Back to Journals » International Journal of Nanomedicine » Volume 20
Stomatobaculum longum-Derived Extracellular Vesicles Enhance Oral Squamous Cell Carcinoma Malignancy Through BRCA1/EXO1/TP53BP1 Modulation
Authors Yan L, Wu F , Jiao J, Maimaiti A, Li Y, Shao L, Liang Q , Xiong X, Qin Z
Received 19 September 2024
Accepted for publication 24 April 2025
Published 25 May 2025 Volume 2025:20 Pages 6659—6674
DOI https://doi.org/10.2147/IJN.S491473
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Jie Huang
Lingjian Yan,1,2,* Fan Wu,1,2,* Jiuyang Jiao,1,* Abudusaimi Maimaiti,3 Yachong Li,1,2 Libin Shao,1,2 Qixiang Liang,4 Xinxin Xiong,5 Zeman Qin1,2
1Department of Stomatology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangdong-Hong Kong Joint Laboratory for RNA Medicine, Medical Research Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 3Department of Stomatology, The First People’s Hospital of Kashi Area, Xinjiang Uygur Autonomous Region, People’s Republic of China; 4Department of Stomatology, The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 5The Second Affiliated Hospital, Guangdong Provincial Key Laboratory of Allergy & Clinical Immunology, Guangzhou Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xinxin Xiong, Email [email protected] Zeman Qin, Email [email protected]
Background: Oral squamous cell carcinoma (OSCC) is a common malignancy among Asian populations, and emerging evidence suggests that oral microbiota dysbiosis may play a role in its pathogenesis. This study investigates the role of the oral microbiome, particularly Stomatobaculum longum (S. longum), in OSCC progression and explores the underlying molecular mechanisms involving bacterial extracellular vesicles (EVs).
Methods: 16S rRNA sequencing was conducted on tumor and adjacent non-tumor tissues from OSCC patients to identify microbial composition. In vitro and in vivo experiments were used to evaluate the effects of S. longum on OSCC proliferation and cell cycle progression. GW4869, an inhibitor of EVs’ release, was applied to validate the EVs-mediated mechanisms. Extracellular vesicles from S. longum (SBL-EVs) were isolated using ultracentrifugation and characterized by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). High-throughput sequencing and functional assays were performed to identify signaling pathways regulated by SBL-EVs.
Results: Tumor tissues exhibited a significant enrichment of S. longum compared to non-tumor tissues. S. longum promoted OSCC proliferation and cell cycle progression both in vitro and in vivo, which was reversed by GW4869 treatment. SBL-EVs were successfully isolated and characterized, and they were found to promote OSCC progression by activating the BRCA1/EXO1/TP53BP1 signaling axis.
Conclusion: This study demonstrates the oncogenic role of S. longum in OSCC, mediated through EVs-dependent regulation of the BRCA1/EXO1/TP53BP1 pathway. Targeting bacterial EVs or their downstream signaling may represent a novel therapeutic strategy for OSCC.
Keywords: oral squamous cell carcinoma, Stomatobaculum longum, bacterial vesicles, BRCA1
Introduction
Oral squamous cell carcinoma (OSCC) is a malignant tumor that develops from keratinocyte differentiation and affects the lips and oral mucosa. It is the most common type of malignant tumor in this anatomical region, accounting for 90–95% of cases.1 OSCC has a high incidence among Asian populations, with China having the highest incidence rate, which has resulted in a significant social and economic burden. Risk factors associated with the development of OSCC include alcohol or tobacco intake, chewing betel nut, micronutrient deficiency, chronic injury, and HPV infection.2 It is important to note that other potential causes may contribute to the occurrence, and further exploration of additional risk factors is necessary. Recent research has shown that the composition and imbalance of oral microbiota play a significant role in oral tumors.3 However, the relationship between oral microbiota and the development of OSCC is still unclear. Further investigation in this area can provide valuable insights into the pathogenesis of OSCC and pave the way for novel preventive and therapeutic strategies.
Several investigations have shown a significant correlation between the bacterial and the incidence of OSCC.4,5 The toxins produced by these microorganisms associated with tumors can damage host cell DNA directly or indirectly by generating ROS.6 Additionally, these microbes can stimulate cellular growth and proliferation by activating signaling pathways such as β-catenin and NF-κB.7 Porphyromonas, Fusobacterium, and Streptococcus are among the species that have been identified as differentially represented in the microbiota associated with OSCC.8 For example, Porphyromonas gingivalis induces the expression of B7-H1 in OSCC cells, which may promote an immunosuppressive microenvironment by fostering regulatory T cells (Tregs) and inhibiting effector T cells.9 Al-Hebshi et al reported an association between Clostridium nucleatum and Pseudomonas aeruginosa and the presence of OSCC, further supporting the link between microbial activity and cancer development.10 Although these studies have confirmed the association of specific microbial populations with OSCC, there is still a lack of research identifying bacteria that could serve as prognostic indicators for different stages of OSCC.
Bacterial vesicles, as membrane-bound entities secreted by both Gram-positive and Gram-negative bacteria, span a size spectrum from 20 to 300 nanometers.11 These vesicles serve as transporters for an extensive variety of biomolecules, encompassing membranous and cytoplasmic proteins, peptidoglycan, nucleic acids, and toxins. Their capacity to be internalized into cells and subsequently release their payload positions them as potential vehicles for targeted molecular delivery within biological systems, suggesting novel therapeutic applications.12 Recent investigations have revealed that bacterial vesicles can preferentially accumulate in the tumor microenvironment.13 A study by Kim has showcased through in vivo fluorescence imaging that bacterial vesicles derived from Escherichia coli are predominantly localized in various tumor tissues, including those of colon adenocarcinoma, melanoma, and breast cancer.14 Additionally, vesicles produced by Porphyromonas gingivalis have been found to contain small RNAs that down-regulate DSC2, a member of the cadherin family, thereby fostering tumor invasion and metastasis in OSCC.15 Furthermore, vesicles originating from Fusobacterium species have been reported to activate autophagy pathways in OSCC cells, potentially facilitating epithelial–mesenchymal transition and the subsequent development of lung metastases.16 Despite these advancements, the underlying mechanisms by which bacterial vesicles contribute to tumor invasion and metastasis in OSCC remain incompletely understood.
In this investigation, 16S rRNA sequencing was employed to elucidate the correlation between the oral microbiome and the progression of OSCC in patient samples. Notably, a significant enrichment of S. longum was observed within tumor tissues. Both in vivo and in vitro experiments indicated that the presence of S. longum significantly enhanced the proliferation and cell cycle progression of OSCC cells, which was counteracted by GW4869, an inhibitor of extracellular vesicle synthesis and release. SBL-EVs were isolated through ultra-high speed centrifugation, and their morphology was characterized using TEM and NTA. Subsequent high-throughput sequencing, coupled with in vitro and in vivo assays, confirmed that SBL-EVs modulate the progression of OSCC via the regulation of BRCA1/EXO1/TP53BP1 signaling pathways. Our findings pave the way for future exploration into the potential therapeutic implications of bacterial vesicles in OSCC treatment.
Methods and Materials
Bacterial Culture Preparation
The study commenced after obtaining approval from the Ethics Committee of Sun Yat-sen University and written informed consent. Tissue samples, including normal tissues, peri-tumor tissues, and tumor tissues from patients with OSCC, were collected and homogenized. The samples comprised tumor tissues (3 mm³) obtained from the deep invasive regions of the tumor, normal mucosal tissues (2–3 mm³) collected at least 5 cm away from the tumor site, and peritumoral tissues (2–3 mm³) harvested within 1 cm of the tumor margin. The supernatant of these samples was then added into anaerobic culture bottles and aerobic culture bottles containing Brain-Heart Infusion (BHI) medium. The anaerobic culture bottles were incubated in an anaerobic chamber with shaking, while the aerobic culture bottles were incubated in a constant temperature incubator with shaking. After 72 hours, 100 μL of the cultured medium was transferred to a blood agar plate, and the entire surface of the blood agar was spread using an inoculation loop. All inoculated culture plates were incubated inside a humidified 5% CO2 incubator at 37°C.
16S rRNA Sequencing
Tumor (n = 14) and adjacent normal tissues (n = 9) were homogenized in liquid nitrogen-chilled disposable grinding tubes for microbial 16S rRNA gene sequencing, which was conducted by Guangzhou IGE Biotechnology LTD. Genomic DNA was extracted using the FastPure Blood/Cell/Tissue/Bacteria DNA Isolation Mini Kit (Vazyme, China) following the manufacturer’s protocol. The V3-V4 hypervariable region of the bacterial 16S rRNA gene was amplified with specific primer pairs, and the amplicons were quantified using the Qubit fluorometric quantitation system (Thermo Fisher Scientific, USA). Subsequently, the DNA library was sequenced on the Illumina® MiSeq platform (Illumina, Inc., San Diego, CA, USA). Raw sequence data were processed using Qiime2 software. Samples were demultiplexed based on index and barcode information, and both barcode sequences and PCR primers were removed. Denoising of the sequence data was performed according to the DADA2 analysis pipeline within Qiime2 to obtain Amplicon Sequence Variants (ASVs). The taxonomic annotations and abundance information for all samples across various classification levels were utilized to select the top 35 most abundant taxa. A heatmap was then generated based on the abundance of these taxa within each sample.
qPCR Assay
The cultured medium containing bacteria was collected and centrifuged, and then the pellet was used for RNA isolation with TRIZOL regent (ThermoFisher, USA). mRNAs were subjected to reverse transcription using a cDNA Reverse Transcription Kit according to the manufacturer’s instructions (Vazyme, China), and the mRNA expression was examined by using SYBR® Green PCR Master Mix (TAKARA, China) with a real-time PCR Detection System (Bio-rad CFX96, USA). The primer sequences were as follows:
S. longum-16S rRNA-F: 5′-CGCGTTCGATTAGCCAGTTG-3′
S. longum-16S rRNA-R: GCCACCGGTGTTCTTCCTAA
E. coil-16S rRNA-F: 5′-TGCCTGATGGAGGGGGATAA-3′
E. coli-16S rRNA-R: 5′-TGGAGTTAGCCGGTGCTTCTT-3′
Cell Culture
Human OSCC CAL27 and SCC9 cell lines were purchased from ATCC and cultured in Dulbecco’s modified Eagles’ medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin/streptomycin (Invitrogen, USA) at 37°C in a humidified atmosphere containing 5% CO2.
CCK8 Assay
The Cell Counting Kit-8 (CCK-8) assay was employed to assess cell proliferation. 1×103 cells were seeded into each well of a 96-well plate, then 10 µL of CCK-8 solution was added to the culture medium in each well at designated time points followed by incubation for an additional hour. Subsequently, the absorbance at 450 nm was measured using a microplate reader (BioTeck, USA) to determine the extent of cell proliferation.
Stomatobaculum longum-Derived Extracellular Vesicles (SBL-EVs) Isolation
S. longum (DSM 24645, purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ) by the Guangdong Institute of Microbiology, Guangzhou, Guangdong, P.R. China) was cultivated in BHI medium at 37°C under anaerobic conditions for 72 hours until the optical density at 600 nm (OD600) reached 2–3. The culture underwent centrifugation at 10,000 × g for 10 minutes to pellet the cells. The supernatant was then filtered sequentially through a 0.45-µm pore size filter followed by a 0.22-µm pore size filter to remove larger particles and contaminants. The filtrate underwent sequential centrifugations at 300 g for 10 minutes, 2,000 g for 20 minutes, and 10,000 g for 30 minutes. The resulting supernatant was then processed by ultracentrifugation at 130,000 × g for 70 minutes at 4°C to pellet the membrane vesicles. After ultracentrifugation, the supernatant was discarded, and the vesicle pellet was resuspended in 5 mL of sterile Phosphate Buffered Saline (PBS). The suspension was filtered through a 0.22-µm pore size filter to ensure purity and stored at −80°C. The size distribution of the SBL-EVs was determined using NanoSight nanoparticle characterization system (NanoSight, UK). Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Invitrogen, USA).
Transmission Electron Microscopy (TEM)
A 20 μL aliquot of the SBL-EVs suspension was deposited onto a carbon-coated perforated film pre-mounted on a transmission electron microscopy (TEM) grid. After a 2-minute adsorption period, excess fluid was carefully removed by blotting twice from the reverse side of the grid using filter paper. This procedure resulted in the formation of an ultrathin film of the vesicles, optimal for imaging. The HT7800 TEM (120 kV, Hitachi Co. Ltd., Japan) was used to examine the specimens.
HPLC-MS Analysis of SBL-EVs
Samples of S. longum were prepared by adding 100 μL of deionized water and 500 μL of a methanol-acetonitrile mixture (1:1), followed by immersion in liquid nitrogen for 1 minute and sonication for 10 minutes. In parallel, 200 μL of SBL-EVs was combined with 600 μL of the methanol-acetonitrile mixture (1:1). Subsequently, all samples underwent centrifugation at 17,000 g for 15 minutes, and the supernatants were vacuum-centrifuged at 35°C. The pellets were then reconstituted in 100 μL of 50% methanol in water and once more centrifuged at 17,000 g for 15 minutes. For HPLC analysis, 60 μL of the supernatant was analyzed using System A (0.1% formic acid/H2O) and System B (0.1% acetonitrile formate) for positive ion mode, and System A (2mM ammonium acetate) and System B (acetonitrile) for negative ion mode. The Agilent 6545A QTOF mass spectrometer was operated using the LC/MS Data Acquisition software, Version B.08.00. The mass spectrometric acquisition rates were as follows: 6 spectra per second for MS, and 12 spectra per second for MS2 Collection. The collision energies for the second-order impact were set to 10 V and 40 V, respectively, selecting 12 ions from the first-order spectra for second-order scanning. The primary mass scan range was m/z 50–1300, and the secondary mass scan range was m/z 20–1300. The parameters for the electrospray ionization (ESI) source were as follows: ion source drying temperature, 320°C; nitrogen flow, 8 L/min; sheath gas flow, 12 L/min; sheath gas temperature, 350°C. Capillary voltages were set to 4000 V for positive ion mode and 3500 V for negative ion mode.
Uptake of SBL-EVs by OSCC Cells
To detect the internalization of extracellular vesicles, SBL-EVs were labelled with PKH26 red fluorescent dye (Sigma, USA) according to the manufacturer’s instructions. Specifically, SBL-EVs were suspended in 500 μL of Diluent C containing 5 μL of PKH26 dye. The mixture was then incubated for 5 minutes at 37°C. The staining reaction was stopped by adding 250 μL of 5% bovine serum albumin (Beyotime, China). The PKH26-labelled SBL-EVs were subsequently co-cultured with OSCC cells for 6 h. The cells were fixed using 4% paraformaldehyde, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime, China). Images were captured using fluorescence microscopy.
Flow Cytometry Analysis of Cell Cycle
OSCC cells were plated at a density of 3×105 cells per well in six-well plates and incubated overnight. The next day, the cells were harvested by trypsinization and fixed with 70% (v/v) ethanol at 4°C for 2 hours. After fixation, the cells were washed with PBS and incubated in PBS containing 0.1 mg/mL RNaseA and 50 µg/mL Propidium Iodide (PI) for 30 minutes at 37°C in the dark. Finally, the cells were immediately analyzed using flow cytometry.
Mouse Experiments
BALB/c nude mice, aged 6–8 weeks, were sourced from Beijing Vitalstar Biotechnology Co., Ltd. and housed under specific-pathogen-free (SPF) conditions at a controlled temperature of 22 ± 2°C. The animal care and experimental protocols were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University, with all studies conducted in accordance with the approved guidelines. For the subcutaneous tumor model, BALB/c nude mice were inoculated with CAL27 and SCC9 cells. Post-tumor implantation, the animals were randomly assigned to five treatment groups: a negative control group; a S. longum supernatant group (100 mg/mL, 100 μL); a GW4869 group (2.5 µg/g); a S. longum + GW4869 group (100 mg/mL, 100 μL, where the cultured S. longum was pre-treated with 10 µM GW4869); and an SBL-EVs group (100 mg/mL, 100 μL). All treatments were administered intraperitoneally every five days. Tumor dimensions were measured alternating days using caliper measurements, and the tumor volume was calculated. Three weeks after the initiation of treatment, the mice were euthanized, and the tumors were excised for further analysis.
Tracking of SBL-EVs’ Biodistribution
SBL-EVs were labelled with DiI dye following the manufacturer’s protocol. After washing with PBS, they were centrifuged at 100,000×g for 1 hour at 4°C. Next, 100 μg of DiL-SBL-EVs and 100 μL of PBS-DiL were administered via intraperitoneal injection to mice with subcutaneous tumors. The mice were euthanized by cervical dislocation eight hours after injection. Subsequently, the tumors and various organs, such as the liver, heart, kidney, spleen, and lungs, were harvested for in vivo imaging using an IVIS Spectrum In vivo Imaging System.
Western Blotting
Protein extraction was performed using RIPA lysis buffer supplemented with PMSF (Beyotime, China). The extracted proteins were separated via SDS-PAGE and subsequently electrotransferred onto PVDF membranes (Millipore, USA). The membranes were then blocked with 5% nonfat milk for 1 hour and incubated overnight at 4°C with the appropriate primary antibodies. Following this, the membranes were washed three times with TBST and incubated for 1 hour at room temperature with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies. Protein detection was achieved using ECL plus reagent (Millipore, USA) on a ChemiDoc™ System (Bio-Rad, USA).
Statistical Analysis
Statistical analyses were conducted using Prism 8 software (GraphPad). All data are presented as the mean ± standard deviation (SD). For pairwise comparisons, Student’s t-tests were employed. Multiple comparisons among groups were assessed using one-way analysis of variance (ANOVA) or two-way ANOVA as appropriate. A p-value less than 0.05 was considered to indicate a statistically significant difference.
Results
Stomatobaculum longum Induces the Progression of OSCC
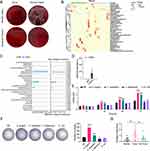
The analysis of bacterial clones showed a significant difference between tumor and normal tissues after cultivation in both anaerobic and aerobic conditions using BHI medium. It is worth noting that the size and number of bacterial clones in tumor tissues were considerably lower than those in normal tissues (Figure 1A). After analyzing the microbial diversity between anaerobic and aerobic flora in five pairs of clinical tissue samples using 16S rRNA gene sequencing, the heatmap showed that the top 25 differentially abundant taxa were statistically significant, including Peptostreptococcus, Prevotella, and Stomatobaculum (Figure 1B). Additionally, a correlation analysis showed that Mesorhizobium, Stomatobaculum, Prevotella and Peptostreptococcus had high microbial abundance (Figure 1C). The Wilcoxon rank-sum test revealed that the abundance of S. longum was significantly higher in tumor tissues than in normal tissues, among the ten bacteria with high abundance (Figure 1D). The effects of S. longum, Escherichia coli (E. coli; DSM 1103), Lactobacillus rhamnosus (L. rhamnosus; HN001), and Fusobacterium nucleatum (F. nucleatum; DSM 20482) on cell proliferation were further elucidated using CCK8 assay and colony formation assay. Our findings indicate that S. longum and F. nucleatum significantly enhance the proliferative capacity (Figure 1E) and colony formation (Figure 1F) of CAL27 cells, with S. longum exhibiting a more pronounced effect. Conversely, L. rhamnosus was found to suppress the proliferation and clonogenic potential of CAL27 cells. Moreover, the expression of S. longum was highly upregulated in tumor tissues compared with normal and peri-tumor tissues (Figure 1G). These data indicated that S. longum induce the progression of OSCC.
Stomatobaculum longum Promote the Progression of OSCC via Extracellular Vesicles
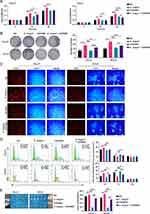
To investigate whether the modulation of OSCC development by S. longum is mediated through bacterial exosomes, we employed the non-competitive neutral sphingomyelinase (N-SMase) inhibitor GW4869, which is known to effectively block the synthesis and release of extracellular vesicles.17 CAL27 and SCC9 OSCC cell lines were cultured with supernatants from S. longum cultures that were either supplemented with GW4869 or remained untreated. The impact on cell viability was subsequently evaluated using the CCK8 assay (Figure 2A), colony formation assay (Figure 2B), and EDU cell proliferation assay (Figure 2C). The findings suggest that the culture supernatant devoid of exosomal vesicles from S. longum cultures does not significantly enhance the proliferation and colony formation of OSCC cells. However, the addition of GW4869 appears to slightly suppress the proliferation and colony formation of OSCC cells (Figure 2A–C).
Additional cell cycle analyses were performed to determine the modulatory effect of culture supernatants with and without extracellular vesicles. According to Figure 2D, the supernatant containing extracellular vesicles from S. longum effectively increased cell accumulation in the G1 phase while impeding the progression through the S phase. When extracellular vesicle release was inhibited by GW4869, the resulting culture supernatant showed a significant decrease in G1 phase accumulation and an augmented number of cells in the S phase. In vivo experiment also demonstrated that the supernatant containing extracellular vesicles from S. longum significantly promoting the tumor growth of CAL27 and SCC9 in nude mice, which was also inhibited by GW4869 (Figure 2E). These observations above imply that S. longum promote the progression of OSCC via extracellular vesicles.
Extracellular Vesicles Derived from Stomatobaculum longum Isolation and Characterization
Extracellular vesicles originating from S. longum (SBL-EVs) were meticulously isolated employing an intricate sequence of filtration steps culminating in ultracentrifugation. The resultant SBL-EVs underwent comprehensive characterization pertaining to their morphological attributes and dimensions. An exemplary Cryo-TEM micrograph corroborates the presence of these vesicles within the ultracentrifuged pellet, whilst elucidating their structural morphology (Figure 3A). Furthermore, the size distribution profile of the SBL-EVs was ascertained utilizing NanoSight technology, revealing a preponderance of vesicles with an average diameter of approximately 88 nm (Figure 3B). SBL-EVs underwent analytical scrutiny for their molecular content through nucleic acid electrophoresis to visualize RNA constituents (Figure 3C). Additionally, protein components were discerned using silver staining (Figure 3D). HPLC analyses were also conducted to elucidate the presence of small molecules within the vesicles (Figure 3E). The cumulative data revealed that SBL-EVs encapsulate RNA, proteins, and a variety of small molecules. Notably, the composition of these substances mirrors that found within S. longum, albeit with variations in concentration and relative enrichment. Six hours after the competitive uptake experiment, SBL-EVs were mainly found within CAL27 and SCC9 cells, as shown by PKH26 staining (Figure 3F). In addition, tumor-bearing mice were injected intraperitoneally with Dil-labeled SBL-EVs. The presence of Dil-labeled SBL-EVs was observed in tumor tissues, indicating a sustained accumulation of the fluorescent signal up to 24 hours after injection. Upon collection of various organs, it was noted that the majority of the fluorescence signals were concentrated in the liver and kidney (Figure 3G). In summary, successful isolation and detailed characterization of SBL-EVs have been achieved.
Extracellular Vesicles Derived from Stomatobaculum longum Promote the Malignant Phenotype of OSCC Cells
The above findings provide evidence that S. longum regulates the development of OSCC through extracellular vesicles. It has been shown that SBL-EVs can be isolated using ultra-high speed centrifugation methods. The impact of SBL-EVs on cellular proliferation and the cell cycle was investigated due to their capacity to be internalized by OSCC cells and concentrate within tumors in vivo. The results demonstrated that SBL-EVs increased the proliferative potential and clonogenic ability of CAL27 and SCC9 cells, as measured by CCK8 assay (Figure 4A), colony formation assay (Figure 4B), and EDU staining (Figure 4C). Furthermore, SBL-EVs were shown to significantly increase the proportion of cells in the G1 phase while decreasing the population in the S phase for both CAL27 and SCC9 cells (Figure 4D). Additionally, the tumor volumes in nude mice injected with CAL27 and SCC9 cells and treated with SBL-EVs were significantly larger compared to the control group (Figure 4E), highlighting the strong impact of SBL-EVs on tumor progression.
SBL-EVs Transport Oncogenic Signaling Molecules
Subsequently, we conducted a bioinformatics analysis on the RNA sequencing data of SBL-EVs co-cultured with CAL27 cells to elucidate the underlying mechanisms. Our analysis revealed a total of 16,285 differentially expressed genes (DEGs), with 9,756 genes being down-regulated and 6,529 genes up-regulated post SBL-EVs treatment (Figure 5). In the Gene Ontology (GO) analysis of these DEGs, the most enriched terms were associated with DNA repair (BP), microtubule organizing center (CC), and cadherin binding (MF) (Figure 5B). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the ubiquitin-mediated proteolysis, nuclear transcription, bacterial invasion, and cell cycle pathways were significantly enriched (Figure 5C). To further investigate the functional relevance of these DEGs within the GO pathways, we constructed a protein–protein interaction (PPI) network using the STRING database and visualized it with the Cytoscape tool. This PPI network was subjected to topological analysis using the Degree (Deg, Figure 5D), Maximal clique centrality (MCC, Figure 5E), and Betweenness (BC, Figure 5F) algorithms from the cytoHubba plugin. By intersecting the top 50 results from these three analytical methods, we identified 11 core genes, including BRCA1, EXO1, TOP2A, BRCA2, ATRX, ASPM, TP53BP1, MCM10, PRKDC, CENPJ, and TPR (Figure 5G). These core genes were then mapped onto the STRING database to construct a refined PPI network, which demonstrated that BRCA1 had the highest number of interactions among the 11 core genes (Figure 5H), suggesting its potential role as a key target gene modulated by SBL-EVs.
SBL-EVs Induced the Progression of OSCC via Regulating BRCA1/EXO1/TP53BP1 Signaling Pathway
To elucidate the molecular mechanisms underlying the development of OSCC cells regulated by BRCA1, we established a stable transfection model with BRCA1 knockdown for transcriptome sequencing (Supplemental Figure 1). A total of 2,781 differentially expressed genes were identified in CAL27, including 1,252 down-regulated genes and 1,529 up-regulated genes (Figure 6A). The Go analysis of the DEGs showed that the BP, CC, and MF categories were enriched with immune response activation, cell-substrate junction, and cytokine activity, respectively (Figure 6B). The KEGG pathway analysis revealed significant enrichment in the TNF signaling pathway, NOD-like receptor signaling pathway, and cell cycle pathway (Figure 6C). By intersecting the top 15 hub genes with 11 core genes identified in the administration of SBL-EVs, we obtained BRCA1, EXO1, TP53BP1, ASPM, ATRX, and TOP2A genes (Figure 6D). The PPI network suggested that the BRCA1/EXO1/TP53BP1 signaling pathway may be the targeted pathways (Figure 6E).
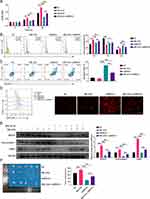
The CCK8 assay (Figure 7A) and cell cycle analysis (Figure 7B) revealed that SBL-EVs enhanced the proliferative capacity of SCC9 cells, which was reversed upon BRCA1 suppression. The knockdown of BRCA1 induced the apoptosis in SCC9 cells, which was counteracted by SBL-EVs administration (Figure 7C). The BRCA1 gene encodes a protein that is integral to DNA repair mechanisms, facilitating the repair of damaged DNA.18 Figure 7D shows that ROS accumulation was intensified upon BRCA1 suppression, which was reversed by SBL-EVs treatment. SBL-EVs significantly increased the expression of BRCA1, EXO1, and TP53BP1 proteins, and this upregulation was nullified by targeted BRCA1 knockdown (Figure 7E). Furthermore, the ablation of BRCA1 significantly impeded the tumorigenic growth of SCC9 cells in NOD mice (Figure 7F). The data indicate that SBL-EVs contribute to the progression of OSCC by regulating the BRCA1/EXO1/TP53BP1 signaling pathway.
Discussion
OSCC is one of the most common malignant tumors, with high malignancy and poor prognosis.19 The study of the relationship between bacteria and OSCC is an important research field because bacteria can trigger a cascade of biological responses such as infection and inflammatory responses that ultimately lead to the development of tumors. Bacterial extracellular vesicles (EVs) play an important role in the genesis, development, and metastasis of tumors.13 In this study, we found that S. longum was highly enriched in OSCC sections and can effectively promote the progression of OSCC through bacterial EVs. Additionally, we discovered that EVs derived from S. longum induced tumor malignancy by regulating the BRCA1/EXO1/TP53BP1 signaling pathway. Therefore, the specific mechanism and function of bacterial EVs in tumors are discussed, as well as how to use these EVs as biomarkers and targets for early diagnosis and treatment, which may provide more effective treatment options for OSCC patients.
The Human Oral Microbiome Database (eHOMD, http://www.homd.org) indicates the presence of over 770 identified bacterial strains within the oral cavity, highlighting the complexity of the microbial communities residing in this environment.20 The oral and maxillofacial region, with its consistent temperature of 37°C and salivary pH ranging between 6.5 and 7.5, provides a stable ecosystem conducive to the proliferation of both aerobic and anaerobic bacteria.21 These factors underscore a potentially significant relationship between the treatment of OSCC and the composition of oral microbiota. To elucidate the connection between the oral microenvironment and OSCC, clinical tissue samples were meticulously collected and subsequently cultured under both anaerobic and aerobic conditions. Our findings revealed notable disparities in the quantity of bacterial colonies present in tumor tissues compared to adjacent by blood agar plate cultivation. Intriguingly, the 16S rRNA sequence analysis indicated an enrichment of S. longum within the tumor tissues. This observation aligns with previous research by E. Babikow, who reported dynamic shifts in the abundance of S. longum during fixed orthodontic treatment, suggesting its role in maintaining oral health in conjunction with the gingival microbiota.22 Additionally, the study conducted by Weihua Shi et al implicated the fluctuations in the prevalence of S. longum in the incidence of dental caries among adolescents.23 Collectively, these findings underscore a potential correlation between the levels of S. longum and various oral diseases, including OSCC.
EVs are defined as small vesicles ranging from 50 to 250 nm in diameter, which are released by bacteria and have the capacity to transport a broad spectrum of bioactive molecules, such as proteins, lipids, DNA, and RNA.24 The significance of EVs in oncology has been increasingly recognized, with numerous studies underscoring their involvement in the initiation, progression, and metastasis of tumors.25 Shuang Qing’s research elucidated that bacterial EVs are efficiently taken up and concentrated within tumor cells.13 Mukeng Hong et al discovered that bacteria within the intestinal microbiota could modulate the inflammatory process of osteoarthritis through the secretion of bacterial vesicles, thus participating in the regulatory mechanisms underlying osteoarthritis.26 In our current study, we have uncovered that S. longum promotes the proliferation of OSCC cells and contributes to tumor growth through the release of EVs. We successfully isolated and characterized SBL-EVs using TEM and NTA. Further experiments revealed that SBL-EVs are effectively internalized by OSCC cells, playing a biological role in promoting cell proliferation and regulating the cell cycle. To gain a deeper understanding of the in vivo distribution of SBL-EVs, we conducted small animal imaging studies. Our findings indicated that SBL-EVs exhibit a pronounced enrichment in tumor tissues and are also concentrated in the liver and kidneys, suggesting a systemic distribution pattern with potential implications in tumor. Unfortunately, this study did not further analyze the components of SBL-EVs and the metabolomics of OSCC cells, future studies will explore the mechanism of SBL-EVs in tumor microenvironment and provide a new theoretical basis for tumor treatment.
High-throughput sequencing analysis identified BRCA1, EXO1, and TP53BP1 as hub genes following administration with SBL-EVs. These proteins play crucial roles in cellular proliferation, DNA repair, and cell cycle regulation. BRCA1 is a DNA repair gene that inhibits apoptosis and ROS accumulation, thus protecting cells from oxidative stress damage.27–29 The expression of the BRCA1 gene has a certain correlation with the occurrence and development of OSCC. Mutations in the BRCA1 gene can increase the risk of OSCC progression.30 However, the molecular mechanism of action between BRCA1 and oral cancer remains unclear, and the regulation between oral flora and BRCA1 has not been reported. EXO1 is an exonuclease involved in processes such as DNA damage repair, DNA replication, and recombination. Aberrant expression and activity of the EXO1 gene are related to the occurrence of various tumors.31 TP53BP1 (also known as 53BP1) is a protein that interacts with the p53 protein, which plays a key role in DNA damage and cell cycle regulation.32 Abnormal expression of these proteins in OSCC may lead to uncontrolled cell proliferation, which in turn promotes the occurrence and development of tumors. Our results confirmed that SBL-EVs can significantly promote the expression of BRCA1, EXO1, and TP53BP1, thereby promoting cell proliferation and inhibiting cell apoptosis. Furthermore, SBL-EVs could also inhibit apoptosis and ROS accumulation caused by BRCA1 knockdown, indicating their role in protecting cells from oxidative stress damage. These findings provide new ideas and targets for the treatment of OSCC.
In conclusion, this study revealed the role and mechanism by which SBL-EVs regulate the progression of OSCC via BRCA1. Future studies can investigate the role and molecular mechanism of SBL-EVs in the treatment of OSCC, providing novel insights and therapeutic targets for the treatment of OSCC.
Data Sharing Statement
Sequence files and metadata for all samples used in this study have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.25827688.v2).
Ethical Approval and Consent to Participate
The present study was approved by The Sun Yat-sen University Committee for Ethical Review of Research Involving Human Subjects (SYSKY-2024-214-03) and the Animal Ethics Committee of Sun Yat-sen University (AP20220040) in accordance with the provisions of the “Experimental Animal Care and Use Guide”. All experiments were conducted in Sun Yat-sen Memorial Hospital. Written informed consent was obtained from all patients. The principles outlined in the Declaration of Helsinki were follow. Subjects’ right have been protected by an appropriate Institutional Review Board.
Acknowledgments
This study was supported by the technical and equipment support by the Department of Oral & Maxillofacial Surgery and Department of General Dentistry of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. We thank all the patients who consented to participate in the study as well as all nurses and physicians who performed the procedures: their enthusiasm and support as co-researchers in this project was indispensable.
Funding
This study was supported by Sun Yat-sen Pilot Scientific Research Fund (Grant No. YXQH202412), National Nature Science Foundation in China (NSFC) (Grant No. 82273179), and Science and Technology Program of Guangzhou (Grant No. 202201010916).
Disclosure
The authors declare that they have no competing interests.
References
1. Tan Y, Wang Z, Xu M, et al. Oral squamous cell carcinomas: state of the field and emerging directions. Int J Oral Sci. 2023;15(1):44. doi:10.1038/s41368-023-00249-w
2. Chamoli A, Gosavi AS, Shirwadkar UP, et al. Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics. Oral Oncol. 2021;121:105451. doi:10.1016/j.oraloncology.2021.105451
3. Sukmana BI, Saleh RO, Najim MA, et al. Oral microbiota and oral squamous cell carcinoma: a review of their relation and carcinogenic mechanisms. Front Oncol. 2024;14:1319777. doi:10.3389/fonc.2024.1319777
4. Stasiewicz M, Karpinski TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 2022;86(Pt 3):633–642. doi:10.1016/j.semcancer.2021.11.002
5. Lamont RJ, Fitzsimonds ZR, Wang H, Gao S. Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontol. 2022;89(1):154–165. doi:10.1111/prd.12425
6. Bakhti SZ, Latifi-Navid S. Oral microbiota and Helicobacter pylori in gastric carcinogenesis: what do we know and where next? BMC Microbiol. 2021;21(1):71. doi:10.1186/s12866-021-02130-4
7. Tuganbaev T, Yoshida K, Honda K. The effects of oral microbiota on health. Science. 2022;376(6596):934–936. doi:10.1126/science.abn1890
8. Gholizadeh P, Eslami H, Yousefi M, Asgharzadeh M, Aghazadeh M, Kafil HS. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother. 2016;84:552–558. doi:10.1016/j.biopha.2016.09.082
9. Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology. 2011;216(12):1302–1310. doi:10.1016/j.imbio.2011.05.005
10. Al-Hebshi NN, Nasher AT, Maryoud MY, et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep. 2017;7(1):1834. doi:10.1038/s41598-017-02079-3
11. Sartorio MG, Pardue EJ, Feldman MF, Haurat MF. Bacterial outer membrane vesicles: from discovery to applications. Annu Rev Microbiol. 2021;75:609–630. doi:10.1146/annurev-micro-052821-031444
12. Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17(1):13–24. doi:10.1038/s41579-018-0112-2
13. Qing S, Lyu C, Zhu L, et al. Biomineralized bacterial outer membrane vesicles potentiate safe and efficient tumor microenvironment reprogramming for anticancer therapy. Adv Mater. 2020;32(47):e2002085. doi:10.1002/adma.202002085
14. Kim OY, Park HT, Dinh NTH, et al. Bacterial outer membrane vesicles suppress tumor by interferon-gamma-mediated antitumor response. Nat Commun. 2017;8(1):626. doi:10.1038/s41467-017-00729-8
15. Liu D, Liu S, Liu J, Miao L, Zhang S, Pan Y. sRNA23392 packaged by Porphyromonas gingivalis outer membrane vesicles promotes oral squamous cell carcinomas migration and invasion by targeting desmocollin-2. Mol Oral Microbiol. 2021;36(3):182–191. doi:10.1111/omi.12334
16. Chen G, Gao C, Jiang S, et al. Fusobacterium nucleatum outer membrane vesicles activate autophagy to promote oral cancer metastasis. J Adv Res. 2024;56:167–179. doi:10.1016/j.jare.2023.04.002
17. Essandoh K, Yang L, Wang X, et al. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim Biophys Acta. 2015;1852(11):2362–2371. doi:10.1016/j.bbadis.2015.08.010
18. Wu J, Lu LY, Yu X. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1(2):117–123. doi:10.1007/s13238-010-0010-5
19. Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Semin Cancer Biol. 2020;61:71–83. doi:10.1016/j.semcancer.2019.09.011
20. Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. 2018;200(4):525–540. doi:10.1007/s00203-018-1505-3
21. Galeano Nino JL, Wu H, LaCourse KD, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611(7937):810–817. doi:10.1038/s41586-022-05435-0
22. Babikow E, Ghaltakhchyan N, Livingston T, et al. Longitudinal microbiome changes in supragingival biofilm transcriptomes induced by orthodontics. JDR Clin Trans Res. 2023:23800844231199393.
23. Shi W, Tian J, Xu H, Qin M. Microbial relationship of carious deciduous molars and adjacent first permanent molars. Microorganisms. 2023;11(10):2461. doi:10.3390/microorganisms11102461
24. Gupta D, Zickler AM, El Andaloussi S. Dosing extracellular vesicles. Adv Drug Deliv Rev. 2021;178:113961. doi:10.1016/j.addr.2021.113961
25. Marar C, Starich B, Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat Immunol. 2021;22(5):560–570. doi:10.1038/s41590-021-00899-0
26. Hong M, Li Z, Liu H, et al. Fusobacterium nucleatum aggravates rheumatoid arthritis through FadA-containing outer membrane vesicles. Cell Host Microbe. 2023;31(5):798–810e797. doi:10.1016/j.chom.2023.03.018
27. Wang J, Nan N, Shi L, et al. Arabidopsis BRCA1 represses RRTF1-mediated ROS production and ROS-responsive gene expression under dehydration stress. New Phytol. 2020;228(5):1591–1610. doi:10.1111/nph.16786
28. Li Y, Wang Y, Zhang W, Wang X, Chen L, Wang S. BKM120 sensitizes BRCA-proficient triple negative breast cancer cells to olaparib through regulating FOXM1 and Exo1 expression. Sci Rep. 2021;11(1):4774. doi:10.1038/s41598-021-82990-y
29. Oda T, Gotoh N, Kasamatsu T, Handa H, Saitoh T, Sasaki N. DNA damage-induced cellular senescence is regulated by 53BP1 accumulation in the nuclear foci and phase separation. Cell Prolif. 2023;56(6):e13398. doi:10.1111/cpr.13398
30. Oliveira-Costa JP, Oliveira LR, Zanetti R, et al. BRCA1 and gammaH2AX as independent prognostic markers in oral squamous cell carcinoma. Oncoscience. 2014;1(5):383–391. doi:10.18632/oncoscience.47
31. Sertic S, Quadri R, Lazzaro F, Muzi-Falconi M. EXO1: a tightly regulated nuclease. DNA Repair. 2020;93:102929. doi:10.1016/j.dnarep.2020.102929
32. Mirman Z, de Lange T. 53BP1: a DSB escort. Genes Dev. 2020;34(1–2):7–23. doi:10.1101/gad.333237.119
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.