Back to Journals » Journal of Inflammation Research » Volume 18
Shared Genes and Pathways in Ulcerative Colitis and Ankylosing Spondylitis: Functional Validation and Implications for Diagnosis
Authors Li L , An G, Li F , Zhang D, Zhu X, Liang C, Zhao Y, Xie K, Zhou P, Zhu H, Jin X, Du L
Received 15 October 2024
Accepted for publication 24 January 2025
Published 4 February 2025 Volume 2025:18 Pages 1657—1678
DOI https://doi.org/10.2147/JIR.S497201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Lin Li,* Guangqi An,* Fuzhen Li,* Donghui Zhang, Xinyue Zhu, Chunyu Liang, Yu Zhao, Kunpeng Xie, Pengyi Zhou, Haiyan Zhu, Xuemin Jin,* Liping Du*
Department of Ophthalmology, The First Affiliated Hospital of Zhengzhou University, Henan Province Eye Hospital, Henan International Joint Research Laboratory for Ocular Immunology and Retinal Injury Repair, Zhengzhou, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xuemin Jin; Liping Du, Department of Ophthalmology, The First Affiliated Hospital of Zhengzhou University, No. 1 Jianshe East Road, Zhengzhou, 450000, People’s Republic of China, Tel +8613603983193 ; +8613838191481, Email [email protected]; [email protected]
Background: Associations between ulcerative colitis (UC) and ankylosing spondylitis (AS) have been reported in multiple studies, but the common etiologies of UC and AS remain unknown. Thus, in the current study, we aimed to investigate the shared genes and relevant mechanisms in UC and AS.
Methods: Using datasets for UC (GSE113079) and AS (GSE1797879), we initially identified differentially expressed genes (DEGs) through differential expression analysis. The DEGs from both datasets were intersected to identify common DEGs, relevant to both UC and AS, which were used in receiver operating characteristic (ROC) curve analysis to confirm key genes in the shared pathway. Gene set enrichment analysis (GSEA) was used to obtain information on key gene pathways and interactions with UC or AS-related diseases, followed by immune infiltration analysis. Finally, peripheral blood samples of AS and UC were used to verify the mRNA expression of the eight key genes using reverse transcription-polymerase chain reaction (RT-PCR).
Results: Our results revealed that GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, S100A12 and DGKQ are potential diagnostic biomarkers of AS and UC. Rimegepant, eptinezumab, methotrexate, atogepant, and ubrogepant were identified as potential drugs for S100A12 and S100A8 in patients with UC and AS. GSEA showed that these key genes were associated with antigen processing and presentation, natural killer cell mediated cytotoxicity and the T cell receptor signaling pathway in AS and UC, and were significantly associated with immune cells in various immune-related pathways. Subsequent functional experiments revealed significant increases in the mRNA expressions of S100A12 and VAMP5 in patients with AS and UC. Additionally, CLEC4D mRNA expression was notably higher in patients with UC than in healthy controls.
Conclusion: Key genes and shared pathways were identified in UC and AS, which may improve understanding of their relationship and guide diagnosis and treatment strategies.
Keywords: ulcerative colitis, ankylosing spondylitis, diagnosis, etiology
Background
Ulcerative colitis (UC), classified as an inflammatory bowel disease (IBD), affects the digestive system and causes persistent inflammation within the inner lining of the large intestine.1 In recent years, many studies have been conducted to gain further insights and understandings into and understand this debilitating condition. UC symptoms vary from person to person, but common signs and symptoms include diarrhea, rectal bleeding, abdominal cramps and pain, unintended weight loss, and anemia.2 In addition, some people with UC experience joint pain, skin rashes, and eye irritation.3 Between 20 and 250 cases of UC are believed to occur in every 100,000 people, depending on the population and geographic region. Several studies have revealed that the prevalence of UC has doubled in the last two decades, indicating that the condition is becoming more common worldwide.3 The pathogenesis of UC involves a complex interplay of multiple factors, including microbial infections, abnormal immune responses, endocrine disorders, and genetic susceptibility.2,4 The treatment of UC primarily focuses on reducing inflammation, relieving symptoms, and maintaining remission.5 However, due to the lack of a comprehensive understanding of the etiology of UC, existing therapies often fail to fully meet treatment goals, resulting in limited therapeutic efficacy.5 Therefore, in-depth exploring the molecular mechanisms of acute flare-ups and recurrences in UC and identifying potential targets for treatment sensitivity is of great significance for better understanding the etiology and symptoms of UC, developing more effective treatment strategies, and improving patient prognosis.
Ankylosing spondylitis (AS) is a type of chronic and inflammatory arthritis that mainly targets the spine and sacroiliac joints.6 The condition is distinguished by progressive stiffness and fusion of the spine, which causes severe back pain, limited mobility, and disability.7 Other associated symptoms include fever, weight loss, joint swelling, and eye inflammation. Studies have shown that the prevalence of AS is 0.1–0.2% in the general population, with a greater frequency in specific populations.8 AS is a complex disease that significantly impacts the quality of life of patients. Although genetic and environmental factors may play a role, the exact etiology remains unclear, making it difficult to develop preventive measures and fundamental treatment strategies targeting the underlying causes.8 Currently, the management of this disease is confined to symptomatic treatment, which aims to alleviate pain and stiffness through pharmacological interventions, physical activity, and lifestyle modifications. However, the therapeutic outcomes are highly individualized, and these approaches fail to address the underlying pathophysiological mechanisms of the disease.9 Therefore, further research is required to better understand the pathogenesis of AS and to develop strategies to improve its management.
Both AS and UC involve abnormal immune system responses in their pathogenesis. At the molecular level, cross-linking studies of UC and AS have focused primarily on common inflammatory pathways and cytokine networks. Tumor necrosis factor-alpha (TNF-α) is a significant inflammatory mediator in both diseases. Additionally, the IL-23/IL-17 axis plays a crucial role in the pathogenesis of both UC and AS.10,11 Genetic studies have indicated that HLA-B27 is highly associated not only with AS but also with an increased risk of IBD, suggesting a genetic overlap in susceptibility between these two diseases. Several studies have shown a link between AS and UC. Indeed, 5–10% of patients with AS are also diagnosed with IBD, implying that AS may be an independent risk factor for IBD.12 Furthermore, UC can often lead to sacroiliac arthritis, which is characterized by lumbosacral pain and limited mobility. The prevalence of sacroiliac arthritis in individuals with UC is estimated to be around 20%.13 The symptoms of sacroiliac arthritis may improve or disappear along with UC. However, when UC is combined with AS, patients usually experience severe joint symptoms, and inadequate treatment may lead to deformity and spinal rigidity.14 In summary, UC and AS are interconnected through immune dysregulation, cytokine networks, genetic factors, and inflammatory pathways. Genetic research has revealed a shared susceptibility between the two, contributing to a more complex clinical presentation when both conditions coexist. Due to the diverse and overlapping clinical symptoms that complicate diagnosis, investigating shared biomarkers and mechanisms is of critical importance.
This study hypothesizes that there may be similar pathogenic mechanisms between UC and AS, particularly in terms of immune responses and inflammatory pathways. Exploring the common pathogenesis of AS and UC and identifying shared biomarkers are crucial for improving diagnostic accuracy, enhancing patient prognosis, and developing new treatment strategies for these two diseases. This approach will help establish the basis for a more comprehensive understanding of the connection between AS and UC.
Methods
Data Source
The flow of the experimental elements of this study can be seen in Figure S1. In this study, the gene expression profiles of 21 control and 87 UC samples in the GSE87466 dataset based on the GPL13158 platform were derived from data obtained from mRNA microarray studies performed on mucosal biopsy samples. Additionally, 16 control and 16 AS samples in the GSE25101 dataset based on the GPL6947 platform were acquired from the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and used as training sets. Finally, the gene expression profiles of 22 control and 25 UC samples from the GSE94648 dataset and 20 control and 52 AS samples from the GSE73754 dataset were obtained and used as external validation sets. These are genome-wide transcriptional analysis data from peripheral whole blood (PAXgene tubes) involving patients with endoscopically active and inactive UC and non-inflammatory controls. The datasets were all derived from publicly accessible microarray databases and web-based data mining platforms. By checking the upper and lower limits of expression of the expression spectra with the max(probes_expr) and min(probes_expr) functions, as well as plotting the dataset’s expression box line graphs, it is possible to understand that the data for the expression spectra of both datasets are standardized and have good inter-sample consistency (Figure S2).
Identification and Functional Enrichment Analysis of Differentially Expressed Genes (DEGs)
To identify the DEGs between the different sample groups, and further explore the functions of these genes, the limma R package was employed to detect DEGs between control and UC subjects in GSE87466 and DEGs between control and AS samples in GSE25101 with |log2FC| >0.5 and adjusted p-value <0.05 for Standardized processing. The findings were displayed as volcano plots and heatmaps generated by ggplot (version 3.3.5) and the pheatmap R package (version 1.0.12), respectively. The clusterProfiler R package (version 4.0.5) was used for enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in DEGs. In the GO analysis, biological process (BP), cellular component (CC) and molecular function (MF) were included. A p-value <0.05 was significantly enriched. By overlapping DEGs in GSE87466 and GSE25101, common DEGs involved in both UC and AS were identified and used for downstream analysis.
Comprehensive Analysis of DEGs, miRNAs, Transcription Factors, and Potential Drug Targets
To identify potential diagnostic biomarkers, ROC curves were generated to evaluate the performance of common DEGs in classifying AS or UC samples from control samples. Common DEGs with an AUC ≥0.8 were defined as potential diagnostic biomarkers for AS or UC. Then, we overlapped the diagnostic biomarkers for AS and UC and identified them as key genes connecting these two conditions. The miRNAs of key genes were predicted through the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/), with the score value set to 1. Subsequently, the transcription factors (TFs) of key genes were predicted in the Cistrome database (http://cistrome.org/), with the threshold set as RP_score > 0.6. The TF-miRNA relationship pairs were predicted in the TransmiR database (http://www.cuilab.cn/transmir). Then, the miRNA-mRNA, mRNA-TF and miRNA-TF-mRNA regulatory networks were drawn by using Cytoscape software (version 3.9.1).15 By integrating information from different databases, the network of interactions between miRNAs, TFs, and key genes in the context of UC and AS was revealed, aiming to understand the complex structure of the regulatory network and its role in the disease process. Additionally, potential drugs interacting with key genes were predicted using the DGIdb database (https://www.dgidb.org/). This analysis was conducted to forecast drug-gene interactions, evaluate the therapeutic potential of these drugs as targets, and provide a scientific foundation for the development and clinical trials of novel treatments.
Integrative Analysis of Gene Expression Profiles and Publicly Available Data
Genes in GSE87466 and GSE25101 were ranked according to the correlations with each key gene, and GSEA was performed to detect the pathways that those genes were significantly enriched into. KEGG pathway gene sets were obtained from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb) and were used as reference gene sets. AS and UC related pathways were also downloaded from the CTD database (http://ctdbase.org/) using the keywords “ulcerative colitis” and “ankylosing spondylitis”. By intersecting the pathways identified by GSEA and CTD, key gene-related pathways involved in UC and AS were identified. Meanwhile, using the CTD database, we also investigated the interaction of each key gene with diseases associated with UC or AS.
Exploring Immune Cell Variations and Biomarker Relationships in Patients with UC and AS
In contrast to GO and KEGG enrichment analyses, which are based on all differential genes while ignoring upper downregulated expression by super geometric distribution calculated enrichment, the GSEA basic method employs pre-defined gene sets to determine whether the genes in the gene set are randomly distributed or clustered at the top or bottom of the gene set. To further explore the possible signaling pathways and biological functions of key genes in UC and AS, we used single-gene GSEA to calculate each target gene separately in AS versus UC. Correlation of all genes and rank all genes from high to low, ranking the sorted base. The KEGG signaling pathway was used as a pre-defined gene set to detect the enrichment of the KEGG signaling pathway in the gene sets (GSE87466 and GSE25101). The proportions of 28 immune cells between the control and UC subjects in GSE87466 and between the control and AS samples in GSE25101were calculated by using the “single sample gene set enrichment analysis (ssGSEA)” algorithm and compared using the “Wilcoxon” test. Moreover, the correlations between biomarkers and immune cells were further studied using Spearman correlation.
Differential Expression of Key Genes in Peripheral Blood Mononuclear Cells (PBMCs)
In this study, a total of 12 healthy individuals (6 male, 6 female, mean age=29.83± 7.17), 7 patients (5 male, 2 female, mean age=32.14±4.14) diagnosed with active AS, and 12 patients (7 male, 5 female, mean age=31.25 ± 12.25) diagnosed with active UC were included (Table 1). AS was diagnosed using the 1984 New York Diagnostic Criteria. Whereas the diagnosis of UC was based on clinical, endoscopic, radiological, and histological criteria. The research was conducted in accordance with the principles outlined in the Declaration of Helsinki, and ethical approvals were obtained from the research committees of Zhengzhou University (2021-KY-0246-001). Blood was drawn from participants to obtain PBMCs, which were subsequently used for RNA extraction. All participants provided written informed consent to take part in the study. RT-PCR was performed to evaluate the mRNA expression of eight key genes. The primers used for RT-PCR are listed in Table 2. Beta-actin was used as an internal reference to standardize the data (forward: 5′GGATGCAGAAGGAGATCACTG3′; reverse: 5′CGATCCACACGGAGTACT3′). The 2−ΔΔCt method was used to calculate gene expression levels. Statistical analyses were performed using the SPSS 23.0 software, employing t-test for two-group comparisons. The p value was calculated, and a p value < 0.05 was considered statistically significant.
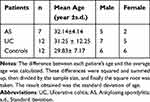 |
Table 1 Demographic Characteristics of Patients with AS, UC, and Healthy Controls |
 |
Table 2 Primers Used for mRNA Expression Analysis of GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, S100A12 and DGKQ |
Results
DEGs Involved in UC are Associated with Immune and Extracellular Matrices
The analysis conducted in GSE87466 revealed a set of 3,123 DEGs (1,749 up-regulated genes and 1,374 down-regulated genes) between controls and UC samples (Figure 1a and Table S1, see Additional file 1). The expressions of these DEGs are shown in a heatmap (Figure 1b). GO and KEGG pathway analyses were conducted to detect the biological effects of the DEGs. A total of 1,463 BPs, 76 CCs, 146 MFs (Table S2, see Additional file 2), and 67 KEGG pathways (Table S3, see Additional file 3) were significantly enriched (Table S3, see Additional file 3). As demonstrated in Figure 1c and d, DEGs exhibited significant enrichment in immune- and extracellular matrix-related biological functions, including extracellular matrix organization and positive regulation of cell adhesion in terms of BPs, external side of the plasma membrane and collagen-containing extracellular matrix in terms of CCs, and extracellular matrix structural constituent and cytokine activity in terms of MFs. The DEGs were also enriched in KEGG pathways, including cell adhesion molecules and cytokine-cytokine receptor interactions.
DEGs Involved in AS are Associated with Protein Biogenesis, Oxidative Stress and Neurodegeneration
The analysis conducted in GSE25101 revealed a set of 137 DEGs (100 up-regulated genes and 37 down-regulated genes) between the control and AS samples (Figure 1e and Table S4, see Additional file 4). The expressions of these DEGs are shown in a heatmap (Figure 1f). To detect the biological effects of the DEGs, GO and KEGG pathway analyses were conducted. In total, 80 BPs, 28 CCs, 18 MFs (Table S5, see Additional file 5), and 16 KEGG pathways (Table S6, see Additional file 6) were significantly enriched. As demonstrated in Figure 1g and h, DEGs exhibited significant enrichment in protein biogenesis-, oxidative stress-, and neurodegeneration- related biological functions, including co-translational protein targeting to the membrane and establishment of protein localization to the endoplasmic reticulum in terms of BPs, ribosomal subunit and respiratory chain complex in terms of CCs, structural constituent of ribosome, and receptors of advanced glycation end-products (RAGE) binding in terms of MFs. The DEGs were also enriched in KEGG pathways, including ribosome, Parkinson’s disease and oxidative phosphorylation.
Eight Key Common Genes in UC and AS Have Diagnostic Value
By overlapping those DEGs, 20 common DEGs were found in UC and AS (Figure 2a), including DGKQ, PPBP, GLRX, CKLF, RGS18, S100A8, TMEM158, CLEC4D, C19orf59, CMTM2, S100A12, GNG11, GMFG, LY96, TSTA3, SLPI, S100P, ANKRD22, VAMP5, and HDC, most of which were significantly upregulated in UC and AS samples (Figure 2b and c). Based on the 20 critical DEGs AU selected in the previous step, to evaluate the break efficiency of the selected patients with UC and AS, the area under the single gene curve was analyzed by constructing a ROC curve between healthy controls and patients with UC / AS (AUC). As shown in Figure 2d, our findings suggest that DGKQ, PPBP, GLRX, S100A8, TMEM158, CLEC4D, CMTM2, S100A12, GNG11, GMFG, LY96, TSTA3, SLPI, S100P, ANKRD22, and VAMP5 may serve as diagnostic markers for UC in GSE87466. As shown in Figure 2e, GMFG, GNG11, CKLF, CLEC4D, CMTM2, VAMP5, S100A8, S100A12 and DGKQ performed better, with an AUC greater than 0.8, in distinguishing AS samples in GSE25101. The intersections of those genes, including GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, S100A12, and DGKQ, were then identified as common genes in UC and AS (Figure 2f).
Construction of TF-miRNA-mRNA Regulatory Networks and Identification of Potential Therapeutics for UC and AS
The miRNAs that target common key genes were predicted using the miRWalk database, which led to the construction of miRNA-mRNA network composed of 52 nodes and 45 edges (Figure 3a). TFs regulating the expression of key genes were predicted using the Cistrome database, leading to the establishment of a TF-mRNA network comprising 167 nodes and 230 edges (Figure 3b). The TransmiR database was then used to identify relationships between TFs and miRNAs. By integrating miRNA-mRNA, TF-mRNA, and TF-miRNA pairs, a TF-miRNA-mRNA regulatory network with 212 nodes and 721 edges was created (Figure 3c). In addition, the DGIdb database was used to identify potential medicines that target S100A12 and S100A8 for the treatment of UC and AS (rimegepant, eptinezumab, methotrexate, atogepant and ubrogepant) (Figure 3d).
Key Immune Pathways Regulated by Gene Expression in UC and AS Identified via GSEA and CTD Database Analysis
GSEA was conducted to investigate the potential mechanisms underlying the regulation of UC and AS. As shown in Tables S7 and S8 (see additional File 7 and 8), the analysis revealed that these key genes were linked with various immune- related pathways in both AS and UC, including antigen processing and presentation, natural killer cell- mediated cytotoxicity and the T cell receptor signaling pathway. The top nine enriched KEGG pathways associated with each key gene in AS and UC are displayed in Figure 4, respectively. Pathways associated with AS and UC were also searched in the CTD database (Tables S9 and 10, see Additional files 9 and 10). The top nine pathways related to AS included the NOD-like receptor signaling pathway, RIG-I-like receptor signaling pathway, JAK/STAT signaling pathway, HTLV-I infection, IBD, Herpes simplex infection, amoebiasis, allograft rejection, and cytokine-cytokine receptor interaction (Figure 5a), and the top nine pathways conferred to UC included the TNF signaling pathway, pathways in cancer, chemokine signaling pathway, amoebiasis, AGE-RAGE signaling pathway in diabetic complications, toxoplasmosis, IBD, tuberculosis and cytokine-cytokine receptor interaction (Figure 5b). Intersecting the results from GSEA and CTD revealed key pathways associated with the regulation of UC and AS, including the Toll-like receptor signaling pathway, antigen processing and presentation, allograft rejection, natural killer cell mediated cytotoxicity, pathways in cancer, graft- versus host- disease, and viral myocarditis (Figure 5c). In addition, the key genes involved in regulation had high interaction scores with AS- related nephritis, and UC- related systemic lupus erythematosus (SLE), and hypersensitivity (Figure 6).
GNG11 had the Highest Positive Correlation with Effector Memory CD4 T Cells in AS
Following the finding that the biomarkers were implicated in immune-related pathways, we further found that activated dendritic cells (DCs), effector memory CD4 T cells, gamma delta T cells, macrophages, regulatory T cells and type 17 T helper cells were significantly increased, and effector memory CD8 T cells, natural killer T cells, and T follicular helper cells were significantly decreased in the AS group (Figure 7a and b). Among them, GNG11 had the highest positive correlation with effector memory CD4 T cells. All of the key genes were correlated with natural killer T cells and gamma delta T cells (Figure 7c). For UC, we further found that the proportion of all 27 kinds of immune cells except memory B cells, was remarkably changed in the UC group compared to the healthy control group (Figure 7d and e). All of these key genes were notably correlated with almost all immune- infiltrating cells (Figure 7f).
Validation of Key Gene Expression Patterns in UC and AS Across Different Patient Cohorts
In the UC training set GSE87466, GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, and S100A12 were highly expressed in UC, whereas DGKQ was lowly expressed in UC compared to the control (Figure 8a and Table S11, see Additional files 11). In the UC external validation set GSE94648, the changes in the expression of GMFG, CLEC4D, CMTM2, VAMP5, S100A8, and S100A12 were consistent with those in the training set (Figure 8b and Table S12, see Additional files 12). In the AS training set, GSE25101, GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, and S100A12 were highly expressed in AS, whereas DGKQ was lowly expressed in AS compared to the control (Figure 8c and Table S13, see Additional files 13). In the AS external validation set GSE73754, the expression changes of S100A8 and S100A12 were consistent with those in the training set (Figure 8d and Table S14, see Additional files 14).
The mRNA expression levels of GMFG, DGKQ, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, and S100A12 in PBMCs were extracted from 31 participants (12 healthy controls, 12 patients with active UC and 7 patients with active AS). Significant associations were observed between S100A12 and VAMP5 mRNA expression levels and patients (both AS and UC) (P<0.05, Figure 9a and b). Additionally, CLEC4D mRNA expression levels were increased in patients with UC compared to healthy controls (P<0.05, Figure 9c).
Discussion
The interrelationship between AS and UC has been widely reported. The Combination attacks of the two diseases can result in severe symptoms and poor prognosis, which are frequently misdiagnosed and undertreated.14 We conducted this experiment to establish a theoretical foundation for early diagnosis and treatment, and to investigate the genes and pathways that are shared in the development of both diseases. Our study identified eight key genes (GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, S100A12 and DGKQ) involved in AS and UC. By intersecting the results from GSEA and CTD, the Toll-like receptor signaling pathway, antigen processing and presentation, allograft rejection, viral myocarditis, pathways in cancer, GVHD, and natural killer cell- mediated cytotoxicity were identified as key pathways associated with genes that regulate UC and AS. In addition, the result of functional experiments validated the elevated levels of S100A12 and VAMP5 mRNA expression in both patients with UC and AS, whereas CLEC4D mRNA expression was significantly increased in patients with UC.
Glia maturation factor gamma (GMFG) protein is predominantly expressed in immune cells, and it has been implicated in actin reorganization, neutrophil chemotaxis, and T cell adherence, among others.16 A recent study demonstrated a positive connection between GMF-γ expression and the inflammatory process of UC.17 The involvement of GMFG in the TLR4 signaling pathway (lipopolysaccharide-induced) was investigated in THP-1 and human primary macrophages.18 GMFG has been revealed to negatively regulate the MAPK, NF-κB, and IFN regulatory factor 3 signaling pathways.19 Knocking down the GMFG gene in macrophages led to elevated levels of mitochondrial reactive oxygen species (mtROS) which was linked to reduced levels of several mitochondrial respiration chain components, including antioxidant enzymes (SOD1 and SOD2) and the iron-sulfur cluster assembly scaffold protein ISCU.20 It has been revealed that oxidative stress and the ROS signaling pathway contribute to both AS and UC by enhancing the release of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α).21 Our study revealed that GMFG is a common critical gene in both AS and UC, and the aberrant linkages between GMFG and the ROS signaling pathway may contribute to the development of UC and AS.
GNG11, a member of the gamma subunit family of G proteins, is expressed in macrophages and DCs. In a gene array study, GNG11 was shown to be associated with psoriatic arthritis (PsA) compared to healthy controls.22 GNG11 appears to respond to oxidative stress by inducing its gene expression. GNG11 also plays a role in transmitting external signals to internal effectors, and its overexpression is firmly linked to the onset of cellular senescence through activation of the extracellular signal-regulated kinase (ERK)1/2 pathway.23 The ERK signaling pathway is one of the best-characterized MAPK signaling pathways. Through the phosphorylation of transcription factors like c-Fos and c-Jun, activated ERK can directly influence cell division and proliferation.23 Moreover, it has been demonstrated that ERK1/2 activation promotes osteoclast formation and bone resorption, which may aid in the development of spinal abnormalities and bone erosion in AS.24,25 The etiology of UC is closely tied to the traditional mechanism of the ERK pathway. After activation, ERK may regulate its downstream targets, such as NF-κB and Bcl-2, which can influence the inflammatory response and cell death during the development of UC.25 According to the aforementioned statements, our research indicates that GNG11 is a crucial gene shared by both AS and UC, and it may contribute to the copathogenesis of AS and UC by influencing the concentrations of associated transcription factors via the ERK1/2 pathway.
CLEC4D (also known as Dectin-3/ MCL), which is a member of myeloid C-type lectin receptors (CLRs), can recognize microbial components and subsequently activate intracellular signaling pathways while modulating immune responses.26,27 CLRs exhibit diverse functions, such as inducing endocytic, phagocytic, antimicrobial, pro‐inflammatory and anti‐inflammatory responses, and these functions are dependent on specific signaling motifs found within the cytoplasmic domains of CLRs. The CLRs transmit signals via spleen tyrosine kinase (Syk), which then combines with the CARD9/ B-cell CLL/ BCL-10/ MALT1 complex, resulting in the release of pro-inflammatory cytokines and activation of adaptive T-cell immunity.26 CLEC4D is predominantly expressed in macrophages, neutrophils in the peripheral circulation, classical monocytes, and certain subsets of DCs. CLEC4D plays a role in endocytosis and interacts with mycobacteria by recognizing TDM on the cell walls.28 Binding to the TDM upregulates Mincle expression, leading to enhanced cellular responses. Downstream signaling is mediated by the CARD9/BCL10/MALT1 and Syk kinase pathways. These pathways induce numerous intracellular responses, including the activation of NF-κβ, phagocytosis, and release of inflammatory cytokines.29,30 Moreover, the interaction of CLEC4D with TDM promotes the maturation of DCs and priming of T cells within the body.31 A higher level of CLEC4D mRNA was detected in patients with AS using whole blood transcriptional profiling.32 It was also found that CLEC4D-deficient mice were more vulnerable to colitis than wild-type mice (induced by DSS).33 CLEC4D gene polymorphisms are related to TB prevalence, making the CLR a key component of anti-mycobacterial defense.34 The association between Syk and the initiation of inflammation in osteoarthritis (OA) has been revealed, resulting in the secretion of inflammatory factors such as IL-1β that hasten the degradation of cartilage and bone tissue. The severity of the inflammatory response can be decreased by using Syk inhibitors.35,36 As a clinical focus of UC, Syk participates in the initial stages of B-cell receptor and Fc receptor signaling.37 In a mouse model of colitis induced by acetic acid, the selective Syk inhibitor (fostamatinib) reduced mucosal damage.38 In the present study, CLEC4D was elevated in UC and AS by overlapping DEGs. The functional experiments confirmed a significant increase in CLEC4D mRNA expression in patients with UC, consistent with the results observed in the validation dataset. Although there are limited studies on the association between the CLEC4D gene and AS and UC, the aforementioned research allows us to speculate on the potential mechanisms of CLEC4D in AS and UC, especially in UC. CLEC4D may accelerate the development of UC by stimulating the Syk kinase/CARD9/BCL10/MALT1 pathway and Mincle transcription, which in turn activates NF-κβ and pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α).
The chemokine-like factor (CKLF) superfamily (CKLFSF), also known as CMTM, is a critical component in pathological and physiological processes. In recent years, CMTM has gained the interest of researchers due to its potential effects on a wide range of disorders. CMTM2, a subtype of CMTM, is primarily found in bone marrow and CD4+ T cells.39 Multiple studies have suggested that CMTM2 plays a role in suppressing HIV-1 LTR activity by blocking the CREB and AP-1 pathways.40 According to a study in a Chinese cohort, individuals with HBV-related liver disorders exhibited notably lower serum CMTM2 expression compared to healthy subjects. It has also been demonstrated that serum CMTM2 levels correlate with the DNA load of HBV and may be used to distinguish between patients with HBV and healthy individuals.41 A comprehensive histologic and microarray analysis of patients with OA revealed reduced CMTM2 expression in patients with OA compared to healthy controls.42 Furthermore, the use of whole-genome microarray revealed that patients with AS exhibited CMTM2 overexpression in their peripheral blood.32 Our study found that CTMT2 was upregulated in both UC and AS, based on the overlap of DEGs. CMTM2 was enriched in signaling pathways such as “Allograft rejection” and “Cell adhesion molecules (CAMs)”, suggesting that in the development of AS and UC, CMTM2 may regulate the activation and migration of cells during the rejection response. Additionally, CMTM2 is involved in regulating adhesion molecules between cells, playing a critical role in the migration and signaling of immune cells.43 Therefore, CMTM2 may act as a molecular bridge, connecting different immune cells and signaling pathways, thus contributing to immune responses and inflammatory diseases. Despite the limited studies on the association between the CTMT2 gene and AS and UC, our findings provide insights into the potential pathways of CTMT2 in these two conditions.
VAMP5 is an important member of the Soluble N-Ethylmaleimide Sensitive Factor (SNARE) protein family,44 which plays a role in vesicle docking and fusion by interacting with other SNAREs. Vesicle-mediated protein transportation, which is the primary means of transporting proteins along the secretory and endocytic pathways, regulates the secretion of proteins and neurotransmitter release.45 This process is essential for the proper functioning of cells, as it allows proteins to be delivered to the correct locations in the cell and regulates the release of neurotransmitters. VAMP5 is an important gene that facilitates intracellular transport functions such as exocytosis, internal recycling and endocytosis. Mice lacking VAMP5 have a low birth rate and respiratory system defects, indicating its functional role in developing the respiratory system.46 The elevated expression of the genes in patients with tuberculosis indicates their crucial roles in the immunological mechanisms of the disease. Research has demonstrated that the levels of VAMP5 are relatively higher in CD14+ and CD16+ monocytes than in other cell types. This is consistent with the role of monocytes in tuberculosis infection, as they participate in vesicle transport, which requires VAMP5 for this process.46,47 Furthermore, the elimination of tuberculosis could involve the complement activation pathway, and the predicted role of the discovered protein in tuberculosis is closely linked to its sequence-structure-function relationship.48 Multiple genetic variation studies have suggested that VAMP5 confers susceptibility to Hirschsprung disease (HSCR), demonstrating the involvement of VAMP5 in the physiological processes of the digestive system.49 Through validation using external datasets and functional experiments, we identified VAMP5 as a common key gene in UC and AS. Given the paucity of data on this topic, further research is required to confirm the underlying link between the VAMP5 gene and AS with UC illness.
Diacylglycerol kinases (DGKs) are a class of enzymes that catalyze the phosphorylation of diacylglycerol (DAG) to form phosphatidic acid (PA), which is a vital intermediate of lipid metabolism and signaling pathways within cells.50 DGKQ, a critical member of the DGK family, consists of three cysteine-rich domains (CRDs), a proline/glycine-rich region, and a pleckstrin homology (PH) domain that overlaps with a Ras-binding domain.51 Previous studies have shown that DGKQ might be linked to several autoimmune diseases, including SLE, type 1 diabetes (T1D), Rheumatoid arthritis (RA), idiopathic inflammatory myopathies (IIM), primary biliary cirrhosis (PBC), and Sjögren’s syndrome (SS), among others.52–54 The high-risk SNP alleles were linked to increased DGKQ expression in fibroblasts, lymphocytes, and the lungs. DGKQ serves as a critical mediator of cell signal transduction and is expressed at higher levels in the vasculature of patients with systemic sclerosis and pulmonary involvement.55 This protein can indirectly stimulate the epidermal growth factor receptor (EGFR) pathway,56 which is crucial for regulating cell proliferation and migration. The GSEA results of this study showed that DGKQ was enriched in pathways such as “ADHERENS JUNCTION”, suggesting that it may regulate intestinal epithelial barrier function by influencing the stability of adherens junctions. This indicated that DGKQ played an important role in immune regulation and cell death, thereby affecting disease progression.57,58 Thus, the potential role of DGKQ in modulating immune responses and inflammation suggests that it could serve as a new therapeutic target for AS and UC. Our study identified DGKQ as a crucial gene shared by UC and AS. Additional research is required to confirm this result.
The S100 family comprises 25 recognized members that exhibit considerable structural and sequence similarity. Interacting with signal transducers and various receptors, the S100 family modulates processes involved in inflammation, energy consumption, calcium homeostasis, apoptosis, cell cytoskeletal processes, microbial resistance, cell differentiation and proliferation.59 In our research, both S100A12 and S100A8 were identified as common key genes for AS and UC. S100A8, a type of Ca2+-binding protein, is highly expressed in a variety of infectious and inflammatory disorders, including RA, IBD, pseudomonas aeruginosa keratitis psoriasis, and acute anterior uveitis (AAU).60,61 Additionally, S100A8 (also known as MRP8) primarily engages with S100A9 (also known as MRP14) and forms a heterodimer.62,63 By combining with the RAGE and TLR4, extracellular S100A8/S100A9 is also hypothesized to affect several processes in leukocyte recruitment. As a result, the transcription factor NF-kB is activated, leading to the release of proinflammatory cytokines (IL-1β, IL-6, and TNFα).64 Moreover, the activation of S100A8/S100A9 in monocytes may promote ROS to enhance the production of proinflammatory cytokines and the expression of the NLRP3 inflammasome.65,66 An increased level of S100A8 has been observed in patients with arthritis and IBD, indicating that it could play a role in AS and UC by affecting the aforementioned pathways and cytokines.
S100A12, which is an alarm signal that specifically targets granulocytes, binds to RAGE and TLR4.67 According to reports, RAGE-dependent activation of NF-κB has been shown to result in the release of pro-inflammatory cytokines, ultimately leading to the recruitment of monocytes.68 S100A12 is involved in the recruitment of inflammatory cells in mouse models and has been reported to be overexpressed in inflamed tissues of patients with a variety of conditions, including IBD, PsA, JIA, and RA.69–71 S100A12 is considered a reliable biomarker of IBD and systemic-onset JIA. Both JIA and AS are considered autoimmune illnesses in which the immune system mistakenly targets the body’s own tissues, resulting in joint inflammation and damage.72 Our previous research identified S100A12 as a disulfidptosis-related gene that may be involved in the pathogenesis of AS, CD, and UC. However, subsequent Mendelian randomization analysis confirmed a causal relationship only between S100A12 and IBD.73 In this study, both initial bioinformatics analysis and RT-PCR experiments emphasized the potential role of S100A12 in the development of UC and AS. However, additional functional studies are required to explore the underlying mechanisms.
Therapeutic agents that serve as functional inhibitors of receptors and ligands have been widely used to manage diverse conditions, including immunological disorders. Studies have shown that anti-allergic drugs that bind to S100A12 effectively block downstream RAGE signaling and NF-κB activation.74 According to research conducted on mouse models, the use of inhibitors and antibodies to limit the activity of S100A8/A9 can improve pathological conditions.60 Certainquinoline-3-carboxamides, which have been widely studied for the treatment of human autoimmune and inflammatory illnesses, interact with S100A9 and the S100A8/A9 complex, thereby reducing their interaction with TLR4 or RAGE and TNF-αproduction in vivo.75 A recent study indicated that blocking S100A8/A9 reduces inflammation in mouse arthritis models.75 It has been suggested that targeting S100A8 represents an effective strategy for addressing the chronic inflammation caused by obesity. Based on the DGIdb database, we found that rimegepant, eptinezumab, methotrexate, atogepant and ubrogepant may serve as potential drugs for treating UC and AS by targeting S100A12 and S100A8. Given the potential significance of the S100L8 and S100L12 in AS and UC, it is reasonable to assume that these medications could serve as research topics for disease treatment in the future. Furthermore, we constructed an integrated transcription factor-miRNA-S100L8/S100L12 regulatory network. This complex network not only revealed the intricate regulatory mechanisms in UC and AS but also opened new avenues for therapeutic interventions.
In the present study, GSEA was performed to identify pathways where the key genes exhibited significant enrichment. GMFG, CLEC4D, S100A8, and S100A12 exhibited high levels of enrichment in various processes related to the activation and function of neutrophils, including neutrophil activation involved in the immune response, neutrophil mediated immunity, neutrophil degranulation, and neutrophil activation pathways (Tables S5 and S6). Neutrophils have long been recognized as important players in the immune system, both for innate and adaptive immunity. Recent research has revealed that neutrophils exhibit pronounced phenotypic and functional abnormalities in various systemic autoimmune disorders. These abnormalities can lead to inappropriate immune responses and organ damage. Both GMFG and S100A8 were shown to be related to the Toll-like receptor pathway which has been identified as playing a crucial role in numerous autoimmune diseases. In our study, the Toll-like receptor pathway was also identified as a common process underlying AS and UC. We revealed natural killer cell mediated cytotoxicity, antigen processing and presentation, allograft rejection, cancer, viral myocarditis, and GVHD as key pathways associated with the aforementioned genes in regulating UC and AS. These pathways have provided new ideas for future research.
In the pathogenesis of AS and UC, changes in the number or proportion of immune cells and the correlation between key genes and immune cells are of great significance. In AS, the patterns of immune cell changes are complex and multifaceted. The increase in activated DCs, for example, leads to excessive antigen presentation, which perpetuates immune system activation and triggers a sustained inflammatory response.76 Similarly, the accumulation of effector memory CD4 T cells is associated with the persistence of chronic inflammation, as these cells continuously release inflammatory mediators, exacerbating joint tissue damage.77 Conversely, a reduction in effector memory CD8 T cells, natural killer T cells, and T follicular helper cells disrupts immune homeostasis, impairing the body’s ability to monitor self-antigens and regulate immune responses, thereby facilitating disease progression.78,79 Notably, the strong positive correlation between GNG11 and effector memory CD4 T cells suggests that GNG11 may play a pivotal role in the functional regulation, proliferation, and differentiation of these cells,80 offering new insights into AS pathogenesis and providing potential therapeutic targets. Furthermore, the correlations of all key genes with natural killer T cells and γδT cells suggest that these genes modulate the function of these immune cells in AS, influencing disease progression. In UC, the alteration of immune cell proportions, with the notable exception of memory B cells, reflects a profound imbalance within the intestinal immune system.81 This imbalance is likely the result of multiple interacting factors, including gut microbiota, genetic predisposition, and environmental influences.82 The disruption of immune cell proportions in the gut can compromise the intestinal mucosal barrier, increasing susceptibility to pathogen invasion and triggering autoimmune responses and inflammatory damage.83,84 The significant correlation between key genes and nearly all immune infiltrating cells underscores their central role in the immune pathogenesis of UC. These genes may influence the intestinal immune microenvironment by regulating immune cell activity, migration, or cytokine secretion.34,85 This provides promising targets for precision medicine in UC, with the potential to restore intestinal immune balance by modulating the interaction between key genes and immune cells. In summary, the study of immune cell alterations and the correlation between key genes and immune cells offers critical insights into the pathogenesis of AS and UC. This research aids in constructing a more comprehensive disease model, unveiling the immune regulatory networks involved, and highlights the potential for therapeutic interventions targeting the interactions between immune cells and key genes, which may lead to the development of novel treatment strategies.
Our study thoroughly explored the common genes and pathways associated with both AS and UC. In terms of diagnosis, monitoring these genes may serve as preliminary indicators for the occurrence of AS combined with UC, or be combined with existing diagnostic methods to improve diagnostic accuracy and efficiency. In terms of treatment, the identification of common genes and pathways provides targets for the development of new therapeutic approaches. Developing drugs targeting these shared key genes or pathways may allow for the simultaneous treatment of both diseases, improving treatment efficacy, alleviating patient suffering, and enhancing quality of life. Although AS and UC are considered distinct disease entities, their connection suggests that identifying common genes and pathways can help us understand them from a broader perspective, explore shared pathogenic mechanisms, break traditional disease classification boundaries, and build a more comprehensive and systematic disease knowledge framework, advancing medical research toward a more in-depth and precise direction. However, this study has several limitations. First, the number of genes identified is limited, and further studies are needed to validate their diagnostic efficiency. Second, power analysis was not conducted during the differential expression analysis, which may lead to insufficient comprehensiveness and precision in detecting gene expression differences. The small sample size may also limit the generalizability of the results. While we validated expression using external datasets and RT-PCR, further validation at the protein level or through cellular and animal models is required, as well as clinical trials to confirm the reliability of these findings. Simultaneous diagnosis of acute AS and UC is very rare. Therefore, our validation was conducted on patients experiencing acute UC and AS, using the PBMCs we collected. The validation dataset (GSE94648) includes 17 active UC samples and 8 UC samples in remission to compare and analyze the differences in biomarker expression across different disease states. In the future, expanding the sample size is crucial. We plan to broaden the sample range to include diverse populations across different ages, genders, regions, and disease states, in order to comprehensively and extensively validate the role of genes in UC and AS. We will also explore how gene expression varies during different disease phases and symptoms to ensure these genes can distinguish UC from other diseases. Further functional studies are needed to understand the mechanisms behind these genes and pathways, providing stronger evidence for their role as biomarkers in AS combined with UC. We also plan to use larger datasets and additional validation steps, which may uncover more potential biomarkers. We will validate biomarkers at the protein level through Western blot (WB) and immunohistochemistry (IHC), as protein-based validation provides an important extension to mRNA expression studies. Additionally, we will integrate cellular and animal models to perform both in vitro and in vivo validations to offer a more comprehensive evaluation of biomarker efficacy. Furthermore, we will utilize single-cell technologies and multi-omics approaches to gain deeper insights into the pathogenesis of complex diseases and to precisely identify potential biomarkers. By comparing patients with AS combined with UC to those with UC alone, we can further identify specific genetic and molecular biomarkers, enabling the development of more precise and effective diagnostic and therapeutic strategies. We plan to address these limitations in future research.
Conclusions
Our study highlighted eight common key genes (GMFG, GNG11, CLEC4D, CMTM2, VAMP5, S100A8, S100A12, and DGKQ) and seven common pathways (Toll-like receptor signaling pathway, antigen processing and presentation, allograft rejection, viral myocarditis, pathways in cancer, graft versus host disease and natural killer cell mediated cytotoxicity) shared by UC and AS. Functional experiments confirmed the distinct expression of S100A12 and VAMP5 in both AS and UC, as well as CLEC4D in UC. The shared molecular mechanisms between UC and AS were uncovered, offering deeper insights into the pathophysiology of these diseases. This discovery not only held the potential to improve outcomes for patients with both conditions but also emphasized the need for interdisciplinary collaboration between UC and AS research, providing a solid foundation for more precise diagnosis.
Abbreviations
UC, ulcerative colitis; AS, ankylosing spondylitis; IBD, inflammatory bowel disease; DEGs, differentially expressed genes; GSEA, Gene Set Enrichment Analysis; BP, Biological process; CC, cellular component; MF, molecular function; GMFG, Glia maturation factor gamma; mtROS, mitochondrial reactive oxygen species; PsA, psoriatic arthritis; ERK, extracellular signal-regulated kinase; CLRs, C-type lectin receptors; Syk, spleen tyrosine kinase; DCs, dendritic cells; OA, osteoarthritis; CKLF, Chemokine-like factor; CKLFSF, Chemokine-like factor superfamily; VAMP, Vesicle-mediated protein; DGKs, Diacylglycerol kinases; PA, phosphatidic acid; CRDs, cysteine-rich domains; PH, pleckstrin homology; SLE, systemic lupus erythematosus; T1D, type 1 diabetes; IIM, idiopathic inflammatory myopathies; PBC, primary biliary cirrhosis; SS, Sjögren’s syndrome; RA, Rheumatoid arthritis; AAU, acute anterior uveitis. RT-PCR, reverse transcription-polymerase chain reaction; TNF-α, Tumor necrosis factor-alpha; GEO, gene expression omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ROC, receiver operating characteristic; TFs, transcription factors; CAMs, Cell adhesion molecules; SNARE, Soluble N-Ethylmaleimide Sensitive Factor; HSCR, Hirschsprung disease; DAG, diacylglycerol; EGFR, epidermal growth factor receptor; WB, Western blot; IHC, immunohistochemistry.
Data Sharing Statement
The datasets used and/or analysed during the current study are available online. GSE87466, GSE25101, GSE94648 and GSE73754 datasets were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/). miRNA-key gene, TF-key gene and TF-miRNA relationships were obtained from the miRWalk (http://mirwalk.umm.uni-heidel berg.de/), Cistrome (http://cistrome.org/) and TransmiR (http://www.cuilab.cn/transmir). Potential drugs for key genes were predicted by DGIdb database (Figure 4d; https://www.dgidb.org/). KEGG pathway gene sets were downloaded from MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb). AS and UC related pathways were also downloaded from the CTD database (http://ctdbase.org/).
Ethics Approval and Consent to Participate
Ethical approvals were obtained from the research committees of Zhengzhou University (2021-KY-0246-001).
Acknowledgments
We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript. This paper has been uploaded to Biorxiv as a preprint: https://www.biorxiv.org/content/10.1101/2023.04.20.537616v1.full.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by National Natural Science Foundation of China [82301271, 81970792, 82171040, 82101108].
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
2. Du L, Ha C. Epidemiology and pathogenesis of ulcerative colitis. Gastroenterol Clin North Am. 2020;49(4):643–654. doi:10.1016/j.gtc.2020.07.005
3. Ordas I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380(9853):1606–1619. doi:10.1016/S0140-6736(12)60150-0
4. Patra R, Dey AK, Mukherjee S. Identification of genes critical for inducing ulcerative colitis and exploring their tumorigenic potential in human colorectal carcinoma. PLoS One. 2023;18(8):e0289064. doi:10.1371/journal.pone.0289064
5. Kucharzik T, Koletzko S, Kannengiesser K, Dignass A. Ulcerative colitis-diagnostic and therapeutic algorithms. Dtsch Arztebl Int. 2020;117(33–34):564–574. doi:10.3238/arztebl.2020.0564
6. Braun J, Sieper J. Ankylosing spondylitis. Lancet. 2007;369(9570):1379–1390. doi:10.1016/S0140-6736(07)60635-7
7. Zhu W, He X, Cheng K, et al. Ankylosing spondylitis: etiology, pathogenesis, and treatments. Bone Res. 2019;7(22). doi:10.1038/s41413-019-0057-8
8. Ebrahimiadib N, Berijani S, Ghahari M, Pahlaviani FG. Ankylosing Spondylitis. J Ophthalmic Vis Res. 2021;16(3):462–469. doi:10.18502/jovr.v16i3.9440
9. Garcia-Montoya L, Gul H, Emery P. Recent advances in ankylosing spondylitis: understanding the disease and management. F1000Res. 2018;7:1512. doi:10.12688/f1000research.14956.1
10. Jang DI, Lee AH, Shin HY, et al. The role of tumor necrosis factor alpha (TNF-alpha) in autoimmune disease and current TNF-alpha inhibitors in therapeutics. Int J mol Sci. 2021;22(5):2719. doi:10.3390/ijms22052719
11. Sieper J, Poddubnyy D, Miossec P. The IL-23-IL-17 pathway as a therapeutic target in axial spondyloarthritis. Nat Rev Rheumatol. 2019;15(12):747–757. doi:10.1038/s41584-019-0294-7
12. Wang S, Tsou HK, Chiou JY, Wang YH, Zhang Z, Wei JC. Increased risk of inflammatory bowel disease among patients with Ankylosing Spondylitis: a 13-year population-based cohort study. Front Immunol. 2020;11:578732. doi:10.3389/fimmu.2020.578732
13. De Vos M. Review article: joint involvement in inflammatory bowel disease. Aliment Pharmacol Ther. 2004;20:36–42. doi:10.1111/j.1365-2036.2004.02044.x
14. Cui Z, Hou G, Meng X, Feng H, He B, Tian Y. Bidirectional causal associations between inflammatory Bowel Disease and Ankylosing Spondylitis: a two-sample Mendelian randomization analysis. Front Genet. 2020;11:587876. doi:10.3389/fgene.2020.587876
15. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
16. Lippert DN, Wilkins JA. Glia maturation factor gamma regulates the migration and adherence of human T lymphocytes. BMC Immunol. 2012;13:1.
17. Li K, Strauss R, Ouahed J, et al. Molecular comparison of adult and pediatric ulcerative colitis indicates broad similarity of molecular pathways in disease tissue. J Pediatr Gastroenterol Nutr. 2018;67(1):45–52. doi:10.1097/MPG.0000000000001898
18. Husebye H, Halaas O, Stenmark H, et al. Endocytic pathways regulate toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25(4):683–692. doi:10.1038/sj.emboj.7600991
19. Aerbajinai W, Chin K, Lee HW, Zhu J, Rodgers GP. Glia maturation factor-gamma negatively modulates TLR4 signaling in macrophages induced by lipopolysaccharide (LPS). Blood. 2010;116(21):1482. doi:10.1182/blood.V116.21.1482.1482
20. Aerbajinai W, Ghosh MC, Liu J, et al. Glia maturation factor-gamma regulates murine macrophage iron metabolism and M2 polarization through mitochondrial ROS. Blood Adv. 2019;3(8):1211–1225. doi:10.1182/bloodadvances.2018026070
21. Baeten D, De Keyser F, Mielants H, Veys EM. Ankylosing spondylitis and bowel disease. Best Pract Res Clin Rheumatol. 2002;16(4):537–549. doi:10.1053/berh.2002.0249
22. Szodoray P, Dolcino M, Ottria A, et al. Gene expression profiling in peripheral blood cells and synovial membranes of patients with psoriatic arthritis. PLoS One. 2015;10(6):e0128262.
23. Moens U, Kostenko S, Sveinbjornsson B. The role of mitogen-activated protein kinase-activated protein kinases (MAPKAPKs) in inflammation. Genes. 2013;4(2):101–133. doi:10.3390/genes4020101
24. Liao HT, Tsai CY, Lai CC, et al. The potential role of genetics, environmental factors, and gut dysbiosis in the aberrant non-coding RNA expression to mediate inflammation and osteoclastogenic/osteogenic differentiation in ankylosing spondylitis. Front Cell Dev Biol. 2021;9:748063. doi:10.3389/fcell.2021.748063
25. Gao W, Wang C, Yu L, et al. Chlorogenic acid attenuates dextran sodium sulfate-induced ulcerative colitis in mice through MAPK/ERK/JNK pathway. Biomed Res Int. 2019;2019:6769789. doi:10.1155/2019/6769789
26. Marakalala MJ, Ndlovu H. Signaling C-type lectin receptors in antimycobacterial immunity. PLoS Pathog. 2017;13(6):e1006333. doi:10.1371/journal.ppat.1006333
27. Del Fresno C, Iborra S, Saz-Leal P, Martinez-Lopez M, Sancho D. Flexible signaling of myeloid C-type lectin receptors in immunity and inflammation. Front Immunol. 2018;9:804. doi:10.3389/fimmu.2018.00804
28. Miyake Y, Toyonaga K, Mori D, et al. C-type lectin MCL is an FcRg-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38(5):1050–1062. doi:10.1016/j.immuni.2013.03.010
29. Miyake Y, Masatsugu OH, Yamasaki S. C-type lectin receptor MCL facilitates mincle expression and signaling through complex formation. J Immunol. 2015;194(11):5366–5374. doi:10.4049/jimmunol.1402429
30. Zhao XQ, Zhu LL, Chang Q, et al. C-type lectin receptor dectin-3 mediates trehalose 6,6’-dimycolate (TDM)-induced Mincle expression through CARD9/Bcl10/MALT1-dependent nuclear factor (NF)-kappaB activation. J Biol Chem. 2014;289(43):30052–30062. doi:10.1074/jbc.M114.588574
31. Geijtenbeek TB, Gringhuis SI. C-type lectin receptors in the control of T helper cell differentiation. Nat Rev Immunol. 2016;16(7):433–448. doi:10.1038/nri.2016.55
32. Pimentel-Santos FM, Ligeiro D, Matos M, et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res Ther. 2011;13(2):R57. doi:10.1186/ar3309
33. Wang T, Pan D, Zhou Z, et al. Dectin-3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PLoS Pathog. 2016;12(6):e1005662. doi:10.1371/journal.ppat.1005662
34. Wilson GJ, Marakalala MJ, Hoving JC, et al. The C-type lectin receptor CLECSF8/CLEC4D is a key component of anti-mycobacterial immunity. Cell Host Microbe. 2015;17(2):252–259. doi:10.1016/j.chom.2015.01.004
35. Novikov FN, Panova MV, Titov IY, Stroylov VS, Stroganov OV, Chilov GG. Inhibition of SYK and cSrc kinases can protect bone and cartilage in preclinical models of osteoarthritis and rheumatoid arthritis. Sci Rep. 2021;11(1):23120. doi:10.1038/s41598-021-02568-6
36. Geahlen RL. Getting Syk: spleen tyrosine kinase as a therapeutic target. Trends Pharmacol Sci. 2014;35(8):414–422. doi:10.1016/j.tips.2014.05.007
37. Lowell CA. Src-family and Syk kinases in activating and inhibitory pathways in innate immune cells: signaling cross talk. Cold Spring Harb Perspect Biol. 2011;3(3):a002352–a002352. doi:10.1101/cshperspect.a002352
38. Can G, Ayvaz S, Can H, et al. The Syk inhibitor Fostamatinib decreases the severity of colonic mucosal damage in a rodent model of colitis. J Crohn's Colitis. 2015;9(10):907–917. doi:10.1093/ecco-jcc/jjv114
39. Li M, Luo F, Tian X, Yin S, Zhou L, Zheng S. Chemokine-like factor-like MARVEL transmembrane domain-containing family in hepatocellular carcinoma: latest advances. Front Oncol. 2020;10:595973. doi:10.3389/fonc.2020.595973
40. Song HS, Shuang SH, Lu XZ, et al. Intracellular CMTM2 negatively regulates human immunodeficiency virus type-1 transcription through targeting the transcription factors AP-1 and CREB. Chin Med J. 2010;123(17):2440–5.
41. Chen J, Zhou J, Kelly M, Holbein BE, Lehmann C. Iron chelation for the treatment of uveitis. Med Hypotheses. 2017;103:1–4. doi:10.1016/j.mehy.2017.03.029
42. Chen S, Hu Q, Chen H, et al. Identification of serum CMTM2 as a potential biomarker for HBV-related disorders. Dis Markers. 2020;2020:2032056. doi:10.1155/2020/2032056
43. Heeger PH, Jordan MC, Jordan S. S: translating B cell immunology to the treatment of antibody-mediated allograft rejection. Nat Rev Nephrol. 2024;20:218–232. doi:10.1038/s41581-023-00791-0
44. Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(2):120–144. doi:10.1016/j.bbamcr.2005.03.014
45. Tajika Y, Takahashi M, Khairani AF, Ueno H, Murakami T, Yorifuji H. Vesicular transport system in myotubes: ultrastructural study and signposting with vesicle-associated membrane proteins. Histochem Cell Biol. 2014;141(4):441–454. doi:10.1007/s00418-013-1164-z
46. Ikezawa M, Tajika Y, Ueno H, Murakami T, Inoue N, Yorifuji H. Loss of VAMP5 in mice results in duplication of the ureter and insufficient expansion of the lung. Dev Dyn. 2018;247(5):754–762. doi:10.1002/dvdy.24618
47. Gong Z, Gu Y, Xiong K, et al. The evaluation and validation of blood-derived novel biomarkers for precise and rapid diagnosis of tuberculosis in areas with high-TB burden. Front Microbiol. 2021;12:650567. doi:10.3389/fmicb.2021.650567
48. Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi:10.1038/nature09247
49. Zhao J, Xie X, Yao Y, et al. Association of VAMP5 and MCC genetic polymorphisms with increased risk of Hirschsprung disease susceptibility in Southern Chinese children. Aging. 2018;10(4):689.
50. Cai J, Abramovici H, Gee SH, Topham MK. Diacylglycerol kinases as sources of phosphatidic acid. Biochimica et Biophysica Acta. 2009;1791(9):942–948. doi:10.1016/j.bbalip.2009.02.010
51. Houssa B, Schaap D, van der Wal J, et al. Cloning of a novel human diacylglycerol kinase (DGKθ) containing three cysteine-rich domains, a proline-rich region, and a Pleckstrin homology domain with an overlapping ras-associating domain. J Biol Chem. 1997;272(16):10422–10428. doi:10.1074/jbc.272.16.10422
52. Jia X, Shi N, Feng Y, et al. Identification of 67 pleiotropic genes associated with seven autoimmune/autoinflammatory diseases using multivariate statistical analysis. Front Immunol. 2020;11. doi:10.3389/fimmu.2020.00030
53. Robertson CC, Inshaw JRJ, Onengut-Gumuscu S, et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nature Genet. 2021;53(7):962–971. doi:10.1038/s41588-021-00880-5
54. Rothwell S, Amos CI, Miller FW, et al. Genome-wide imputation identifies novel associations and localises signals in idiopathic inflammatory myopathies. Arthritis Rheumatol. 2022;74:342–352. doi:10.1002/art.41929
55. Overbeek MJ, Boonstra A, Voskuyl AE, et al. Platelet-derived growth factor receptor-beta and epidermal growth factor receptor in pulmonary vasculature of systemic sclerosis-associated pulmonary arterial hypertension versus idiopathic pulmonary arterial hypertension and pulmonary veno-occlusive disease: a case-control study. Arthritis Res Ther. 2011;13(2):R61. doi:10.1186/ar3315
56. van Baal J, de Widt J, Divecha N, van Blitterswijk WJ. Diacylglycerol kinase θ counteracts protein kinase C-mediated inactivation of the EGF receptor. Int J Biochem Cell Biol. 2012;44(11):1791–1799. doi:10.1016/j.biocel.2012.06.021
57. Kageyama T, Ito T, Tanaka S, Nakajima H. Physiological and immunological barriers in the lung. Semin Immunopathol. 2024;45(4–6):533–547. doi:10.1007/s00281-024-01003-y
58. Lechuga S, Marino-Melendez A, Naydenov NG, Zafar A, Braga-Neto MB, Ivanov AI. Regulation of epithelial and endothelial barriers by molecular chaperones. Cells. 2024;13(5):370. doi:10.3390/cells13050370
59. Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X. S100 proteins as an important regulator of macrophage inflammation. Front Immunol. 2017;8:1908. doi:10.3389/fimmu.2017.01908
60. Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J: S100A8/A9 in Inflammation. Front Immunol 2018, 9:1298.
61. Schonthaler HB, Guinea-Viniegra J, Wculek SK, et al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39(6):1171–1181. doi:10.1016/j.immuni.2013.11.011
62. Narumi K, Miyakawa R, Ueda R, et al. Proinflammatory proteins S100A8/S100A9 activate NK cells via Interaction with RAGE. J Immunol. 2015;194(11):5539–5548. doi:10.4049/jimmunol.1402301
63. Donato R. RD: S100 a multigenic family of calcium modulated proteins of the EF hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33(7):637–68.
64. Kessel C, Holzinger D, Foell D. Phagocyte-derived S100 proteins in autoinflammation: putative role in pathogenesis and usefulness as biomarkers. Clin Immunol. 2013;147(3):229–241. doi:10.1016/j.clim.2012.11.008
65. Wilson EH, Simard J-C, Cesaro A, et al. S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-κB1. PLoS One. 2013;8(8):e72138.
66. Liu Y, Kong X, You Y, et al. S100A8-mediated NLRP3 inflammasome-dependent pyroptosis in macrophages facilitates liver fibrosis progression. Cells. 2022;11(22):3579. doi:10.3390/cells11223579
67. Vogl T, Pröpper C, Hartmann M, et al. S100A12 is expressed exclusively by granulocytes and acts independently from MRP8 and MRP14. J Biol Chem. 1999;274(36):25291–25296. doi:10.1074/jbc.274.36.25291
68. Lira-Junior R, Holmstrom SB, Clark R, et al. S100A12 expression is modulated during monocyte differentiation and reflects periodontitis severity. Front Immunol. 2020;11(86). doi:10.3389/fimmu.2020.00086.
69. Foell D, Kane D, Bresnihan B, et al. Expression of the pro-inflammatory protein S100A12 (EN-RAGE) in rheumatoid and psoriatic arthritis. Rheumatology. 2003;42(11):1383–1389. doi:10.1093/rheumatology/keg385
70. Foell D, Kucharzik T, Kraft M, Vogl T, Sorg C, Domschke W, Roth J. Neutrophil derived human S100A12 (EN-RAGE) is strongly expressed during chronic active inflammatory bowel disease. Gut. 2003;52:847–853.
71. Orczyk K, Smolewska E. A granulocyte-specific protein S100A12 as a potential prognostic factor affecting aggressiveness of therapy in patients with juvenile idiopathic arthritis. J Immunol Res. 2018;2018:5349837. doi:10.1155/2018/5349837
72. Wittkowski H, Frosch M, Wulffraat N, et al. S100A12 is a novel molecular marker differentiating systemic-onset juvenile idiopathic arthritis from other causes of fever of unknown origin. Arthritis Rheum. 2008;58(12):3924–3931. doi:10.1002/art.24137
73. Li L, Fang H, Li F, et al. Regulation mechanisms of disulfidptosis-related genes in ankylosing spondylitis and inflammatory bowel disease. Front Immunol. 2024;15:1326354. doi:10.3389/fimmu.2024.1326354
74. Chiou JW, Fu B, Chou RH, Yu C. Blocking the interactions between calcium-bound S100A12 protein and the V domain of RAGE using tranilast. PLoS One. 2016;11(9):e0162000. doi:10.1371/journal.pone.0162000
75. Austermann J, Zenker S, Roth J. S100-alarmins: potential therapeutic targets for arthritis. Expert Opin Ther Targets. 2017;21(7):739–751. doi:10.1080/14728222.2017.1330411
76. Zhang P, Zhao WL, Li JK, Tong JY. [RNA m (6)A modification and its roles in immune function regulation]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2022;53(6):1118–1126. Danish. doi:10.12182/20221160511
77. Hoeks C, Duran G, Hellings N, Broux B. When helpers go above and beyond: development and characterization of cytotoxic CD4(+) T cells. Front Immunol. 2022;13:951900. doi:10.3389/fimmu.2022.951900
78. Poggi A, Zocchi MR. Role of bone marrow stromal cells in the generation of human CD8+ regulatory T cells. Hum Immunol. 2008;69(11):755–759. doi:10.1016/j.humimm.2008.08.278
79. Kim YJ, Choi J, Choi YS. Transcriptional regulation of Tfh dynamics and the formation of immunological synapses. Exp Mol Med. 2024;56(6):1365–1372. doi:10.1038/s12276-024-01254-7
80. Liu X, Xu X, Hilger D, et al. Structural insights into the process of GPCR-G protein complex formation. Cell. 2019;177(5):1243–1251.e1212. doi:10.1016/j.cell.2019.04.021
81. Chao K, Zhong BH, Zhang SH, Gong XR, Yao JY, Chen MH. [Imbalance of CD4(+) T cell subgroups in ulcerative colitis]. Zhonghua Yi Xue Za Zhi. 2011;91(23):1605–1608. Danish
82. Zhang Y, Liu Y, Yang S, Yan S. Mechanism of Nrf2 in the treatment of ulcerative colitis via regulating macrophage polarization. Zhong Nan da Xue Xue Bao Yi Xue Ban. 2023;48(11):1746–1752. doi:10.11817/j.issn.1672-7347.2023.230281
83. Hou X, Zhu F, Zheng W, et al. Protective effect of Schistosoma japonicum eggs on TNBS-induced colitis is associated with regulating Treg/Th17 balance and reprogramming glycolipid metabolism in mice. Front Cell Infect Microbiol. 2022;12:1028899. doi:10.3389/fcimb.2022.1028899
84. Tajima M, Wakita D, Noguchi D, et al. IL-6-dependent spontaneous proliferation is required for the induction of colitogenic IL-17-producing CD8+ T cells. J Exp Med. 2008;205(5):1019–1027. doi:10.1084/jem.20071133
85. Graham LM, Gupta V, Schafer G, et al. The C-type lectin receptor CLECSF8 (CLEC4D) is expressed by myeloid cells and triggers cellular activation through Syk kinase. J Biol Chem. 2012;287(31):25964–25974. doi:10.1074/jbc.M112.384164
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Pediatric Stroke from Bench to Bedside: A Single-Center Experience in Saudi Arabia
Al-Sharydah AM, Al-Arfaj HK, Al-Suhibani SS, Al-Safran FS, Al-Abdulwahhab AH, Al-Jubran SA, AlSaflan AA
Vascular Health and Risk Management 2022, 18:529-540
Published Date: 13 July 2022










