Back to Journals » International Journal of Nanomedicine » Volume 19
The Advances in the Development of Epigenetic Modifications Therapeutic Drugs Delivery Systems
Received 12 July 2024
Accepted for publication 14 October 2024
Published 19 October 2024 Volume 2024:19 Pages 10623—10637
DOI https://doi.org/10.2147/IJN.S480095
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Tingyi Li,1,2 Yanwei Chen,1 Shuai Li1
1Department of Pharmacy, The First Affiliated Hospital of Dalian Medical University, Dalian, People’s Republic of China; 2Dalian Medical University, Dalian, People’s Republic of China
Correspondence: Shuai Li; Yanwei Chen, Department of Pharmacy, The First Affiliated Hospital of Dalian Medical University, Dalian, People’s Republic of China, Email [email protected]; [email protected]
Abstract: Epigenetic dysregulation can significantly trigger the onset and progression of various diseases, epigenetic therapy is a new treatment strategy by changing DNA methylation, histone modification, N6-methyladenosine, chromatin modification and other epigenetic modifications to regulate gene expression levels for therapeutic purposes. However, small-molecule epigenetic drugs face challenges in disease treatment, such as lack of selectivity, limited therapeutic efficacy, and insufficient safety. Nanomedicine delivery systems offer significant advantages in addressing these issues by enhancing drug targeting, improving bioavailability, and reducing nonspecific distribution. This help minimize side effects while increasing both therapeutic effectiveness and safety of epigenetic drugs. In this review, we focus on the mechanism and role of epigenetic regulatory factors in diseases, as well as the challenges faced by small molecule inhibitors in treatment strategies, especially the research advancements in epigenetic drug delivery systems, review and discuss the therapeutic potential and challenges of using nanotechnology to develop epigenetic drug delivery systems.
Keywords: epigenetic therapy, DNA methylation, N6-methyladenosine, histone modification, nanomedicine delivery systems, nanotechnology
Introduction
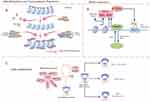
Epigenetics investigates alterations in gene expression that occur independently of changes in the DNA sequence. Instead of modifying the DNA sequence itself, it involves processes such as altering nucleotide residues, modifying chromatin structure, or degrading already translated messenger RNA.1 Researchers have pinpointed several distinct mechanisms involved in epigenetic regulation. These mechanisms encompass DNA methylation, histone modification, and gene regulation mediated by modifications of non-coding RNA. In mammals, DNA methylation is mediated by DNA methyl transferases (DNMT), which methylate by transferring methyl groups from S-adenosyl methionine (SAM) to the 5′ site of cytosine residues on the DNA molecule. This causes the C site on the cytosine residue to bind to a methyl group to form 5-methylcytosine (5mC).2 This 5mC usually occurs on CpG dinucleotide sequences. Histones are major components of chromatin, which form nucleosomes in the nucleus that wrap around DNA sequences. Chemical modifications of histones, including acetylation, methylation, phosphorylation, and ubiquitination, regulate the compactness of chromatin and the accessibility of genes. These modifications influence how tightly or loosely DNA is packaged around histone proteins, thus controlling gene expression. RNA methylation serves as a method for regulating gene expression after transcription has occurred. To date, researchers have identified over 150 types of RNA modifications, highlighting the intricate landscape of post-transcriptional regulatory mechanisms. N6-methyladenosine (m6A) stands out as the most abundant and significant modification found in mRNA. It plays a crucial role in various biological processes and is implicated in both normal physiological functions and disease states. For the regulation of RNAs, in addition to the RNA molecules coding for proteins, there exists a large number of non-coding RNAs (ncRNAs), which are not directly translated into proteins, and which perform a regulatory function within the cell.3 Some ncRNAs can interact with DNA or RNA to affect gene expression and cellular functions by altering chromatin structure, post-transcriptional modifications and transcriptional regulation. BET (bromodomain and extraterminal) proteins, including BRD2, BRD3, BRD4, play significant roles in human disease research (Figure 1B). These proteins feature two highly similar bromodomains (BD1 and BD2), which serve as potential targets for small molecule inhibitors. Each BET protein is involved in distinct functions related to gene regulation, biological processes, and various human diseases, with BD1 and BD2 in each protein having specific contributions to these functions.
Epigenetic drugs have shown significant therapeutic effects for many diseases, especially cancer and neurological disorders. DNA methyltransferase inhibitors (DNMTi) and histone deacetylase (HDAC) inhibitors are frequently utilized drug classes in the treatment of cancer. DNMTi inhibitors inhibit the methylation process and reply to the expression of methylated oncogenes, and HDAC inhibitors regulate histone modification status and affect chromatin results and gene expression.4 In neurological disorders, HDAC inhibitors can improve neurodegenerative diseases, such as alleviating symptoms of Parkinson’s and Alzheimer’s disease. For cardiovascular diseases, researchers have found that DMNT inhibitors can inhibit inflammatory responses and increase vascular endothelial function in atherosclerosis, promising to reduce the risk of atherosclerosis.
As with many chemotherapeutic drugs, the side effects of epigenetic drugs need to be minimized. The therapeutic effect of single epigenetic drugs is not obvious, and the existing epigenetic drugs have problems such as weak targeting ability, toxic side effects and drug resistance. The development of new combination therapeutic strategies and the realization of targeted, controlled release of drugs are urgent problems. Epigenetically modified nanocarrier-based drug delivery systems (NDDSs) can effectively improve drug targeting, delivery efficiency and therapeutic efficacy. NDDSs, which are composed of nanomaterials smaller than 1000 nm, show great promise for biomedical applications due to their ability to load drugs effectively, deliver them to specific targets, and release them in a controlled manner. This article reviews the mechanisms and role of epigenetics in disease, advances in epigenetic nanodelivery systems, focusing on their principles, technological developments, clinical applications, and challenges.
Mechanisms of Epigenetics and Their Role in Disease Development
The Mechanisms and Roles of Epigenetic Modifications
DNA Methylation
DNA methylation, along with various regulatory elements, exerts significant influence over gene expression as a pivotal epigenetic factor. Within mammalian genomes, DNA methylation primarily transpires at CpG islands and is facilitated by a group of enzymes called DNA methyltransferases (DNMTs). These enzymes facilitate the transfer of a methyl group from S-adenosyl methionine (SAM) to the fifth carbon position of a cytosine base, resulting in the formation of 5-methylcytosine (5mC).5 Methylation to form 5mC recruits proteins that repress gene expression or inhibit the binding of transcription factors to DNA, maintaining cellular homeostasis, DNA integrity, genome-associated gene stability, and promoting the formation of functionally inert heterochromatin (Figure 1A). Hypomethylation causes chromosomal instability and the activation of oncogenes or genes that are normally silenced. Methylation abnormalities have a significant impact on the development of cancer, neurological disorders, and cardiovascular disease. In typical cellular conditions, CpG islands situated at gene promoters tend to remain unmethylated. Nevertheless, when hypermethylation occurs abnormally, it results in the silencing of gene transcription.6 In cancer, there is a widespread reduction in methylation across the genome, coupled with targeted increases in methylation at specific sites. Additionally, mutations in tumors tend to cluster within regions that are methylated at CpG sites.7 The process of active DNA demethylation is enhanced by the oxidation of 5-methylcytosine, facilitated by enzymes such as ten-eleven translocation (TET) proteins, which are dependent on α-ketoglutarate (αKG) as a cofactor for their catalytic activity.8
Histone Modifications
Histone modification is a critical epigenetic mechanism that plays a fundamental role in cancer development. Histone proteins form the core of nucleosomes, fundamental units of chromatin structure. The addition of chemical marks, known as post-translational modifications, to histones plays a crucial role in regulating chromatin dynamics and function.9 Histones, including H1, H2A, H2B, H3, H4, and H5, are proteins rich in positively charged amino acids like lysine and arginine. These proteins are remarkably similar across different eukaryotic organisms, with nearly identical amino acid sequences, underscoring their fundamental role in chromatin structure and function. This conservation underscores the vital role these histones play in maintaining chromatin structure and function across different species. The nucleosome serves as the basic unit of chromatin structure. It consists of a tetramer made up of two copies each of four core histone proteins: H2A, H2B, H3, and H4. Together, these histones constitute the nucleosome core, which serves as the structural foundation for organizing and packaging DNA within the chromatin fiber.10 Histone H1 or, in some cases, H5 (predominantly found in the inactive nuclei of avian erythrocytes), serves to link the octameric units of nucleosomes together. The linker histones are essential for maintaining the stability of chromatin structure and aiding in the organization of chromatin into more complex arrangements. The nucleosomes, along with the linker histones, are further compacted into 30 nm fibers. These fibers are then organized into various structures such as loops, solenoids, helices, and chromatin beads, contributing to the overall organization and packaging of DNA within the nucleus.11 The modifications of linker histones are closely linked to the formation of heterochromatin and the regulation of transcriptional activation. A wide range of histone modifications have been identified to intricately control the chromatin state (as depicted in Figure 1A). These modifications are pivotal for controlling transcriptional activities and maintaining the correct chromatin state as cells divide. They encompass various post-translational alterations such as methylation, acetylation, phosphorylation, glycosylation, cooperation, poly ADP-ribosylation, and ubiquitination. These modifications play crucial roles in regulating gene expression and chromatin structure. Disruptions or dysregulations of these processes have been linked to the development and progression of cancer. Such modifications can lead to the upregulation of oncogenes or the downregulation of tumor suppressor genes, both of which play a role in the development of cancer.
Non-Coding RNAs
Even though the genome has the capacity to encode roughly 20,000 proteins, this represents just a fraction, approximately 2%, of the entire genome. Put differently, not all RNA molecules are translated into functional proteins.12 Increasingly, research suggests that non-coding RNAs (NcRNAs), a type of RNA that does not encode proteins, are instrumental in controlling gene expression and influencing the development of various human diseases.13 Take, for instance, how NcRNAs facilitate post-transcriptional gene silencing, which results in the degradation of messenger RNA and prevents the translation of proteins.14 Furthermore, NcRNAs play a role in restructuring the chromatin framework by guiding the assembly of heterochromatin.15 Moreover, NcRNAs are implicated in modulating the activity of nearby (cis) and distant (trans) genes, either amplifying or dampening their expression levels.16 These regulatory mechanisms are vital for influencing processes such as cell growth, specialization, tissue renewal, and programmed cell death in humans, thereby playing a crucial role in maintaining balance within the body.17
m6A Modification
N6-methyladenosine (m6A) stands out as the predominant internal modification found in both messenger RNAs (mRNAs) and non-coding RNAs within eukaryotic cells. Its significance spans across various biological functions, notably impacting processes like tumor advancement, through its regulatory role in RNA processing and metabolism.18,19 The levels and functional impact of m6A in RNA are governed by the intricate interplay of methyltransferases (known as “Writers”), demethylases (“Erasers”), and m6A-binding proteins (“Readers”), reflecting a dynamic regulatory mechanism. M6A methylation represents a dynamic and reversible phenomenon (Figure 1C). Primarily, the METTL3/METTL14 methyltransferase complex, along with the WTAP cofactor, orchestrates m6A methylation, while the demethylation process primarily involves the FTO and ALKBH520–23 methylases. Among mammals, the roster of m6A reader proteins primarily encompasses members of the YTH protein family, comprising YTHDF1, YTHDF2, YTHDF3, as well as YTHDC1 and YTHDC2. Additionally, representatives of the heterogeneous nuclear ribonucleoprotein family such as HNRNPA2B1, HNRNPC, and HNRNPG also play significant roles as m6A readers. Numerous human ailments are intricately linked to m6A modifications, spanning the spectrum from immune function regulation to lipid metabolism, viral replication and susceptibility to infection, as well as neurodevelopmental processes and diseases. Dysregulation of m6A regulators affects the expression, stability and function of transcripts targeting m6A modification, resulting in cell cycle abnormalities, apoptosis evasion, epithelial-mesenchymal transition and angiogenesis. The m6A modification can lead to the generation of tumors, leading to tumorigenesis, progression, metastasis and resistance to anticancer therapy.24
Chromatin Modification
Chromatin remodeling plays a crucial role in diseases, primarily involving dynamic changes in chromatin structure and positioning, which influence gene transcription by altering chromatin accessibility, thus affecting cellular and tissue phenotypes without changing the DNA sequence.25 Chromatin remodeling complexes (CRCs) refer to the macromolecular complexes involved in the chromatin remodeling process, which possess ATPase activity, using energy from ATP hydrolysis to drive changes in chromatin structure and positioning. In humans, four ATP-dependent chromatin remodeling factors have been identified: SWI/SNF (BRG1-associated factor, BAF), ISWI, INO80, and CHD. Among them, the SWI/SNF complex has garnered attention due to its close association with various cancer tissues and cell lines. Studies have found that the SWI/SNF complex frequently undergoes mutations in cancer, particularly with biallelic deletions of SMARCA4 and ARID1A, leading to a specific enhanced dependency on another subunit (SMARCA2 or ARID1B). Based on this enhanced dependency, scientists have proposed a synthetic lethality treatment strategy, where simultaneous inactivation of two genes leads to cell death, while inactivation of only one allows for cell survival. In this context, targeting the subunits that exhibit specific enhanced dependency in cancer cells (such as SMARCA2 or ARID1B) could kill cancer cells without harming normal cells. This finding not only highlights the significant role of chromatin remodeling in cancer but also provides new potential therapeutic targets for cancer treatment.26
The Role of Epigenetics in Disease Causation and Progression
In most diseases, disease arises due to a combination of epigenetic changes and genetic mutations. One of the most studied is the methylation status of cancer. In cancer cells, there is a notable widespread decrease in DNA methylation,27 typically ranging from a 20% to 60% reduction in overall 5-methylcytosine levels. Simultaneously, it’s commonly noted that certain promoter CpG islands undergo distinctive hypermethylation patterns. In general, the prevalent hypomethylation primarily targets repetitive DNA sequences, fostering chromosomal instability, translocations, disruption of genes, and reawakening of dormant parasitic sequences within the genome.28–30 An illustrative instance is the LINE family member L1, which research has demonstrated to undergo hypermethylation across various cancer types, such as breast, lung, bladder, and liver cancers.31 Modification via m6A methylation stands out as among the most prevalent and abundant epigenetic alterations in eukaryotic RNAs. This modification has been linked to the onset and progression of various diseases, including acute lymphoblastic leukemia (AML), atherosclerosis, and more. The m6A demethylase, FTO, serves as a pivotal regulator in the dynamics of leukemia. In hypertrophied and failing hearts, deviations in m6A levels occur regardless of transcriptional and translational regulatory processes. These irregular modifications drive pathological changes in cardiac structure and function, leading to conditions such as heart failure, atherosclerosis, and congenital heart disease. Furthermore, they may exacerbate risk factors associated with cardiovascular diseases, such as obesity, inflammation, hypertension, and type 2 diabetes.
Until now, much of the emphasis in epigenetic research has been on cancer. However, as the field progresses, it is uncovering fresh perspectives on a broader spectrum of diseases, with particular attention to neurological and autoimmune disorders. The human central nervous system stands as one of the most intricate systems in the body. Recent investigations have unveiled a connection between changes in epigenetic markers and the onset or progression of neurodegenerative and neurological conditions. The bulk of the evidence centers on DNA methylation and modifications to histones. In numerous neurological disorders, there seems to be a disruption in DNA methylation patterns, resulting in the formation of both hypermethylated and hypomethylated regions. As an illustration, individuals with Fragile X syndrome exhibit heightened methylation in the promoter region of the FMR1 gene. This syndrome arises from the expansion of CGG trinucleotide repeats within the 5′-untranslated region of FMR1. When the CGG repeat sequence surpasses 200 copies, it triggers methylation of FMR1, leading to the gene’s suppression and subsequent transcriptional silencing.32 Additionally, instances of hypomethylation have been documented. In Parkinson’s disease patients, for instance, the substantia nigra shows increased expression of tumor necrosis factor alpha (TNFα) because its promoter undergoes hypomethylation. This alteration contributes to neuronal apoptosis, or programmed cell death.33 In neurological disorders, changes in histone markings are also observed, with a prevalent alteration being histone hypoacetylation. A notable instance of histone hypoacetylation occurs in amyotrophic lateral sclerosis (ALS). Within ALS patients, cytoplasmic deposits of misfolded proteins contain aggregates of the FUS protein. FUS interacts with CBP, significantly impeding its histone acetyltransferase (HAT) activity and downregulating certain CREB target genes. Consequently, elevated FUS levels lead to reduced histone acetylation.34
Enhancing the Therapeutic Potential of Epigenetic Drugs Through the Delivery System
Despite the increasing progress, epigenetic medicine still encounters numerous challenges. At present, epigenetic drugs approved by the FDA lack precision in targeting specific sites and are not selective in inhibiting various DNMT and HDAC enzyme variants. The absence of specificity may cause unforeseen off-target effects, resulting in elevated drug toxicity and an inability to sustain long-term responses.35 Furthermore, the limited solubility and permeability of these epigenetic drugs, along with their suboptimal pharmacokinetic characteristics, including instability and low bioavailability, pose substantial hurdles to their broader clinical utilization. Hence, refining targeting precision and enhancing drug stability and delivery efficacy are crucial for maximizing the clinical utility of these medications. Nanoscale drug delivery systems and prodrugs offer promise in addressing certain clinical challenges associated with existing epigenetic drugs. They can prevent premature breakdown, enhance bioavailability, facilitate intracellular uptake, and improve targeted delivery to tumors.36,37 Commonly utilized nanoparticles (NPs) encompass various types such as biodegradable polymer NPs, dendritic macromolecules, solid lipid NPs, liposomes, inorganic materials like metal NPs and quantum dots, as well as biological materials such as viral and albumin NPs. Numerous nanoparticle platforms have received approval for cancer therapy, with several others undergoing clinical investigations.38,39 Nanoparticle systems hold promise in surpassing the drawbacks of traditional delivery methods. They can enhance the pharmacological characteristics of the compound, such as stability, solubility, circulation duration, and tumor localization. Additionally, they can reduce nonspecific dispersion, enable precise cancer targeting, mitigating off-target effects, and enhance intracellular penetration to combat drug resistance.
Nanoscale delivery systems hold the potential to enhance the effectiveness of epigenetic medications. By improving permeability and precision targeting while reducing adverse effects, these systems offer promise. Various biodegradable materials, whether synthetic or natural, can serve as carriers for demethylating agents. As an example, researchers have developed a nanodelivery system employing a PLGA-PEG diblock structure to enhance the stability of AZA conjugation (Figure 2).40 In a mouse model of breast cancer where tumors were transplanted, the AZA in its coupled form demonstrated superior therapeutic outcomes. These included enhanced solubility and bioavailability of the drug compared to its free form. Research indicates that lipid nanocarriers offer protective benefits against acidic degradation of drugs, facilitate enhanced permeability across the intestinal barrier, and bolster oral absorption. Notably, there’s strong evidence supporting the effectiveness of nanostructured lipid carriers in administering decitabine orally.41 Nanogels (NGs) provide an alternative approach for drug delivery. They can be customized to release the enclosed medication in response to the patient’s physiological conditions or specific stimuli, such as changes in temperature, pH levels, or molecular recognition.42,43 Utilizing nanogels loaded with decitabine has proven effective in overcoming resistance mechanisms observed in cancer cell lines.44 Moreover, a synergistic approach involving a combination of epigenetic drugs encapsulated within biodegradable nanogels (DAC + SAHA) demonstrated superior efficacy in overcoming resistance compared to conventional drug therapies. Research has demonstrated that utilizing nanogel-mediated delivery enhances drug stability, promotes increased cellular uptake, and extends the duration of drug effects compared to administering unmodified drugs. This advancement leads to heightened efficacy in treatment outcomes.45 Innovative nanoparticles incorporating gelatinase, polyethylene glycol (PEG), and poly-ε-caprolactone (PCL) have been successfully employed for delivering DAC. Promising results have been noted, suggesting their capacity to overcome epigenetically induced multidrug resistance in experimental settings, including laboratory (in vitro) studies and animal (in vivo) models.46 Consequently, utilizing nanotechnology for the delivery of epigenetic drugs holds promise as a viable strategy to augment both the effectiveness and safety profiles of these medications, ultimately leading to improved treatment outcomes.
 |
Figure 2 Vehicles for transport and delivery of iDNMTs in poly(lactic-co-glycolic acid) (PLGA)- and poly(ethylene glycol) (PEG)-based nanomicelles, in gelatinase with PEG and poly-ε-caprolactone (PCL), and in alendronate-PEG-2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE). Note: Adapted from Roberti A, Valdes AF, Torrecillas R, Fraga MF, Fernandez AF. Epigenetics in cancer therapy and nanomedicine. Clin Epigenetics. 2019;11(1):81. Creative Commons.40 |
DNA Methyltransferase Inhibitors (DNMTi) and Their Delivery for Treatment
DNA methylation is a vital mechanism that regulates various fundamental biological and cellular processes via epigenetic alterations. Orchestrating this process are enzymes collectively referred to as DNA methyltransferases (DNMTs), including DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L.2,47 DNMT share a common catalytic structural domain called AdoMet-dependent MTase fold.48 DNMT1 is the key enzyme in charge of maintaining DNA methylation patterns during DNA replication, thereby preserving important epigenetic marks. In contrast, DNMT3A, DNMT3B, along with their regulatory factor DNMT3L, primarily initiate the process of de novo DNA methylation during the early embryonic stages of mammalian development. Additionally, DNMT2 has a unique function in cytoplasmic RNA methylation.49 Dysregulation of DNA methylation, marked by alterations in DNA methylation patterns such as hypomethylation and hypermethylation, is strongly associated with numerous medical conditions. These include cancer, hereditary diseases, and disorders impacting the neurological and autoimmune systems. Consequently, DNMT enzymes have garnered significant attention as potential targets for therapeutic intervention. In response, numerous small molecule inhibitors that specifically target the DNMT family have been documented and studied extensively.50,51 Two prominent examples of DNMT inhibitors are 5-azacitidine (Azacitidine or Vidaza) and 5-aza-2′-deoxycytidine (Decitabine or Dacogen), which belong to the cytidine chemotype class of nucleoside-based DNMT inhibitors. These inhibitors have gained FDA approval for the treatment of myelodysplastic syndromes, a group of disorders affecting the production of healthy blood cells. These drugs have shown efficacy in helping to restore normal blood cell formation by targeting and inhibiting DNMT enzymes involved in aberrant DNA methylation patterns. These inhibitors function by inserting themselves between the base pairs of DNA, which effectively impedes the process of DNA methylation, particularly in CpG islands. These regions are often densely populated with CpG dinucleotides and are highly significant in controlling gene expression as they frequently correspond to promoter regions involved in gene regulation. By blocking DNMT activity, these inhibitors prevent the methylation of CpG islands, thereby preserving the transcriptionally relevant regulatory sequences that are crucial for proper gene expression.52,53 Given the challenges associated with DNMTi cytidine analogs, including their chemical instability and non-specific target association profiles, there is a growing need for the development of more precise and selective DNMT inhibitors.54–56
In investigating the therapeutic potential of DNMT inhibitors for autosomal dominant polycystic kidney disease (ADPKD), it was discovered that Aza exhibits synergistic effects when combined with the ADPKD medication tolvaptan (MT). Administering the ADPKD medications metformin and tolvaptan (MT) along with the DNMT inhibitor 5-aza-20-deoxycytidine (Aza), either as standalone drugs or encapsulated within nanoparticles, decreased cell viability and inhibited cystic growth in both 2D and 3D cultures of cystic Pkd1 heterozygous renal epithelial cells (PKD1-Het cells) (Figure 3).57 To prepare for potential use in nanoparticle-based drug delivery systems, the researchers integrated combinations of drugs into micelle formulations optimized for accumulation in renal tissues.58 In short, the researchers anchored hydrophilic metformin to the outer layer of the micelles, while hydrophobic Aza and tolvaptan were encapsulated within the micellar core through hydrophobic interactions. This approach was validated to mitigate off-target effects and enhance the bioavailability of the drugs.57
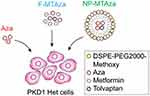 |
Figure 3 Illustration depicting experimental drug conditions. PKD1-Het cells were treated with Aza, free MT and Aza (F-MTAza), or MT and Aza delivered within a nanoparticle (NP-MTAza). Note: Adapted from Trinh A, Huang Y, Shao H et al. Targeting the ADPKD methylome using nanoparticle-mediated combination therapy. APL Bioeng. 2023;7(2):026111. Creative Commons.57 |
Two commonly used drugs that focus on DNA methylation, AZA and 5-aza-20-deoxycytidine (decitabine, DAC), are now established as standard treatments for myelodysplastic syndromes, a fatal type of leukemia.59–61 Both drugs function by incorporating into replicating DNA and forming covalent adducts, which subsequently inhibit DNMT, effectively trapping the enzyme within rapidly dividing cells. This leads to a gradual reduction in DNA methylation as DNMT activity diminishes throughout successive cancer cell divisions. However, AZA and DAC exhibit instability both in laboratory settings and within living organisms, undergoing degradation through processes such as hydrolysis, deformation of the triazine ring, and isomerization of the ribose ring under neutral to alkaline conditions. The researchers engineered a chemical fusion between AZA and the diblock backbone of PLGAPEG to improve drug stability and enable pH-responsive drug release. Aliphatic polyesters like PLGA have been blended with hydrophilic PEG chain segments to create A-B type diblock copolymers. These copolymers can pass through the loosely arranged junctions of endothelial cells in capillaries surrounding tumor tissues. Subsequently, they gather within tumor cells via the “enhanced permeation and retention” (EPR) phenomenon. Furthermore, incorporating PEG onto the surface of PLGA particles has demonstrated the ability to extend their presence in the bloodstream, enhance cellular absorption, and mitigate degradation by metabolic enzymes.62 The findings indicated that AZA in its bound state exhibited heightened therapeutic effectiveness in contrast to its free form. This suggested that the proposed method of delivering epigenetic drugs could expand the repertoire of nanomedicine tools, offering novel avenues for developing cancer treatments.
In cancer immunotherapy, oncolytic viruses (OVs) have garnered significant attention for their ability to induce immunogenic cell death (ICD). Research has shown that OVs can drive anti-tumor immunity and alleviate immune suppression in the tumor microenvironment (TME) by recruiting T cells through the release of tumor-associated antigens (TAAs) into immune-excluded or immune-desert TME.63 However, monotherapy with OVs or their combination with immune checkpoint inhibitors (ICIs) has not achieved satisfactory clinical results, making it urgent to explore the mechanisms behind the suboptimal efficacy of OVs and optimize OV treatment strategies. DNMT inhibitors (DNMTi) can target epigenetic changes, induce gene demethylation, and potentially reactivate tumor suppressor genes. Additionally, DNMTi have been shown to convert immunosuppressive “cold” tumors into immune-permissive “hot” tumors, inhibit the differentiation of regulatory T cells (Tregs), and reverse T cell exhaustion. Therefore, the combination of engineered oncolytic herpes simplex virus (oHSV) and DNMTi appears to be a promising strategy.64 Researchers proposed a ROS/pH dual-responsive DNMTi prodrug nanocarrier, ACNPs, which can specifically release the drug 5-Aza in the TME, thereby reducing off-target effects and significantly alleviating drug toxicity. There is a synergistic effect between ACNPs and oHSV, leading to an increased expression of Gasdermin E (GSDME), which enhances GSDME-induced pyroptosis in tumor cells and the release of immunogenic substances. This combination presents a promising strategy for tumor immunotherapy.65
Zebularine (Zeb), known for its demethylating properties, has shown substantial advantages and improved therapeutic effectiveness in the realm of tumor immunotherapy. However, Zeb therapy alone requires administration of higher doses due to the lack of targeting capabilities. This study introduced a pioneering nanomedical formulation comprising Zeb, a DNA methyltransferase inhibitor, and pH-responsive chitosan (CS). This formulation, termed CS-Zeb nanoparticles (NPs), was specifically crafted for the research. The study findings revealed that CS-Zeb nanoparticles demonstrated increased drug release in acidic conditions (pH 5.5) compared to neutral conditions (pH 7.4). In addition, in vivo studies conclusively demonstrated that the nanocomplexes significantly reduced tumor load and prolonged survival in a B16F10 tumor-bearing mouse model compared to equivalent amounts of isolated Zeb. Furthermore, CS-Zeb nanoparticles led to elevated levels of CD8+ T cells and tumor-infiltrating lymphocytes (TIL) in the peripheral bloodstream of mice. Importantly, the dosage of CS-Zeb nanoparticles was notably reduced by a factor of 70 compared to standalone Zeb administration.43 In conclusion, DNMTis in combination with the nanomedicine delivery system showed superior efficacy than when used alone.
Histone Deacetylase Inhibitors (HDACi) and Their Delivery for Treatment
So far, researchers have identified 18 different species of histone deacetylases (HDACs) in mammals. These enzymes can be categorized into four distinct classes based on their structural and phylogenetic characteristics. Class I HDACs share similarities with the Rpd3 deacetylase found in yeast, whereas classes IIA and IIB are akin to the yeast Hda1 deacetylase. Class III HDACs, on the other hand, are similar to yeast Sir2. It’s notable that HDAC11 displays resemblances to both class I and class II enzymes, yet it’s categorized as a distinct class IV enzyme. Class I, II, and HDAC11 enzymes function as zinc-dependent hydrolases, whereas class III HDACs are reliant on the coenzyme NAD for their activity, making them NAD-dependent enzymes. These enzymes are vital players in various biological functions, encompassing apoptosis (cell death), differentiation (cell specialization), proliferation (cell growth and division), and senescence (cellular aging).66,67 Scientists have developed various types of compounds known as HDAC inhibitors (HDACi) for therapeutic purposes. These inhibitors can be broadly categorized into various groups based on their chemical structures. Instances of the initial chemotype comprise short-chain fatty acids such as sodium butyrate, phenylbutyrate, pivanex, and valproic acid, along with cyclic tetrapeptides and naturally existing compounds. The second chemotype comprises newer and more selective classes of HDAC inhibitors, such as isohydroxamic acids including vorinostat, belistat, pabilostat, and dacilostat. Another class is the benzamides, represented by entrestat and mocilostat, while the third class consists of bicyclic phenyl peptides, with romidepsin as a notable example. These diverse classes of HDAC inhibitors offer potential targets for therapeutic interventions in various diseases.68 In particular, most of the compounds in clinical trials were isohydroxamic acid analogs.69 The FDA-approved drug vorinostat first demonstrated clinical success with isohydroxamic acid analogs.70 The ability of these compounds to inhibit HDAC stems from their essential polar isohydroxamic group. This group hinders deacetylation by either interacting with zinc-binding proteins or chelating zinc ions situated within the enzyme pocket’s catalytic site.70 HDAC inhibitors, when combined with other therapeutic agents, have demonstrated preclinical potential.
Additionally, utilizing cutting-edge drug delivery techniques, such as liposomal encapsulation, can enhance the clinical development and efficacy of epigenetic drugs in cancer treatment.71 Surface functionalization of liposomes improves their stability and enables targeted delivery of drugs, genes, or imaging agents, even across biological and physiological barriers. They are biocompatible, exhibit low immunogenicity, and enhance the solubility of various chemotherapeutics. Several liposomal formulations have received EMA and/or FDA approval for the treatment of various cancer types.72 To enhance the effectiveness of histone deacetylase inhibitors (HDACIs) in solid tumors, PEGylated liposomes were developed for the encapsulation of trichostatin A (TSA), CG1521 (CG), and PXD101 (PXD).73 These liposome formulations serve as promising nanocarriers, demonstrating stability in size, charge, and biological activity for one month when stored at 4°C.
To achieve optimal inhibition of intracellular HDAC activity and induce tumor cell inhibition or apoptosis, it is necessary to maintain a sustained circulation of the HDAC inhibitors within the body and ensure prolonged exposure of the drugs. This continuous drug exposure is crucial for attaining significant clinical responses in treating tumors. Various factors can diminish the effectiveness of HDAC inhibitors against tumors by decreasing their availability and antitumor activity. These factors include rapid elimination from the body, high affinity for proteins, swift metabolism-induced breakdown, or rapid deactivation of reactive functional groups such as epoxy groups. A recent study investigated the viability of delivering the HDAC inhibitor LAQ824 via liposomes. In a pilot study lasting three weeks, where liposomal LAQ824 was administered once weekly, noteworthy inhibition of growth was observed in rapidly proliferating human breast tumor xenografts.74 Similar to findings in prior research utilizing diverse tumor model systems, administering free LAQ824 typically demands daily injections of higher doses to impede tumor growth. Furthermore, liposomes offer the advantage of either passively targeting tumor cells or being customized for active targeting, enhancing their potential in therapeutic interventions.75 In this context, three HDAC inhibitors from classes I and II (7-phenyl-2,4,6-heptatrienoylisohydroxamic acid, belinostat [PXD101], and trichostatin A [TSA]) were incorporated into polyethylene glycol-coated liposomes. It has been noted that the creation of “multilayered” liposomes effectively facilitates the loading of each HDAC inhibitor. However, while the encapsulation efficiencies for 7-phenyl-2,4,6-heptatrienoylisohydroxamic acid and belinostat remain above 90%, TSA achieves an encapsulation efficiency of only 75%. Remarkably, the ideal drug concentration for effective loading is 1 mm. Certain nanoparticles within belinostat and TSA liposomes exhibited a size of 150 nm, whereas liposomes loaded with CG1521 were sized at 100 nm.73 This size spectrum holds promise for effectively targeting tumors and facilitating drug delivery. In a separate investigation, the HDAC inhibitor octanediylaniline isohydroxamic acid (SAHA), alternatively known as vorinostat (Vor), was co-encapsulated within cationic liposomes alongside an angiogenesis inhibitor, the low-molecular-weight heparin taurocholate concatenate (LHT7). Initially, the liposomes were formulated to contain encapsulated SAHA. Subsequently, the negatively charged LHT7 was bound to the cationic liposomes through electrostatic interactions, resulting in a favorable particle size of 117.6 nm.76 Scientists further investigated the creation of specialized liposomes tailored for active targeting objectives. For example, they engineered liposomes containing a combination of trastuzumab-functionalized mannosylated gefitinib and Vor. These liposomes were crafted to selectively home in on tumor-associated macrophages characterized by heightened expression of mannose receptors. The objective was to repolarize these macrophages and overcome resistance to gefitinib, thus enhancing the effectiveness of the treatment. The liposomes utilized in the study had a particle size of 180 nm and demonstrated excellent stability. Additionally, they achieved a high encapsulation rate of over 80% for both drugs, indicating efficient loading of the therapeutic agents into the liposomes.77 Nanocarriers present considerable advantages for delivering HDAC inhibitors in cancer therapy, such as heightened effectiveness, minimized side effects, improved stability, enhanced water solubility, targeted delivery, slowed breakdown in the body, and prolonged duration of action.78
Delivery of Bromodomain Inhibitors and Degraders for Treatment
BRD2, BRD3, and BRD4 are part of the extensively researched group of proteins known as the bromodomain and extra-terminal (BET) family. These proteins interact with acetylated lysine residues on histones and have been associated with various conditions spanning from cancer and inflammation to cardiovascular diseases.79 Glioblastoma (GBM) is a type of brain tumor with a grim outlook, mainly because it quickly becomes resistant to single chemotherapy agents and faces challenges in targeted delivery within the brain. Recent research indicates that combining temozolomide (TMZ) with the epigenetic bromodomain inhibitor OTX015, packaged within a nanodrug delivery system, can significantly enhance treatment effectiveness. Due to OTX’s restricted ability to penetrate the blood-brain barrier (BBB) and its limited targeting of glioblastoma (GBM), researchers employed a laboratory-developed technique involving cell membrane coating to simultaneously encapsulate TMZ and OTX. This approach aims to enhance the delivery of both drugs to the brain and improve their targeting specifically to GBM. The nanodrugs were deliberately enveloped in red blood cell membranes (RBCm) and adorned with apolipoprotein E peptide (ApoE) to facilitate penetration of the BBB and uptake by glioblastoma (GBM) cells. Internally, we used pH-responsive polymers that degrade at low pH to facilitate drug release triggered by the GBM micro- and intracellular environment.42 Collectively, these design features result in the creation of ABNM@TMZ/OTX nanomedicines, offering a promising solution to the challenge of delivering drugs to the brain, thereby enhancing clinical effectiveness.
Research has demonstrated that heightened levels of BRD4 are associated with unfavorable outcomes in lung cancer. Conversely, suppressing BRD4 expression triggers apoptosis, resulting in tumor reduction. Researchers have engineered a new versatile nano-PROTAC (CRV-LLC membrane/DS-PLGA/dBET6), which involves incorporating a BRD4-targeting PROTAC (DBET6) into a pH/GSH-responsive polymer (DS-PLGA) and camouflaging it with lung cancer cell membranes engineered for dual-targeting abilities. Remarkably, CREATE exhibited a pronounced capacity to target both lung cancer cells and tumor-associated macrophages (TAM). The pH/GSH-responsive structure notably enhanced the release of the dBET6 payload from the nanoparticles, effectively inducing apoptosis in both cell types. This synergistic effect significantly impeded tumor growth in both subcutaneous and in situ hormonal mouse models.80 Altogether, these results shed light on the fact that the combination of epigenetic drugs with nanodelivery systems opens up new avenues for the effective treatment of diseases.
Inhibition of N6-Methyladenosine and Delivery for Treatment
The regulation of N6-methyladenosine (m6A) modulators plays a pivotal role in dictating the destiny of m6A-modified transcripts, thereby influencing cancer progression. Targeting m6A modulators through RNA interference holds significant potential as a novel therapeutic approach in combating cancer. YTH N6-methyladenosine RNA-binding protein 1 (YTHDF1), belonging to the YT521-B (YTH) homology structural domain family, serves as a crucial m6A reader protein. It exhibits specificity in binding to m6A-modified transcripts, thereby influencing their translation efficiency.81 YTHDF1 has been identified as a promoter of tumor advancement across various cancer types such as gastric,82 ovarian,83 hepatocellular, and cervical cancers.84 Its role as both a prognostic indicator and a potential target for cancer therapy has garnered significant attention. Various strategies, including the use of small molecule inhibitors and RNA interference (RNAi), have been devised to target dysregulated m6A modulators. These approaches have demonstrated notable efficacy in impeding tumor advancement85 and reinstating anti-tumor immune responses.85 The researchers opted for a specific type of macrophage-derived extracellular vesicle (EV) with elevated CD47 expression86,87 (RAW-264.7), denoted as RAW-SEV or RsEV. Additionally, they enhanced the efficacy of these EVs by functionalizing them with cyclic arginine-glycine-aspartate (c(RGDyC)) peptides, resulting in what they termed PRsEVs. This modification aimed to enhance the retention of nanocarriers in circulation and improve their ability to target tumors.88 Furthermore, the PRsEVs were engineered to serve as gadolinium-based magnetic contrast agents, labeled as PGRsEVs (Figure 4A).24 This design allows for precise cancer therapy guided by magnetic resonance imaging (MRI), enhancing the accuracy of treatment delivery. Recombinant rsEVs demonstrate superior performance in evading phagocytosis through CD47 signaling and enhancing tumor targeting via c(RGDyC) interactions. They effectively deliver siYTHDF1 to the tumor location, leading to YTHDF1 depletion in an m6-dependent manner. Consequently, they hinder FZD7 protein translation and curb tumor advancement and metastasis by suppressing the Wnt/β-catenin pathway. Deletion of YTHDF1 triggers the upregulation of interferon-gamma receptor 1 (IFNGR1) in tumor cells. This elevation in IFNGR1 enhances the response to interferon-gamma, resulting in increased expression of major histocompatibility complex class I (MHC-I) molecules on the tumor cell surface. As a result, tumor cells can present themselves as immunogenic targets to CD8+ T cells, stimulating a potent cytotoxic T lymphocyte response. The heightened expression of CD47 on engineered SEVs facilitates increased phagocytosis by competitively binding to Sirpα receptors on tumor-associated macrophages (TAMs) (Figure 4B).24 This adaptable nanotheranostic platform offers a safe and effective approach for precisely delivering siYTHDF1, which targets m6A modulators, with minimal toxicity.24
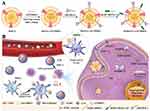 |
Figure 4 (A) Preparation of PGRsEV-siYTHDF1. (B) PGRsEV-siYTHDF1 inhibits gastric cancer progression by targeting FZD7 and modulating its translation through an m6A-dependent mechanism. This action disrupts the Wnt/β-catenin signaling pathway. Additionally, it enhances immunotherapy by promoting the self-presentation of immunogenic tumor antigens and employing a CD47 blockade strategy, which intensifies the cytotoxic effects of cytotoxic T lymphocytes (CTLs) and tumor-associated macrophages (TAMs). Note: Adapted from You Q, Wang F, Du R et al m(6) A reader ythdf1-targeting engineered small extracellular vesicles for gastric cancer therapy via epigenetic and immune regulation. Adv Mater. 2023;35(8):e2204910. ©2022 Wiley-VCH GmbH.24 |
In summary, RNA interference aimed at m6A regulators holds significant potential as a burgeoning cancer treatment strategy. Presently, various studies are leveraging nanotechnology to address challenges such as inadequate tumor targeting and excessive systemic toxicity.
Challenges and Prospects
In recent years, extensive research into epigenetic changes in various disease has highlighted new treatment options that focus on reversible modifications of the epigenome. Significant progress has been achieved in understanding the epigenetic modifications observed in cancer and their association with tumorigenesis. Yet, the effective utilization of epigenetic medications has been constrained, mainly attributable to their limited specificity and the onset of unintended broad-ranging effects. Challenges such as poor tolerability, limited effectiveness, and off-target effects of many epigenetic drugs continue to pose significant hurdles in their application to solid tumors. Investigating lower doses and targeted delivery could significantly enhance the therapeutic index. Nanotechnology for precise delivery and controlled release of epigenetic medications, heralds a notable advancement toward personalized and precisely targeted therapies. Nanotechnology has transformed drug delivery methods for epigenetic therapies aimed at solid tumors. Various formulations of nanocarriers have been used to enhance the stability, solubility, and specificity of DNMT and histone inhibitors. Moreover, conventional chemotherapy agents have been effectively co-loaded with epigenetic drugs in nanocarriers for combination therapy. Preclinical and clinical trial data for nano-based epigenetic drugs in breast cancer treatment show decreased systemic toxicity and enhanced efficacy compared to traditional free-drug formulations.
Although nano-delivery of epigenetic drugs is promising, nano-based delivery systems must adhere to “safety by design” principles and be thoroughly tested to mitigate potential health risks and environmental impacts. Research into the epigenetic effects caused by nanomaterials has gradually gained attention. For example, nanoparticles like silver and zinc oxide can alter the overall methylation levels in cell genomes, and they also influence histone modifications, such as acetylation.40,89 The impact of nanomaterials on epigenetics depends on factors like the type of nanomaterial, its size, charge, and crystal structure. When using nanomaterials to deliver epigenetic drugs, it’s important to carefully consider the potential toxicity that these materials could introduce. Mitigating the epigenetic toxicity of nanomaterials can be achieved by using drugs to counteract their effects, avoiding inorganic nanomaterials, and opting for highly biocompatible biological nanomaterials like liposomes and albumin, as they might effectively reduce the epigenetic toxicity associated with nanomaterials. In summary, the development of nano-delivery systems and the advancement of epigenetic drugs and biomarkers will significantly promote personalized targeted therapy and improve the effectiveness and safety of epigenetic therapy.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13(12):877–919. doi:10.1007/s13238-021-00846-7
2. Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021;37(11):1012–1027. doi:10.1016/j.tig.2021.05.002
3. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–639. doi:10.1042/ebc20200032
4. Moran B, Davern M, Reynolds JV, Donlon NE, Lysaght J. The impact of histone deacetylase inhibitors on immune cells and implications for cancer therapy. Cancer Lett. 2023;559:216121. doi:10.1016/j.canlet.2023.216121
5. Mattei AL, Bailly N, Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38(7):676–707. doi:10.1016/j.tig.2022.03.010
6. Hogg SJ, Beavis PA, Dawson MA, Johnstone RW. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov. 2020;19(11):776–800. doi:10.1038/s41573-020-0077-5
7. Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat Rev Cancer. 2019;19(3):151–161. doi:10.1038/s41568-019-0109-9
8. He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi:10.1126/science.1210944
9. Chen Q, Yang B, Liu X, Zhang XD, Zhang L, Liu T. Histone acetyltransferases CBP/p300 in tumorigenesis and CBP/p300 inhibitors as promising novel anticancer agents. Theranostics. 2022;12(11):4935–4948. doi:10.7150/thno.73223
10. He S, Wu Z, Tian Y, et al. Structure of nucleosome-bound human BAF complex. Science. 2020;367(6480):875–881. doi:10.1126/science.aaz9761
11. Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2018;19(3):192–206. doi:10.1038/nrm.2017.94
12. Zhang K, Hocker JD, Miller M, et al. A single-cell atlas of chromatin accessibility in the human genome. Cell. 2021;184(24):5985–6001. doi:10.1016/j.cell.2021.10.024
13. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. 2024;25(3):211–232. doi:10.1038/s41576-023-00662-1
14. Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188491. doi:10.1016/j.bbcan.2020.188491
15. Wang K, Liu H, Hu Q, et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Target Ther. 2022;7(1):374. doi:10.1038/s41392-022-01211-8
16. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37(3):270–288. doi:10.1016/j.ccell.2020.02.004
17. Zhan Y, Chen Z, He S, et al. Correction: long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer. 2023;22(1):115. doi:10.1186/s12943-023-01822-x
18. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20(6):303–322. doi:10.1038/s41568-020-0253-2
19. Pomaville MM, He C. Advances in targeting RNA modifications for anticancer therapy. Trends Cancer. 2023;9(7):528–542. doi:10.1016/j.trecan.2023.04.003
20. Liu P, Li F, Lin J, et al. m(6)A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat Cell Biol. 2021;23(4):355–365. doi:10.1038/s41556-021-00656-3
21. Yankova E, Blackaby W, Albertella M, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601. doi:10.1038/s41586-021-03536-w
22. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13(1):117. doi:10.1186/s13045-020-00951-w
23. Azzam SK, Alsafar H, Sajini AA. FTO m6A Demethylase in Obesity and Cancer: implications and Underlying Molecular Mechanisms. Int J Mol Sci. 2022;23(7):3800. doi:10.3390/ijms23073800
24. You Q, Wang F, Du R, et al. m(6) A Reader YTHDF1-Targeting Engineered Small Extracellular Vesicles for Gastric Cancer Therapy via Epigenetic and Immune Regulation. Adv Mater. 2023;35(8):e2204910. doi:10.1002/adma.202204910
25. Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer. 2021;21(7):413–430. doi:10.1038/s41568-021-00357-x
26. Wang L, Tang J. SWI/SNF complexes and cancers. Gene. 2023;870:147420. doi:10.1016/j.gene.2023.147420
27. Guo H, Vuille JA, Wittner BS, et al. DNA hypomethylation silences anti-tumor immune genes in early prostate cancer and CTCs. Cell. 2023;186(13):2765–2782.e28. doi:10.1016/j.cell.2023.05.028
28. Besselink N, Keijer J, Vermeulen C, et al. The genome-wide mutational consequences of DNA hypomethylation. Sci Rep. 2023;13(1):6874. doi:10.1038/s41598-023-33932-3
29. Li Y, Fan Z, Meng Y, Liu S, Zhan H. Blood-based DNA methylation signatures in cancer: a systematic review. Biochim Biophys Acta Mol Basis Dis. 2023;1869(1):166583. doi:10.1016/j.bbadis.2022.166583
30. Switzer CH, Cho HJ, Eykyn TR, Lavender P, Eaton P. NOS2 and S-nitrosothiol signaling induces DNA hypomethylation and LINE-1 retrotransposon expression. Proc Natl Acad Sci U S A. 2022;119(21):e2200022119. doi:10.1073/pnas.2200022119
31. de Cubas AA, Dunker W, Zaninovich A, et al. DNA hypomethylation promotes transposable element expression and activation of immune signaling in renal cell cancer. JCI Insight. 2020;5(11):137569. doi:10.1172/jci.insight.137569
32. Faulkner S, Maksimovic I, David Y. A chemical field guide to histone nonenzymatic modifications. Curr Opin Chem Biol. 2021;63:180–187. doi:10.1016/j.cbpa.2021.05.002
33. Pieper HC, Evert BO, Kaut O, Riederer PF, Waha A, Wüllner U. Different methylation of the TNF-alpha promoter in cortex and substantia nigra: implications for selective neuronal vulnerability. Neurobiol Dis. 2008;32(3):521–527. doi:10.1016/j.nbd.2008.09.010
34. Shanker OR, Kumar S, Dixit AB, Banerjee J, Tripathi M, Sarat Chandra P. Epigenetics of neurological diseases. Prog Mol Biol Transl Sci. 2023;198:165–184. doi:10.1016/bs.pmbts.2023.01.006
35. Ghosh A, Himaja A, Biswas S, Kulkarni O, Ghosh B. Advances in the Delivery and Development of Epigenetic Therapeutics for the Treatment of Cancer. Mol Pharm. 2023;20(12):5981–6009. doi:10.1021/acs.molpharmaceut.3c00610
36. Magro M, Venerando A, Macone A, Canettieri G, Agostinelli E, Vianello F. Nanotechnology-Based Strategies to Develop New Anticancer Therapies. Biomolecules. 2020;10(5):735. doi:10.3390/biom10050735
37. Urbanova M, Cihova M, Buocikova V, et al. Nanomedicine and epigenetics: new alliances to increase the odds in pancreatic cancer survival. Biomed Pharmacother. 2023;165:115179. doi:10.1016/j.biopha.2023.115179
38. Fan D, Cao Y, Cao M, Wang Y, Cao Y, Gong T. Nanomedicine in cancer therapy. Signal Transduct Target Ther. 2023;8(1):293. doi:10.1038/s41392-023-01536-y
39. Sun Q, Bai X, Sofias AM, et al. Cancer nanomedicine meets immunotherapy: opportunities and challenges. Acta Pharmacol Sin. 2020;41(7):954–958. doi:10.1038/s41401-020-0448-9
40. Roberti A, Valdes AF, Torrecillas R, Fraga MF, Fernandez AF. Epigenetics in cancer therapy and nanomedicine. Clin Clin Epigenet. 2019;11(1):81. doi:10.1186/s13148-019-0675-4
41. Neupane YR, Srivastava M, Ahmad N, Kumar N, Bhatnagar A, Kohli K. Lipid based nanocarrier system for the potential oral delivery of decitabine: formulation design, characterization, ex vivo, and in vivo assessment. Int J Pharm. 2014;477(1–2):601–612. doi:10.1016/j.ijpharm.2014.11.001
42. Liu Y, Wang W, Zhang D, et al. Brain co-delivery of first-line chemotherapy drug and epigenetic bromodomain inhibitor for multidimensional enhanced synergistic glioblastoma therapy. Exploration. 2022;2(4):20210274. doi:10.1002/exp.20210274
43. Lai J, Liang J, Zhang Y, et al. A drug-delivery depot for epigenetic modulation and enhanced cancer immunotherapy. Biomed Pharmacother. 2023;168:115687. doi:10.1016/j.biopha.2023.115687
44. Vijayaraghavalu S, Labhasetwar V. Efficacy of decitabine-loaded nanogels in overcoming cancer drug resistance is mediated via sustained DNA methyltransferase 1 (DNMT1) depletion. Cancer Lett. 2013;331(1):122–129. doi:10.1016/j.canlet.2012.12.009
45. Vijayaraghavalu S, Labhasetwar V. Nanogel-mediated delivery of a cocktail of epigenetic drugs plus doxorubicin overcomes drug resistance in breast cancer cells. Drug Deliv Transl Res. 2018;8(5):1289–1299. doi:10.1007/s13346-018-0556-y
46. Hong YD, Zhang J, Zhuang M, et al. Efficacy of decitabine-loaded gelatinases-stimuli nanoparticles in overcoming cancer drug resistance is mediated via its enhanced demethylating activity to transcription factor AP-2 epsilon. Oncotarget. 2017;8(70):114495–114505. doi:10.18632/oncotarget.21274
47. Weisenberger DJ, Lakshminarasimhan R, Liang G. The Role of DNA Methylation and DNA Methyltransferases in Cancer. Adv Exp Med Biol. 2022;1389:317–348. doi:10.1007/978-3-031-11454-0_13
48. Gasiulė L, Stankevičius V, Kvederavičiu Tė K, et al. Engineered Methionine Adenosyltransferase Cascades for Metabolic Labeling of Individual DNA Methylomes in Live Cells. J Am Chem Soc. 2024;146(27):18722–18729. doi:10.1021/jacs.4c06529
49. Yan L, Geng Q, Cao Z, et al. Insights into DNMT1 and programmed cell death in diseases. Biomed Pharmacother. 2023;168:115753. doi:10.1016/j.biopha.2023.115753
50. Zhang L, Li HT, Shereda R, et al. DNMT and EZH2 inhibitors synergize to activate therapeutic targets in hepatocellular carcinoma. Cancer Lett. 2022;548:215899. doi:10.1016/j.canlet.2022.215899
51. Mehdipour P, Chen R, De Carvalho DD. The next generation of DNMT inhibitors. Nat Cancer. 2021;2(10):1000–1001. doi:10.1038/s43018-021-00271-z
52. Hoang NM, Rui L. DNA methyltransferases in hematological malignancies. J Genet Genomics. 2020;47(7):361–372. doi:10.1016/j.jgg.2020.04.006
53. Yu J, Xie T, Wang Z, et al. DNA methyltransferases: emerging targets for the discovery of inhibitors as potent anticancer drugs. Drug Discov Today. 2019;24(12):2323–2331. doi:10.1016/j.drudis.2019.08.006
54. Ma L, Li C, Yin H, et al. The Mechanism of DNA Methylation and miRNA in Breast Cancer. Int J Mol Sci. 2023;24(11):9360. doi:10.3390/ijms24119360
55. Wang J, Yang J, Li D, Li J. Technologies for targeting DNA methylation modifications: basic mechanism and potential application in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188454. doi:10.1016/j.bbcan.2020.188454
56. Wang T, Li P, Qi Q, et al. A multiplex blood-based assay targeting DNA methylation in PBMCs enables early detection of breast cancer. Nat Commun. 2023;14(1):4724. doi:10.1038/s41467-023-40389-5
57. Trinh A, Huang Y, Shao H, et al. Targeting the ADPKD methylome using nanoparticle-mediated combination therapy. APL Bioeng. 2023;7(2):026111. doi:10.1063/5.0151408
58. Jiang K, Huang Y, Chung EJ. Combining Metformin and Drug-Loaded Kidney-Targeting Micelles for Polycystic Kidney Disease. Cell Mol Bioeng. 2023;16(1):55–67. doi:10.1007/s12195-022-00753-9
59. Patnaik MM, Lasho T. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: a focused review. Hematol Am Soc Hematol Educ Program. 2020;2020(1):460–464. doi:10.1182/hematology.2020000163
60. Gurion R, Vidal L, Gafter-Gvili A, et al. 5-azacitidine prolongs overall survival in patients with myelodysplastic syndrome--a systematic review and meta-analysis. Haematologica. 2010;95(2):303–310. doi:10.3324/haematol.2009.010611
61. Chowdhury B, McGovern A, Cui Y, et al. The hypomethylating agent Decitabine causes a paradoxical increase in 5-hydroxymethylcytosine in human leukemia cells. Sci Rep. 2015;5:9281. doi:10.1038/srep09281
62. Laurent G, Benbalit C, Chrétien C, et al. Characterization and biodistribution of Au nanoparticles loaded in PLGA nanocarriers using an original encapsulation process. Colloids Surf B. 2021;205:111875. doi:10.1016/j.colsurfb.2021.111875
63. Zhu Z, McGray AJR, Jiang W, Lu B, Kalinski P, Guo ZS. Improving cancer immunotherapy by rationally combining oncolytic virus with modulators targeting key signaling pathways. Mol Cancer. 2022;21(1):196. doi:10.1186/s12943-022-01664-z
64. Ehrlich M, Bacharach E. Oncolytic Virotherapy: the Cancer Cell Side. Cancers. 2021;13(5):939. doi:10.3390/cancers13050939
65. Forbes NE, Abdelbary H, Lupien M, Bell JC, Diallo JS. Exploiting tumor epigenetics to improve oncolytic virotherapy. Front Genet. 2013;4:184. doi:10.3389/fgene.2013.00184
66. Mirzaei H, Ghorbani S, Khanizadeh S, Namdari H, Faghihloo E, Akbari A. Histone deacetylases in virus-associated cancers. Rev Med Virol. 2020;30(1):e2085. doi:10.1002/rmv.2085
67. Ramaiah MJ, Tangutur AD, Manyam RR. Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 2021;277:119504. doi:10.1016/j.lfs.2021.119504
68. Ononye SN, van Heyst M, Falcone EM, Anderson AC, Wright DL. Toward isozyme-selective inhibitors of histone deacetylase as therapeutic agents for the treatment of cancer. Pharm Pat Anal. 2012;1(2):207–221. doi:10.4155/ppa.12.21
69. Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Mol Oncol. 2012;6(6):657–682. doi:10.1016/j.molonc.2012.09.004
70. West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30–39. doi:10.1172/jci69738
71. Gurunathan S, Kang MH, Qasim M, Kim JH. Nanoparticle-Mediated Combination Therapy: two-in-One Approach for Cancer. Int J Mol Sci. 2018;19(10):3264. doi:10.3390/ijms19103264
72. Buocikova V, Rios-Mondragon I, Pilalis E, et al. Epigenetics in Breast Cancer Therapy-New Strategies and Future Nanomedicine Perspectives. Cancers. 2020;12(12):3622. doi:10.3390/cancers12123622
73. Urbinati G, Marsaud V, Plassat V, Fattal E, Lesieur S, Renoir JM. Liposomes loaded with histone deacetylase inhibitors for breast cancer therapy. Int J Pharm. 2010;397(1–2):184–193. doi:10.1016/j.ijpharm.2010.06.046
74. Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi:10.1146/annurev.pharmtox.45.120403.095825
75. García-Pinel B, Porras-Alcalá C, Ortega-Rodríguez A, et al. Lipid-Based Nanoparticles: application and Recent Advances in Cancer Treatment. Nanomaterials. 2019;9(4):638. doi:10.3390/nano9040638
76. Kim JY, Shim G, Choi HW, et al. Tumor vasculature targeting following co-delivery of heparin-taurocholate conjugate and suberoylanilide hydroxamic acid using cationic nanolipoplex. Biomaterials. 2012;33(17):4424–4430. doi:10.1016/j.biomaterials.2012.02.066
77. Peng H, Chen B, Huang W, et al. Reprogramming Tumor-Associated Macrophages To Reverse EGFR(T790M) Resistance by Dual-Targeting Codelivery of Gefitinib/Vorinostat. Nano Lett. 2017;17(12):7684–7690. doi:10.1021/acs.nanolett.7b03756
78. Talaei S, Mellatyar H, Asadi A, Akbarzadeh A, Sheervalilou R, Zarghami N. Spotlight on 17-AAG as an Hsp90 inhibitor for molecular targeted cancer treatment. Chem Biol Drug Des. 2019;93(5):760–786. doi:10.1111/cbdd.13486
79. Prachayasittikul V, Prathipati P, Pratiwi R, et al. Exploring the epigenetic drug discovery landscape. Expert Opin Drug Discov. 2017;12(4):345–362. doi:10.1080/17460441.2017.1295954
80. Zhang HT, Peng R, Chen S, et al. Versatile Nano-PROTAC-Induced Epigenetic Reader Degradation for Efficient Lung Cancer Therapy. Adv Sci. 2022;9(29):e2202039. doi:10.1002/advs.202202039
81. Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24(12):1490–1492. doi:10.1038/cr.2014.153
82. Chen XY, Liang R, Yi YC, et al. The m(6)A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m(6)A-Dependent Manner. Front Cell Dev Biol. 2021;9:647702. doi:10.3389/fcell.2021.647702
83. Liu T, Wei Q, Jin J, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816–3831. doi:10.1093/nar/gkaa048
84. Wang H, Luo Q, Kang J, et al. YTHDF1 Aggravates the Progression of Cervical Cancer Through m(6)A-Mediated Up-Regulation of RANBP2. Front Oncol. 2021;11:650383. doi:10.3389/fonc.2021.650383
85. Su R, Dong L, Li C, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172(1–2):90–105. doi:10.1016/j.cell.2017.11.031
86. Jang SC, Kim OY, Yoon CM, et al. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7(9):7698–7710. doi:10.1021/nn402232g
87. Nie W, Wu G, Zhang J, et al. Responsive Exosome Nano-bioconjugates for Synergistic Cancer Therapy. Angew Chem Int Ed Engl. 2020;59(5):2018–2022. doi:10.1002/anie.201912524
88. Zhu Q, Ling X, Yang Y, et al. Embryonic Stem Cells-Derived Exosomes Endowed with Targeting Properties as Chemotherapeutics Delivery Vehicles for Glioblastoma Therapy. Adv Sci. 2019;6(6):1801899. doi:10.1002/advs.201801899
89. Lu X, Miousse IR, Pirela SV, Melnyk S, Koturbash I, Demokritou P. Short-term exposure to engineered nanomaterials affects cellular epigenome. Nanotoxicology. 2016;10(2):140–150. doi:10.3109/17435390.2015.1025115
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.