Back to Journals » International Journal of Nanomedicine » Volume 19
Strategies for Targeting Peptide-Modified Exosomes and Their Applications in the Lungs
Authors Qiu M, Zou J, Yang Z, Yang D, Wang R, Guo H
Received 3 April 2024
Accepted for publication 2 August 2024
Published 12 August 2024 Volume 2024:19 Pages 8175—8188
DOI https://doi.org/10.2147/IJN.S472038
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Eng San Thian
Min Qiu,1,2,* Jinru Zou,2,* Zheng Yang,3 Dan Yang,2 Rui Wang,1 Haie Guo1,4
1College of Veterinary Medicine, Inner Mongolia Agricultural University, Inner Mongolia, People’s Republic of China; 2College of Pharmacy, Baotou Medical College, Baotou, People’s Republic of China; 3The First Affiliated Hospital, Baotou Medical College, Baotou, People’s Republic of China; 4Agriculture, Animal Husbandry and Science and Technology Bureau of Liangcheng County, Ulanqab, Inner Mongolia, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Rui Wang; Haie Guo, Email [email protected]; [email protected]
Abstract: Exosomes belong to a subgroup of extracellular vesicles secreted by various cells and are involved in intercellular communication and material transfer. In recent years, exosomes have been used as drug delivery carriers because of their natural origin, high stability, low immunogenicity and high engineering ability. However, achieving targeted drug delivery with exosomes remains challenging. In this paper, a phage display technology was used to screen targeted peptides, and different surface modification strategies of targeted peptide exosomes were reviewed. In addition, the application of peptide-targeted exosomes in pulmonary diseases was also summarised.
Keywords: exosome, targeted peptide modification, targeted delivery, lung
Introduction
Exosomes are extracellular vesicles that are approximately 30–150 nm in diameter, can be produced by various types of cells and are widely found in biological fluids, such as blood, urine, saliva and milk.1–3 The main roles of exosomes are to carry and transmit intercellular information and participate in cell communication and regulation. They contain a variety of bioactive molecules, such as proteins, mRNAs, microRNAs and lipids, which can be transported as metabolites or nutrients to other cells or tissues.4 Exosomes are involved in signal transduction and interactions among cells, affecting cell fate and tissue function.5 To date, research on exosomes is focused on medical diagnosis, therapy and drug development. For example, exosomes have been used as biomarkers for cancer diagnosis and specific protein or RNA detection in exosomes.6 In addition, exosomes are used as drug carriers to deliver drugs and genetic material to target cells or tissues, improving the efficacy of drugs and reducing side effects.7 Exosomes have a natural biological origin and are thus generally considered more biocompatible than synthetic nanoparticles. Owing to the natural targeting of exosomes, they have been extensively explored as drug delivery systems. The targeting ability of exosomes can be improved through surface modification methods, including antibody or ligand modification, polymer modification (particularly with polyethylene glycol), physical modification (such as electroporation), biotinisation, genetic engineering, membrane fusion and magnetic nanoparticle modification.8 In this review, we mainly discuss the different methods for the targeted peptide modification of exosomes and the application of exosomes to the lungs.
Exosomes
Biogenesis of exosomes
The biogenesis of exosomes is complex. Some bioactive substances, such as miRNA, proteins, lipids and other components, and cell surface proteins can enter cells through endocytosis and fuse with the membranes of the endoplasmic reticulum and Golgi apparatus to form plasma membrane buds outside and inside cells.9–11 This budding process leads to the formation of early endosomes, which gradually transform into late endosomes (LSEs). The secondary invagination of LSEs produces intracavicular vesicles (ILVs), and cytoplasmic components enter newly formed ILVs with secondary invagination. ILVs were discovered by Pan,12 who studied the vesicular secretion of transferrin receptors in mature reticulocytes. ILVs form multivesicular bodies (MVBs), which can fuse with autophagosomes or lysosomes. The final contents can be degraded in the lysosomes. MVBs that do not follow this pathway can be transported to the plasma membrane via the cytoskeleton and microtubule networks and released as exosomes by exocytosis after fusion with the cell surface (Figure 1).
 |
Figure 1 (A) Biogenesis of exosomes, (B) Main pathways for exosomes to enter cells; (a) endocytosis; (b) membrane fusion; (c) receptor-mediated endocytosis. |
Main Pathways for Exosomes to Enter Cells
The entry of exosomes into cells usually involves interactions and fusion among cell membranes (Figure 1). This process can be achieved through different mechanisms:13,14 (a) endocytosis, where exosomes can bind to receptors on the cell membrane, prompting the cell membrane to form vesicles that enclose the exosomes in the cell;15 (b) membrane fusion, where exosomes directly fuse with the cell membrane, and the exosome contents are released into the cytoplasm; this fusion may depend on factors, such as membrane fusion proteins and lipid environment regulation;16 (c) receptor-mediated endocytosis, where some exosomes interact with specific receptors and are then taken into the cell through receptor-mediated endocytosis;17,18 the details of this process are still being studied extensively. Understanding the internal communication of exosomes has important implications for the fields of cell communication, immune regulation and disease research.
Exosomal Structure
Exosomes are mainly composed of lipids, proteins and nucleic acids. Exosomal membranes are rich in sphingolipids, cholesterol, phosphatidylserine and ceramides (Figure 2).19 Proteomic studies have shown that tetraspanins (CD9, CD63, CD81 and CD82) exist in exosome membranes. The four transmembrane domains of tetraspanins are the unequal inner ring of amino acid residues (EC1), outer ring (EC2), cytoplasmic carboxyl group (C-terminal) and amino tail (N-terminal).20 These proteins regulate the formation, stabilisation and release of exosomes and form complexes on the exosome membrane to participate in the biosynthesis and directed secretion of exosomes. In addition, they are involved in the regulation of the fusion between exosomes and cell membranes. Integrins,21,22 such as integrins αVβ5 and β3, are involved in adhesion and interactions between exosomes and target cells. These proteins can bind to ligands on the cell membrane, mediate adhesion between exosomes and target cells and promote the uptake or phagocytosis of exosomes. Major histocompatibility complexes (MHCs), including MHCI and MHCII, play a role in immune regulation.23,24 MHC molecules on exosomes can interact with immune cells and mediate the recognition of exosomes by the immune system, thus affecting the immune response. Heat shock proteins25 (HSPs, eg Hsp 70 and Hsp 90) play a role in cellular stress and antigen presentation. HSPs on exosomes can interact with target cells, regulate signalling inside and outside cells and affect exosome uptake. Annexins, such as annexins A1 and A2, regulate the adhesion and membrane binding of exosomes.26 Annexins bind to phospholipids on the cell membrane, promoting the membrane fusion of exosomes and interaction with target cells.27 Glycosyl phosphatidyl inositol (GPI)–anchored proteins, such as CD55 and CD59, play a role in anti-haemolysis and regulation of immune response.28 These proteins are anchored to the exosome membrane through GPI and participate in the regulation of exosome function.29,30 HSPs,31 such as Hsp70 and Hsp90, are involved in the uptake of exosomes and signalling inside and outside cells.32 Peripheral surface proteins, including transforming growth factor β, tumour necrosis factor (TNF) and cytokines, are involved in signalling.20 Lysosomal associated protein 2 (Lamp-2), specifically its variants Lamp-2a, Lamp-2b and Lamp-2c,33 is highly expressed on the surfaces of exosomes and consists of a long carbon chain, in which the highly glycosylated N end is located on the lumen side of the membrane and the C end is exposed to the cytoplasm. Therefore, peptides fusing with the N-terminal of Lamp-2b are highly expressed on the surfaces of exosomes.34 Cells with plasmid vectors encoding Lamp-2b fusion peptides are transfected for the production of peptide-modified exosomes.35 Exosomes have been identified to contain the following types of RNA, such as microRNA and messenger RNA, as well as long non-coding RNAs.36
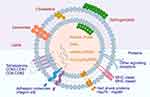 |
Figure 2 Exosome structure diagram. Exosomes are mainly composed of lipids, proteins and nucleic acids. |
Function of Exosomes
Exosomes are essential to intercellular and intra-organ communication systems, transferring signalling molecules,37 such as proteins, DNA, mRNA and miRNA,38 across cells and thereby facilitating intercellular communication.39 Exosomes can regulate immune responses40 by transferring miRNAs and can present antigens to activate the adaptive immune system.41 Given that exosomes are present in all biological fluids and secreted by all cells, a small number of miRNAs and surface proteins in exosomes can be used as markers for disease diagnosis. Exosomes are applied to the diagnosis of cancer and diseases of the cardiovascular and central nervous systems, liver, kidney and lungs. Exosomes can be designed to deliver different therapeutic payloads, including siRNAs, antisense oligonucleotides, chemotherapeutic agents and immunomodulators, to specific targets.9 Compared with traditional vectors, such as liposomes, nanospheres, micelles, microemulsions and conjugates, exosomes are biologically derived in nature, thus showing excellent physicochemical stability, low immunostimulatory activity, low toxicity, biocompatibility and biobarrier permeability.42–44 Owing to these features, exosomes are promising vectors for targeted drug delivery. However, conventional vectors and exosomes are prone to be trapped in non-specific organs,45 and thus, they are unsuitable for targeting specific organs. Nevertheless, their targeting ability for specific tissues or organs can be enhanced by modifying their surface proteins.
Strategies for Modifying Exosomes by Targeting Peptides
Peptides are the most widely explored targeted ligands because of their small molecular weight,46 strong binding affinity and specificity for target cells or tissues, low immunostimulatory activity, and low toxicity.47 In addition, they can be easily modified. Biomimetic peptides can be easily synthesised on the basis of natural ligand–receptor interactions or by screening a large library of peptides. However, peptides are easily degraded by intracellular lysosomal proteases during the formation of exosomes. Thus, peptide synthesis is challenging. To improve the stability of peptides on exosomes’ surfaces, some studies bound glycosylation sequences (GNSTM) to specific peptides and then fused them to the N-terminal of Lamp-2b to prevent specific degradation.48 The three transmembrane proteins widely used to target exosomes are Lamp-2b, lactomucin, and platelet-derived growth factor receptor (PDGF-R).19 A targeting peptide is a short peptide with a specific sequence that binds to a specific cell surface receptor for a targeted effect. When the encoding sequence of the targeted peptide is fused with the exosome membrane protein and transferred to the cell for expression, the secreted exosome will display the targeted peptide on its surface. By modifying targeted peptides to the surfaces of exosomes, the ability of exosomes to target cells and targeted drug delivery efficiency can be improved.49 The screening of targeted peptides can be realised by the phage display technology.
Phage Display Technology
The phage display technology is based on phage DNA gene modification for expressing foreign peptides, antibody fragments or proteins on phage shells.50 Cells that specifically bind to a specific organ or tissue can be screened by phage display and genetically modified on the N-terminal of Lamp-2b for a targeted effect. The British scientist Smith invented this technology in the early 1985s, using a phage surface display technology to identify phages that can bind to fixed peptides,51 laying the foundation for phage display technology. Subsequently, some scientists fused foreign peptides with phage surface proteins to further develop the phage display technology.52
This technology clones a coding gene or target gene fragment of a polypeptide or protein into a specific position in the structure gene of a phage coat protein (Figure 3).53 When the reading frame is correct, and the normal functions of other coat proteins are unaffected, a foreign polypeptide or protein fuses with a coat protein, and the resulting fusion protein is displayed on the surface of the phage during the reassembly of progeny phage.54,55 The peptides or proteins can maintain independent spatial structures and biological activity, thereby facilitating the identification and binding of target molecules. In the process of display, the peptide library is incubated with target protein molecules on the solid phase within a certain period, washes off unbound free phages and removes phages bound to and adsorbed by target molecules with competitive receptors or pickling.56 Eluted phages are propagated and amplified, and then the next round of elution is performed. After 3–5 rounds of “adsorption–elution–amplification”, the phages are eluted. Bacteriophages that bind specifically to target molecules are highly enriched.57 The phage preparation can be used for further enrichment of target bacteria with desired binding properties.58 The enriched peptides are fused with exosome membrane proteins and then displayed on the exosome surface by genetic engineering.
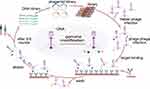 |
Figure 3 Flowchart of phage display technology. |
Genetic Engineering Modification
Proteins on exosome membranes, such as Lamp, GPI and CD63, can fuse with targeted ligands (such as peptides, antibodies and aptamers).In genetic engineering modification, peptides with specific targeting properties are identified first. These peptides typically have a high affinity for target cell surface receptors and can enable exosomes to precisely reach target cells. Then, according to the amino acid sequences of the targeted peptides, corresponding DNA sequences that will encode the peptides are synthesised using a gene synthesis technology. The DNA sequences of the targeted peptides are fused with gene sequences encoding exosome membrane proteins (such as CD63 and Lamp-2b) with molecular cloning techniques (PCR amplification and DNA ligation). Appropriate genetically engineered vectors (such as plasmids and viral vectors) are then selected and stably transfected into host cells, which will carry the fusion genes and secrete exosomes with the targeted peptides (Figure 4). Kim et al59 transfected HEK293 cells with a plasmid encoding cell-targeting peptide (CTP)–flag–Lamp-2b–HA to produce a heart-targeting exosome (CTP-EXO). CTP is a heart-targeting peptide with the sequence APWHLSSQYSRT. Some researchers genetically engineered ischaemic myocardial targeting peptide (IMTP; peptide sequence CSTSMLKAC) into Lamp-2b, which is an exosomal membrane protein derived from mesenchymal stem cells (MSCs); the resulting IMTP–exosome complexes specifically targeted ischaemic myocardial muscles.60 Liang et al61 designed a chondrocyte affinity peptide (CAP; peptide sequence DWRVIIPPRPSA) exosome based on the CAP–Lamp-2b plasmid, which specifically delivers miRNA-140 to chondrocytes in joints and attenuates the progression of osteoarthritis in a rat model. Wang et al62 constructed an EGFP-C1-iRGD-Tyr7-Lamp-2b lentiviral vector containing the exosome membrane protein Lamp-2b,63 tyrosine and iRGD peptide (αv integrin–specific peptide that can specifically target tumours) for HEK-293T cell transfection. A novel exosome (iRGD-Exos) was obtained by loading doxorubicin (Dox) and radioactive iodine-131 into an exosome. The results showed efficient targeting and Dox delivery to integrin αvβ3-positive thyroid anaplastic carcinoma cells.
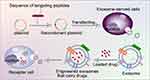 |
Figure 4 Targeted peptide modified exosomes-Genetic engineering. |
Anchoring CP05 Peptide
The CP05 peptide (specific sequence: CRHSQMTVTSRL) has high affinity and specificity for CD63 on an exosome’s surface and is thus often used in exosome modification. CP05 does not alters the characteristics and distribution of exosomes in vivo and functionalises different parts of exosomes simultaneously, facilitating the delivery of targeted phosphodiamide morpholino oligomer (PMO) to the muscles of anti-muscular dystrophin deficiency mice by anchoring a muscle-targeting peptide and PMO to the same exosome with CP05 (Figure 5).64 Thus, the expression of anti-muscular dystrophy protein and muscle function increased in a muscular dystrophy mouse model. In addition, CP05 can specifically capture exosomes from patient sera. Siming Yu et al65 used human umbilical cord mesenchymal cell–derived exosomes as carriers to deliver anti-Mir-146B-5PASo (PMO-146b) and then covalently coupled with anchor-peptide CP05 to assemble on the surface of the exosomes and form a conjugate named ePPMO-146b. The drug exhibited anti-tumour effects in a mouse model of colon cancer. Exosomal proteins and nucleic acids have also been proved to be potential biomarkers for the diagnosis and prognosis of lung cancer, such as epidermal growth factor receptor (EGFR), CD151, CD171, placental alkaline phosphatase and dsDNA.
 |
Figure 5 Targeted peptide modified exosomes-Anchoring CP05 peptide. |
Chemical Modification
Loading targeted peptides onto exosome surfaces can be achieved by chemical methods, such as click-through chemistry (cycloaddition, affinity ring opening, carbonyl chemistry and carbon–carbon multi-bond addition).66 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide-n-hydroxysuccinimide is first used to activate amino groups on the surfaces of exosomes through a condensation reaction, providing active sites for subsequent click chemical reactions. The end of each targeted peptide is then modified with a group that can react with an active site on an exosome’s surface, such as an azide group. This modification ensures the specific bonding of targeted peptides to exosomes’ surfaces. Finally, an azide–acetylene cycloaddition reaction is performed and catalysed using copper and the azide groups on the targeted peptides are combined with the alkyne groups on the exosomes’ surfaces to form stable triazole bonds.67,68 An advantage of click chemistry is that it can quickly and effectively connect biomolecules and drug molecules to exosomes (Figure 6), facilitating the connection of a large number of molecules and showing high efficiency. By contrast, conventional chemical reactions usually require long reaction times and high reaction conditions, which may adversely affect the structure and function of exosomes. Jia et al69 first used the electroporation technology to load superparamagnetic iron oxide nanoparticles and curcumin (Cur) into exosomes and then coupled the exosome membrane with RGE (neurociliin-1 targeting peptide; peptide sequence RGERPPR) according to the click chemistry principle. To obtain therapeutically targeted glioma exosomes, other researchers have used bioorthogonal cupric azide-free alkyne cycloaddition (click chemistry) to couple the functional ligand ring (Arg-Gly-Asp-D-Tyr-Lys) peptide (c (RGDyK)) to the exosome surface. The resulting RGD-exos targets ischaemic cerebral vascular endothelial cells after intravenous administration.70 Some researchers have used the same method to construct and prepare exosome cRGD-Exo-PTX modified with c (RGDyK) and paclitaxel (PTX) loaded, showing good targeted therapy for glioblastoma (GBM).71 Exosomes modified with c (RGDyK) have targeting ability to tumours and are overexpressed in tumour vascular endothelial cells.72
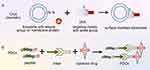 |
Figure 6 (A) Targeted peptide modified exosomes-click chemistry. (B) Targeting peptide–drug coupling. |
Polypeptide-Drug Conjugates
In addition to modifying exosomes with targeted peptides for the targeting of specific sites (Figure 6), peptides can be directly coupled with drugs to exhibit a targeting function. Polypeptide-drug conjugates (PDCs) are novel targeted drugs that can improve the physicochemical properties and the targeting and therapeutic effects of drugs.73 PDCs are mainly composed of cytotoxic drugs, peptides and binding bonds.74 Peptides used in PDCs are divided into two categories: cell-penetrating peptides (CPP) and cell-targeting peptide.75 A stable bond ensures the integrity of a PDC before it reaches a specific site and prevents premature drug release. Inside a target cell, a binding bond ensures effective drug release. Drugs commonly used in PDCs are doxorubicin, paclitaxel and radionuclide.76 The core advantages of PDC are enhanced tissue permeability, easy chemical synthesis, low production cost and remarkable therapeutic effect. The US Food and Drug Administration has approved two PDCs for the treatment of cancer: 177Lu-dotatate (lutathera) and melflufen.77
Progress in the Application of Targeted Peptide Modified Exosomes in Lung
Necessity of Exosomes for Targeted Delivery to the Lungs
Targeted delivery to the lungs is currently achieved mainly through inhalation delivery, local injection and nanomaterial delivery (eg liposomes, polymer nanoparticles and extracellular vesicles). Compared with these strategies, using exosomes, which show excellent biocompatibility, efficient intercellular communication and loading efficiency, have the advantages of having low immunogenicity and causing only mild local trauma.78 Therefore, exosomes as targeted delivery systems for the lungs have attracted considerable interest.
After intravenously injected exosomes enter the blood circulation, they are quickly eliminated from the blood vessels and enter parenchymal organs.79 Most exosomes are captured by the liver and spleen, which are the primary sites of exosome metabolism. Only a small amount of exosomes are captured by other organs, such as the lungs, kidneys and bone marrow, and thus enhancing the ability of exosomes to target the lungs is critical. The first step is to select exosomes of suitable cell origin. Currently, exosomes used in lung injury treatment are mainly derived from stem cells, especially mesenchymal stem cells. MSC-derived exosomes are rich in bioactive molecules, including growth factors, cytokines and miRNAs, which can participate in intercellular signal transduction, mediate immune regulation and promote cell proliferation and differentiation, playing an important role in lung injury repair. MSC-derived exosomes are highly efficient in treating lung injury. For example, in acute lung injury caused by COVID-19, the nebulised inhalation of MSC-derived exosomes can greatly improve lung function and prognosis.80 Second, a peptide-modified exosome can be selected to improve lung targeting. The RGD peptide and its derivatives specifically target the lungs.81 Other peptides, such as GE11, MOTS-c, GFE, F3 and tLyP-1, also show lung-targeting capability to varying degrees.82–85
In the treatment of lung diseases, especially lung infections, oral drugs often do not enter the lungs directly but circulate in the body, resulting in high potential toxicity and limited efficacy. To improve the targeting and therapeutic effects of drugs, researchers have developed exosomal drug delivery systems based on targeted peptide modifications.
Anti-Inflammatory Effects of Peptides Targeting Exosomes in the Lungs
Inflammatory diseases are the key features of acute lung injury (ALI)/acute respiratory distress syndrome (ARDS), a multi-factor respiratory disease with a high mortality rate and numerous pathologic features, such as increased alveolar capillary permeability, increased lung weight and loss of lung tissue inflation. Exosomes have been gradually used in ALI/ARDS treatment in recent years due to their anti-inflammatory, apoptosis-inhibiting and cell-regenerating effects.86 MicroRNAs in exosomes can participate in intercellular communication and play an immunomodulatory role in ALI/ARDS disease models.87 Kim et al88 engineered a RAGE binding peptide (RBP) and linked it to the exosome membrane protein Lamp-2b to prepare RBP-EXO exosomes, which were then loaded with Cur to enhance anti-inflammatory effects; the results showed that RBP-EXO–Cur complex reduced proinflammatory cytokines in ALI models more effectively than Cur, RBP-EXO or unmod-exo/Cur, suggesting that the complex is beneficial for ALI treatment. Klinger et al89 designed a model of Sugen5416 and hypoxia-induced pulmonary hypertension in rats; they showed that MSCs-EXO transformed the macrophage phenotype from a classical inflammatory pathway (M1) to an anti-inflammatory pathway (M2) and reduced the levels of Interleukin-6 (IL-6), macrophage inflammatory protein 2 (MIP-2) and tumor necrosis factor-α (TNF-α).
Exosomes of different cell origins may vary in composition and function, and some exosomes have no anti-inflammatory effects. Mammalian cells, such as macrophages, dendritic cells, B cells, T cells, platelets, epithelial cells, reticulocytes, mast cells, neurons, oligodendrocytes, tumor cells, and Schwann cells, have been demonstrated to be able to release exosomes.23,90–92 Plant exosomes are mainly used for exosome extraction and drug delivery.93–95 The source of an exosome may determine the type of bioactive molecules it carries, which in turn affect its therapeutic effects against lung diseases. For example, exosomes from tumour cells release exosomes carrying carcinogenic and apoptotic factors,96 which may further promote the development of tumours rather than exert anti-inflammatory effects. Exosomes may be detrimental to anti-inflammatory processes because they promote the apoptosis of immune cells and help tumour cells escape an immune response or drug clearance.97 In a state of disease or stress, non-physiological cells may secrete exosomes that function abnormally, which may no longer have normal biological functions, including anti-inflammatory effects. In addition, some activated immune cells may secrete exosomes that exert pro-inflammatory effects.
Peptide-Targeted Exosomes in Lung Gene Therapy
Gene therapy is the therapeutic delivery of genetic material (DNA or RNA) into a patient’s cells for the treatment of a disease.98 This mode of delivery can be achieved by gene replacement, correction, addition and suppression.99 Over the past few years, many nucleic acid delivery methods have been developed for therapeutic purposes, mainly including viral and non-viral vectors (liposomes and nanoparticles).100 However, viral vectors have toxic, immunogenic and carcinogenic effects, and non-viral vectors have low transduction efficiency, induce immune response and are quickly cleared by the reticuloendothelial system. Therefore, the emerging class: exosomes are ideal candidates for the delivery of gene drugs due to their low immunogenicity, good biocompatibility, low toxicity and easy engineering modification.101 Therefore, exosomes as carriers to carry drugsto specific tissues and organs for treatment may become a promising strategy of future research.
Bai et al102 developed a novel engineered tLyp-1 exosome for efficient delivery of siRNA, which is commonly used to mediate gene silencing in cells, primarily for viral infections and cancer, to human cancer cells.103 We designed a plasmid vector encoding the fusion protein tLyp-1–Lamp-2b for transfection into human embryonic kidney HEK293T tool cells, where the tLyp-1 peptide (amino acid sequence CGNKRTR) selectively targets neurociliin 1 and neurociliin 2. The obtained targeted tLyp-1-exo was used as a gene delivery vector to encapsulate siRNA through electroporation and then applied to human non-small cell lung cancer (NSCLC) cells. The results showed that the transfection of lung cancer and tumour stem cells with targeted siRNAtLyp-1 exosomes is efficient and can knock down the target gene of cancer cells, which can be used for gene therapy. Zhou et al104 constructed and expressed two plasmas in A549 cells on the basis of the principle underlying specific binding between TAT peptide and a stem-ring containing trans-activation reaction (TAR). One plasmid contains a membrane localisation protein ADC linked to a TAT peptide, the other plasmid contains a precursor of miR-449a linked to a TAR element, and ADC-TAT is loaded into an exosome membrane. Through this approach, Zhou collected the engineered exosomes secreted by A549 cells (miR-449aExo), which exhibited strong anti-tumour ability in vitro and in vivo. Thus, the method is a promising method for NSCLC treatment. mRNAs and proteins loaded into lung-derived exosomes and administered to the lungs by inhalation showed high translation and expression levels in the lung tissues of mice that had received lung-EXO.105 Liu et al106 loaded IL-12 mRNA into human embryonic kidney cell–derived exosomes by electroporation. The resulting IL-12 mRNA–loaded exosomes facilitated the direct treatment of lung tumours through inhalation administration.
Application Potential of Peptide-Targeted Exosomes in Pulmonary Fibrosis
Pulmonary fibrosis (PF) is a chronic, progressive and destructive fibrotic interstitial lung disease characterised by alveolar epithelial damage, fibroblast proliferation, excessive extracellular matrix deposition and decreased lung function.107 However, the exact pathogenesis of the disease is unknown and effective treatment is lacking. Thus, patients with PF often have poor prognoses. The role of exosomes in intercellular communication in PF has attracted interest. miR-17-5p derived from human embryonic stem cell exosomes alleviate bleomycin-induced PF in mice by directly binding to thrombospondin-2.108 Macrophage-derived exosomes alleviate fibrosis in airway epithelial cells and lung fibroblasts by delivering miR-142-3p.109 Exosomes derived from bone marrow mesenchymal stem cells can up-regulate the mesenchymal cells of type II alveolar epithelial cells and regulate the transformation of fibroblasts into myofibroblasts.110 Therefore, exosomes are promising tools for cell therapy for PF. Strategies for treating the lungs with targeted peptides have advanced. Although research on peptide-targeted exosomes in the treatment of PF is still in the preliminary stage, using peptide-targeted exosomes is a potential strategy for PF treatment.
Conclusion
Under physiological conditions, exosomes can act as signalling molecules, regulating information exchange between cells and cell fate. Under pathological conditions, exosomes can be involved in tumour cell metastasis, infectious disease transmission and immune response. In addition, exosomes transport substances.111 By modifying exosomes with targeted peptides, the precise targeted delivery of exosomes to specific sites can be achieved.112 This approach improves drug bioavailability, reduces side effects and enhances diagnostic accuracy.Although exosomes show promising application prospects for basic research, exosome-based drug delivery systems still face many challenges in clinical settings. More clinical trials are needed to verify its safety and efficacy; At the same time, it is also necessary to solve the standardization of exosomal preparation and purification, the selection of targeted peptides and modification methods, etc., in the click-chemistry method, there is likely to be chemical residues in the final product, which may affect the safety of exosomes. Genetic engineering methods cannot be used to label/target molecules other than genetically coded proteins/peptides, and we need to select suitable targeting ligands or labeled parts without generating any adverse immunogenic activity. However, with continuous advancements in technologies and in-depth research, these challenges can be addressed. Targeted peptide-modified exosomes are essential for disease treatment and diagnosis, offering prospects for effective and personalised medical programmes.
Abbreviations
LSEs, late endosomes; ILVs, intracavicular vesicles; MVBs, multivesicular bodies; MHCs, major histocompatibility complexes; HSPs, heat shock proteins; GPI, glycosyl phosphatidyl inositol; TNF, tumour necrosis factor; CTP, cell-targeting peptide; IMTP, ischaemic myocardial targeting peptide; CAP, chondrocyte affinity peptide; Dox, doxorubicin; PMO, phosphodiamide morpholino oligomer; Cur, curcumin; PDCs, Polypeptide-drug conjugates; MSCs, mesenchymal stem cells; ALI, acute lung injury; ARDS, acute respiratory distress syndrome; NSCLC, non-small cell lung cancer; TAR, trans-activation reaction; PF, Pulmonary fibrosis.
Acknowledgments
This work was supported by the National Natural Science Foundation of China under grant no. 82260808; Natural Science Foundation of Inner Mongolia, China, under grant nos. 2020MS08071, 2020MS08077, 2021MS08126 and 2022LHMS08011; Supported By Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region no. NJYT23112; Scientific research project of universities in Inner Mongolia Autonomous Region no. NJZZ23104.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kimiz-Gebologlu I, Oncel SS. Exosomes: large-scale production, isolation, drug loading efficiency, and biodistribution and uptake. J Control Release. 2022;347:533–543. doi:10.1016/j.jconrel.2022.05.027
2. Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31:1059–1062. doi:10.1248/bpb.31.1059
3. Admyre C, Johansson SM, Qazi KR, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi:10.4049/jimmunol.179.3.1969
4. Liang Y, Duan L, Lu J, Xia J. Engineering exosomes for targeted drug delivery. Theranostics. 2021;11:3183–3195. doi:10.7150/thno.52570
5. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19:47. doi:10.1186/s12964-021-00730-1
6. Zhang Y, Bi J, Huang J, et al. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int j Nanomed. 2020;15:6917–6934. doi:10.2147/ijn.S264498
7. Shao J, Zaro J, Shen Y. Advances in exosome-based drug delivery and tumor targeting: from tissue distribution to intracellular fate. Int j Nanomed. 2020;15:9355–9371. doi:10.2147/ijn.S281890
8. Ming-Kun C, Zi-Xian C, Mao-Ping C, et al. Engineered extracellular vesicles: a new approach for targeted therapy of tumors and overcoming drug resistance. Can Communic. 2024;44:205–225. doi:10.1002/cac2.12518
9. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. doi:10.1126/science.aau6977
10. Heusermann W, Hean J, Trojer D, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol. 2016;213:173–184. doi:10.1083/jcb.201506084
11. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi:10.1038/nrm.2017.125
12. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi:10.1016/0092-8674(83)90040-5
13. Krylova SV, Feng D. The machinery of exosomes: biogenesis, release, and uptake. Int J Mol Sci. 2023;25:24. doi:10.3390/ijms24021337
14. Yue B, Yang H, Wang J, et al. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020;53:e12857. doi:10.1111/cpr.12857
15. Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinf. 2015;13:17–24. doi:10.1016/j.gpb.2015.02.001
16. Yao Z, Qiao Y, Li X, et al. Exosomes exploit the virus entry machinery and pathway to transmit alpha interferon-induced antiviral activity. J Virol. 2018:92. doi:10.1128/jvi.01578-18
17. Arya SB, Collie SP, Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2024;34:90–108. doi:10.1016/j.tcb.2023.06.006
18. Sung BH, von Lersner A, Guerrero J, et al. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat Commun. 2020;11:2092. doi:10.1038/s41467-020-15747-2
19. Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi:10.1016/j.pharmthera.2017.02.020
20. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
21. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi:10.1038/nature15756
22. Grigoryeva ES, Savelieva OE, Popova NO, Cherdyntseva NV, Perelmuter VM. Do tumor exosome integrins alone determine organotropic metastasis? Mol Biol Rep. 2020;47:8145–8157. doi:10.1007/s11033-020-05826-4
23. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi:10.1038/nri855
24. Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15:203–216. doi:10.1038/nri3818
25. Regimbeau M, Abrey J, Vautrot V, et al. Heat shock proteins and exosomes in cancer theranostics. Semin Cancer Biol. 2022;86:46–57. doi:10.1016/j.semcancer.2021.07.014
26. Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e418. doi:10.1016/j.cell.2019.02.029
27. He B, Cai Q, Qiao L, et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nature Plants. 2021;7:342–352. doi:10.1038/s41477-021-00863-8
28. Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60:9–18. doi:10.1194/jlr.R084343
29. López-Cobo S, Campos-Silva C, Valés-Gómez M. Glycosyl-phosphatidyl-inositol (GPI)-anchors and metalloproteases: their roles in the regulation of exosome composition and NKG2D-mediated immune recognition. Front Cell Develop Biol. 2016;4:97. doi:10.3389/fcell.2016.00097
30. Tutanov OS, Glass SE, Coffey RJ. Emerging connections between GPI-anchored proteins and their extracellular carriers in colorectal cancer. Extracell Vesicl Circul Nucl Aci. 2023;4:195–217. doi:10.20517/evcna.2023.17
31. Taha EA, Ono K, Eguchi T. Roles of extracellular HSPs as biomarkers in immune surveillance and immune evasion. Int J Mol Sci. 2019;21:20. doi:10.3390/ijms20184588
32. Eguchi T, Sogawa C, Ono K, et al. Cell stress induced stressome release including damaged membrane vesicles and extracellular HSP90 by prostate cancer cells. Cells. 2020;9:755. doi:10.3390/cells9030755
33. Leone DA, Peschel A, Brown M, et al. Surface LAMP-2 is an endocytic receptor that diverts antigen internalized by human dendritic cells into highly immunogenic exosomes. J Immunol. 2017;199:531–546. doi:10.4049/jimmunol.1601263
34. Salunkhe S, Basak M, Chitkara D, Mittal A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: strategies and significance. J Control Release. 2020;326:599–614. doi:10.1016/j.jconrel.2020.07.042
35. Xu X, Liang Y, Li X, et al. Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid-derived mesenchymal stem cells and cartilage regeneration. Biomaterials. 2021;269:120539. doi:10.1016/j.biomaterials.2020.120539
36. Kim H, Kim EH, Kwak G, et al. Exosomes: cell-derived nanoplatforms for the delivery of cancer therapeutics. Int J Mol Sci. 2020;22:22. doi:10.3390/ijms22010014
37. Kim SB. Function and therapeutic development of exosomes for cancer therapy. Arch Pharm Res. 2022;45:295–308. doi:10.1007/s12272-022-01387-1
38. Li B, Cao Y, Sun M, Feng H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021;35:e21916. doi:10.1096/fj.202100294RR
39. Lai JJ, Chau ZL, Chen SY, et al. Exosome processing and characterization approaches for research and technology development. Advan Sci. 2022;9:e2103222. doi:10.1002/advs.202103222
40. Tavasolian F, Hosseini AZ, Rashidi M, et al. The impact of immune cell-derived exosomes on immune response initiation and immune system function. Curr Pharm Des. 2021;27:197–205. doi:10.2174/1381612826666201207221819
41. Sun YF, Pi J, Xu JF. Emerging role of exosomes in tuberculosis: from immunity regulations to vaccine and immunotherapy. Front Immunol. 2021;12:628973. doi:10.3389/fimmu.2021.628973
42. Chinnappan M, Srivastava A, Amreddy N, et al. Exosomes as drug delivery vehicle and contributor of resistance to anticancer drugs. Cancer Lett. 2020;486:18–28. doi:10.1016/j.canlet.2020.05.004
43. Rehman FU, Liu Y, Zheng M, Shi B. Exosomes based strategies for brain drug delivery. Biomaterials. 2023;293:121949. doi:10.1016/j.biomaterials.2022.121949
44. Familtseva A, Jeremic N, Tyagi SC. Exosomes: cell-created drug delivery systems. Mol Cell Biochem. 2019;459:1–6. doi:10.1007/s11010-019-03545-4
45. Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. doi:10.1016/j.jconrel.2015.07.030
46. Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290:8166–8172. doi:10.1074/jbc.M114.621383
47. He J, Ren W, Wang W, et al. Exosomal targeting and its potential clinical application. Drug Deliv Transl Res. 2022;12:2385–2402. doi:10.1007/s13346-021-01087-1
48. Liu X, Yang X, Sun W, et al. Systematic evolution of ligands by exosome enrichment: a proof-of-concept study for exosome-based targeting peptide screening. Adv Biosyst. 2019;3:e1800275. doi:10.1002/adbi.201800275
49. Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9:1015–1028. doi:10.7150/thno.30853
50. Sioud M. Phage display libraries: from binders to targeted drug delivery and human therapeutics. Mol Biotechnol. 2019;61:286–303. doi:10.1007/s12033-019-00156-8
51. Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi:10.1126/science.4001944
52. Parmley SF, Smith GP. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi:10.1016/0378-1119(88)90495-7
53. Jaroszewicz W, Morcinek-Orłowska J, Pierzynowska K, Gaffke L, Węgrzyn G. Phage display and other peptide display technologies. FEMS Microbiol Rev. 2021;46. doi:10.1093/femsre/fuab052
54. Saw PE, Song EW. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein and Cell. 2019;10:787–807. doi:10.1007/s13238-019-0639-7
55. Zhang K, Liu Y, Tang Y. Screening of TNFR1 binding peptides from deinagkistrodon acutus venom through phage display. Toxins. 2022;14. doi:10.3390/toxins14020155
56. Zhang K, Tang Y, Chen Q, Liu Y. The screening of therapeutic peptides for anti-inflammation through phage display technology. Int J Mol Sci. 2022;24:23. doi:10.3390/ijms23158554
57. Newman MR, Benoit DSW. In vivo translation of peptide-targeted drug delivery systems discovered by phage display. Bioconjug Chem. 2018;29:2161–2169. doi:10.1021/acs.bioconjchem.8b00285
58. Pierzynowska K, Morcinek-Orłowska J, Gaffke L, et al. Applications of the phage display technology in molecular biology, biotechnology and medicine. Crit. Rev. Microbiol. 2023;50:450–490. doi:10.1080/1040841X.2023.2219741
59. Kim H, Yun N, Mun D, et al. Cardiac-specific delivery by cardiac tissue-targeting peptide-expressing exosomes. Biochem Biophys Res Commun. 2018;499:803–808. doi:10.1016/j.bbrc.2018.03.227
60. Wang X, Chen Y, Zhao Z, et al. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc. 2018;7:e008737. doi:10.1161/jaha.118.008737
61. Liang Y, Xu X, Li X, et al. Chondrocyte-targeted MicroRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Appl Mater Interfaces. 2020;12:36938–36947. doi:10.1021/acsami.0c10458
62. Wang C, Li N, Li Y, et al. Engineering a HEK-293T exosome-based delivery platform for efficient tumor-targeting chemotherapy/internal irradiation combination therapy. J Nanobiotechnology. 2022;20:247. doi:10.1186/s12951-022-01462-1
63. Xin L, Yuan YW, Liu C, et al. Preparation of internalizing RGD-modified recombinant methioninase exosome active targeting vector and antitumor effect evaluation. Dig Dis Sci. 2021;66:1045–1053. doi:10.1007/s10620-020-06262-x
64. Gao X, Ran N, Dong X, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. 2018:10. doi:10.1126/scitranslmed.aat0195
65. Yu S, Liao R, Bai L, et al. Anticancer effect of hUC-MSC-derived exosome-mediated delivery of PMO-miR-146b-5p in colorectal cancer. Drug Deliv Transl Res. 2023;14:1352–1369. doi:10.1007/s13346-023-01469-7
66. Wang M, Altinoglu S, Takeda YS, Xu Q. Integrating protein engineering and bioorthogonal click conjugation for extracellular vesicle modulation and intracellular delivery. PLoS One. 2015;10:e0141860. doi:10.1371/journal.pone.0141860
67. Porte K, Riomet M, Figliola C, Audisio D, Taran F. Click and bio-orthogonal reactions with mesoionic compounds. Chem Rev. 2021;121:6718–6743. doi:10.1021/acs.chemrev.0c00806
68. Choi H, Choi Y, Yim HY, et al. Biodistribution of exosomes and engineering strategies for targeted delivery of therapeutic exosomes. Tissue Eng and Regener Med. 2021;18:499–511. doi:10.1007/s13770-021-00361-0
69. Jia G, Han Y, An Y, et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. doi:10.1016/j.biomaterials.2018.06.029
70. Zhang H, Wu J, Wu J, et al. Exosome-mediated targeted delivery of miR-210 for angiogenic therapy after cerebral ischemia in mice. J Nanobiotechnology. 2019;17:29. doi:10.1186/s12951-019-0461-7
71. Zhu Q, Ling X, Yang Y, et al. Embryonic stem cells-derived exosomes endowed with targeting properties as chemotherapeutics delivery vehicles for glioblastoma therapy. Advan Sci. 2019;6:1801899. doi:10.1002/advs.201801899
72. Zhang M, Hu S, Liu L, et al. Engineered exosomes from different sources for cancer-targeted therapy. Signal Transduct Target Therap. 2023;8:124. doi:10.1038/s41392-023-01382-y
73. Wang M, Liu J, Xia M, et al. Peptide-drug conjugates: a new paradigm for targeted cancer therapy. Eur J Med Chem. 2024;265:116119. doi:10.1016/j.ejmech.2023.116119
74. Wang L, Chen H, Wang F, Zhang X. The development of peptide-drug conjugates (PDCs) strategies for paclitaxel. Expert Opin Drug Deliv. 2022;19:147–161. doi:10.1080/17425247.2022.2039621
75. Fu C, Yu L, Miao Y, et al. Peptide-drug conjugates (PDCs): a novel trend of research and development on targeted therapy, hype or hope? Acta Pharm Sin B. 2023;13:498–516. doi:10.1016/j.apsb.2022.07.020
76. Rizvi SFA, Abbas N, Zhang H, Fang Q. Identification of a pH-responsive peptide-paclitaxel conjugate as a novel drug with improved therapeutic potential. J Med Chem. 2023;66:8324–8337. doi:10.1021/acs.jmedchem.3c00382
77. Lindberg J, Nilvebrant J, Nygren P, Lehmann F. Progress and future directions with peptide-drug conjugates for targeted cancer therapy. Molecules. 2021;26. doi:10.3390/molecules26196042
78. Xi XM, Xia SJ, Lu R. Drug loading techniques for exosome-based drug delivery systems. Pharmazie. 2021;76:61–67. doi:10.1691/ph.2021.0128
79. Koh HB, Kim HJ, Kang SW, Yoo TH. Exosome-based drug delivery: translation from bench to clinic. Pharmaceutics. 2023;16:15. doi:10.3390/pharmaceutics15082042
80. Zhu YG, Shi MM, Monsel A, et al. Nebulized exosomes derived from allogenic adipose tissue mesenchymal stromal cells in patients with severe COVID-19: a pilot study. Stem Cell Res Ther. 2022;13:220. doi:10.1186/s13287-022-02900-5
81. Yadav B, Chauhan M, Shekhar S, et al. RGD-decorated PLGA nanoparticles improved effectiveness and safety of cisplatin for lung cancer therapy. Int J Pharm. 2023;633:122587. doi:10.1016/j.ijpharm.2023.122587
82. Cheng L, Huang FZ, Cheng LF, et al. GE11-modified liposomes for non-small cell lung cancer targeting: preparation, ex vitro and in vivo evaluation. Int j Nanomed. 2014;9:921–935. doi:10.2147/ijn.S53310
83. Xinqiang Y, Quan C, Yuanyuan J, Hanmei X. Protective effect of MOTS-c on acute lung injury induced by lipopolysaccharide in mice. Int Immunopharmacol. 2020;80:106174. doi:10.1016/j.intimp.2019.106174
84. Oh Y, Mohiuddin I, Sun Y, et al. Phenotypic diversity of the lung vasculature in experimental models of metastases. Chest. 2005;128:596s–600s. doi:10.1378/chest.128.6_suppl.596S
85. Zhang X, Huang Y, Song H, et al. Inhibition of growth and lung metastasis of breast cancer by tumor-homing triple-bioresponsive nanotherapeutics. J Control Release. 2020;328:454–469. doi:10.1016/j.jconrel.2020.08.066
86. Fan E, Brodie D, Slutsky AS. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA. 2018;319:698–710. doi:10.1001/jama.2017.21907
87. Liu C, Xiao K, Xie L. Advances in the use of exosomes for the treatment of ALI/ARDS. Front Immunol. 2022;13:971189. doi:10.3389/fimmu.2022.971189
88. Kim G, Lee Y, Ha J, Han S, Lee M. Engineering exosomes for pulmonary delivery of peptides and drugs to inflammatory lung cells by inhalation. J Control Release. 2021;330:684–695. doi:10.1016/j.jconrel.2020.12.053
89. Klinger JR, Pereira M, Del Tatto M, et al. Mesenchymal stem cell extracellular vesicles reverse sugen/hypoxia pulmonary hypertension in rats. Am J Respir Cell Mol Biol. 2020;62:577–587. doi:10.1165/rcmb.2019-0154OC
90. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi:10.1083/jcb.201211138
91. Marbán E. The secret life of exosomes: what bees can teach us about next-generation therapeutics. J Am Coll Cardiol. 2018;71:193–200. doi:10.1016/j.jacc.2017.11.013
92. Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:186–192. doi:10.1161/atvbaha.112.300139
93. Mondal J, Pillarisetti S, Junnuthula V, et al. Hybrid exosomes, exosome-like nanovesicles and engineered exosomes for therapeutic applications. J Control Release. 2023;353:1127–1149. doi:10.1016/j.jconrel.2022.12.027
94. Dad HA, Gu TW, Zhu AQ, Huang LQ, Peng LH. Plant exosome-like nanovesicles: emerging therapeutics and drug delivery nanoplatforms. Mol Ther. 2021;29:13–31. doi:10.1016/j.ymthe.2020.11.030
95. Cao M, Diao N, Cai X, et al. Plant exosome nanovesicles (PENs): green delivery platforms. Mater Horizons. 2023;10:3879–3894. doi:10.1039/d3mh01030a
96. Zhou Y, Zhang Y, Gong H, Luo S, Cui Y. The role of exosomes and their applications in cancer. Int J Mol Sci. 2021;23:22. doi:10.3390/ijms222212204
97. Lakshmi S, Hughes TA, Priya S. Exosomes and exosomal RNAs in breast cancer: a status update. Eur. J. Cancer. 2021;144:252–268. doi:10.1016/j.ejca.2020.11.033
98. Zhang Y, Liu Q, Zhang X, et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J Nanobiotechnology. 2022;20:279. doi:10.1186/s12951-022-01472-z
99. Tang R, Xu Z. Gene therapy: a double-edged sword with great powers. Mol Cell Biochem. 2020;474:73–81. doi:10.1007/s11010-020-03834-3
100. Soltani F, Parhiz H, Mokhtarzadeh A, Ramezani M. Synthetic and biological vesicular nano-carriers designed for gene delivery. Curr Pharm Des. 2015;21:6214–6235. doi:10.2174/1381612821666151027153410
101. Sun M, Zhang H, Liu J, et al. Extracellular vesicles: a new star for gene drug delivery. Int j Nanomed. 2024;19:2241–2264. doi:10.2147/ijn.S446224
102. Bai J, Duan J, Liu R, et al. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J Pharm Sci. 2020;15:461–471. doi:10.1016/j.ajps.2019.04.002
103. Mainini F, Eccles MR. Lipid and polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules. 2020;25:2692. doi:10.3390/molecules25112692
104. Zhou W, Xu M, Wang Z, Yang M. Engineered exosomes loaded with miR-449a selectively inhibit the growth of homologous non-small cell lung cancer. Cancer Cell Int. 2021;21:485. doi:10.1186/s12935-021-02157-7
105. Popowski KD, López de Juan Abad B, George A, et al. Inhalable exosomes outperform liposomes as mRNA and protein drug carriers to the lung. Extracell Vesicle. 2022;1:100002. doi:10.1016/j.vesic.2022.100002
106. Liu M, Hu S, Yan N, Popowski KD, Cheng K. Inhalable extracellular vesicle delivery of IL-12 mRNA to treat lung cancer and promote systemic immunity. Nat Nanotechnol. 2024;19:565–575. doi:10.1038/s41565-023-01580-3
107. Koudstaal T, Funke-Chambour M, Kreuter M, Molyneaux PL, Wijsenbeek MS. Pulmonary fibrosis: from pathogenesis to clinical decision-making. Trends Mol Med. 2023;29:1076–1087. doi:10.1016/j.molmed.2023.08.010
108. Liu Q, Bi Y, Song S, et al. Exosomal miR-17-5p from human embryonic stem cells prevents pulmonary fibrosis by targeting thrombospondin-2. Stem Cell Res Ther. 2023;14:234. doi:10.1186/s13287-023-03449-7
109. Guiot J, Cambier M, Boeckx A, et al. Macrophage-derived exosomes attenuate fibrosis in airway epithelial cells through delivery of antifibrotic miR-142-3p. Thorax. 2020;75:870–881. doi:10.1136/thoraxjnl-2019-214077
110. Xie L, Zeng Y. Therapeutic potential of exosomes in pulmonary fibrosis. Front Pharmacol. 2020;11:590972. doi:10.3389/fphar.2020.590972
111. Kim HI, Park J, Zhu Y, et al. Recent advances in extracellular vesicles for therapeutic cargo delivery. Exp Mol Med. 2024;56:836–849. doi:10.1038/s12276-024-01201-6
112. Donoso-Quezada J, Ayala-Mar S, González-Valdez J. State-of-The-art exosome loading and functionalization techniques for enhanced therapeutics: a review. Crit Rev Biotechnol. 2020;40:804–820. doi:10.1080/07388551.2020.1785385
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

