Back to Journals » Infection and Drug Resistance » Volume 18
Staphylococcal Drug Resistance: Mechanisms, Therapies, and Nanoparticle Interventions
Authors Shao K, Yang Y, Gong X, Chen K, Liao Z, Ojha SC
Received 3 December 2024
Accepted for publication 6 February 2025
Published 19 February 2025 Volume 2025:18 Pages 1007—1033
DOI https://doi.org/10.2147/IDR.S510024
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Kunyu Shao,1,* Yuxun Yang,1,* Xuankai Gong,1,* Ke Chen,1,2 Zixiang Liao,1 Suvash Chandra Ojha2
1School of Clinical Medicine, Southwest Medical University, Luzhou, 646000, People’s Republic of China; 2Department of Infectious Diseases, the Affiliated Hospital of Southwest Medical University, Luzhou, 646000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Suvash Chandra Ojha, Email [email protected]
Abstract: The increasing incidence of antibiotic resistance in Staphylococcus aureus (S. aureus) poses a substantial threat to global public health. In recent decades, the evolution of bacteria and the misuse of antibiotics have led to a progressive development in drug resistance of S. aureus, resulting in a worldwide rise in methicillin-resistant S. aureus (MRSA) infection rates. Understanding the molecular mechanisms underlying staphylococcal drug resistance, the treatments for staphylococcal infections, and the efficacy of nanomaterials in addressing multi-drug resistance is crucial. This review explores the resistance mechanisms, which include limiting drug uptake, target modification, drug inactivation through the production of degrading enzymes, and active efflux of drugs. It also examines the current therapeutic strategies, such as antibiotic combination therapy, phage therapy, monoclonal antibody therapy, and nanoparticle therapy, with a particular emphasis on the role of silver-based nanomaterials. Nanoparticles possess the ability to overcome multi-drug resistance, offering a novel avenue for the management of drug-resistant bacteria. The nanomaterials have demonstrated potent antibacterial activity against S. aureus through various mechanisms, including cell membrane disruption, generation of reactive oxygen species (ROS), and inhibition of essential cellular processes. It also highlights the need for further research to optimize nanoparticle design, enhance their antibacterial potency, and ensure their biocompatibility and biodegradability. The review ultimately concludes by emphasizing the importance of a multifaceted approach to treatment, including the development of new antibiotics, investment in stewardship programs to prevent antibiotic misuse, and the exploration of natural compounds and bacteriocins as potential antimicrobial agents.
Keywords: Staphylococcus aureus, MRSA, drug resistance mechanism, antimicrobial therapy, nanomaterials’ antistaphylococcal mechanism
Background
Bacterial resistance is at an alarming level, leading to both life-threatening bloodstream infections and an increase in resistance for several bacteria in communities. Notable among the plethora of bacteria is S. aureus, a genus of Gram-positive bacteria of the family Staphylococcaceae.1 The Lancet systematic analysis of the global burden of antimicrobial resistance in 2019 states that S. aureus is the second leading pathogen of death associated with resistance.2 In 2019, methicillin-resistant S. aureus (MRSA) alone caused more than 100,000 deaths.2 The 2024 WHO Bacterial Priority Pathogens List updates and refines the prioritization of antibiotic-resistant bacterial pathogens in order to address the evolving challenges posed by antibiotic resistance.3 The list categorizes these pathogens into critical, high, and medium priority groups to guide research and development (R&D) and public health interventions, reflecting the urgent need for new antibiotics, with S. aureus designated as a high priority.
Typically, the antibiotics utilized for the treatment of S. aureus infections fall into several categories, including β-lactam antibiotics, glycopeptide antibiotics, oxazolidinone antibiotics, lipopeptide antibiotics, and others (Table 1). Among these, the penicillin, methicillin, and vancomycin resistance mechanisms have received the utmost attention. Many S. aureus strains have developed resistance through the production of β-lactamase enzymes, which hydrolyze the β-lactam ring, thereby rendering the antibiotic ineffective.4 The presence of PBP2a (encoded by mecA) reduces the affinity of the bacteria for β-lactam antibiotics, allowing them to survive in the presence of methicillin.5 Vancomycin resistance in S. aureus is rare but has been reported.6 The resistance is primarily due to the acquisition of resistance genes from vancomycin-resistant enterococci (VRE). The most common resistance mechanism involves the substitution of D-Ala-D-Ala with D-Ala-D-Lac or D-Ala-D-Ser in the peptidoglycan precursors, which reduces the binding affinity of vancomycin.6 Due to the increasing resistance of S. aureus, the total number of antibiotics effective against S. aureus is declining, leaving us with fewer and fewer effective antibiotics to use in the future.
 |
Table 1 Antibiotics Employed to Combat S. aureus |
A variety of treatments for Staphylococcus have been proposed, including antibiotic combination therapy, phage therapy, antibody therapy, vaccine therapy, natural compound therapy, and nanoparticle therapy.7,8 Of these, nanoparticles (NPs) are increasingly being employed to target bacteria as an alternative to antibiotics and appear to have high potential in addressing the emergence of multi-resistant bacteria.9–14 Studies have shown that NPs are less likely than antibiotics to promote resistance in bacteria.13,14 Therefore, novel NP-based materials exhibiting antibacterial properties have attracted peoples’ attention, particularly silver-based nanoparticles, which are of particular interest.
At present, some literature has described the resistance mechanism of S. aureus to specific antibiotics, while other articles have addressed antibiotic resistance in a general manner.4–6 However, there is a lack of comprehensive summaries regarding the resistance mechanism of S. aureus to currently used antibiotics.8 Simultaneously, certain literatures outlines new treatment schemes and relevant action targets for drug-resistant bacteria in the future.7 However, there is an absence of a comprehensive overview about the potential advantages and downsides of these treatment schemes. Therefore, we consolidate the information pertaining to this subject and examine the molecular basis of staphylococcal drug resistance, the current therapeutics utilized in staphylococcal treatment, and elucidate the mechanisms through which nanoparticles counteract multidrug resistance, thereby facilitating the application of nanomaterials in the management of drug-resistant bacteria.
Drug Resistance Mechanism
Antibiotics are among the most significant medical innovations of the twentieth century, clearly benefiting humanity in the battle against bacteria and saving millions of lives.15 However, the excessive use of antibiotics has led to the emergence of multidrug-resistant strains (MDR), with S. aureus serving as a prominent example. The drug resistance mechanism of S. aureus is mainly divided into limiting drug uptake, target modification, drug inactivation (degrading enzymes), and active efflux of drugs. This section examines the molecular processes of antibiotic resistance in S. aureus, with an emphasis on the main targets of antibiotics, which are the cell envelope, ribosomes, and nucleic acids (Figure 1).
 |
Figure 1 Therapeutic targets of antibiotics against S. aureus. |
Antibiotics Targeting Cell Envelope
β-Lactam Antibiotics
The primary inhibitory target of S. aureus against β-lactam antibiotics is the bifunctional transglycolytic enzyme-transpeptidase PBP2 (Figure 2). The structure of β-lactam antibiotics is similar to the D-Ala-D-Ala terminal pentapeptide of lipid II. The latter serves as the natural substrate for the transpeptidase penicillin-binding proteins (PBPs).16 β-lactam antibiotics competitively inhibit the transpeptidase, thus affecting bacterial cell wall synthesis and ultimately leading to bacterial cell death. However, penicillin and other β-lactam antibiotics are also abused. A large number of S. aureus bacteria were found to have a plasmid that encodes the penicillium enzyme braZ. This plasmid is capable of hydrolyzing the β-lactam ring, rendering it unable to bind to PBPs17 (Figure 2a). Consequently, S. aureus acquires pharmacological resistance, exemplified as penicillin-resistant S. aureus (PRSA).
To treat the resistant bacteria, researchers eventually turned to methicillin (MET), which was susceptible to penicillinase action. However, the identification of a low-affinity PBP (PBP2A/PBP2′) indicates that S. aureus has acquired resistance to MET and certain cephalosporins (Figure 1). This low-affinity PBP is encoded by the mecA gene in the staphylococcal chromosome cassette mec (SCCmec). The resistant strain to the modified β-lactam is referred to as MRSA.
Vancomycin and Other Glycopeptides
The widespread occurrence of MRSA has led to the global use of glycopeptides like vancomycin (VAN), which were previously regarded as “last-resort” antimicrobial agents. Glycopeptides similarly inhibit cell wall synthesis. These antibiotics bind to the D-Ala-D-Ala residue of lipid II, resulting in the cessation of the transpeptidase reaction due to the depletion of the substrate required for the reaction. However, the emergence of vancomycin intermediate-type S. aureus (hVISA) has created a novel challenges for individuals. The hVISA is a precursor of vancomycin intermediate-type S. aureus (VISA) with a minimum inhibitory concentration (MIC) of 48 μg/mL.
The emergence of VISA is closely related to genetic mutations, with the most prominent manifestation being its impact on UDP-MurNAc-L-Ala-D-iso-Gln-L-Lys-D-Ala-D-Ala, the main component of the bacterial cell wall. The vanA operon associated with vanA-type vancomycin resistance is influenced by the genes vanA, vanH, vanX, vanS, vanR, vanY, and vanZ.18 The vanA operon-mediated resistance requires two key events: 1) Hydrolysis of the dipeptide D-Ala-D-Ala peptidoglycan precursor binding to vancomycin; 2) Synthesis of D-Ala-D-lactate peptidoglycan precursor that does not bind to vancomycin. The latter characteristic is to reduce its vulnerability to vancomycin by altering the cell wall constituents (Figure 3).
Naturally, there are alternative methods for producing VISA. Mutations in genes that stimulate cell wall stress, including walRK, yycH, vraRS, graRS, rpoB, and tcaA may also result in the emergence of VISA. These mutations reduce the level of crosslinks within the cell wall, resulting in an increased number of free D-Ala-D-Ala residues, hence generating a certain number of “false targets”. Thus, VAN is sequestered within the cell wall rather than reaching lipid II targets on the membrane.19 This mechanism of resistance is called “drug catch” or “blockage” (Figure 4).
Studies have pointed out that the underlying molecular basis of insensitivity extends beyond alterations in cell wall structure, with the increased thickness of the cell wall also affects the bactericidal effect of vancomycin.20 The thickening of the bacterial cell membrane necessitates that the drug traverse a greater distance to engage with lipid II. The presence of the bound “sham target” significantly diminishes the binding affinity of VAN to the pertinent target site. Furthermore, large bulk glycopeptides aggregate bound to the enlarged outer layer of the cell wall can hinder drug diffusion. In some VISA strains, the increased level of D-Ala substitution on phosphonic acid altered the charge of the cell envelope, resulting in the repulsion of positively charged glycopeptides.
Daptomycin (DAP)
DAP exerts its bactericidal effect by interacting with cell membranes and phospholipids, a mechanism that has not been fully elucidated.21,22 It is established that DAP requires Ca2+ ions to maintain antimicrobial activity. The positive charge of the Dap-Ca2+ complex binds to the negatively charged phosphatidylglycerol (PG) headgroup, and this induced tension is thought to facilitate the insertion of the antibiotic into the bacterial cell membrane22 (Figure 2c).
Resistance to DAP typically results from a point mutation in MprF. The MrpF protein functions as an integral membrane protein that incorporates positively charged lysine residues into PG to form lysine-phosphatidylglycerol (L-PG). This compound is synthesized in the cytoplasm and subsequently transferred to the outer membrane surface. They cause altered gain-of-function of proteins that increase the levels of L-PG in the membrane. This increases the charge on the outer surface of the membrane, thereby rejecting Dap-Ca2+ and a reduction in the PG level.23,24 It has been suggested that MprF-mediated DAP resistance can increase the cell wall thickness. This is very similar to the “drug capture” mechanism of vancomycin.
Antibiotics Targeting the Ribosomes
Antibiotic binding sites on the ribosome are concentrated at two major sites: one on the 50S subunit near the polypeptide exit channel around the peptidyl transferase center (PTC); and the other on the 30S subunit at site A, where the incoming aminoacyl-tRNA attaches. All antibiotics targeting the ribosome have been demonstrated to bind to rRNA (16S or 23S).
Antibiotics That Inhibit Protein Synthesis in the 30S Subunit
Drugs that inhibit the 30S subunit of the ribosome, include aminoglycosides and tetracyclines.
Aminoglycosides (AGs)
AGs are the only ribosome-targeting antibiotics with bactericidal activity, primarily through disrupting bacterial protein synthesis.25 They bind to the 30S ribosomal subunit, inducing translation errors by increasing the amino acid misincorporation rate from less than 1 in 1,000 to approximately 1 in 100. This results in the production of functionally impaired proteins, particularly membrane proteins, which compromises cell membrane integrity and leads to increased permeability and leakage of essential cellular components. Additionally, AGs induce clusters of translation errors, amplifying downstream error rates and further contributing to proteotoxic stress. Collectively, these mechanisms, which include increased translation errors, membrane protein malfunction, and compromised membrane integrity, all contribute to AGs’ bactericidal effect.
Resistance mechanism to AGs antibiotics in pathogenic bacteria include aminoglycoside-modifying enzymes (AMEs), mutations and modifications of ribosomal targets, and efflux pumps.26,27 The predominant mechanism of resistance to aminoglycoside antibiotics is the inactivation by modifying enzymes. Among the AMEs are acetyltransferases, phosphotransferases, and nucleotidyl transferases. These modifications reduce the binding affinity of the drug to the target and result in a reduction of antimicrobial potency. Adenosine transferases are enzymes associated with drug resistance that hold clinical significance.
The resistance of AGs to S. aureus is due to the acquisition of cytoplasmic AMEs. This enzyme can modify AGs antibiotic molecules by adding, altering, or removing chemical functional groups, thus reducing their bactericidal capacity and bacteriostatic effect on target cells, resulting in drug resistance. Aminoglycoside resistance is dependent on the acquisition of AMEs including aac(6′)-Ie+aph(2″), ant(4′)-Ia, aph(3′)-IIIa, and ant(6)-Ia genes, which inactivate the antibiotic.28,29
Modification of ribosomal targets is another resistance mechanism, specifically the methylation of 16S ribosomal RNA by methyltransferase. These enzymes alter specific rRNA nucleotides, inhibiting the efficient binding of the AGs to their targets (Figure 5). Based on the nucleotide position altered at 16S rRNA, the methyltransferases associated with AGs resistance can be categorized into N7-G1405 (methylation at the N7 position of guanine-1405) 16S rRNA methyltransferase and N1-A1408 (N1 position methylation of adenine-1408) 16S rRNA methyltransferase. Members of the drug-nodule-division (RND) family of efflux systems have been shown to contribute to in the intrinsic aminoglycoside resistance of various pathogens.
Tetracycline (TET)
TET prevents the binding of aminoacyl tRNA via interaction with the 16S rRNA A site. TET resistance occurs through at least four mechanisms: binding site mutations, ribosome-protective proteins, efflux pumps, and enzyme inactivation. S. aureus uses two primary ways to evade its destruction.30 The initial mechanism involves the active expulsion of the antibiotic via the plasmid-mediated TetK and TetK MFS efflux pumps (Figure 6a). The active removal of TET from S. aureus cells is regulated by membrane proteins Tet (K), Tet (L), Tet (38), Tet (42), Tet (43), Tet (45), and Tet (63), utilizing energy from a proton pump and categorized as major facilitator superfamily (MFS). TetK is usually located on a plasmid within the MRSA strain SCCmecIII. The second mechanism involves the expression of TetO and TetM, two ribosome-protecting proteins (RPPs) that bind to the ribosome and obstruct TET from accessing its targets. Tet (M) protein determines resistance to TET, particularly minocycline, and is usually responsible for TET resistance in S. aureus. The tet (M) gene is locate inside the chromosomes of numerous S. aureus strains. Notably, these proteins, despite possessing GTPase activity, do not serve as elongation factors.
Antibiotics That Inhibit Protein Biosynthesis at the 50S Subunit
Chloramphenicol (CHL)
In S. aureus, resistance manifests through several mechanisms: acquisition of the chloramphenicol-florfenicol resistance cfr gene, which encodes a ribosomal RNA methyltransferase that methylates an adenine residue at position 2503 of the 23S rRNA V domain, hence inhibiting CHL activity.31,32
A contributing factor to CHL resistance is the synthesis of chloramphenicol acetyltransferases CATA 7, CATA 8, and CATA 9.33,34 Alternatively, CHL functions by reversible binding to the L16 protein of the 50S ribosomal subunit. Since L16 can directly interact with 23S rRNA in the 50S peptidyl transferase center, CHL can specifically disrupt peptide bond formation. The resistance of S. aureus to CHL occurs through several ways: 1. The acquisition of resistance to CHL and florfenicol; 2. The cfr gene encodes CHL acetyltransferase, which facilitates the transfer of an acetyl group from acetyl-CoA to CHL, resulting in drug inactivation (Figure 5b).
Antibiotics That Target the Nucleic Acids
Quinolones
Staphylococcus can develop resistance to fluoroquinolones by antibiotic efflux. The overexpression of chromosomally encoded efflux pumps (NorA, NorB, and NorC) is a key step; NorA processes hydrophilic molecules, while NorB and NorC target hydrophobic molecules (Figure 6b). The small transcriptional regulator, MgrA, is the primary factor implicated, as it may directly bind to the promoter of the nor gene.35
Moreover, several studies have identified topoisomerase mutations as one of the mechanisms underlying the development of fluoroquinolone resistance.36 These mutations may induce alterations in the amino acid residues at the drug-binding site, a phenomenon referred to by some studies as modifications within the quinolone resistance-determining region (QRDR).37 In staphylococci, ParC is the most sensitive topological enzyme, rendering it the principal target.
Sulfa-Drugs
Sulfonamides can exhibit bactericidal properties by interfering with folate metabolism in bacteria. Sulfonamides inhibit dihydrophosphonate synthase (DHPS), which condenses dihydrophosphonate and p-aminobenzoic acid (pABA) to form dihydrophosphonate, a precursor of folic acid essential for prokaryotes. The investigation of sulfonamide resistance in S. aureus clinical isolates has indicated that drug resistance is associated with several different amino acid substitutions in DHPS targets.38
Trimethoprim (TMP)
The 2,4-diaminopyridyl trimethoprim targets dihydrofolate reductase (DHFR), which catalyzes the conversion of dihydrofolate into tetrahydrofolate. Both sulfonamides and TMP can affect bacterial folic acid synthesis. Therefore, TMP is generally used in combination with sulfamethoxazole in clinical settings, referred to as the compound neoxazole.
Resistance to TMP in clinical isolates can be achieved with a single point mutation, F98Y, in the chromosomal dfrB gene that encodes the DHFR enzyme.39 Another approach could involve obtaining genes that encode DHFR enzymes that are not easily repressed, so enabling the bypass of the chromosomal DHFR blockage. DHFR activity is required for DNA synthesis, and the inhibition of this enzyme prevents DNA replication. The most common change in TMP-resistant DHFR in S. aureus is the single amino acid substitution of F98Y in the DfrB-resistant phenotype.
Rifampicin (RIF)
RIF is a class of spectral bactericidal antibiotics directed against RNA polymerases. It can deactivate RNA polymerase and affect bacterial RNA synthesis, with its efficacy being dose-dependent. It binds to subunit B about 12 angstroms away from the active site of the enzyme. It does not prevent the RNA polymerase from binding to the promoter and initiating transcription.40 RIF resistance arises from target-site modification; mutations in the rpoB gene occur in a hotspot region known as the rifampin resistance-determining region. It suppresses the formation of phosphodiester bonds of the first ribonucleotide, thereby halting mRNA synthesis. The most common rpoB resistance mutations were H481Y and L466S, while some rpoB mutations were associated with increased cell wall thickness.41
Therapeutics
The advent of innovative therapy and short-term prophylactic strategies has expanded treatment choices for staphylococcal infections. A table delineating the advantages and disadvantages of each treatment has been included (Table 2).
 |
Table 2 An Overview of the Benefits and Drawbacks of Various Treatment Approaches |
Combination of Antibiotics
Due to genetic mutations or the acquisition of exogenous drug-resistant genes in bacteria, the efficacy of a single antibiotic is often suboptimal; thus, combination therapy has become an important treatment strategy for drug-resistant S. aureus.42–48 Combination therapy usually involves the use of two or more antibiotics at the same time, using the synergies between them to improve the antibacterial efficacy. Additionally, the likelihood of a pathogen acquiring resistance to the combination of drugs is significantly lower than that associated with a monotherapy approach.8 This treatment is designed to kill the bacteria to the fullest extent, minimizing the chances of the bacteria adaptation and resistance development.45–48
The investigation of the mechanism of action of different antibiotics reveals three main approaches to combination therapy: 1. Increase membrane permeability; 2. Reduce efflux pump activity; 3. Inhibit kinase activity and intrinsic antibiotic resistance.7 The majority of in vitro anti-staphylococcal combination therapies utilize daptomycin or vancomycin in combination with other antibiotics. A recent study by Saravolatz and Pawlak (2023) discovered that most MRSA strains were effectively treated when fosfomycin was coupled with linezolid or daptomycin.42 Combination therapy is not only a combination of antibiotics, but also a combination of antibiotics and non-antibiotic drugs.8 For example, a research has demonstrated that pyridines re-sensitize MRSA to beta-lactam antibiotics.7 Thus, the combination of pyridine compounds and β-lactam antibiotics is used to treat staphylococcal infections. The study conducted by Peris et al (2022) is the first of its kind to demonstrate the synergistic effect of icariin in combination with key antibiotics used in the treatment of MRSA infections.43 The formation of biofilms is a crucial factor in the MRSA resistance. Biofilms are formed from a community of microorganisms embedded in a self-produced extracellular polymeric substance, which include polysaccharides, extracellular DNA, proteins, and lipids, adhering to both living or nonliving surfaces.49 A study found that ranbezolid could completely eradicate MRSA biofilms at clinically relevant concentrations and also demonstrated the feasibility of ranbezolid in combination with C-TEMPO.44 However research indicates that the administration of more drugs is not always better, prolonged usage of multiple drugs may be less efficacious than a single drug, thereby limiting long-term drug clearance and increase the risk of adverse reactions.46 This indicates that the implementation of combination therapy needs to be adjusted according to the actual situation and cannot be generalized.
There are still some problems and challenges in combination therapy; first of all, combining multiple drugs increases the financial burden of patients as well as the incidence of adverse reactions. Secondly, long-term use of antibiotics may lead to increased bacterial resistance. Therefore, while using combination therapy, it is vital to treat strictly in accordance with the prescriptions, regulate the doctor’s usage habits, minimize the large use of antibiotics, and ensure the safety and rationality of medication.
Phage Therapy
The emergence of many drug-resistant strains has led to increased attention in phage research.50 Phage therapy, as an alternative to traditional antibiotic therapy, has garnered widespread attention in recent years for addressing drug-resistant staphylococcal infections. Bacteriophage is a kind of virus that specifically targets and lyses bacteria. Its unique targeting and bactericidal action offer a novel approach for addressing drug-resistant bacteria.8 Phage therapy advantages lie in its high host specificity and targeted action, enabling it to eliminate pathogenic bacteria without damaging the normal microbiota of the body, hence minimizing treatment-related side effects, similar to antibiotics.8
Phages, biologically distinct from small-molecule antibiotics, possess several theoretical advantages as therapeutics, including their environmental abundance, minimal off-target effects owing to host specificity, and ability to replicate through the attainment of bacterial clearance.51 The potential of phages to exhibit resistance against biofilms is also evident. This mechanism may be executed through depolymerase enzymes which bind polysaccharide in capsule, lipopolysaccharide, or extrapolymeric substance, or through endolysins which bind peptidoglycan.51 Many studies demonstrate that phage therapy is highly effective against different strains of staphylococcal strains that produce biofilms, including MRSA.52–57 The efficacy of a monophage to control a specific bacterial infection is somewhat determined by the extent of its host range.57 A general means of enhancing spectrum of activity breadth beyond that achievable by a monophage is to combine treatments. A phage cocktail is a therapeutic approach that uses a combination of multiple bacteriophages to fight bacterial infections.
Bacteriophage therapy operates by modulating the expression of host virulence and drug resistance genes during the infection process, thereby killing pathogenic bacteria to achieve therapeutic outcomes. Concurrently, phages exhibit robust proliferation, enabling them to grow and generate sufficient quantities to attain optimal therapeutic efficacy against the rapidly proliferating drug-resistant staphylococcal strains.57 Phages are utilized in animal models, therapeutic applications, and clinical trials. It is worth mentioning that phage-derived endolysins are also a treatment method for MRSA.58 Therefore, we assert that phage therapy is a promising antimicrobial strategy that may provide novel solutions for infections caused by drug-resistant Staphylococcus.
Albeit phage therapy seems to be an ideal approach to treat drug-resistant bacteria, it also has numerous challenges and limitations. First of all, the host range of phages is limited, allowing a phage to infect a few bacteria; therefore, it is essential to screen and prepare different phages for distinct types of S. aureus. In addition, owing to the specificity of phages, their application necessitates a more personalized design, therefore presenting a challenge in developing a phage map for each patient. Secondly, there is a lack of in-depth studies on the safety and effectiveness of phage therapy.59 Previous studies lacked established and standardized phage extraction and purification protocols, resulting in discrepancies in reported findings across several investigations.58 There is a paucity of authentic clinical studies regarding staphylococcal infections, and no randomized double-blind trials have been conducted. Established treatments have predominantly depended on case reports or limited clinical trials. The available clinical case reports also indicate that certain treatments may have limited effectiveness, as evidenced by their small sample sizes. The efficacy of phage therapy in treating S. aureus-induced osteomyelitis is limited, often resulting in the need for amputation.52 Combination therapy with phages and antibiotics does not necessarily yield favourable outcomes, for example, when administered simultaneously with bactericidal antibiotics, PYOSa demonstrates diminished efficacy in killing S. aureus compared to its effectiveness in the absence of these drugs.54 The antibiotics reduce the bacterial densities, thereby reducing the phage’s replication capacity. Finally, the proliferation of therapeutic phages requires the use of strains from pathogenic species that amplify the secretion of virulence factors produced by the host (such as toxins, immune escape proteins, etc)., thereby contaminating phage lysates, which may adversely affect patients during phage administration.
Antibody Therapy
Antibody therapy mainly refers to monoclonal antibody therapy. Monoclonal antibodies are highly uniform antibodies produced from a single B cell clone that target specific epitopes with a high degree of specificity.60 Compared with small molecules, monoclonal antibodies (mAbs) exhibit exquisite target selectivity, resulting in reduced toxicity due to diminished binding to non-targets.61 At present, human monoclonal antibodies are majorly utilized in antiviral, anticancer, and autoimmune therapies; however, their applications in bacterial infections remain limited. Numerous studies have reported the use of monoclonal antibodies in the treatment of drug-resistant S. aureus.61–64 Its high specificity and affinity enable precise targeting of specific antigens associated with drug-resistant S. aureus. A study shows that mAbs targeting common surface components of S. aureus can recognize clinically relevant biofilm types.63 Monoclonal antibodies targeting drug-resistant S. aureus can effectively neutralize bacterial virulence, inhibit growth and reproduction, thereby facilitating treatment outcomes. It should be noted that multivalent antibodies are also an alternative way to treat S. aureus. A study suggests that S. aureus produces multiple virulence factors, complicating the identification of specific targets for vaccine or monoclonal therapy development.65 A human-derived anti-S. aureus monoclonal antibody (mAb)-centyrin fusion protein has been described, which targets multiple bacterial adhesins concurrently.65
Monoclonal antibodies treat S. aureus by facilitating interaction between antibodies and bacteria, such as the anti-SRAP L-lectin module-mediated monoclonal antibodies block the ability of S. aureus to recognize the salivary host receptor on the host cell, thereby reducing the adhesion and invasion of bacteria to the host cell.66 Monoclonal antibodies can treat S. aureus through activation and regulation of the immune system, exemplified by monoclonal antibody 3F6, which blocks SpA activity in human cord blood and promotes complement-dependent cell-mediated S. aureus phagocytosis.67
However, monoclonal antibody therapy also has several challenges and shortcomings. First of all, the treatment of drug-resistant S. aureus using monoclonal antibodies is intricate and necessitates the integration of additional therapeutic modalities to optimize efficacy. Secondly, monoclonal antibodies targeting bacterial antigens may cross-react with host tissue components, resulting in the formation of immune complexes that can induce undesirable side effects. Consequently, additional research and clinical studies are necessary to ascertain their efficacy and safety. At the same time, the preparation of high-quality monoclonal antibodies also necessitates continuous optimization and improvement of technology.
Nanoparticle Therapy
Nanoparticles mainly include lipid nanoparticles, polymer nanoparticles, and metal nanoparticles, characterized by distinctive physical and chemical properties, including small size and large specific surface area, facilitating effective drug loading and precise delivery.11 Targeted therapy and drug release for resistant S. aureus can be achieved by encapsulating antibacterial drugs or other therapeutic agents within the nanoparticles or by attaching them to their surfaces.9,10 Additional therapeutic methods using nanoparticles include photothermal therapy, photodynamic therapy, and transcription factor trapping to facilitate the synergistic treatment of S. aureus.11 Nanoparticles can also act as immunomodulators to enhance the body’s immune response to drug-resistant S. aureus.14 By stimulating the immune system, nanoparticles can promote the body’s production of specific antibodies and cytotoxic T cells to clear the bacteria at the site of infection.
In recent years, the continuous development of nanoparticle technology has led to the design of several nanoparticles for the treatment of drug-resistant S. aureus. These particles exhibit not only high antistaphylococcal activity but also commendable biocompatibility and stability. Diverse metal nanoparticles have been used to study their efficacy on MRSA, as these nanoparticles possess inherent antibacterial activity that can destroy bacterial cell membranes and inhibit bacterial growth and reproduction. Silver nanoparticles (AgNPs) have been extensively studied and possess significant potential value.9–12
Nanoparticles can infiltrate and precisely target drug-resistant bacterial biofilms by delivering a concentrated dose of medication at the infection site, potentially reducing undesirable effects associated with increased drug concentrations.12 Despite its potential, several factors contribute to the failure of NP techniques in clinical settings. Challenges remain in transferring excellent in vitro outcomes to in vivo applications, with long-term NPs safety being a notable concern, alongside hurdles in scaling up NPs production to an industrial batch scale.
Natural Compounds
Natural compounds mainly include the following categories. 1. Biological; usually a large peptide (>50 residues) or protein either isolated from an organism or cell line or produced by biotechnological means in a surrogate host. 2. Natural product. 3. Derivative of a natural product, it is typically a semisynthetic modification. 4. A totally synthetic drug, often found by random screening or modification of an existing agent. 5. Prepared by total synthesis, albeit the pharmacophore originated from a natural product.68 Natural compounds exhibit a multitude of biological activities, including antimicrobial and antibiofilm activity.69,70 They may inhibit S. aureus by interfering with bacterial metabolic processes, altering cell membrane integrity, or inhibiting bacterial protein synthesis.71 Some studies indicate that specific active ingredients in plant extracts show significant inhibition of drug-resistant S. aureus, offering new prospects for treating drug-resistant strains.72
In addition, the antimicrobial mechanisms of natural compounds are often different from those of conventional antibiotics, which gives them a unique advantage in certain instances. It is worth noting that while certain natural compounds exhibit effective inhibition of S. aureus, the cytotoxic effects of these compounds and their possible adverse reactions must be taken into account. Some studies have identified that triterpenes inhibit biofilm formation and exhibit antibacterial activity against reference staphylococcal strains; however, the compound has demonstrated cytotoxicity to osteoblasts.70 Simultaneously, while several natural compounds have antibacterial or synergistic effects, their antibacterial activity is limited or difficult to obtain, resulting in a lack of further development. Additionally, the development of novel biotechnologies is crucial.
Vaccine Treatment
Vaccines have great potential for the prevention and control of drug-resistant S. aureus.73 Vaccination can stimulate the body’s immune system to produce a targeted immune response against S. aureus, thereby preventing or reducing the occurrence of infection. Bacterial vaccines have significantly reduced morbidity and mortality caused by several common pathogens, including Haemophilus influenzae type B, Streptococcus pneumoniae, Neisseria meningitidis, Corynebacterium diphtheria, Bordetella pertussis, and Clostridium tetani.74 However, there is currently no vaccine that is widely applicable to S. aureus. Therefore, the development of a vaccine against S. aureus has great potential. Due to the presence of numerous fluctuating infection-related factors, particularly in staphylococcal spp., one study advocates for the development of multivalent vaccines consisting of multiple antigens associated with different infection stages.75
S. aureus vaccine treatment is mainly categorized into three directions: extracellular vesicles (EVs), whole-cell or attenuated live vaccines, and nucleic acid vaccines.76 Whole-cell vaccines guarantee a wide array of antigens; however, adverse reactions cannot be ruled out compared to subcellular vaccines. In contrast to nucleic acid vaccines, it is widely reported that S. aureus can survive within cells, and research indicates that RNA-based vaccines promote the production of antiviral immune responses, including CD8+ T cells, so RNA vaccine may be effective in treating S. aureus infections.76 Currently, RNA-based vaccines have not been explicitly studied against S. aureus; nonetheless, studies indicate that rRNA vaccines drive a protective immune response against Streptococcus in mouse experiments and provide cross-generational humoral protection.
However, the development of S. aureus vaccine still needs to consider its complex pathogenic mechanism and numerous pathogenic factors, indicating that multiple challenges remain in vaccine development. The genome of different staphylococcal strains exhibit significant variation, resulting in different antigenic properties, which complicates the development of S. aureus vaccine. Also, the safety and efficacy of vaccines must be verified through rigorous clinical trials. Additionally, more research is required to determine the duration of the protective effect of immunization.
Bacteriocin Treatment
Bacteriocins are proteins or peptides with antibacterial activity produced by certain bacteria. They usually exert an inhibitory effect on neighboring or closely similar bacteria while remaining safe to the human body and exhibiting great specificity.49,77–79 It is currently regarded as having strong potential for the development of new antibiotics. Bacteriocins can inhibit pathogens through specific mechanisms, such as destroying cell membranes, inhibiting cell wall synthesis, or interfering with DNA replication. Compared with traditional antibiotics, they possess a narrower antibacterial spectrum, allowing for more precise action against target bacteria while minimizing impact on the body’s normal microbiota. It is also less probable that bacteria will evolve a resistance to bacteriocins because of their diverse action mechanisms.
Recent investigations have demonstrated that bacteriocin exerts a substantial inhibitory impact on S. aureus. Certain microcins can directly damage the cell membrane of S. aureus, resulting in leakage of cellular contents and subsequent bacterial death. Bacteriocin concurrently inhibits the cell wall synthesis of S. aureus, hence preventing its normal growth and reproduction. A study discovered that the potent activity on staphylococcal biofilms indicated that bacitracin may prevent or treat biofilm-associated infections.49 These findings provide a solid theoretical basis for bacteriocin as a potential therapeutic agent for S. aureus infections. In addition to their direct antibacterial effects, bacteriocins also have the ability to regulate immune responses. They can stimulate the body’s immune system and enhance the body’s ability to clear S. aureus. The immunomodulatory effect enhances the efficacy of bacteriocin in treating S. aureus infections, aiding in disease management and minimizing consequences.78
Although bacteriocins show great potential in the treatment of S. aureus infections, there are still some challenges in their clinical application. Firstly, the stability, bioavailability, and safety of bacteriocins need to be further investigated. Secondly, it is necessary to screen out bacteriocins with high efficiency and specificity for different types of S. aureus infection. Finally, the production process and cost of bacteriocins are important factors affecting their wide application.
Antibacterial Mechanism of Nanomaterials
Nanomaterials exhibit significant potential for antibacterial applications owing to their distinctive physicochemical properties.80 They exhibit diverse structures, and those with antistaphylococcal properties can be classified into inorganic nanomaterials (metal nanomaterials, metal oxide nanomaterials), organic nanomaterials (chitosan, liposomes), carbon-based nanomaterials (carbon nanotubes, fullerenes), and composite nanomaterials.81 Studies have revealed that nanomaterials operate through many concurrent antibacterial pathways, making it difficult for bacteria to develop resistance.82 Their remarkable efficacy is achieved through their tiny size and substantial surface-to-volume ratio. The properties of the materials (type, shape, size, charge, surface coating, concentration, etc.) and environmental factors also impact their effectiveness.83 Among the various nanomaterials, silver and gold are considered the most effective in combating staphylococcal infections. In addition to metallic nanomaterials, other nano-based materials have also been investigated. This section summarizes the antibacterial mechanisms of different types of nanomaterials against staphylococci (Table 3).
 |
Table 3 A Summary of the Antistaphylococcal Properties of Nanomaterials |
Inorganic Nanomaterials
Metal nanoparticles are at the forefront of research due to their applications in disease diagnosis and treatment.116 The specific antibacterial mechanisms can be summarized as follows: (1) destruction of cell membranes and cell walls; (2) damage to DNA; (3) disruption of proteins; (4) generation of reactive oxygen species (ROS); (5) disruption of electron transport chains; (6) inhibition of ribosomal subunits (Figure 7).
AgNPs
Silver nanoparticles (AgNPs) are among the most studied nanoparticles in recent years. Their mechanism of action against staphylococci involves the inhibition of bacterial DNA replication, degradation of bacterial membranes, or modification of intracellular triphosphate phosphate levels to generate ROS that induce oxidative stress. AgNPs readily adhere to the surface of bacterial cells due to the electrostatic attraction between the positive surface charge of AgNPs and the negatively charged cell membranes of microorganisms, together with the interaction among the AgNPs themselves.84 The ions released by AgNPs interact with thiol groups on cysteines, altering their three-dimensional structures, thereby blocking active binding sites and inhibiting protease activity.85 Given that sulfur and phosphorus are important components of DNA, the interaction between silver ions and sulfur and phosphorus in DNA may impede DNA replication.86
AgNPs destroy cell membranes through direct contact, leading to intracellular permeation. The silver ions released from AgNPs react rapidly with sulfhydryl groups on bacterial cell membranes. Stable S-SAG bonds are formed through the exchange of terminal hydrogen atoms, which completely block the respiratory chain, electron transfer, protein secretion, and lipid biosynthesis.87,88 Confocal laser scanning microscopy (CLSM) study of biofilms from infected wounds, specifically MRSA and methicillin-resistant S. epidermidis (MRSE), demonstrates definitive proof of AgNPs ability to inhibit bacterial growth.117
Upon entering cells, free silver ions deactivate respiratory enzymes, generating ROS that interrupt ATP production and affect respiratory chains. Studies evaluating the ROS-mediated antibacterial activity of AgNPs against multidrug-resistant S. aureus have found that ROS generation significantly contributes to the antibacterial effect.89 Similarly, AgNPs immobilized on nanoscale silicate plates (AgNP/NSPs) exhibit significant antibacterial activity against MRSA through ROS generation.90 Research indicates that ROS generated by SNP treatment of MRSA may cause significant covalent modifications to proteins, leading to cell protein inactivation and impaired DNA replication, resulting in intense oxidative stress and bacterial cell death.90
The antibacterial activity of AgNPs is influenced by several factors. According to Ayala-Núñez et al (2010), AgNPs with smaller diameters exhibit better antibacterial activity and lower toxicity to human cells.118 Furthermore, prism-shaped (truncated) AgNPs with sharp peaks and edges approximately 20 nm in size demonstrate superior antibacterial activity. AgNPs can be combined with stabilizers to enhance their antibacterial capabilities,119 with different stabilizers (glucose, lactose, fructose, sucrose) yield varying inhibition zones against S. aureus.
AuNPs
Research demonstrates that citrate-capped AuNPs with a particle size of 12 nm synthesized through the Turkevich chemical method exhibit effective staphylococcal inhibitory activity.120 The antibacterial action of AuNPs occurs in two stages. First, they alter membrane potential, reducing the activity of ATP synthases and metabolic processes. High concentrations of AuNPs (>50 mg/L) have shown a significant reduction in S. aureus biofilm formation.91 In an in vitro experimental study, polyethylene (PE) discs coated with gold nanoparticles (AuNPs) on one side (experimental group) and PE discs without coating on the other side (control group) were placed in a CDC biofilm reactor and then inoculated with S. aureus for culturing. The results showed that the biofilm formation of S. aureus in the control group increased.92 Secondly, AuNPs diminish the ribosomal subunits, thereby impairing their biological functions.93
AuNPs basically react with sulfur- or phosphorus-containing soft groups. Therefore, proteins containing sulfur and DNA molecules containing phosphorus are favorable sites for GNPS to attack. AuNPs bind to thiol groups, including NADH dehydrogenase, release ROS to destroy the respiratory chain, induce oxidative stress, severely damage the cell structure, and ultimately lead to cell death.
Fe2O3NPs
Magnetic iron oxide nanoparticles utilize direct and alternating magnetic fields to disrupt MRSA biofilms.94 Following MRSA infection, Fe2O3NPs are implanted into the bone marrow cavity of mice (at high temperatures) to control S. aureus responsible for osteomyelitis.121 Similar to AgNPs, Fe2O3NPs can destroy macromolecules,95 including DNA and proteins, through the generation of ROS.
CuNPs
In a recent study, CuNPs were prepared using medicinal plants ginger and turmeric, which exhibited higher antistaphylococcal activity than traditional antibiotics, including penicillin, methicillin, and ampicillin.122 Studies indicate that the enhanced inhibitory activity of copper and copper oxide nanoparticles is attributed to copper ions,96 which can alter the local pH and conductivity at the cellular level, disrupt cell membranes, and affect respiratory enzymes.
TiO2NPs
Positively charged TiO2NPs exhibit strong interactions with bacterial cells, leading to bacterial membrane permeabilization and ensuring oxidative damage. Another crucial mechanism for their bactericidal effect lies in their photocatalytic activity, which enables the killing of MRSA through ROS generated by electron-hole pairs under UV light excitation.97 Notably, even without UV exposure, TiO2NPs encapsulated within self-assembled tripeptide hydrogels, forming hgel-TiO2NP composites, significantly enhance the growth inhibition of S. aureus due to their remarkable photo-antibacterial properties.98 Furthermore, the photocatalytic activity of TiO2NPs can be enhanced by doping with transition metals or forming titanium nanotubes, thereby improving their bactericidal performance.
ZnONPs
In a mouse model experiment, the use of ZnONPs to reduce skin infections related to MRSA has demonstrated efficacy.123 The main antibacterial mechanism of ZnONPs is the release of zinc ions in a bacterial medium containing ZnONPs. The released Zn2+ subsequently inhibits protein synthesis, amino acid metabolism, and disrupts the enzyme system.99 ZnONPs exert antibacterial effects through direct contact with bacterial cell walls, leading to cell integrity disruption, the release of antibacterial zinc ions, and the formation of ROS.100 Nano-ZnO can enhance the antibacterial activity of incorporated antimicrobials. When combined with oxacillin, neomycin, and ampicillin/sulbactam antibiotics against Staphylococcus, ZnONPs significantly reduce the bacterial load compared to antibiotic use alone.101 This mechanism may involve inhibiting biofilm formation by S. aureus.102
Organic Nanomaterials
Chitosan
Chitosan nanoparticles, which carry a positive charge on their surface, can bind to negatively charged cell membranes, resulting in cell leakage. Due to their excellent permeability, chitosan nanoparticles can penetrate biofilms and hinder their formation.124 Additionally, chitosan, as a well-studied polymer, serves as a drug carrier, capable of loading antibiotics and metal nanoparticles to achieve synergistic antibacterial effects. Studies indicate that chitosan nanoparticles loaded with daptomycin require only 4 hours to achieve high drug delivery and anti-MRSA activity.103 Chitosan loaded with silver nanoparticles exhibits potent antibacterial activity against S. aureus, with the mechanism involving the decomposition of silver/chitosan nanoparticles (Ag-CHNps) into silver nanoparticles (AgNps) and chitosan nanoparticles (CHNps), each contributing distinct antibacterial activities.104
Liposomes
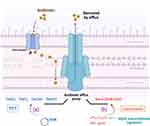
Liposomes are vesicular structures composed of phospholipids (PL) and cholesterol (Chol), demonstrating successful antibacterial effects against MRSA and staphylococcal biofilms.105 Liposomes, as nano- or micro-structured closed spherical vesicles, are typically formed by one or more layers of phospholipids. The charge of liposomes imparts stability and promotes electrostatic repulsion, hence facilitating their interaction with cell surfaces. This may be a crucial factor in the interaction between cationic liposomes and the negatively charged cell walls of S. aureus106 (Figure 8). Studies indicate that the combination of cationic photosensitizers with cationic liposomes can induce potent anti-MRSA activity.125 Levofloxacin liposomes exhibit prolonged and improved anti-biofilm and antibacterial effects in the treatment of S. aureus infections.107 Liposomes prolong the therapeutic plasma drug concentration time, enhance the stability of cinnamon oil, and potentiate its activity against MRSA biofilms.108 Antibiotic encapsulation within liposomes can evade enzymatic hydrolysis associated with bacterial resistance, including hydrolysis by staphylococcal β-lactamases,109 suggesting that liposomes may serve as an effective antibiotic carrier against staphylococcal infections.
 |
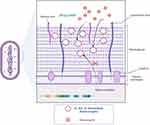
Figure 8 Antibacterial mechanism of liposomes. It serves as an efficient antibiotic carrier, interacting with the negatively charged cell walls of S. aureus to boost antistaphylococcal activity. |
Carbon-Based Nanomaterials
Carbon-based materials are regarded as a valuable resource due to the existence of various carbon allotropes.81 Their diverse forms make them an important material for various biomedical applications.126
Carbon Nanotubes (CNTs)
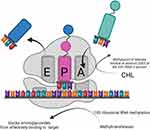
CNTs are cylindrical or tubular structures. In recent years, CNTs have garnered significant scientific attention due to their fundamental structure and unparalleled characteristics, including size-dependent electronic coupling, high electrical conductivity, and mechanical strength with small radii. Their antibacterial mechanisms against Staphylococcus can be summarized as follows: (1) CNTs attach to microbial surfaces, causing damage to cell walls and membranes; (2) CNTs penetrate bacterial cells, leading to DNA disruption and protein dysfunction; (3) generation of ROS110 (Figure 9).
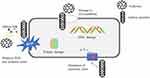 |
Figure 9 Antibacterial mechanism of Fullerene and CNT. They exert their effects on multiple locations, including the cell wall, cell membrane, and DNA, to effectively eliminate S. aureus. |
Fullerenes
Fullerenes are photochemically active carbon-based NPs that generate ROS under light exposure, disrupting respiratory chains and inhibiting bacterial growth, thereby exhibiting antibacterial activity, particularly against Gram-positive bacteria such as S. pyogenes.111 Among these mechanisms, oxidative stress is considered the most effective for NPS against staphylococcal activity.112 Fullerenes disrupt cell membranes, degrade DNA, alter metabolic pathways, and render microorganisms inactive. It is suggested that their antibacterial activity is induced by inhibiting energy metabolism after nanoparticle internalization into bacteria111 (Figure 9).
Graphene-Based Nanomaterials
Graphene, a prominent representative of two-dimensional nanomaterials (2D NBGs), holds notable importance in antibacterial research, particularly against pathogens like S. aureus. Its antibacterial mechanisms encompass physical membrane disruption, oxidative stress-induced damage to bacterial structures, membrane stress effects, photothermal ablation, and the synergistic improvement of antibacterial activity through multicomponent integration. These mechanisms collectively underscore graphene’s superior performance in antibacterial applications.127,128 Nevertheless, prior to practical implementation, a rigorous biosafety assessment is imperative to ensure no unintended adverse effects.129 Research indicates that graphene-coated zinc oxide, in conjunction with curcumin, demonstrates substantial antibacterial and disinfection properties against MRSA.130 In another research study, Elias et al (2019) explored a nanotechnological approach that utilizes graphene oxide (GO) or carbon nanofibers (CNFs) in conjunction with light-emitting diode (LED) irradiation as innovative nano-tools to combat two clinically significant Gram-positive multidrug-resistant pathogens: MRSA and methicillin-resistant S. epidermidis (MRSE).131
Graphene oxide-silver nanocomposite (GO-Ag), a pivotal branch of graphene-based composites, integrates GO with AgNPs. Its antibacterial mechanisms encompass the potent toxicity stemming from the release of Ag+, specifically targeting thiol groups within bacterial cells, membrane disruption resulting from direct physical interaction between GO nanosheets and bacterial cells, the activation of oxidative stress pathways, and augmented multi-component synergies facilitated by the extensive surface area of GO nanosheets.132 The concerted action of these mechanisms bestows GO-Ag composites with swift and potent antimicrobial capabilities in tests against recalcitrant pathogens, including MRSA.133
Recent years have witnessed notable advancements in the research of 2D NBGs, beyond graphene, for antibacterial applications. These materials exhibit diverse antibacterial mechanisms, including membrane disruption via physical contact, oxidative stress-induced cellular damage through ROS generation, light-induced strategies (photothermal effects, photocatalytic reactions, photodynamic therapy), controlled release of drugs/metal ions, and multi-mode synergistic effects.134,135 These mechanisms significantly enhance antibacterial efficiency, providing a robust foundation and practical support for high-efficiency, multifunctional antibacterial applications.
Composite Nanomaterials
Nanomaterials do not merely function in isolation; different nanomaterials can be combined for use. The currently developed bimetallic nanoparticle systems have proven to be a reliable, reproducible, and effective source of nanomaterials with biomedical potential to combat multidrug-resistant pathogens.113 When combined in the form of bimetallic NPS, their antibacterial properties are enhanced. Consequently, AuNPs are often used in conjunction with other nanometals, such as ZnONPs, to increase their antibacterial effects.
Similarly, nanomaterials can also be combined with antibiotics. When AuNPs are functionalized with ampicillin, they transform into effective bactericides with unique properties that interfere with the resistance mechanisms of various drug-resistant bacteria, particularly inhibiting MRSA. Kuo et al combined synthetic peptides with arginine, tryptophan, and cysteine termini with an aqueous solution of tetrachloroauric acid to generate peptide-immobilized AuNPs,136 which demonstrated efficacy as antibacterial agents against Staphylococcus.
MXene-based composites exhibit remarkable antibacterial potential, particularly against Gram-positive and Gram-negative bacteria, including MRSA, due to their exceptional features, including superior photothermal effects, highly reactive sites, substantial interlayer spacing, distinctive chemical structures, and hydrophilicity.137 Their antibacterial mechanism is rooted in their unique nanostructure: the sharp edges of their nanosheets physically disrupt bacterial cell membranes, resulting in the release of cellular contents.138 Furthermore, under near-infrared (NIR) light exposure, MXenes exhibit a pronounced photothermal effect, converting light energy into heat to further damage bacterial cells.139 Notably, MXenes of different sizes exhibit varying degrees of temperature elevation under NIR light, with smaller MXenes demonstrating the highest antibacterial activity due to their enhanced photothermal conversion efficiency, leading to higher localized temperatures and more effective bacterial eradication.139 Furthermore, MXene nanomaterial incorporated into the non-crosslinked chitosan hydrogel exhibits significant synergistic anti-MRSA efficacy and has considerable promise for the treatment of MRSA-infected conditions.140 Nonetheless, despite extensive research on MXene-based materials, their superior properties cannot match the standards for a variety of applications. After surface modification, MXene-based materials are widely used in biomedical, energy, and environmental applications.138,141 Although substantial progress has been made in surface modification and biomedical applications of MXene-based materials, there are still numerous gaps in their development, warranting further research.
Metal-organic frameworks (MOFs) possess high loading capacities, and their inherent microporosity or mesoporosity enables them to encapsulate multiple drugs simultaneously as nanocarriers, leading to synergistic therapeutic effects. Researchers have developed LL-37@MIL-101-Van nanoparticles that achieve a synergistic effect of ROS generation alongside the antibacterial actions of vancomycin and LL-37 peptides,114 significantly inhibiting biofilm formation and promoting healing in MRSA-infected wounds. While nanogels are still in the early stages of exploration for combating biofilm infections, research indicates that nanogels derived from natural polymers like chitosan (CS), hyaluronic acid (HA), and alginates effectively deliver anti-biofilm drugs, demonstrating efficacy against S. aureus.115
Moreover, iron-based MOFs (Fe-MOFs) exhibit distinct advantages in antibacterial mechanisms.142 Their antibacterial activity stems primarily from the release of Fe³⁺ metal ions and the sustained release of bioactive agents (eg, drug molecules) following MOFs degradation. Additionally, Fe-MOFs can serve as carriers for several antibacterial agents, including silver ions, demonstrating broad-spectrum antibacterial properties against diverse bacterial strains. In practice, Fe-MOFs have been widely used in the fabrication of surface antibacterial coatings, antibacterial drug delivery systems, and composite antibacterial materials, providing innovative avenues and perspectives for the research and application of antibacterial materials.
Challenges and Limitations in the Application of Nanomaterials
Albeit nanomaterials exhibit enormous potential for combating staphylococcal drug resistance, there are numerous challenges and limitations to overcome in real-world scenarios while deploying them. The principal concern involves the biosafety of nanomaterials, which may induce considerable cytotoxicity and tissue toxicity, adversely impacting red blood cells, blood coagulation mechanisms, and vital organ functions.143,144 In addition, the long-term effects and safety of nanomaterials in the human body are still ambiguous, thus constraining potential therapeutic applications.80
Secondly, the biodegradation and clearance mechanisms of nanomaterials are not yet well established, potentially leading to their prolonged retention and accumulation of toxicity in living organisms, thereby increasing health risks.143 The environmental risks of nanomaterials cannot be ignored either, as they may facilitate the horizontal transfer of antibiotic resistance genes, posing potential threats to ecosystems and amplifying toxic effects throughout the food chain, with long-term and far-reaching impacts on human health and environmental safety.80
At the preparation and application levels, nanomaterials face multiple technical obstacles, including complex preparation processes, high costs, material stability, and storage condition control, all of which restrict their large-scale production and widespread application.80 In particular, ensuring the stability of nanomedicines in vivo and their expected therapeutic efficacy is a critical technical challenge that urgently needs to be addressed in the field of nanomedicine.
Additionally, the intricate regulations surrounding nanomaterials, the protracted investigation of commercialization routes, and the emergence of bacterial resistance to nanoparticles provide significant obstacles to their broader utilization.143 Ethical, social, and legal considerations in nanomedicine necessitate comprehensive discourse, particularly about treatment affordability and the efficacy of high-risk management, which are focal points for forthcoming research and practice in the field144 Consequently, while advocating for the utilization of nanomaterials in medicine, it is imperative to thoroughly evaluate and mitigate the previously mentioned problems and limitations to guarantee their safe, effective, and sustainable advancement.
Future Prospects of Nanomaterials
Despite encountering an array of hurdles in its current development phase, nanomaterials continue to exhibit vast and promising prospects for future applications. In the realm of antistaphylococcal therapy, especially for the treatment of refractory infections like MRSA, nanomaterials have shown remarkable potential. They enhance the sensitivity of diagnostic techniques, enable precise modulation of drug delivery processes, and effectively inhibit biofilm formation, thereby significantly boosting treatment efficiency and outcomes. Additionally, research on the feasibility of using extracellular vesicles as a carrier for staphylococcal vaccines is being explored.145 These vesicles possess the ability to augment antigen-specific immune responses and may not necessitate the use of additional adjuvants.
The utilization of nanoparticles in drug delivery systems has enabled precise targeting and controlled release of drugs, offering innovative strategies for the management of chronic illnesses.144 Future research in this domain will concentrate on: the advanced development of novel nano-antimicrobials through precise modulation of their microstructure and chemical composition to optimize drug loading and release efficacy while effectively overcoming bacterial resistance; the integration of nanotechnology with other sophisticated medical modalities, such as photothermal therapy and immunotherapy, to establish multimodal synergistic treatment systems for targeted and efficient management of staphylococcal infections; the active investigation and implementation of environmentally sustainable chemical synthesis methods to minimize the ecological impact of nanomaterial fabrication processes and foster their green and sustainable advancement; and the expedited clinical translation and application of nano-antimicrobial technologies to offer patients a broader array of effective treatment alternatives, comprehensively addressing the major hurdles posed by staphylococcal infections.
Conclusions
This comprehensive review has examined the molecular basis of staphylococcal drug resistance, existing therapeutic approaches, and the emerging role of nanoparticles in addressing multi-drug resistance. Our analysis demonstrates that S. aureus employs a multifaceted approach to evade the effects of antibiotics, including limiting drug uptake, target modification, drug inactivation, and active efflux of drugs. Therapeutic strategies such as combination antibiotic therapy, phage therapy, bacteriocin therapy, monoclonal antibody therapy, and nanoparticle therapy have shown promise in enhancing antibacterial efficacy and reducing resistance development. Nanoparticles, especially silver-based nanomaterials, hold great promise in controlling staphylococcal infections due to their small size, rapid penetration, and ability to deposit within cells for extended durations. However, production costs, scalability, cytotoxicity, stability, viscosity differences, skin penetration, and biocompatibility remain barriers to nanoparticles’ widespread application. Future studies should focus on optimizing nanoparticle design to enhance their antibacterial potency, biocompatibility, and biodegradability. Additionally, rigorous toxicological assessments are imperative to ensure the safety of these nanomaterials for human use.
Abbreviations
S. aureus, Staphylococcus aureus; MRSA, Methicillin-Resistant S. aureus; WHO, World Health Organization; PBP2a, Penicillin-Binding Protein 2a; VRE, Vancomycin-Resistant Enterococci; NPs, Nanoparticles; AgNPs, Silver nanoparticles; MDR, Multi-Drug Resistance; PRSA, Penicillin-Resistant S. aureus; VISA, Vancomycin-Intermediate S. aureus; hVISA, Heterogeneous Vancomycin-Intermediate S. aureus; DAP, Daptomycin; CM, Cytoplasmic Membrane; ROS, Reactive Oxygen Species; ATPase, Adenosine Triphosphatase; CNTs, Carbon Nanotubes; MOF, Metal-Organic Frameworks; CS, Chitosan; HA, Hyaluronic Acid.
Data Sharing Statement
The manuscript includes all data generated during this study.
Funding
This work was funded in part by grants from the Sichuan Provincial Undergraduate Innovation Training Project “Feasibility Study of Silver Nanoparticles for Inhibiting Staphylococcus aureus in Humans” (S202310632170) and the National Natural Science Fund of China (Grant No. 82150410452) to S.C.O.
Disclosure
The authors declare no competing interests in this work.
References
1. Lowy FD. Staphylococcus aureusInfections. N Engl J Med. 1998;339(8):520–532. doi:10.1056/NEJM199808203390806
2. Murray CJL, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi:10.1016/S0140-6736(21)02724-0
3. WHO. Bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Available from: https://www.who.int/publications/i/item/9789240093461.
4. Cassat JE, Thomsen I. Staphylococcus aureus infections in children. Curr Opin Infect Dis. 2021;34(5):510–518. doi:10.1097/QCO.0000000000000752
5. Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4). doi:10.1128/CMR.00020-18
6. Liu C, Bayer A, Cosgrove SE, et al. Clinical practice guidelines by the infectious diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children. Clinl Infect Dis. 2011;52(3):e18–e55. doi:10.1093/cid/ciq146
7. Wang C-H, Hsieh Y-H, Powers ZM, Kao C-Y. Defeating antibiotic-resistant bacteria: exploring alternative therapies for a post-antibiotic era. Int J Mol Sci. 2020;21(3):1061.
8. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.00539
9. Zarenezhad E, Abdulabbas HT, Marzi M, et al. Nickel nanoparticles: applications and antimicrobial role against methicillin-resistant staphylococcus aureus infections. Antibiotics. 2022;11(9):1208. doi:10.3390/antibiotics11091208
10. Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int j Nanomed. 2017;Volume 12:1227–1249. doi:10.2147/IJN.S121956
11. Vanamala K, Tatiparti K, Bhise K, et al. Novel approaches for the treatment of methicillin-resistant Staphylococcus aureus: using nanoparticles to overcome multidrug resistance. Drug Discovery Today. 2021;26(1):31–43. doi:10.1016/j.drudis.2020.10.011
12. Swolana D, Wojtyczka RD. Activity of silver nanoparticles against Staphylococcus spp. Int J Mol Sci. 2022;23(8).
13. Kaiser KG, Delattre V, Frost VJ, et al. Nanosilver: an old antibacterial agent with great promise in the fight against antibiotic resistance. Antibiotics. 2023;12(8). doi:10.3390/antibiotics12081264
14. Bruna T, Maldonado-Bravo F, Jara P, Caro N. Silver nanoparticles and their antibacterial applications. Int J Mol Sci. 2021;22(13):7202. doi:10.3390/ijms22137202
15. Ribeiro da Cunha B, Fonseca LP, Calado CR. Antibiotic discovery: where have we come from, where do we go? Antibiotics. 2019;8(2):45. doi:10.3390/antibiotics8020045
16. Sharkey LK, Edwards TA, O’Neill AJ. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio. 2016;7(2):e01975. doi:10.1128/mBio.01975-15
17. Hammes WP, Neuhaus FC. On the mechanism of action of vancomycin: inhibition of peptidoglycan synthesis in Gaffkya homari. Antimicrob Agents Chemother. 1974;6(6):722–728. doi:10.1128/AAC.6.6.722
18. Kirby WM. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science. 1944;99(2579):452–453. doi:10.1126/science.99.2579.452
19. Nieto M, Perkins HR. Physicochemical properties of vancomycin and iodovancomycin and their complexes with diacetyl-L-lysyl-D-alanyl-D-alanine. Biochem J. 1971;123(5):773–787. doi:10.1042/bj1230773
20. Cui J, Zhang H, Mo Z, Yu M, Liang Z. Cell wall thickness and the molecular mechanism of heterogeneous vancomycin‐intermediateStaphylococcus aureus. Lett Appl Microbiol. 2021;72(5):604–609. doi:10.1111/lam.13456
21. Hiramatsu K, Kayayama Y, Matsuo M, et al. Vancomycin-intermediate resistance in Staphylococcus aureus. J Global Antimicrob Resist. 2014;2(4):213–224. doi:10.1016/j.jgar.2014.04.006
22. Tran TT, Munita JM, Arias CA. Mechanisms of drug resistance: daptomycin resistance. Ann NY Acad Sci. 2015;1354:32–53. doi:10.1111/nyas.12948
23. Miller WR, Bayer AS, Arias CA. Mechanism of action and resistance to Daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harbor Perspectives Med. 2016;6(11):a026997. doi:10.1101/cshperspect.a026997
24. Mehta S, Cuirolo AX, Plata KB, et al. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(1):92–102. doi:10.1128/AAC.00432-10
25. Serio AW, Keepers T, Andrews L, Krause KM. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal Plus. 2018;8(1). doi:10.1128/ecosalplus.ESP-0002-2018
26. Douglas EJA, Alkhzem AH, Wonfor T, et al. Antibacterial activity of novel linear polyamines against Staphylococcus aureus. Front Microbiol. 2022;13:948343. doi:10.3389/fmicb.2022.948343
27. Huang YT, Liao CH, Chen SY, et al. Characterization of rifampin-resistant Staphylococcus aureus nasal carriage in patients receiving rifampin-containing regimens for tuberculosis. Infect Drug Resist. 2018;11:1175–1182. doi:10.2147/IDR.S163634
28. Loss G, Simões PM, Valour F, et al. Staphylococcus aureus Small Colony Variants (SCVs): news From a Chronic Prosthetic Joint Infection. Front Cell Infect Microbiol. 2019;9:363. doi:10.3389/fcimb.2019.00363
29. Rahimi F. Characterization of Resistance to Aminoglycosides in Methicillin-Resistant Staphylococcus aureus Strains Isolated From a Tertiary Care Hospital in Tehran, Iran. Jundishapur J Microbiol. 2016;9(1):e29237. doi:10.5812/jjm.29237
30. Zhang Y, Zhang N, Wang M, et al. The prevalence and distribution of aminoglycoside resistance genes. Biosaf Health. 2023;5(1):14–20. doi:10.1016/j.bsheal.2023.01.001
31. Weigel LM, Donlan RM, Shin DH, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51(1):231–238. doi:10.1128/AAC.00576-06
32. Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67(3):419–431. doi:10.1007/s00018-009-0172-6
33. Gould IM. Frontiers in Antimicrobial Resistance: a Tribute to Stuart B. Levy David G. White, Michael N. Alekshun and Patrick F. McDermott, Eds. ASM Press, USA, 2005.ISBN 1-55581-329-1. $119.95, 598 pp. J Antimicrob Chemother. 2006;58(1):232. doi:10.1093/jac/dkl194
34. Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. Tetracycline antibiotics and resistance mechanisms. Biol Chem. 2014;395(5):559–575. doi:10.1515/hsz-2013-0292
35. Yu JL, Grinius L, Hooper DC. NorA functions as a multidrug efflux protein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J Bacteriol. 2002;184(5):1370–1377. doi:10.1128/JB.184.5.1370-1377.2002
36. Hooper DC, Jacoby GA. Topoisomerase Inhibitors: fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harbor Perspectives Med. 2016;6(9):a025320. doi:10.1101/cshperspect.a025320
37. Pitondo-Silva A, Martins VV, Silva C, Stehling EG. Conjugation between quinolone-susceptible bacteria can generate mutations in the quinolone resistance-determining region, inducing quinolone resistance. Int J Antimicrob Agents. 2015;45(2):119–123. doi:10.1016/j.ijantimicag.2014.07.018
38. Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol. 2005;187(7):2395–2405. doi:10.1128/JB.187.7.2395-2405.2005
39. Hampele IC, D’Arcy A, Dale GE, et al. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol. 1997;268(1):21–30. doi:10.1006/jmbi.1997.0944
40. Dale GE, Broger C, D’Arcy A, et al. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J Mol Biol. 1997;266(1):23–30. doi:10.1006/jmbi.1996.0770
41. Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104(6):901–912. doi:10.1016/S0092-8674(01)00286-0
42. Saravolatz LD, Pawlak J. In vitro activity of fosfomycin alone and in combination against Staphylococcus aureus with reduced susceptibility or resistance to methicillin, vancomycin, daptomycin or linezolid. J Antimicrob Chemother. 2023;78(1):238–241. doi:10.1093/jac/dkac380
43. Peris MC, Martínez A, Ortíz MP, Sheth CC, Veses V. Icariin in Combination with Amoxycillin-Clavulanate and Ampicillin, but Not Vancomycin, Increases Antibiotic Sensitivity and Growth Inhibition against Methicillin-Resistant Staphylococcus aureus. Antibiotics. 2022;11(2):233. doi:10.3390/antibiotics11020233
44. Ndukwe ARN, Qin J, Wiedbrauk S, Boase NRB, Fairfull-Smith KE, Totsika M. In Vitro Activities of Oxazolidinone Antibiotics Alone and in Combination with C-TEMPO against Methicillin-Resistant Staphylococcus aureus Biofilms. Antibiotics. 2023;12(12):1706. doi:10.3390/antibiotics12121706
45. Ledger EVK, Sabnis A, Edwards AM. Polymyxin and lipopeptide antibiotics: membrane-targeting drugs of last resort. Microbiology. 2022;168(2). doi:10.1099/mic.0.001136
46. Lázár V, Snitser O, Barkan D, Kishony R. Antibiotic combinations reduce Staphylococcus aureus clearance. Nature. 2022;610(7932):540–546. doi:10.1038/s41586-022-05260-5
47. Duan L, Zhang J, Chen Z, et al. Antibiotic Combined with Epitope-Specific Monoclonal Antibody Cocktail Protects Mice Against Bacteremia and Acute Pneumonia from Methicillin-Resistant Staphylococcus aureus Infection. J Inflamm Res. 2021;Volume 14:4267–4282. doi:10.2147/JIR.S325286
48. Ahmad-Mansour N, Loubet P, Pouget C, et al. Staphylococcus aureus Toxins: an Update on Their Pathogenic Properties and Potential Treatments. Toxins. 2021;13(10):677. doi:10.3390/toxins13100677
49. Liu S, Deng S, Liu H, et al. Four Novel Leaderless Bacteriocins, Bacin A1, A2, A3, and A4 Exhibit Potent Antimicrobial and Antibiofilm Activities against Methicillin-Resistant Staphylococcus aureus. Microbiol Spectrum. 2022;10(5). doi:10.1128/spectrum.00945-22
50. Teng F, Xiong X, Zhang S, et al. Efficacy Assessment of Phage Therapy in Treating Staphylococcus aureus-Induced Mastitis in Mice. Viruses. 2022;14(3):620. doi:10.3390/v14030620
51. Totten KMC, Patel R. Phage Activity against Planktonic and Biofilm Staphylococcus aureus Periprosthetic Joint Infection Isolates. Antimicrob Agents Chemother. 2022;66(1). doi:10.1128/AAC.01879-21
52. Plumet L, Ahmad-Mansour N, Dunyach-Remy C, et al. Bacteriophage Therapy for Staphylococcus Aureus Infections: a Review of Animal Models, Treatments, and Clinical Trials. Front Cell Infect Microbiol. 2022;12.
53. Feng T, Leptihn S, Dong K, et al. JD419, a Staphylococcus aureus Phage With a Unique Morphology and Broad Host Range. Front Microbiol. 2021;12.
54. Berryhill BA, Huseby DL, McCall IC, Hughes D, Levin BR. Evaluating the potential efficacy and limitations of a phage for joint antibiotic and phage therapy of Staphylococcus aureus infections. Proc Natl Acad Sci. 2021;118(10). doi:10.1073/pnas.2008007118
55. Azam AH, Tanji Y. Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy. Appl Microbiol Biotechnol. 2019;103(11):4279–4289. doi:10.1007/s00253-019-09810-2
56. Atshan SS, Hamat RA, Aljaberi MA, et al. Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections. Antibiotics. 2023;12(2):286. doi:10.3390/antibiotics12020286
57. Abedon ST, Danis-Wlodarczyk KM, Wozniak DJ. Phage Cocktail Development for Bacteriophage Therapy: toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals. 2021;14(10):1019. doi:10.3390/ph14101019
58. Murray E, Draper LA, Ross RP, Hill C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses. 2021;13(4):680. doi:10.3390/v13040680
59. Rai A, Khairnar K. Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Braz J Microbiol. 2021;52(4):2031–2042. doi:10.1007/s42770-021-00566-4
60. Nelson PN. Demystified.: monoclonal antibodies. Mol Pathol. 2000;53(3):111–117. doi:10.1136/mp.53.3.111
61. Shepard HM, Phillips GL, Thanos CD, Feldmann M. Developments in therapy with monoclonal antibodies and related proteins. Clin Med. 2017;17(3):220–232. doi:10.7861/clinmedicine.17-3-220
62. van Dijk B, Hooning van Duyvenbode JFF, de Vor L, et al. Evaluating the Targeting of a Staphylococcus-aureus-Infected Implant with a Radiolabeled Antibody In Vivo. Int J Mol Sci. 2023;24(5):4374. doi:10.3390/ijms24054374
63. van Dijk B, de Vor L, van Kessel K, et al. Human monoclonal antibodies against Staphylococcus aureus surface antigens recognize in vitro and in vivo biofilm. eLife. 2022;11.
64. Speziale P, Pietrocola G. Monoclonal Antibodies Targeting Surface-Exposed and Secreted Proteins from Staphylococci. Vaccines. 2021;9(5):459. doi:10.3390/vaccines9050459
65. Buckley PT, Chan R, Fernandez J, et al. Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections. Cell Host Microbe. 2023;31(5):751–765.e711. doi:10.1016/j.chom.2023.04.004
66. Zhou -T-T, Yue Y, Zheng F, et al. Monoclonal antibody against l-lectin module of SraP blocks adhesion and protects mice against Staphylococcus aureus challenge. J Microbiol Immunol Infect. 2021;54(3):420–428. doi:10.1016/j.jmii.2019.08.019
67. Bernardino PN, Bhattacharya M, Chen X, Jenkins J, Missiakas D, Thammavongsa V. A humanized monoclonal antibody targeting protein a promotes opsonophagocytosis of Staphylococcus aureus in human umbilical cord blood. Vaccine. 2023;41(35):5079–5084. doi:10.1016/j.vaccine.2023.07.018
68. Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Natural Prod. 2020;83(3):770–803. doi:10.1021/acs.jnatprod.9b01285
69. Mastoor S, Nazim F, Rizwan-ul-Hasan S, et al. Analysis of the Antimicrobial and Anti-Biofilm Activity of Natural Compounds and Their Analogues against Staphylococcus aureus Isolates. Molecules. 2022;27(20):6874. doi:10.3390/molecules27206874
70. Guzzo F, Scognamiglio M, Fiorentino A, Buommino E, D’Abrosca B. Plant Derived Natural Products against Pseudomonas aeruginosa and Staphylococcus aureus: antibiofilm Activity and Molecular Mechanisms. Molecules. 2020;25(21):5024. doi:10.3390/molecules25215024
71. Idris FN, Nadzir MM. Multi-drug resistant ESKAPE pathogens and the uses of plants as their antimicrobial agents. Arch. Microbiol. 2023;205(4). doi:10.1007/s00203-023-03455-6
72. Moreno Cardenas C, Çiçek SS. Structure-dependent activity of plant natural products against methicillin-resistant Staphylococcus aureus. Front Microbiol. 2023;14.
73. Henriques-Normark B, Normark S. Bacterial vaccines and antibiotic resistance. Upsala J Med Sci. 2014;119(2):205–208. doi:10.3109/03009734.2014.903324
74. Jahantigh HR, Faezi S, Habibi M, et al. The Candidate Antigens to Achieving an Effective Vaccine against Staphylococcus aureus. Vaccines. 2022;10(2):199. doi:10.3390/vaccines10020199
75. Mirzaei B, Babaei R, Zeighami H, Dadar M, Soltani A. Staphylococcus aureus Putative Vaccines Based on the Virulence Factors: a Mini-Review. Front Microbiol. 2021;12. doi:10.3389/fmicb.2021.704247
76. Clegg J, Soldaini E, McLoughlin RM, Rittenhouse S, Bagnoli F, Phogat S. Staphylococcus aureus Vaccine Research and Development: the Past, Present and Future, Including Novel Therapeutic Strategies. Front Immunol. 2021;12.
77. Wang J, Ma X, Li J, et al. The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus. Int J Mol Sci. 2023;24(7).
78. Teiar R, Pérez-Ramos A, Zgheib H, Cudennec B, Belguesmia Y, Drider D. Anti-adhesion and Anti-inflammatory Potential of the Leaderless Class IIb Bacteriocin Enterocin DD14. Probiotics Antimicrob Proteins. 2022;14(4):613–619. doi:10.1007/s12602-022-09954-0
79. Kranjec C, Kristensen SS, Bartkiewicz KT, et al. A bacteriocin-based treatment option for Staphylococcus haemolyticus biofilms. Sci Rep. 2021;11(1). doi:10.1038/s41598-021-93158-z
80. Bashabsheh RH, AL-Fawares O, Natsheh I, Bdeir R, Al-Khreshieh RO, Bashabsheh HH. Staphylococcus aureus epidemiology, pathophysiology, clinical manifestations and application of nano-therapeutics as a promising approach to combat methicillin resistant Staphylococcus aureus. Pathog Global Health. 2024;118(3):209–231. doi:10.1080/20477724.2023.2285187
81. Alshammari BH, Lashin MMA, Mahmood MA, et al. Organic and inorganic nanomaterials: fabrication, properties and applications. RSC Adv. 2023;13(20):13735–13785.
82. Baptista PV, McCusker MP, Carvalho A, et al. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.01441
83. Hoseinzadeh E, Makhdoumi P, Taha P, et al. A Review on Nano-Antimicrobials: metal Nanoparticles, Methods and Mechanisms. Current Drug Metabolism. 2017;18(2):120–128. doi:10.2174/1389200217666161201111146
84. Abbaszadegan A, Ghahramani Y, Gholami A, et al. The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram‐Positive and Gram‐Negative Bacteria: a Preliminary Study. J Nanomater. 2015;2015(1). doi:10.1155/2015/720654
85. Gordon O, Vig Slenters T, Brunetto PS, et al. Silver Coordination Polymers for Prevention of Implant Infection: thiol Interaction, Impact on Respiratory Chain Enzymes, and Hydroxyl Radical Induction. Antimicrob Agents Chemother. 2010;54(10):4208–4218. doi:10.1128/AAC.01830-09
86. Yin IX, Zhang J, Zhao IS, Mei ML, Li Q, Chu CH. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int j Nanomed. 2020;Volume 15:2555–2562. doi:10.2147/IJN.S246764
87. Mishra YK, Hsueh Y-H, Lin K-S, et al. The Antimicrobial Properties of Silver Nanoparticles in Bacillus subtilis Are Mediated by Released Ag+ Ions. PLoS One. 2015;10(12).
88. Yuan Y-G, Peng Q-L, Gurunathan S. Effects of Silver Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus aureus and Pseudomonas aeruginosa from Mastitis-Infected Goats: an Alternative Approach for Antimicrobial Therapy. Int J Mol Sci. 2017;18(3):569. doi:10.3390/ijms18030569
89. Das B, Dash SK, Mandal D, et al. Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage. Arabian J Chem. 2017;10(6):862–876. doi:10.1016/j.arabjc.2015.08.008
90. Hamida RS, Ali MA, Goda DA, Khalil MI, Al-Zaban MI. Novel Biogenic Silver Nanoparticle-Induced Reactive Oxygen Species Inhibit the Biofilm Formation and Virulence Activities of Methicillin-Resistant Staphylococcus aureus (MRSA) Strain. Front Bioeng Biotechnol. 2020;8. doi:10.3389/fbioe.2020.00433
91. Sathyanarayanan MB, Balachandranath R, Genji Srinivasulu Y, Kannaiyan SK, Subbiahdoss G. The Effect of Gold and Iron-Oxide Nanoparticles on Biofilm-Forming Pathogens. ISRN Microbiol. 2013;2013:1–5. doi:10.1155/2013/272086
92. Kuwabara Y, Okai T, Imanishi Y, et al. Development of Extrauterine Fetal Incubation System Using Extracorporeal Membrane Oxygenator. Artif Organs. 2008;11(3):224–227.
93. Shamaila S, Zafar N, Riaz S, Sharif R, Nazir J, Naseem S. Gold Nanoparticles: an Efficient Antimicrobial Agent against Enteric Bacterial Human Pathogen. Nanomaterials. 2016;6(4):71. doi:10.3390/nano6040071
94. Li J, Nickel R, Wu J, Lin F, van Lierop J, Liu S. A new tool to attack biofilms: driving magnetic iron-oxide nanoparticles to disrupt the matrix. Nanoscale. 2019;11(14):6905–6915. doi:10.1039/C8NR09802F
95. Webster TJ, Leuba K, Durmus NG, Taylor EN. Short communication: carboxylate functionalized superparamagnetic iron oxide nanoparticles (SPION) for the reduction of S. aureus growth post biofilm formation. Int j Nanomed. 2013;731. doi:10.2147/IJN.S38256
96. Strauch BM, Niemand RK, Winkelbeiner NL, Hartwig A. Comparison between micro- and nanosized copper oxide and water soluble copper chloride: interrelationship between intracellular copper concentrations, oxidative stress and DNA damage response in human lung cells. Particle Fibre Toxicol. 2017;14(1). doi:10.1186/s12989-017-0209-1
97. Kubacka A, Diez MS, Rojo D, et al. Understanding the antimicrobial mechanism of TiO2-based nanocomposite films in a pathogenic bacterium. Sci Rep. 2014;4(1). doi:10.1038/srep04134
98. Binaymotlagh R, Hajareh Haghighi F, Di Domenico EG, et al. Biosynthesis of Peptide Hydrogel–Titania Nanoparticle Composites with Antibacterial Properties. Gels. 2023;9(12):940. doi:10.3390/gels9120940
99. Aydin Sevinç B, Hanley L. Antibacterial activity of dental composites containing zinc oxide nanoparticles. J Biomed Mater Res Part B. 2010;94B(1):22–31. doi:10.1002/jbm.b.31620
100. Sirelkhatim A, Mahmud S, Seeni A, et al. Review on Zinc Oxide Nanoparticles: antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015;7(3):219–242. doi:10.1007/s40820-015-0040-x
101. Abo-Shama UH, El-Gendy H, Mousa WS, et al. Synergistic and Antagonistic Effects of Metal Nanoparticles in Combination with Antibiotics Against Some Reference Strains of Pathogenic Microorganisms. Infect Drug Resist. 2020;Volume 13:351–362. doi:10.2147/IDR.S234425
102. Lee J-H, Kim Y-G, Cho MH, Lee J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol Res. 2014;169(12):888–896. doi:10.1016/j.micres.2014.05.005
103. Siddhardha B, Pandey U, Kaviyarasu K, et al. Chrysin-Loaded Chitosan Nanoparticles Potentiates Antibiofilm Activity against Staphylococcus aureus. Pathogens. 2020;9(2). doi:10.3390/pathogens9020115
104. Shi S, Shi W, Zhou B, Qiu S. Research and Application of Chitosan Nanoparticles in Orthopedic Infections. Int j Nanomed. 2024;Volume 19:6589–6602. doi:10.2147/IJN.S468848
105. Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK. Hua S: advances and Challenges of Liposome Assisted Drug Delivery. Front Pharmacol. 2015;6. doi:10.3389/fphar.2015.00286
106. Pinheiro M, Magalhães J, Reis S. Antibiotic interactions using liposomes as model lipid membranes. Chem Phys Lipids. 2019;222:36–46. doi:10.1016/j.chemphyslip.2019.05.002
107. Gupta PV, Nirwane AM, Belubbi T, Nagarsenker MS. Pulmonary delivery of synergistic combination of fluoroquinolone antibiotic complemented with proteolytic enzyme: a novel antimicrobial and antibiofilm strategy. Nanomedicine. 2017;13(7):2371–2384. doi:10.1016/j.nano.2017.06.011
108. Gkartziou F, Giormezis N, Spiliopoulou I, Antimisiaris SG. Nanobiosystems for Antimicrobial Drug-Resistant Infections. Nanomaterials. 2021;11(5).
109. Ferreira M, Ogren M, Dias JNR, et al. Liposomes as Antibiotic Delivery Systems: a Promising Nanotechnological Strategy against Antimicrobial Resistance. Molecules. 2021;26(7).
110. Hada V, Chaturvedi K, Singhwane A, et al. Nanoantibiotic effect of carbon-based nanocomposites: epicentric on graphene, carbon nanotubes and fullerene composites: a review. 3 Biotech. 2023;13(5). doi:10.1007/s13205-023-03552-9
111. Kazemzadeh H, Mozafari M. Fullerene-based delivery systems. Drug Discovery Today. 2019;24(3):898–905. doi:10.1016/j.drudis.2019.01.013
112. Parveen F, Sannakki B, Mandke MV, Pathan HM. Copper nanoparticles: synthesis methods and its light harvesting performance. Solar Energy Mater Solar Cells. 2016;144:371–382. doi:10.1016/j.solmat.2015.08.033
113. Nieto-Argüello A, Medina-Cruz D, Pérez-Ramírez YS, et al. Composition-Dependent Cytotoxic and Antibacterial Activity of Biopolymer-Capped Ag/Au Bimetallic Nanoparticles against Melanoma and Multidrug-Resistant Pathogens. Nanomaterials. 2022;12(5):779. doi:10.3390/nano12050779
114. Qi X, Shen N, Al Othman A, et al. Metal-Organic Framework-Based Nanomedicines for the Treatment of Intracellular Bacterial Infections. Pharmaceutics. 2023;15(5):1521. doi:10.3390/pharmaceutics15051521
115. Ali AA, Al Bostami RD, Al-Othman A. Nanogel-based composites for bacterial antibiofilm activity: advances, challenges, and prospects. RSC Adv. 2024;14(15):10546–10559. doi:10.1039/D4RA00410H
116. Rónavári A, Igaz N, Adamecz DI, et al. Green Silver and Gold Nanoparticles: biological Synthesis Approaches and Potentials for Biomedical Applications. Molecules. 2021;26(4):844. doi:10.3390/molecules26040844
117. Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions onEscherichia coli andStaphylococcus aureus. J Biomed Mater Res. 2000;52(4):662–668. doi:10.1002/1097-4636(20001215)52:4<662::AID-JBM10>3.0.CO;2-3
118. Lara HH, Ayala-Nuñez NV, Ixtepan-Turrent L, Rodriguez-Padilla C. Mode of antiviral action of silver nanoparticles against HIV-1. J Nanobiotechnol. 2010;8(1):1. doi:10.1186/1477-3155-8-1
119. Altaf NUH, Naz MY, Shukrullah S, et al. Statistically Optimized Production of Saccharides Stabilized Silver Nanoparticles Using Liquid–Plasma Reduction Approach for Antibacterial Treatment of Water. Materials. 2021;14(19):5841. doi:10.3390/ma14195841
120. Gouyau J, Duval RE, Boudier A, Lamouroux E. Investigation of Nanoparticle Metallic Core Antibacterial Activity: gold and Silver Nanoparticles against Escherichia coli and Staphylococcus aureus. Int J Mol Sci. 2021;22(4):1905. doi:10.3390/ijms22041905
121. Fang C-H, Tsai P-I, Huang S-W, et al. Magnetic hyperthermia enhance the treatment efficacy of peri-implant osteomyelitis. BMC Infect Dis. 2017;17(1). doi:10.1186/s12879-017-2621-4
122. Varghese B, Kurian M, Krishna S, Athira TS. Biochemical synthesis of copper nanoparticles using Zingiber officinalis and Curcuma longa: characterization and antibacterial activity study. Mater Today Proc. 2020;25:302–306.
123. Rauf MA, Owais M, Rajpoot R, Ahmad F, Khan N, Zubair S. Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals. RSC Adv. 2017;7(58):36361–36373. doi:10.1039/C7RA05040B
124. Alshweiat A, Al-Khresieh RO, Alzarieni KZ, Rashaid AHB. A significant antibiofilm and antimicrobial activity of chitosan-polyacrylic acid nanoparticles against pathogenic bacteria. Saudi Pharm J. 2024;32(1):101918. doi:10.1016/j.jsps.2023.101918
125. Ferro S, Ricchelli F, Monti D, Mancini G, Jori G. Efficient photoinactivation of methicillin-resistant Staphylococcus aureus by a novel porphyrin incorporated into a poly-cationic liposome. Int J Biochem Cell Biol. 2007;39(5):1026–1034. doi:10.1016/j.biocel.2007.02.001
126. González-Gaitán C, Ruiz-Rosas R, Morallón E, Cazorla-Amorós D. Effects of the surface chemistry and structure of carbon nanotubes on the coating of glucose oxidase and electrochemical biosensors performance. RSC Adv. 2017;7(43):26867–26878.
127. Xia M-Y, Xie Y, Yu C-H, et al. Graphene-based nanomaterials: the promising active agents for antibiotics-independent antibacterial applications. J Control Release. 2019;307:16–31.
128. Yu C-H, Chen G-Y, Xia M-Y, et al. Understanding the sheet size-antibacterial activity relationship of graphene oxide and the nano-bio interaction-based physical mechanisms. Colloids Surf B. 2020;191:111009.
129. Kumar P, Huo P, Zhang R, Liu B. Antibacterial properties of graphene-based nanomaterials. Nanomaterials. 2019;9(5).
130. Oves M, Rauf MA, Ansari MO, et al. Graphene Decorated Zinc Oxide and Curcumin to Disinfect the Methicillin-Resistant Staphylococcus aureus. Nanomaterials. 2020;10(5):1004. doi:10.3390/nano10051004
131. Elias L, Taengua R, Frígols B, Salesa B, Serrano-Aroca Á. Carbon nanomaterials and LED irradiation as antibacterial strategies against gram-positive multidrug-resistant pathogens. Int J Mol Sci. 2019;20(14):3603. doi:10.3390/ijms20143603
132. Lu B-Y, Zhu G-Y, Yu C-H, et al. Functionalized graphene oxide nanosheets with unique three-in-one properties for efficient and tunable antibacterial applications. Nano Res. 2021;14:185–190. doi:10.1007/s12274-020-3064-6
133. Moraes A, Araujo Lima B, Fonseca de Faria A, Brocchi M, Luiz Alves O. Graphene oxide-silver nanocomposite as a promising biocidal agent against methicillin-resistant Staphylococcus aureus. Int j Nanomed. 2015;6847. doi:10.2147/IJN.S90660
134. Mei L, Zhu S, Yin W, et al. Two-dimensional nanomaterials beyond graphene for antibacterial applications: current progress and future perspectives. Theranostics. 2020;10(2):757–781. doi:10.7150/thno.39701
135. Kurapati R, Kostarelos K, Prato M, Bianco A. Biomedical uses for 2D materials beyond graphene: current advances and challenges ahead. Adv Mater. 2016;28(29):6052–6074. doi:10.1002/adma.201506306
136. Kuo YL, Wang SG, Wu CY, et al. Functional gold nanoparticle-based antibacterial agents for nosocomial and antibiotic-resistant bacteria. Nanomedicine. 2016;11:2497–2510. doi:10.2217/nnm-2016-0232
137. Iravani S, Varma RS. MXene-based composites against antibiotic-resistant bacteria: current trends and future perspectives. RSC Adv. 2023;13(14):9665–9677. doi:10.1039/D3RA01276J
138. Huang H, Jiang R, Feng Y, et al. Recent development and prospects of surface modification and biomedical applications of MXenes. Nanoscale. 2020;12(3):1325–1338. doi:10.1039/C9NR07616F
139. Gao Y, Dong Y, Yang S, et al. Size-dependent photothermal antibacterial activity of Ti C T MXene nanosheets against methicillin-resistant Staphylococcus aureus. J Colloid Interface Sci. 2022;617:533–541. doi:10.1016/j.jcis.2022.03.032
140. Dong Y, Liu J, Chen Y, et al. Photothermal and natural activity-based synergistic antibacterial effects of Ti3C2Tx MXene-loaded chitosan hydrogel against methicillin-resistant Staphylococcus aureus. Int J Biol Macromol. 2023;240:124482. doi:10.1016/j.ijbiomac.2023.124482
141. Lin X, Li Z, Qiu J, et al. Fascinating MXene nanomaterials: emerging opportunities in the biomedical field. Biomater Sci. 2021;9(16):5437–5471. doi:10.1039/D1BM00526J
142. Peng X, Xu L, Zeng M, Dang H. Application and development prospect of nanoscale iron based metal-organic frameworks in biomedicine. Int j Nanomed. 2023;Volume 18:4907–4931. doi:10.2147/IJN.S417543
143. Chakraborty N, Jha D, Roy I, et al. Nanobiotics against antimicrobial resistance: harnessing the power of nanoscale materials and technologies. J Nanobiotechnol. 2022;20(1). doi:10.1186/s12951-022-01573-9
144. Chakraborty A, Diwan A, Tatake J. Prospect of nanomaterials as antimicrobial and antiviral regimen. AIMS Microbiol. 2023;9(3):444–466. doi:10.3934/microbiol.2023024
145. Hulme J. Application of nanomaterials in the prevention, detection, and treatment of Methicillin-Resistant Staphylococcus aureus (MRSA). Pharmaceutics. 2022;14(4):805. doi:10.3390/pharmaceutics14040805
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.