Back to Journals » International Journal of Nanomedicine » Volume 19
Silver Nanoparticles Exposure Impairs Cardiac Development by Suppressing the Focal Adhesion Pathway in Zebrafish
Authors Lu C, Wu X, Meng X, Liu Y, Yang T, Zeng Y, Chen Y, Huang Y, Fang Z, Yang X , Luo J
Received 30 April 2024
Accepted for publication 4 September 2024
Published 9 September 2024 Volume 2024:19 Pages 9291—9304
DOI https://doi.org/10.2147/IJN.S476168
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Krishna Nune
Chunjiao Lu,* Xuewei Wu,* Xin Meng,* Yi Liu, Ting Yang, Yan Zeng, Yang Chen, Yishan Huang, Zhou Fang, Xiaojun Yang, Juanjuan Luo
Engineering Research Center of Key Technique for Biotherapy of Guangdong Province, Shantou University Medical College, Shantou, 515041, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaojun Yang; Juanjuan Luo, Shantou University Medical College, 22 Xinling Road, Shantou, 515041, People’s Republic of China, Email [email protected]; [email protected]
Introduction: The potential toxic effects of wastewater discharges containing silver nanoparticles (AgNPs) and their release into aquatic ecosystems on aquatic organisms are becoming a major concern for environmental and human health. However, the potential risks of AgNPs to aquatic organisms, especially for cardiac development by Focal adhesion pathway, are still poorly understood.
Methods: The cardiac development of various concentrations of AgNPs in zebrafish were examined using stereoscopic microscope. The expression levels of cardiac development-related genes were analyzed by qRT-PCR and Whole-mount in situ hybridization (WISH). In addition, Illumina high-throughput global transcriptome analysis was performed to explore the potential signaling pathway involved in the treatment of zebrafish embryos by AgNPs after 72 h.
Results: We systematically investigated the cardiac developing toxicity of AgNPs on the embryos of zebrafish. The results demonstrated that 2 or 4 mg/L AgNPs exposure induces cardiac developmental malformations, such as the appearance of pericardial edema phenotype. In addition, after 72 h of exposure, the mRNA levels of cardiac development-related genes, such as myh7, myh6, tpm1, nppa, tbx5, tbx20, myl7 and cmlc1, were significantly lower in AgNPs-treated zebrafish embryos than in control zebrafish embryos. Moreover, RNA sequencing, KEGG (Kyoto Encyclopedia of Genes) and Genomes and GSEA (gene set enrichment analysis) of the DEGs (differentially expressed genes) between the AgNPs-exposed and control groups indicated that the downregulated DEGs were mainly enriched in focal adhesion pathways. Further investigations demonstrated that the mRNA levels of focal adhesion pathway-related genes, such as igf1ra, shc3, grb2b, ptk2aa, akt1, itga4, parvaa, akt3b and vcla, were significantly decreased after AgNPs treatment in zebrafish.
Conclusion: Thus, our findings illustrated that AgNPs could impair cardiac development by regulating the focal adhesion pathway in zebrafish.
Keywords: AgNPs, focal adhesion, cardiac development, zebrafish
Introduction
Nanoparticles (NPs) have a wide range of uses, because of their distinctive characteristics, including size, shape, structure, and surface area.1 Concerns concerning the consequences of unintentional NP exposure on human health have been raised by the extensive usage of NPs.2 NPs can enter organisms through a variety of routes, including through the skin, mouth and respiratory tract, and cause toxic effects.3 The adverse effects of NPs are strongly related to cytotoxicity, oxidative stress, gene expression, proinflammatory responses, alterations in calcium homeostasis and cellular signaling events.3,4
AgNPs are a type of labile metal-based nanoparticle (MeNP), and the metal ions present in AgNPs have the ability to separate from the nanoparticle surface in aqueous solutions and may undergo a variety of physicochemical alterations.5–8 In particular, AgNPs have several attractive properties, such as chemical stability and antifungal and bactericidal properties.9 Therefore, AgNPs are widely used in health products, cosmetics, wound dressings, food packaging films and so on.9,10
The production and extensive application of AgNPs has resulted in their elevated discharge into aquatic ecosystems, and AgNPs can contaminate the groundwater environment.10–13 After entering freshwater, AgNPs typically undergo oxidation, transforming into silver ions (Ag+), which possess toxicity towards aquatic organisms.11 AgNPs can also enter the body via the food chain and accumulate in aquatic organisms.9 In addition, long-term exposure to silver through oral and inhalation routes can result in the development of argyria or argyrosis.9,11 AgNPs can damage many organs and systems in vivo, such as the skin, eyes, hepatobiliary system, nervous system, urinary system, respiratory system, reproductive system and immune system,10 and may cause cell death through necrosis or apoptosis, as well as genetic mutations, and result in toxicity to tissues and organs.14–17
Focal adhesion is an important mechanism of cell communication and plays a key role in cell growth, differentiation, motility, migration, proliferation and survival.18 Renshaw et al indicated that normal cell growth involves the induction of cell adhesion in the presence of growth factors.19 Many focal adhesion proteins, including alpha4 integrin, alpha5 integrin, beta1 integrin, paxillin, vinculin, talin, integrin-like kinase and FAK (focal adhesion kinase), play essential roles in embryonic development.18 In this context, the focal adhesion kinase protein is situated at the cortex of notochord cells and on the notochord-somite boundary. Throughout the process of somitogenesis, FAK accumulates in the basal region of epithelial cells at intersomitic boundaries, and phosphorylated FAK protein is detected at both the notochord-somite boundary and intersomite boundaries.20 FAK plays instrumental roles in cell survival, cell proliferation, cell migration, and the integration of multiple cues in the extracellular environment, and FAK activity can be stimulated by several types of extracellular ligands.21 In postnatal or adult mice, focal adhesion proteins are essential for the functionality of specific tissues and organs.22 FAK phosphorylation is required for somite boundary formation during the somitogenesis of zebrafish embryos.20 FAK deficiency can induce vascular development dysfunction, leading to embryonic lethality.23,24 Endothelial cell-specific deletion of focal adhesion kinase induces vascular defects associated with apoptosis.25,26 Paxillin is a focal adhesion-associated adaptor protein that can recruit FAK and vinculin to focal adhesions to guarantee regular signal reception and transduction.27 Paxillin is very important for the recruitment of vinculin, a pivotal structural component in cardiomyocytes that plays essential part in mechanotransduction, to focal adhesions.27–30 The knockout of the paxillin gene in mice results to an early embryonic lethal phenotype.31 Paxillin and FAK play important roles in the vertebrate heart, and are both required to recruit and stabilize vinculin at sites of force transmission to regulate cardiac contractility.29
Since zebrafish is an excellent animal model for studying the potential ecotoxicity of water pollutants, especially for investigating their biological effects on neurological and cardiovascular development. At early developmental stage, zebrafish embryos are transparent, enabling more easy observation of morphological abnormalities and developmental characteristics.32 In addition, it has been shown that the accumulation of AgNPs in the liver, spleen and lung can cause toxic effects.33,34 AgNPs are toxic to zebrafish embryos and cause death, delayed hatching, physical deformities and heart rate decreases.35 Herein, our study explored the effects of AgNPs on cardiac developmental toxicity and the underlying molecular mechanisms. In addition, the DEGs after exposure to 2 and 4 mg/L AgNPs were analyzed by using transcriptome sequencing techniques to elucidate the potential mechanisms of cardiac developmental toxicity caused by AgNPs exposure. KEGG signaling pathway enrichment analysis of the DEGs were used to further explore the potential downstream signaling pathways in zebrafish with or without AgNPs treatment. qRT‒PCR and Western blotting were also utilized to further analyze the mechanism underlying the potential toxicity of AgNPs exposure in zebrafish cardiac development. Thus, our findings illustrate the mechanism of AgNPs-induced cardiac disruption via regulating the focal adhesion pathway in zebrafish, which provide new insights into the potential risks of the toxic effects of AgNPs exposure to human health.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of Shantou University Medical College. The protocol was approved by the Shantou University Medical College Animal Committee (No: SUMC-2020-015).
AgNPs Solution
Silver nanoparticles powder (CAS No. 7440-22-4, Sigma-Aldrich, Merck, Germany) was suspended in ultrapure water (Millipore, Sigma-Aldrich) and then sonicated for 50 min at 50 W/L and 40 kHz to prepare a 100 mg/L stock solution.36 The morphology and diameter of AgNPs were characterized by transmission electron microscope (TEM) and dynamic light scattering (DLS) according to our previously described.37 The percentages of different diameters of AgNPs particles were 96% (< 62 nm) and 4% (62–100 nm). The AgNPs exposure solutions were prepared every 24 h to maintain fresh and consistent levels of exposure.
Zebrafish Husbandry and Treatment
Wild-type (AB) zebrafish were reared at 28 °C under a constant cycle of 14/10 h light/dark and fed decapsulated freshly hatched brine shrimp twice daily. Healthy adult zebrafish were selected for breeding, and healthy embryos for the exposure experiment were collected at 4 hours postfertilization (hpf).38 A total of 150 embryos (4 hpf) were transferred to 10 cm Petri dishes and supplemented with aqueous solutions of AgNPs at different levels of concentration (0, 1, 2 and 4 mg/L).37,39,40 Three replicates were performed for each concentration group.
For AgNPs treatment, base on our previous report, the median lethal concentration (LC50) assay was performed to evaluate the potential toxic effect of AgNPs in zebrafish embryos.37 Briefly, After treating AgNPs solution with different concentrations in 4 hpf zebrafish embryos for 72 h, the survival rate of 8 mg/L group is 32% with severe developmental disruption, whereas 67% and 74% survival rates in 4 and 6 mg/L groups. Thus, the selected exposure concentrations (0, 1, 2 and 4 mg/L) were utilized in the following experiments.
Zebrafish Morphology
After AgNPs exposure, the phenotypes of the zebrafish embryos at 24–72 h were observed using a stereoscopic microscope. Zebrafish embryo developmental abnormalities, delayed yolk sac absorption and including pericardial edema, were determined at 48–72 h.41,42
Gene Expression Analysis
After zebrafish embryos were exposed to AgNPs for 72 h, 30 larvae were collected for RNA extraction using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. The first-strand cDNAs were transcribed with a PrimeScriptTM RT reagent kit with a gRNA eraser (Takara Bio, Japan) following the manufacturer’s instructions, and 1 μg of total mRNA was utilized for cDNA synthesis process. qRT‒PCR was then performed using an RT‒PCR Kit (Top Green qPCR Super Mix, TransGen, China) in an ABI7500 real-time PCR system (PerkinElmer Applied Biosystems, Foster City, CA). The primers used for the selected genes and their sequences are listed in Table S1. The housekeeping gene β-actin was designated as the internal control. All the samples were run in triplicate. The mRNA expressions of genes were standardized against to those of β-actin utilizing the 2−∆∆CT method.
Whole-Mount In situ Hybridization (WISH)
Zebrafish larvae were collected for WISH experiments after exposure to the highest concentration of AgNPs (4 mg/L) for 72 h as our previously described.43 The ISH antisense probes were designed to target the 3’UTRs of the myh7 (myosin heavy chain 7), myh6 (myosin, heavy chain 6, cardiac muscle, alpha), tpm1 (tropomyosin 1), tnnt2a (troponin T type 2a), nppa (natriuretic peptide A), myl7 (myosin, light chain 7) and cmlc1 (cardiac myosin light chain-1) transcript sequences. The T7 RNA polymerase promoters were included in the probe template. The probe templates were purified with a TAINquick Midi Purification Kit (TIANGEN, China) and then transcribed with a MAXIscript T7 Kit (Ambion, Thermo Fisher Scientific). The sequences of the WISH probes are presented in Table S2.
Global Transcriptome Analysis
After 72 h of AgNPs exposure, zebrafish larvae were sent to the BGI Company (Shenzhen, China) for transcriptome sequencing. The downregulated DEGs were selected and subjected to functional enrichment analysis. KEGG pathway analysis was performed with DAVID (https://david.ncifcrf.gov/gene2gene.jsp). The top 4 pathway with the lowest p values were selected for plotting. The GSEA results were analyzed by Dr. TOM software (https://biosys.bgi.com/#/report/login). The Chiplot website (https://www.chiplot.online) was used to construct a heatmap of the down-regulated DEGs with Log2FC ≤ −0.5, and the KEGG pathway database (https://www.kegg.jp/kegg/pathway.html) was used to analyze the signaling pathways associated with the enriched DEGs.
Western Blot Analysis
After exposure to the AgNPs solution for 72 h, total protein was obtained from the zebrafish larvae using a total protein extraction kit (C500005, Sangon Biotech). Forty micrograms of total protein extract were separated by 10% SDS‒PAGE separation gel followed by transferred to a nitrocellulose membrane (Millipore). Subsequently, the membranes were placed in 5% bovine serum albumin (BSA) in TBST buffer (containing 0.1% Tween-20) for 1 h of incubation. The membranes were then incubated with primary antibodies in the refrigerator at 4 °C overnight. The following day, the primary antibody was removed and after washing with TBST three times, the membranes were incubated with secondary antibodies for 1 h at room temperature. The results were visualized using a Bio-Rad ChemiDoc XRS system. The pixel intensities were measured with ImageJ software (NIH, Bethesda, MA). The sources of the antibodies used are listed in Table S3.
Data Analysis
Statistical analyses were performed using analysis of variance (ANOVA), student’s-t test, and Tukey’s multiple range tests. A value of p < 0.05 was regarded as statistically significant. All values are expressed as the means ± standard errors (SEMs). GraphPad 9.0 software (GraphPad Software, San Diego, CA), and ChiPlot (https://www.chiplot.online/) were used for plotting.
Results
AgNPs Exposure Impairs Cardiac Development in Zebrafish
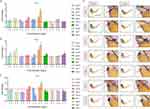
To explore the potential effects of AgNPs on the development of zebrafish, we first examined the morphological phenotypes of zebrafish treated with or without AgNPs. The results indicated that developmental malformations, such as pericardial edema (red broken arrows) and delayed yolk sac absorption (blue broken arrows) (Figure 1A and B), appeared at 48 and 72 h after the embryos were exposed to 2 mg/L AgNPs. Compared with those in the control group, the proportion of embryos with pericardial edema and delayed yolk sac absorption were significantly up-regulated after treatment with 2 or 4 mg/L AgNPs for 48–72 h (Figure 1C–F). Exposure of zebrafish embryos to 2 or 4 mg/L AgNPs affected heart development.
AgNPs Exposure Interferes with the Transcript Levels of Cardiac Development-Related Genes
To study the regulation of cardiac development-related genes after AgNPs exposure, we performed a qPCR assay in zebrafish treated with or without AgNPs at different concentrations (2 and 4 mg/L) and exposure times (24, 48, and 72 h). The results indicated that the mRNA levels of the tnnt2a, nppa and nkx2.5 genes were increased significantly by 1.68-, 1.27- and 2.69-fold, respectively, after treatment with a high concentration (4 mg/L) of AgNPs for 24 h (P < 0.05), whereas the level of tbx20 was relatively decreased compared with that in the control group (Figure 2A). Furthermore, after exposure to 4 mg/L AgNPs for 48 h, the transcript levels of myh7, myh6, nkx2.5, tbx5 and cmlc1 were increased by 1.32-, 1.75-, 2.47-, 1.32- and 1.38-fold, respectively (P < 0.05, Figure 2B). Interestingly, in the 72-h treatment groups, the mRNA levels of myh7, myh6, tpm1, tbx5 and cmlc1 were significantly reduced by 0.87-, 0.63-, 0.74-, 0.88- and 0.85-fold, respectively, in 2 mg/L AgNP-treated zebrafish embryos (P < 0.05, Figure 2C). In the 4 mg/L AgNPs-treated group, the mRNA levels of most cardiac development-related genes, including myh7, myh6, tpm1, nppa, tbx5, tbx20, myl7 and cmlc1, were significantly decreased by 0.87-, 0.61-, 0.70-, 0.46-s, 0.77-, 0.76-, 0.77- and 0.77-fold, respectively (P < 0.05, Figure 2C), except for the mRNA level of the nkx2.5 gene (Figure 2C). Subsequently, we performed WISH analysis on zebrafish larvae treated with 4 mg/L AgNPs for 72 h. We found that the expression trends of cardiac development-related genes, such as myh7, myh6, nppa, myl7 and cmlc1, were broadly consistent with the qPCR analyses (Figure 2D). Taken together, the WISH and qPCR results revealed that AgNPs exposure might induce cardiac abnormalities by interfering with the transcript levels of genes related to cardiac development in zebrafish.
Identification of the Downstream Signaling Pathway Involved in AgNPs-Induced Cardiac Developmental Disruption in Zebrafish
To gain insight into the downstream signaling pathways after AgNPs (2 or 4 mg/L) exposure in zebrafish, KEGG pathway analysis was performed using DAVID. The results indicated that the downregulated DEGs in zebrafish exposed to AgNPs were enriched in the focal adhesion pathway (Figure 3A and B). In addition, GSEA revealed that the most of the DEGs enriched in the focal adhesion pathway were downregulated after AgNPs treatment in zebrafish (Figure 3C and D), suggesting that the expression of genes associated with the focal adhesion pathway was dramatically altered after exposure to AgNPs.
To further investigate the potential roles of 2 or 4 mg/L AgNPs exposure, we next analyzed the downregulated DEGs (Log2FC ≤ −0.5) involved in the focal adhesion pathway using heatmap (https://www.chiplot.online/). The results indicated that the number of DEGs increased after AgNPs treatment in a dose-dependent manner (Figure 4A and B). KEGG signaling pathway website (https://www.kegg.jp/) was used to analyses the downregulated DEGs (Log2FC ≤ −0.5) in the focal adhesion signaling pathway and to explore the underlying mechanisms (Figure 4C). Specifically, the downregulation of randomly selected DEGs, namely, itgb3b, itgb1b, mylpfb, vaspa, and pak6b, was determined by qRT‒PCR (Figure 4D), which were also consistent with those of the KEGG enrichment analyses (Figure 4A and B).
We therefore performed a qPCR assay to analyze the transcript levels of the key genes in the focal adhesion pathway, which were identified by KEGG website (https://www.kegg.jp/), in zebrafish treated with or without AgNPs for 72 h (Figure 5). In this context, we noticed a noticeable alteration in the expression of most of the focal adhesion signaling pathway related genes under the higher concentration groups (2 and 4 mg/L) compared to the lower concentration group (1 mg/L). Specifically, after exposure to 2 mg/L AgNPs, the mRNA levels of shc3, akt1, grb2b, itga4, vcla and parva significantly decreased by 0.65-, 0.61-, 0.63-, 0.73-, 0.62- and 0.5-fold, respectively (P < 0.05). In addition, in the 4 mg/L AgNPs exposure group, the mRNA levels of akt3b, ptk2aa, igf1ra, shc3, akt1, grb2b, itga4, vcla and parvaa were decreased by 0.66-, 0.78-, 0.89-, 0.73-, 0.73-, 0.8-, 0.69-, 0.69- and 0.62-fold, respectively (P < 0.05). Notably, the mRNA level of pik3cd was unexpectedly increased by 1.31-, 1.38- and 1.39-fold after treatment with 1, 2 and 4 mg/L AgNPs, respectively, in zebrafish (Figure 5E), suggesting that AgNPs may affect zebrafish embryonic heart development by interfering with the mRNA expression levels of key genes involved in the focal adhesion signaling pathway.
AgNPs Exposure Suppresses the Expression Levels of Focal Adhesion Signaling Pathway-Related Genes
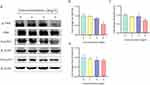
We then determined the expression levels of p-FAK, FAK, Paxillin and Vinculin, which are key focal adhesion pathway genes related to cardiac development, after 72 h of AgNPs exposure (Figure 6A). Western blot analyses indicated that a higher concentration of AgNPs suppressed the expression of genes related to this focal adhesion signaling pathway. Specifically, our findings indicated that the phosphorylation of FAK was dramatically inhibited after AgNPs treatment (P < 0.01, Figure 6B). In addition, the downregulation of paxillin was also detected in zebrafish embryos treated with the high-concentration groups of AgNPs (4 mg/L) (P < 0.05, Figure 6C). Moreover, although not statistically significant, a trend towards the downregulation of Vinculin expression were observed in higher concentrations groups (Figure 6D). Notably, exposure to a lower concentration (1 mg/L) of AgNPs seemed to be insufficient to suppress the focal adhesion signaling pathway, which is consistent with the morphological phenotypes of zebrafish treated with AgNPs at lower concentrations (Figure 1A–F).
Discussion
AgNPs have been shown to have toxic effects, including oxidative stress induction, dysfunction of mitochondria, DNA damage, and apoptosis, both in vivo and in vitro.4,36 In our present study, it was found that the AgNPs exposed could cause cardiac toxicity in zebrafish embryos. The qPCR and WISH analyses revealed that AgNPs interfered with the expression levels of genes related to cardiac development. Transcriptome data showed that AgNPs affected the focal adhesion signaling pathway in zebrafish. The qRT‒PCR analysis showed that the mRNA levels of genes relating to the focal adhesion signaling pathway were regulated after 72 h of AgNPs treatment of zebrafish, suggesting that the focal adhesion signaling pathway was disrupted. In addition, Western blot analysis revealed that AgNPs treatment can affect the protein expression levels of key focal adhesion genes in zebrafish after AgNPs exposure.
AgNPs exposure can cause developmental toxicity in zebrafish embryos. Studies have shown that AgNPs are taken up into embryos via the chorion and distributed throughout developing zebrafish tissues, which could cause an enlarged pore size in the chorion.44 Zebrafish embryos were significantly affected after exposure to 4 nm AgNPs (0.963 mg/L) for 72 hpf, and significant in vivo uptake and delayed yolk sac absorption were evident.41 Our results showed that compared with control zebrafish embryos, exposed zebrafish embryos had growth abnormalities, delayed yolk sac absorption and induced pericardial edema (Figure 1A and B). After exposure to 2 or 4 mg/L AgNPs for 48–72 h, delayed yolk sac absorption and pericardial edema were observed and were significantly different from those in the control group (Figure 1C–F). According to previous studies, exposure to AgNPs can affect the heart rate of zebrafish larvae.37 Xu et al reported that AgNPs could suppress Ca2+ signaling, decrease myogenic locus-specific DNA methylation and reduce heart rate.45
Our results indicated that the transcript levels of zebrafish cardiac development-related genes, such as tnnt2a, nppa and nkx2.5, were increased after 24 h of acute exposure of zebrafish embryos to AgNPs (Figure 2A). As a cardiac contractility-related gene,46 abnormal expression of Tnnt2a can cause hypertrophic cardiomyopathy.47,48 Nppa is known to play an essential part in cardiac development and disease, and has become an important marker for congenital heart defects.48 A recent study indicated that nppa expression is upregulated upon cardiac stress.48 Nkx2.5 has been found to play a key role in determining myocardial cell fate and initiating cardiogenic differentiation.49 Notably, a previous report showed that irregular heartbeats and arrhythmias were mainly due to increased mRNA expression levels of ryr2, tbx5, atp2a2b and nkx2.5.50 Moreover, the upregulation of the transcription of myh7, myh6, nkx2.5, tbx5 and cmlc1 was detected after 48 h of acute AgNPs exposure (Figure 2B). Specifically, Tbx5 is required for heart development and for contacting cardiomyocyte differentiation.51,52 The atrium and the ventricle express the genes myh6 and myh7, respectively; both chambers express myl7.53–55 The role of cardiac myosin light chain-1 (cMLC1) in cardiac development should not be neglected.56 Previous studies indicated that once pathological stress occurs in the heart, the expression of genes related to cardiac development is induced, and this response is thought to play a role in the process of cardiac remodeling and compensation,57–61 which may be orchestrated by a stress-induced regulatory mechanism distinct from that of developmental regulatory programs.62,63 Furthermore, the transcript levels of myh7, myh6, tpm1, nppa, tbx5, tbx20, myl7 and cmlc1 were significantly downregulated after 72 h of AgNPs-treated (Figure 2C), which is similar to previous reports showing that the regulation of the mRNA levels of genes, including myh6, myl7, tnnt2a, nkx2.5 and tbx5, is associated with cardiac developmental and functional defects.64 WISH analysis of zebrafish embryos after 72 h of 4 mg/L AgNPs treatment found that the expression levels of myh7, myh6, nppa, myl7 and cmlc1 were decreased (Figure 2D).
Transcriptome analysis of zebrafish embryos after 72 h of AgNPs treatment revealed that the downregulated DEGs were mainly enriched in focal adhesion signaling pathways under high-concentration treatment (2 and 4 mg/L) (Figure 3A and B). GSEA revealed that the focal adhesion pathway was mainly enriched in the downregulated DEGs in the AgNPs treatment group (Figure 3C and D). In addition, transcriptome expression levels of genes associated with focal adhesion pathway were analyzed, and the DEGs were also validated and analyzed (Figure 4). It was found that the expression levels of focal adhesion pathway-related genes were mostly suppressed under high-concentration (2 and 4 mg/L) of AgNPs treatment (Figure 4). A previous study indicated that AgNPs exposure causes adverse effects on the cell adhesion dynamics and basement membrane.65 Focal adhesion is necessary in cell survival, cell motility, and proliferation.18,66 The lack of function of focal adhesion proteins during embryogenesis could impact the ECM adhesion, cytoskeletal organization, and embryonic survival.67
In order to further investigate the genetic toxicity of AgNPs exposure in zebrafish, we have quantified the mRNA levels of several genes related to the focal adhesion pathway. The mRNA levels of igf1ra, shc3, grb2b, ptk2aa, akt1, itga4, parvaa, akt3b and vcla decreased after 72 h of AgNPs treatment (Figure 5). In this context, the knockdown of SHC expression can inhibit the PI3K-AKT-FoxO pathway through suppressing Akt and FoxO phosphorylation.68 Akt is known to be an essential regulator of cell viability, cell death and the function of mitochondria.69 An inactivating mutation in the grb2b gene can cause severe lymphatic damage.70 Deficiencies of itga5 and itga4 can lead to serious myocardial defects and the morphological defects of the cardia bifida.71 Vcla is known as a vinculin paralog and is associated with epicardial hyperplasia and coronary vessel disorganization.72 Fak1a and fak1b (ptk2aa) deficiency might result in heart failure.29 Integrin β-1 (ITGB1), a member of the integrin family, mediates adhesion to extracellular matrices and facilitates cell-to-cell contact.73,74 The insulin-like growth factor (IGF) system is essential for growth regulation.75 Parvin binds to PINCH proteins and integrin-linked kinases in a protein complex, and this complex is closely associated with heart failure in zebrafish.76 Our findings suggested that AgNPs exposure could cause zebrafish cardiac developmental toxicity by interfering the mRNA levels of the focal adhesion pathway-related genes.
Notably, focal adhesion components also play important roles in cellular migration and differentiation during cardiac development,77,78 which is consistent with our finding that the protein expression levels of p-FAK and paxillin decreased in zebrafish after 72 h of 4 mg/L AgNPs exposure (Figure 6). FAK is a candidate enzyme and its phosphorylation is responsible for integrin-mediated mechanotransduction in cardiomyocyte focal adhesions and costameres.78 In addition, the autophosphorylation site of FAK (Y397), which is a high affinity binding site for the SH2 domain or Src family protein tyrosine kinases,79 can phosphorylate paxillin and other cytoskeletal proteins involved in myofibrillar assembly.78 A previous report showed that inactivation of paxillin’s target gene to progressive reduced cardiac contractility and heart failure in zebrafish.29 FAK and paxillin are both essential for recruiting and stabilizing vinculin to regulate cardiac contractility.37 In summary, our findings indicated that AgNPs exposure may result in cardiac dysgenesis by affecting the phosphorylation of FAK and the expression of paxillin in zebrafish.
Conclusion
In summary, our results demonstrated that AgNPs cause cardiac developmental toxicity in zebrafish embryos. Analysis of zebrafish morphology via qRT‒PCR and WISH revealed that AgNPs can induce abnormal cardiac development and alter the mRNA levels of cardiac development-related genes. RNA-seq analysis revealed that AgNPs exposure disturbs the focal adhesion signaling pathway in zebrafish. Moreover, exposure to AgNPs for 72 h inhibited the focal adhesion pathway by regulating the mRNA levels of several focal adhesion pathway-related genes in zebrafish. Furthermore, the protein levels of focal adhesion pathway-related genes were also affected after AgNP treatment. AgNPs may cause cardiac developmental toxicity by affecting the focal adhesion signaling pathway.
Acknowledgment
This work is supported by the National Natural Science Foundation of China (81872070) and the Natural Science Foundation of Guangdong Province (2022A1515012424, 2023A1515011749), and GuangDong Basic and Applied Basic Research Foundation (2023A1515110234).
Author Contributions
Chunjiao Lu designed the experiment and acquired and analyzed the data. Xuewei Wu, Xin Meng, Yi Liu, Ting Yang, Yan Zeng, Yang Chen, Yishan Huang, Zhou Fang analysed the data. Juanjuan Luo and Xiaojun Yang supervised the project. All the authors reviewed and approved the final manuscript. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int. 2013;2013(942916):1–15. doi:10.1155/2013/942916
2. Shvedova AA, Pietroiusti A, Fadeel B, Kagan VE. Mechanisms of carbon nanotube-induced toxicity: focus on oxidative stress. Toxicol Appl Pharmacol. 2012;261(2):121–133. doi:10.1016/j.taap.2012.03.023
3. Roy R, Kumar S, Tripathi A, Das M, Dwivedi PD. Interactive threats of nanoparticles to the biological system. Immunol Lett. 2014;158(1–2):79–87. doi:10.1016/j.imlet.2013.11.019
4. Jiang X, Lu C, Tang M, et al. Nanotoxicity of Silver Nanoparticles on HEK293T Cells: a Combined Study Using Biomechanical and Biological Techniques. ACS Omega. 2018;3(6):6770–6778. doi:10.1021/acsomega.8b00608
5. Cuillel M, Chevallet M, Charbonnier P, et al. Interference of CuO nanoparticles with metal homeostasis in hepatocytes under sub-toxic conditions. Nanoscale. 2014;6(3):1707–1715. doi:10.1039/C3NR05041F
6. Liu J, Hurt RH. Ion release kinetics and particle persistence in aqueous nano-silver colloids. Environ Sci Technol. 2010;44(6):2169–2175. doi:10.1021/es9035557
7. Wang N, Tong T, Xie M, Gaillard JF. Lifetime and dissolution kinetics of zinc oxide nanoparticles in aqueous media. Nanotechnology. 2016;27(32):324001. doi:10.1088/0957-4484/27/32/324001
8. Behra R, Sigg L, Clift MJ, et al. Bioavailability of silver nanoparticles and ions: from a chemical and biochemical perspective. J Royal Soc Interface. 2013;10(87):20130396. doi:10.1098/rsif.2013.0396
9. Liao C, Li Y, Tjong SC. Bactericidal and Cytotoxic Properties of Silver Nanoparticles. Int J Mol Sci. 2019;20(2):449. doi:10.3390/ijms20020449
10. Xu L, Wang YY, Huang J, Chen CY, Wang ZX, Xie H. Silver nanoparticles: synthesis, medical applications and biosafety. Theranostics. 2020;10(20):8996–9031. doi:10.7150/thno.45413
11. Chernousova S, Epple M. Silver as antibacterial agent: ion, nanoparticle, and metal. Angew Chem. 2013;52(6):1636–1653. doi:10.1002/anie.201205923
12. Mitrano DM, Rimmele E, Wichser A, Erni R, Height M, Nowack B. Presence of nanoparticles in wash water from conventional silver and nano-silver textiles. ACS nano. 2014;8(7):7208–7219. doi:10.1021/nn502228w
13. Syafiuddin A, Salmiati S, Hadibarata T, Kueh ABH, Salim MR, Zaini MAA. Silver Nanoparticles in the Water Environment in Malaysia: inspection, characterization, removal, modeling, and future perspective. Sci Rep. 2018;8(1):986. doi:10.1038/s41598-018-19375-1
14. Ong C, Lee QY, Cai Y, et al. Silver nanoparticles disrupt germline stem cell maintenance in the Drosophila testis. Sci Rep. 2016;6(20632). doi:10.1038/srep20632
15. Asharani PV, Lian Wu Y, Gong Z, Valiyaveettil S. Toxicity of silver nanoparticles in zebrafish models. Nanotechnology. 2008;19(25):255102. doi:10.1088/0957-4484/19/25/255102
16. Botelho DJ, Leo BF, Massa CB, et al. Low-dose AgNPs reduce lung mechanical function and innate immune defense in the absence of cellular toxicity. Nanotoxicology. 2016;10(1):118–127. doi:10.3109/17435390.2015.1038330
17. De Jong WH, Van Der Ven LT, Sleijffers A, et al. Systemic and immunotoxicity of silver nanoparticles in an intravenous 28 days repeated dose toxicity study in rats. Biomaterials. 2013;34(33):8333–8343. doi:10.1016/j.biomaterials.2013.06.048
18. Saeed-Zidane M, Tesfaye D, Mohammed Shaker Y, et al. Hyaluronic acid and epidermal growth factor improved the bovine embryo quality by regulating the DNA methylation and expression patterns of the focal adhesion pathway. PLoS One. 2019;14(10):e0223753. doi:10.1371/journal.pone.0223753
19. Mwpl R, Schwartz MA, Schwartz MA. focal adhesion kinase mediates the integrin signaling requirement for growth factor activation of MAP kinase. J Cell Biol. 1999;147(3):611–618. doi:10.1083/jcb.147.3.611
20. Henry CA, Crawford BD, Yan Y-L, Postlethwait J, Cooper MS, Hille MB. Roles for Zebrafish Focal Adhesion Kinase in Notochord and Somite Morphogenesis. Dev Biol. 2001;240(2):474–487. doi:10.1006/dbio.2001.0467
21. Xu Peng J-LG, Guan J-L. Focal Adhesion Kinase: from In Vitro Studies to Functional Analyses In Vivo. Curr Protein Pept Sci. 2011;12:52–67. doi:10.2174/138920311795659452
22. Wu C. Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh Migr. 2007;1(1):13–18.
23. Ilić D, Furuta Y, Kanazawa S, et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377(6549):539–544. doi:10.1038/377539a0
24. Ilic D, Kovacic B, McDonagh S, et al. Focal adhesion kinase is required for blood vessel morphogenesis. Circ Res. 2003;92(3):300–307. doi:10.1161/01.RES.0000055016.36679.23
25. Braren R, Hu H, Kim YH, Beggs HE, Reichardt LF, Wang R. Endothelial FAK is essential for vascular network stability, cell survival, and lamellipodial formation. J Cell Biol. 2006;172(1):151–162. doi:10.1083/jcb.200506184
26. Shen TL, Park AY, Alcaraz A, et al. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol. 2005;169(6):941–952. doi:10.1083/jcb.200411155
27. Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20(44):6459–6472. doi:10.1038/sj.onc.1204786
28. Pardo JV, Siliciano JD, Craig SW. Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J Cell Biol. 1983;97(4):1081–1088. doi:10.1083/jcb.97.4.1081
29. Hirth S, Buhler A, Buhrdel JB, et al. Paxillin and Focal Adhesion Kinase (FAK) Regulate Cardiac Contractility in the Zebrafish Heart. PLoS One. 2016;11(3):e0150323. doi:10.1371/journal.pone.0150323
30. Zemljic-Harpf AE, Miller JC, Henderson SA, et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol. 2007;27(21):7522–7537. doi:10.1128/MCB.00728-07
31. Hagel M, George EL, Kim A, et al. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22(3):901–915. doi:10.1128/MCB.22.3.901-915.2002
32. Horzmann KA, Freeman JL. Making Waves: new Developments in Toxicology With the Zebrafish. Toxicol Sci. 2018;163(1):5–12. doi:10.1093/toxsci/kfy044
33. Alessandrini F, Vennemann A, Gschwendtner S, et al.: Pro-Inflammatory versus Immunomodulatory Effects of Silver Nanoparticles in the Lung: The Critical Role of Dose, Size and Surface Modification. Nanomaterials 2017, 7(10).
34. Vidanapathirana AK, Thompson LC, Herco M, et al. Acute intravenous exposure to silver nanoparticles during pregnancy induces particle size and vehicle dependent changes in vascular tissue contractility in Sprague Dawley rats. Rep Toxicol. 2018;75:10–22. doi:10.1016/j.reprotox.2017.11.002
35. Massarsky A, Dupuis L, Taylor J, et al. Assessment of nanosilver toxicity during zebrafish (Danio rerio) development. Chemosphere. 2013;92(1):59–66. doi:10.1016/j.chemosphere.2013.02.060
36. Y-B M, C-J L, Junaid M, et al. Potential adverse outcome pathway (AOP) of silver nanoparticles mediated reproductive toxicity in zebrafish. Chemosphere. 2018;207:320–328. doi:10.1016/j.chemosphere.2018.05.019
37. Lu C, Lv Y, Kou G, et al. Silver nanoparticles induce developmental toxicity via oxidative stress and mitochondrial dysfunction in zebrafish (Danio rerio). Ecotoxicol Environ Saf. 2022;243:113993. doi:10.1016/j.ecoenv.2022.113993
38. Gonzalez-Bulnes A, Jia -P-P, Ma Y-B, et al. The Effects of Disturbance on Hypothalamus-Pituitary-Thyroid (HPT) Axis in Zebrafish Larvae after Exposure to DEHP. PLoS One. 2016;11(12). doi:10.1371/journal.pone.0167858
39. Christen V, Capelle M, Fent K. Silver nanoparticles induce endoplasmatic reticulum stress response in zebrafish. Toxicol Appl Pharmacol. 2013;272(2):519–528. doi:10.1016/j.taap.2013.06.011
40. Yang H, Liang X, Zhao Y, et al. Molecular and behavioral responses of zebrafish embryos/larvae after sertraline exposure. Ecotoxicol Environ Saf. 2021;208:111700. doi:10.1016/j.ecoenv.2020.111700
41. Qiang L, Arabeyyat ZH, Xin Q, et al. Silver Nanoparticles in Zebrafish (Danio rerio) Embryos: uptake, Growth and Molecular Responses. Int J Mol Sci. 2020;21(5):1876. doi:10.3390/ijms21051876
42. Shi L, Qian Y, Shen Q, He Y, Jia Y, Wang F. The developmental toxicity and transcriptome analyses of zebrafish (Danio rerio) embryos exposed to carbon nanoparticles. Ecotoxicol Environ Saf. 2022;234:113417. doi:10.1016/j.ecoenv.2022.113417
43. Liu Y, Junaid M, Wang Y, et al. New toxicogenetic insights and ranking of the selected pharmaceuticals belong to the three different classes: a toxicity estimation to confirmation approach. Aquat Toxicol. 2018;201:151–161. doi:10.1016/j.aquatox.2018.06.008
44. Chen ZY, Li NJ, Cheng FY, et al. The Effect of the Chorion on Size-Dependent Acute Toxicity and Underlying Mechanisms of Amine-Modified Silver Nanoparticles in Zebrafish Embryos. Int J Mol Sci. 2020;21(8):2864.
45. Xu QH, Guan P, Zhang T, Lu C, Li G, Liu JX. Silver nanoparticles impair zebrafish skeletal and cardiac myofibrillogenesis and sarcomere formation. Aquat Toxicol. 2018;200:102–113. doi:10.1016/j.aquatox.2018.04.018
46. Staudt DW, Liu J, Thorn KS, Stuurman N, Liebling M, Stainier DY. High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development. 2014;141(3):585–593. doi:10.1242/dev.098632
47. Maron BJ. Hypertrophic CardiomyopathyA Systematic Review. JAMA. 2002;287(10):1308–1320. doi:10.1001/jama.287.10.1308
48. Laing NG. Congenital myopathies. Curr Opin Neurol. 2007;20(5):583–589. doi:10.1097/WCO.0b013e3282ef6e69
49. Balci MM, Akdemir R. NKX2.5 mutations and congenital heart disease: is it a marker of cardiac anomalies? Int J Cardiol. 2011;147(3):e44–45. doi:10.1016/j.ijcard.2009.01.024
50. Wu M, Zuo Z, Li B, Huang L, Chen M, Wang C. Effects of low-level hexabromocyclododecane (HBCD) exposure on cardiac development in zebrafish embryos. Ecotoxicology. 2013;22(8):1200–1207. doi:10.1007/s10646-013-1107-4
51. Hiroi Y, Kudoh S, Monzen K, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28(3):276–280. doi:10.1038/90123
52. Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459(7247):708–711. doi:10.1038/nature08039
53. Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130(24):6121–6129. doi:10.1242/dev.00838
54. England J, Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci. 2013;70(7):1221–1239. doi:10.1007/s00018-012-1131-1
55. Chen X, Li W. Isoflucypram cardiovascular toxicity in zebrafish (Danio rerio). Sci Total Environ. 2021;787(147529):147529. doi:10.1016/j.scitotenv.2021.147529
56. Shiroorkar PN, Afzal O, Kazmi I, et al. Cardioprotective Effect of Tangeretin by Inhibiting PTEN/AKT/mTOR Axis in Experimental Sepsis-Induced Myocardial Dysfunction. Molecules. 2020;25(23):5622. doi:10.3390/molecules25235622
57. Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci USA. 1988;85(2):339–343. doi:10.1073/pnas.85.2.339
58. Chien KR, Knowlton KU, g Z, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5(15):3037–3064. doi:10.1096/fasebj.5.15.1835945
59. Komuro I, Yazaki Y. Control of Cardiac Gene Expression by Mechanical Stress. Ann Rev Physiol. 1993;55(1):55–75. doi:10.1146/annurev.ph.55.030193.000415
60. Kuwahara K, Nishikimi T, Nakao K. Transcriptional regulation of the fetal cardiac gene program. J Pharmacol Sci. 2012;119(3):198–203. doi:10.1254/jphs.12R04CP
61. Man J, Barnett P, Christoffels VM. Structure and function of the Nppa-Nppb cluster locus during heart development and disease. Cell Mol Life Sci. 2018;75(8):1435–1444. doi:10.1007/s00018-017-2737-0
62. Horsthuis T, Houweling AC, Habets PE, et al. Distinct regulation of developmental and heart disease-induced atrial natriuretic factor expression by two separate distal sequences. Circ Res. 2008;102(7):849–859. doi:10.1161/CIRCRESAHA.107.170571
63. Sergeeva IA, Hooijkaas IB, Ruijter JM, et al. Identification of a regulatory domain controlling the Nppa-Nppb gene cluster during heart development and stress. Development. 2016;143(12):2135–2146. doi:10.1242/dev.132019
64. Wu Y, Yang Q, Chen M, Zhang Y, Zuo Z, Wang C. Fenbuconazole exposure impacts the development of zebrafish embryos. Ecotoxicol Environ Saf. 2018;158:293–299. doi:10.1016/j.ecoenv.2018.04.048
65. Martin ME, Reaves DK, Jeffcoat B, et al. Silver nanoparticles alter epithelial basement membrane integrity, cell adhesion molecule expression, and TGF-beta1 secretion. Nanomedicine. 2019;21:102070. doi:10.1016/j.nano.2019.102070
66. Wrighton KH. The ‘ins’ and ‘outs’ of integrin signalling. Nat Rev Mol Cell Biol. 2013;14(12):753. doi:10.1038/nrm3708
67. Bladt F, Aippersbach E, Gelkop S, et al. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol Cell Biol. 2003;23(13):4586–4597. doi:10.1128/MCB.23.13.4586-4597.2003
68. Gong J, Zhang L, Zhang Q, et al. Lentiviral Vector-Mediated SHC3 Silencing Exacerbates Oxidative Stress Injury in Nigral Dopamine Neurons by Regulating the PI3K-AKT-FoxO Signaling Pathway in Rats with Parkinson’s Disease. Cell Physiol Biochem. 2018;49(3):971–984. doi:10.1159/000493228
69. Moore NS, Mans RA, McCauley MK, Allgood CS, Barksdale KA. Critical Effects on Akt Signaling in Adult Zebrafish Brain Following Alterations in Light Exposure. Cells. 2021;10(3):637. doi:10.3390/cells10030637
70. Mauri C, van Impel A, Mackay EW, Schulte-Merker S. The adaptor protein Grb2b is an essential modulator for lympho-venous sprout formation in the zebrafish trunk. Angiogenesis. 2021;24(2):345–362. doi:10.1007/s10456-021-09774-w
71. Schumacher JA, Wright ZA, Owen ML, Bredemeier NO, Sumanas S. Integrin α5 and Integrin α4 cooperate to promote endocardial differentiation and heart morphogenesis. Dev Biol. 2020;465(1):46–57. doi:10.1016/j.ydbio.2020.06.006
72. Cheng F, Miao L, Wu Q, Gong X, Xiong J, Zhang J. Vinculin b deficiency causes epicardial hyperplasia and coronary vessel disorganization in zebrafish. Development. 2016;143(19):3522–3531. doi:10.1242/dev.132936
73. Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268(5208):233–239. doi:10.1126/science.7716514
74. Chen G, Xiong L, Wang Y, et al. ITGB1b-Deficient Rare Minnows Delay Grass Carp Reovirus (GCRV) Entry and Attenuate GCRV-Triggered Apoptosis. Int J Mol Sci. 2018;19(10):3175. doi:10.3390/ijms19103175
75. Nelson SN, Van Der Kraak G. Characterization and regulation of the insulin-like growth factor (IGF) system in the zebrafish (Danio rerio) ovary. General Comparat Endocrinol. 2010;168(1):111–120. doi:10.1016/j.ygcen.2010.04.020
76. Meder B, Huttner IG, Sedaghat-Hamedani F, et al. PINCH proteins regulate cardiac contractility by modulating integrin-linked kinase-protein kinase B signaling. Mol Cell Biol. 2011;31(16):3424–3435. doi:10.1128/MCB.05269-11
77. Fässler R, Rohwedel J, Maltsev V, et al. Differentiation and integrity of cardiac muscle cells are impaired in the absence of beta 1 integrin. J Cell Sci. 1996;109(13):2989–2999. doi:10.1242/jcs.109.13.2989
78. Samarel AM. Focal adhesion signaling in heart failure. Pflugers Arch. 2014;466(6):1101–1111. doi:10.1007/s00424-014-1456-8
79. Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol Cell Biol. 1994;14(3):1680–1688. doi:10.1128/mcb.14.3.1680-1688.1994
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.