPF progression by suppressing the critical PI3K-Akt signaling pathway. These findings bolster the theoretical underpinnings for the clinical utility of HXF in RIPF management.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
Ethical approval to conduct studies using publicly available databases is exempt under the following legislation: Item 1 and 2 of Article 32 of “the Measures for Ethical Review of Life Science and Medical Research Involving Human Subjects”, which was reviewed by the National Science and Technology Ethics Committee, approved by the State Council of China, and jointly promulgated by the National Health Commission, the Ministry of Education, the Ministry of Science and Technology and the State Administration of Traditional Chinese Medicine on Feb. 18, 2023. The animal experiments were approved by the Ethics Committee for Medical Research and New Medical Technology of Sichuan Cancer Hospital (SCCHEC-04-2023-023).
Funding
This work was supported by the Chengdu Technology Bureau (grant number 2024-YF05-02494-SN) and Sichuan Medical Association (grant number 2024HR15).
Disclosure
The authors have no financial conflict of interest.
References
1. Wang K, Tepper JE. Radiation therapy‐associated toxicity: etiology, management, and prevention. CA Cancer J Clin. 2021;71(5):437–454. doi:10.3322/caac.21689
2. Cytlak UM, Dyer DP, Honeychurch J, et al. Immunomodulation by radiotherapy in tumour control and normal tissue toxicity. Nat Rev Immunol. 2022;22(2):124–138. doi:10.1038/s41577-021-00568-1
3. Bucknell NW, Belderbos J, Palma DA, et al. Avoiding toxicity with lung radiation therapy: an IASLC perspective. J Thorac Oncol. 2022;17(8):961–973. doi:10.1016/j.jtho.2022.05.003
4. Wang M, Feng Y, Zhang P, et al. Jiawei Maxing Shigan Tang alleviates radiation-induced lung injury via TGF-beta1/Smad signaling pathway mediated by regulatory T cells. J Ethnopharmacol. 2024;320:117389. doi:10.1016/j.jep.2023.117389
5. Kim KI, Jun JH, Baek H, et al. Oral administration of herbal medicines for radiation pneumonitis in lung cancer patients: a systematic review and meta-analysis. PLoS One. 2018;13(5):e0198015. doi:10.1371/journal.pone.0198015
6. He H, Zhou X, Wang Q, et al. Does the couse of astragalus‐containing Chinese herbal prescriptions and radiotherapy benefit to non‐small‐cell lung cancer treatment: a meta‐analysis of randomized trials. Evid-Based Complementary Altern Med. 2013;2013(1):426207. doi:10.1155/2013/426207
7. Chen J, Zou P, Quan L, et al. Huaxian formula prevents the progression of radiation-induced pulmonary fibrosis by inhibiting the pro-fibrotic effects of macrophages. J Ethnopharmacol. 2025;338(Pt 2):119026. doi:10.1016/j.jep.2024.119026
8. Zhang X, Chen X, Wang L, et al. Review of the efficacy and mechanisms of traditional Chinese medicines as a therapeutic option for ionizing radiation induced damage. Front Pharmacol. 2021;15(12):617559. doi:10.3389/fphar.2021.617559
9. Ma C, Fu Z, Guo H, et al. The effects of Radix Angelica Sinensis and Radix Hedysari ultrafiltration extract on X-irradiation-induced myocardial fibrosis in rats. Biomed Pharmacother. 2019;112:108596. doi:10.1016/j.biopha.2019.01.057
10. Chen J, Wang C, Pan X, et al. Glycyrrhetinic acid mitigates radiation-induced pulmonary fibrosis via inhibiting the secretion of TGF-β1 by treg cells. Int J Radiat Oncol Biol Phys. 2024;118(1):218–230. doi:10.1016/j.ijrobp.2023.08.005
11. Yeh YC, Yang CP, Lee SS, et al. Acute lung injury induced by lipopolysaccharide is inhibited by wogonin in mice via reduction of Akt phosphorylation and RhoA activation. J Pharm Pharmacol. 2016;68(2):257–263. doi:10.1111/jphp.12500
12. Yimam M, Horm T, O’Neal A, et al. UP360, a standardized composition from extracts of Aloe barbadense, Poria cocos, and Rosemary officinalis protected against sepsis and mitigated acute lung injury in murine models. J Med Food. 2023;26(7):489–499. doi:10.1089/jmf.2022.0136
13. Yao L, Sun T. Glycyrrhizin administration ameliorates Streptococcus aureus-induced acute lung injury. Int Immunopharmacol. 2019;70:504–511. doi:10.1016/j.intimp.2019.02.046
14. Ren JL, Dong H, Han Y, et al. Network pharmacology combined with metabolomics approach to investigate the protective role and detoxification mechanism of Yunnan Baiyao formulation. Phytomedicine. 2020;77:153266. doi:10.1016/j.phymed.2020.153266
15. Liu CS, Xia T, Luo ZY, et al. Network pharmacology and pharmacokinetics integrated strategy to investigate the pharmacological mechanism of Xianglian pill on ulcerative colitis. Phytomedicine. 2021;82:153458. doi:10.1016/j.phymed.2020.153458
16. Zhao L, Zhang H, Li N, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J Ethnopharmacol. 2023;309:116306. doi:10.1016/j.jep.2023.116306
17. Jiao X, Jin X, Ma Y, et al. A comprehensive application: molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Comput Biol Chem. 2021;90:107402. doi:10.1016/j.compbiolchem.2020.107402
18. Travis EL, Rachakonda G, Zhou X, et al. NRF2 deficiency reduces life span of mice administered thoracic irradiation. Free Radic Biol Med. 2011;51(6):1175–1183. doi:10.1016/j.freeradbiomed.2011.05.038
19. Curras-Alonso S, Soulier J, Defard T, et al. An interactive murine single-cell atlas of the lung responses to radiation injury. Nat Commun. 2023;14(1):2445. doi:10.1038/s41467-023-38134-z
20. Su L, Dong Y, Wang Y, et al. Potential role of senescent macrophages in radiation-induced pulmonary fibrosis. Cell Death Dis. 2021;12(6):527. doi:10.1038/s41419-021-03811-8
21. Szapiel SV, Elson NA, Fulmer JD, et al. Bleomycin-induced interstitial pulmonary disease in the nude, athymic mouse. Am Rev Respir Dis. 1979;120(4):893–899. doi:10.1164/arrd.1979.120.4.893
22. Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol. 1988;41(4):467–470. doi:10.1136/jcp.41.4.467
23. Gattinoni L, Caironi P, Pelosi P, et al. What has computed tomography taught us about the acute respiratory distress syndrome? Am J Respir Crit Care Med. 2001;164(9):1701–1711. doi:10.1164/ajrccm.164.9.2103121
24. Wong YL, LeBon L, Basso AM, et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife. 2019;8:e42940. doi:10.7554/eLife.42940
25. Paschalis A, Sheehan B, Riisnaes R, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol. 2019;76(4):469–478. doi:10.1016/j.eururo.2019.06.030
26. Jin Y, Cheng S, Liu R, et al. Comprehensive characterization of the chemical composition of Lurong dabu decoction and its absorbed prototypes and metabolites in rat plasma using UHPLC–Q Exactive Orbitrap–HRMS. Food Res Int. 2022;161:111852. doi:10.1016/j.foodres.2022.111852
27. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi:10.1186/1758-2946-6-13
28. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
29. Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38(Web Server issue):W609–614. doi:10.1093/nar/gkq300
30. Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13(4):163. doi:10.1016/s0168-9525(97)01103-7
31. Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: online Mendelian inheritance in man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–798. doi:10.1093/nar/gku1205
32. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
33. Lyu F, Han F, Ge C, et al. OmicStudio: a composable bioinformatics cloud platform with real‐time feedback that can generate high‐quality graphs for publication. Imeta. 2023;2(1):e85. doi:10.1002/imt2.85
34. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45(D1):D362–D368. doi:10.1093/nar/gkw937
35. Missiuro PV, Liu K, Zou L, et al. Information flow analysis of interactome networks. PLoS Comput Biol. 2009;5(4):e1000350. doi:10.1371/journal.pcbi.1000350
36. O’Boyle NM, Banck M, James CA, et al. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi:10.1186/1758-2946-3-33
37. Österberg F, Morris GM, Sanner MF, et al. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46(1):34–40. doi:10.1002/prot.10028
38. Lill MA, Danielson ML. Computer-aided drug design platform using PyMOL. J Comput Aided Mol Des. 2011;25(1):13–19. doi:10.1007/s10822-010-9395-8
39. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
40. Wang Y, Zhang Y, Wang Y, et al. Using network pharmacology and molecular docking to explore the mechanism of Shan Ci Gu (Cremastra appendiculata) against non-small cell lung cancer. Front Chem. 2021;9:682862. doi:10.3389/fchem.2021.682862
41. Li H, Wu H, Gao Y, et al. Effect of Yangyinqingfei decoction on radiation-induced lung injury via downregulation of MMP12 and TIMP-1 expression. Exp Ther Med. 2014;8(1):9–14. doi:10.3892/etm.2014.1686
42. Zhang T, Mi J, Qin X, et al. Rosmarinic acid alleviates radiation-induced pulmonary fibrosis by downregulating the tRNA N7-methylguanosine modification-regulated fibroblast-to-myofibroblast transition through the exosome pathway. J Inflamm Res. 2024;17:5567–5586. doi:10.2147/jir.S458794
43. Drishya S, Dhanisha SS, Raghukumar P, et al. Amomum subulatum mitigates experimental thoracic radiation-induced lung injury by regulating antioxidant status and inflammatory responses. Food Funct. 2023;14(3):1545–1559. doi:10.1039/d2fo03208b
44. Lee SY, Jeong EK, Ju MK, et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16(1):10. doi:10.1186/s12943-016-0577-4
45. Gai X, Tang B, Liu F, et al. mTOR/miR-145-regulated exosomal GOLM1 promotes hepatocellular carcinoma through augmented GSK-3β/MMPs. J Genet Genomics. 2019;46(5):235–245. doi:10.1016/j.jgg.2019.03.013
46. Tzouvelekis A, Ntolios P, Karameris A, et al. Increased expression of epidermal growth factor receptor (EGF‐R) in patients with different forms of lung fibrosis. Biomed Res Int. 2013;2013(1):654354. doi:10.1155/2013/654354
47. Miyake K, Tani K, Kakiuchi S, et al. Epidermal growth factor receptor-tyrosine kinase inhibitor (gefitinib) augments pneumonitis, but attenuates lung fibrosis in response to radiation injury in rats. J Med Invest. 2012;59(1–2):174–185. doi:10.2152/jmi.59.174
48. Huang J, Chen L, Wu J, et al. Targeting the PI3K/AKT/mTOR signaling pathway in the treatment of human diseases: current status, trends, and solutions. J Med Chem. 2022;65(24):16033–16061. doi:10.1021/acs.jmedchem.2c01070
49. Wang J, Hu K, Cai X, et al. Targeting PI3K/AKT signaling for treatment of idiopathic pulmonary fibrosis. Acta Pharm Sin B. 2022;12(1):18–32. doi:10.1016/j.apsb.2021.07.023
50. Zhao DY, Qu HJ, Guo JM, et al. Protective effects of myrtol standardized against radiation-induced lung injury. Cell Physiol Biochem. 2016;38(2):619–634. doi:10.1159/000438655
51. Qu H, Liu L, Liu Z, et al. Blocking TBK1 alleviated radiation-induced pulmonary fibrosis and epithelial-mesenchymal transition through Akt-Erk inactivation. Exp Mol Med. 2019;51(4):1–17. doi:10.1038/s12276-019-0240-4
52. Hu X, Zou M, Ni L, et al. Dec1 deficiency ameliorates pulmonary fibrosis through the PI3K/AKT/GSK-3β/β-Catenin integrated signaling pathway. Front Pharmacol. 2022;13:829673. doi:10.3389/fphar.2022.829673
53. Haspel JA, Choi AM. Autophagy: a core cellular process with emerging links to pulmonary disease. Am J Respir Crit Care Med. 2011;184(11):1237–1246. doi:10.1164/rccm.201106-0966CI
54. Hsu HS, Liu CC, Lin JH, et al. Involvement of ER stress, PI3K/AKT activation, and lung fibroblast proliferation in bleomycin-induced pulmonary fibrosis. Sci Rep. 2017;7(1):14272. doi:10.1038/s41598-017-14612-5
55. Kottmann RM, Kulkarni AA, Smolnycki KA, et al. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-β. Am J Respir Crit Care Med. 2012;186(8):740–751. doi:10.1164/rccm.201201-0084OC
56. Hu X, Xu Q, Wan H, et al. PI3K-Akt-mTOR/PFKFB3 pathway mediated lung fibroblast aerobic glycolysis and collagen synthesis in lipopolysaccharide-induced pulmonary fibrosis. Lab Invest. 2020;100(6):801–811. doi:10.1038/s41374-020-0404-9
57. Lee JH, Ahn J, Kim JW, et al. Flavonoids from the aerial parts of Houttuynia cordata attenuate lung inflammation in mice. Arch Pharm Res. 2015;38(7):1304–1311. doi:10.1007/s12272-015-0585-8
58. Wang B, Wei J, Meng L, et al. Advances in pathogenic mechanisms and management of radiation-induced fibrosis. Biomed Pharmacother. 2020;121:109560. doi:10.1016/j.biopha.2019.109560
59. Shen DY, Juang SH, Kuo PC, et al. Chemical constituents from Andrographis echioides and their anti-inflammatory activity. Int J Mol Sci. 2012;14(1):496–514. doi:10.3390/ijms14010496
60. Wan X, Chen S, Li P, et al. Sinensetin protects against pulmonary fibrosis via inhibiting Wnt/β-Catenin signaling pathway. Tissue Cell. 2022;78:101866. doi:10.1016/j.tice.2022.101866
61. Huang J, Tong X, Zhang L, et al. Hyperoside attenuates bleomycin-induced pulmonary fibrosis development in mice. Front Pharmacol. 2020;11:550955. doi:10.3389/fphar.2020.550955
62. Gao Z, Xu M, Liu C, et al. Structural modification and optimisation of hyperoside oriented to inhibit TGF-β-induced EMT activity in alveolar epithelial cells. Pharmaceuticals. 2024;17(5):584. doi:10.3390/ph17050584
63. Gao Y, Fan X, Gu W, et al. Hyperoside relieves particulate matter-induced lung injury by inhibiting AMPK/mTOR-mediated autophagy deregulation. Pharmacol Res. 2021;167:105561. doi:10.1016/j.phrs.2021.105561
64. Piao MJ, Kang KA, Zhang R, et al. Hyperoside prevents oxidative damage induced by hydrogen peroxide in lung fibroblast cells via an antioxidant effect. Biochim Biophys Acta. 2008;1780(12):1448–1457. doi:10.1016/j.bbagen.2008.07.012
65. Travis EL. The sequence of histological changes in mouse lungs after single doses of x-rays. Int J Radiat Oncol Biol Phys. 1980;6(3):345–347. doi:10.1016/0360-3016(80)90145-5
66. Wang D, Liu Z, Yan Z, et al. MiRNA-155-5p inhibits epithelium-to-mesenchymal transition (EMT) by targeting GSK-3β during radiation-induced pulmonary fibrosis. Arch Biochem Biophys. 2021;697:108699. doi:10.1016/j.abb.2020.108699
67. Liang X, Yan Z, Wang P, et al. Irradiation activates MZF1 to inhibit miR-541-5p expression and promote Epithelial-Mesenchymal Transition (EMT) in Radiation-Induced Pulmonary Fibrosis (RIPF) by upregulating slug. Int J Mol Sci. 2021;22(21):11309. doi:10.3390/ijms222111309
68. Shen L, Fu S, Chen Y, et al. Mannosylated polydopamine nanoparticles alleviate radiation- induced pulmonary fibrosis by targeting M2 macrophages and inhibiting the TGF-β1/Smad3 signaling pathway. Colloids Surf B Biointerfaces. 2023;227:113353. doi:10.1016/j.colsurfb.2023.113353
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.