Back to Journals » Infection and Drug Resistance » Volume 18
The Negative Regulatory Role of Transcriptional Regulator H-NS on the Type VI Secretion System in Acinetobacter baumannii
Authors Zhang Y, Zhou H, Kong J, Hu P, Zhang Y , Cao J, Zhou B
Received 3 February 2025
Accepted for publication 21 May 2025
Published 12 June 2025 Volume 2025:18 Pages 2997—3011
DOI https://doi.org/10.2147/IDR.S512650
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sandip Patil
Yi Zhang,1,2,* Huijing Zhou,3,* Jingchun Kong,1 Panjie Hu,1 Yichi Zhang,1 Jianming Cao,3 Beibei Zhou1
1Department of Clinical Laboratory, the First Affiliated Hospital of Wenzhou Medical University; Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, People’s Republic of China; 2Shaoxing Center for Disease Control and Prevention, Shaoxing, People’s Republic of China; 3Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianming Cao, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China, Email [email protected] Beibei Zhou, Department of Clinical Laboratory, the First Affiliated Hospital of Wenzhou Medical University; Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, 325035, People’s Republic of China, Tel +86-0577-8668-9885, Email [email protected]
Introduction: This study investigates the negative regulatory role of the global transcriptional regulator H-NS (Histone-like Nucleoid Structuring Protein) on the Type VI secretion system (T6SS) in Acinetobacter baumannii (A. baumannii). We explored potential targets of H-NS mediated silencing or activation within the regulation of A. baumannii T6SS, along with the specific regulatory mechanisms involved, thereby providing a theoretical foundation for further research on A. baumannii invasive infections stemming from mixed infections and the development of therapeutic target.
Methods: Using the plasmids pAT04 and pYMAb2-hyg, we constructed A. baumannii ATCC19606 strains with the hns gene knocked out (ABΔhns) and overexpressed (ABhns+). We measured the expression of the T6SS-related gene hcp in wild-type (AB WT), ABΔhns, and ABhns+ strains using RT-qPCR, combined with a mouse sepsis model featuring mixed infections. We assessed their serum resistance, competitive ability against Escherichia coli (E. coli), and blood invasion capability. Proteomic analysis identified differentially expressed proteins, and we further investigated the regulatory role of H-NS on A. baumannii T6SS using electrophoretic mobility shift assays (EMSA).
Results: We successfully constructed both ABΔhns and ABhns+ strains of A. baumannii ATCC19606. RT-qPCR results indicated that H-NS functions as a negative regulator of the T6SS-related gene hcp in A. baumannii. Phenotypic assays for extracellular virulence revealed that the loss of hns enhanced both the competitive ability and serum resistance of ATCC19606. Results from the mouse sepsis infection model demonstrated that knockout of hns significantly increased the bacterium’s blood invasion capability. Bioinformatics analysis of differentially expressed proteins identified elevated levels of T6SS-related proteins in the knockout strain. Furthermore, EMSAs confirmed that H-NS directly binds to multiple sites in the upstream region of hcp.
Conclusion: H-NS inhibits the expression of T6SS-related proteins in A. baumannii by regulating relevant targets associated with the T6SS. This regulation influences the bacterium’s pathogenicity, interspecies competitive ability, and serum resistance.
Keywords: H-NS, Acinetobacter baumannii, mixed infection, type VI secretion system, septicemia
Introduction
A. baumannii is a significant opportunistic pathogen responsible for hospital-acquired infections, including invasive conditions such as sepsis and intra-abdominal infections in critically ill patients and immunocompromised individuals.1 Among these, sepsis ranks as one of the most severe types of infections, often secondary to mixed bacterial infections in other body regions.1 Bloodstream infections caused by Gram-negative bacteria typically result in more severe outcomes, including septic shock and inflammatory responses, compared to those caused by Gram-positive bacteria.2
Bacterial secretion systems are characteristic components on the surface of Gram-negative bacteria, with the Type VI secretion system (T6SS) being closely linked to bacterial pathogenicity.3,4 The T6SS is a widely distributed secretion system that plays a significant role in the ability of pathogenic bacteria to resist environmental stress and evade host immunity,5 often remaining in a “silent” state. Under conditions of environmental stress, the T6SS can function to achieve adaptive equilibrium. For example, in Aeromonas hydrophila, a mutation in the exeA gene weakens the assembly of the Type II secretion system (T2SS) and inhibits biofilm formation, thereby reducing virulence. However, this is compensated by an enhancement of the T6SS, which inhibits further loss of virulence.6 Pseudomonas aeruginosa (P. aeruginosa) employs three evolutionarily distinct T6SS variants to secrete unique effector proteins, which facilitate bacterial competition and host colonization.7 K. pneumoniae also utilizes T6SS to eliminate competing bacterial populations, particularly members of the Betaproteobacteria, in a contact-dependent manner, thereby indirectly promoting its own colonization and pathogenic potential within the gastrointestinal tract.8 The T6SS of A. baumannii mediates the secretion of the bifunctional peptidoglycan-degrading enzyme Tae17 through key amino acids (G1069 and W1075) in the delivery protein VgrG17. The lytic transglycosylase activity of Tae17 plays a dominant role in polymicrobial competition, conferring a competitive advantage to the bacteria.9 Consequently, studies indicate that many Gram-negative bacteria utilize the T6SS to eliminate competing species and influence host responses.
T6SS, which resembles an inverted phage extending outward from the surface of bacterial cells, consists of 13 proteins assembled into three subcomplexes: a caudate tube, a basal baseplate-like structure, and a membrane complex that crosses the inner and outer membranes of the bacteria. These three subcomplexes coordinate through a contractile mechanism to transport effector proteins from the interior to the exterior of the bacteria, facilitating competitive survival.10 The inner tube of the T6SS is formed by hexamers of Hemolysin-coregulated protein (Hcp).11 The tssD/hcp and tssI/vgrG genes represent essential components of the T6SS. Additional copies of these genes can be located outside the main T6SS cluster and are often associated with genes encoding potential effector proteins.12 Adjacent to the paar, vgrG, or hcp genes, numerous other effectors are encoded, implying a secretory correlation between them and adjacent core components. The cases of Serratia marcescens13–15and P. aeruginosa16,17 highlight extensive studies in this area. Hcp is an active marker protein of T6SS, and its presence in the supernatant is indicative of T6SS activity.18–21
Research indicates that the histone-like nucleoid structuring protein (H-NS) regulates T6SS in several clinically significant pathogens such as E. coli and Vibrio parahaemolyticus.22,23 Recent investigations demonstrate that T6SS is also closely associated with the virulence and invasiveness of A. baumannii, particularly during polymicrobial infections.24 The T6SS confers A. baumannii with the ability to outcompete and eliminate both conspecific and heterospecific bacteria, thereby establishing itself as the dominant pathogen within the host. This competitive advantage, combined with its high virulence and multidrug resistance (MDR) profile, enables A. baumannii to cause severe invasive infections, including sepsis.25 It has been reported that MDR strains of A. baumannii isolated from urine carries the large conjugative plasmids (LCP) pAB5, which encodes H-NS. It suppresses biofilm formation by inhibiting exopolysaccharide Poly-N-Acetyl-D-Glucosamine (PNAG) expression. LCP encodes multiple antibiotic resistance genes and negatively regulates T6SS, thereby impacting bacterial virulence.26 Although the functionality and pathogenicity of the T6SS in A. baumannii have been the focus of research, the regulatory role and mechanism of H-NS on T6SS in this bacterium remain to be fully elucidated. This study aims to clarify the regulatory role of H-NS on T6SS in A. baumannii through molecular biology techniques and animal model construction, thereby providing a theoretical foundation for preventing and treating severe invasive infections caused by A. baumannii in the context of mixed bacterial infections, as well as for the development of potential drug targets.
Materials and Methods
Strains and Plasmids
The bacterial strains and plasmids utilized in this study are detailed in Supplementary Table S1. The standard strain, ATCC19606 (designated as AB WT), has been maintained and preserved in our laboratory.
Construction of ATCC19606-Derived Strain
The construction of the ATCC19606 hns gene knockout strain (ABΔhns) followed the previously published protocol.27 In brief, genomic DNA was extracted from the ATCC19606 strain. Using primers containing restriction enzyme sites (Table S2), polymerase chain reaction (PCR) amplification was performed to obtain the upstream and downstream homologous arms of the hns gene. Using the plasmid pKD4 as a template, amplify the kanamycin resistance gene, and construct the linear target fragment by overlapping PCR. The overlap fragment was transformed into ATCC19606 carrying pAT04 through electroporation (1.8 kV, 200 Ω, and 25 μF in a 2 mm cuvette), and the resistance gene and plasmid were discarded to construct the ATCC19606-pAT04 strain, and confirm positive transformants by PCR. Subsequently, the linear target fragment was transformed into competent ATCC19606-pAT04 cells, screen for positive clones on Luria-Bertani (LB) agar plates containing 50 mg/L kanamycin, and successful mutants were confirmed by PCR. The pAT04 plasmid in the hns deletion mutant strain was lost after continuous passage in drug-free LB broth.
The construction of the ATCC19606 hns-overexpressing (ABhns+) strain was performed as previously described, with some modifications.28 Upstream and downstream primers with restriction sites were designed to incorporate for amplifying the full-length hns gene from the wild-type (WT) ATCC19606 strain via PCR. The PCR-amplified hns PCR and the pYMAb2 plasmid were digested with the restriction enzymes BamHI and SalI. The hns gene was then inserted into pYMAb2 using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The resulting pYMAb2-hns plasmid DNA was extracted and electroporated into competent ATCC19606 cells. Positive clones were selected on LB agar plates containing hygromycin. Genomic DNA was extracted from these clones, and the presence of the hns insert was confirmed by PCR and subsequent sequencing.
Quantitative Real-Time PCR (RT-qPCR)
RT-qPCR primers (Table S3) were designed based on the whole-genome sequence of the ATCC19606 strain from the NCBI database. The AB WT, ABΔhns and ABhns+ were inoculated on Columbia blood agar plates and incubated overnight at 37°C for 18–24 h. A single pure colony was picked and inoculated into 3–5 mL of LB liquid medium and cultured at 37°C with shaking at 180 rpm until the OD600 reached 0.6–0.8. The bacterial suspension was then collected, and total RNA was extracted using Trizol reagent. The extracted RNA was reverse transcribed into cDNA using PrimeScript RT reagent Kit (Takara, Japan), and diluted to the same concentration. Real-time quantitative PCR was performed using the TB Green® Premix Ex Taq™ Kit, following the manufacturer’s instructions, on a CFX96 real-time PCR detection system (Bio-Rad, USA). Reaction conditions were set as previously described.29 Post-amplification data analysis was performed using CFX Maestro software (Bio-Rad). The ATCC19606 strain served as the reference, and 16S rRNA was used as the endogenous control. The relative expression levels of the target gene in the ABΔhns and ABhns+ were calculated using the 2−ΔΔCt method.30
Bacterial Competition Assays
The bacterial competition assay was carried out as previously described.31 Single colonies of A. baumannii (AB WT, ABΔhns, and ABhns+) and E. coli DH5α were independently cultured in LB broth and incubated overnight at 37°C with shaking at 180 rpm. The cultures were then subcultured at a 10% ratio and grown for approximately 4 h, the bacterial suspensions were adjusted to a turbidity equivalent to the 0.5 McFarland standard. Next, 40 μL of each A. baumannii strain was thoroughly mixed with 40 μL of E. coli DH5α. Aliquots of 20 μL from the mixed suspension were spread onto LB agar plates and incubated at 37°C for 4 h. Following incubation, colonies from the LB plates were harvested, suspended in 500 μL of phosphate buffer saline (PBS), and thoroughly mixed. Serial dilutions were performed, and samples were plated onto LB agar medium containing tetracycline (for selective enumeration of A. baumannii) and LB medium without antibiotics (for total colony counts). The number of E. coli colonies was determined by subtracting the counts from the antibiotic-containing plates from the total colony counts.
Serum Bactericidal Assays
The serum bactericidal assay was performed as previously described.32 Single colonies of A. baumannii (AB WT, ABΔhns, and ABhns+) were cultured overnight in LB broth at 37°C with shaking at 180 rpm. The cultures were then subcultured at a 10% ratio, and grown for approximately 4 h, the bacterial suspensions were adjusted to match the 0.5 McFarland standard. Then, 1 mL of the bacterial suspension was centrifuged at 12,000 rpm for 2 min, the supernatant was discarded, and the pellet was resuspended in 1 mL of PBS. Next, 50 μL of the resuspended bacterial solution was mixed with 150 μL of normal mouse serum (NMS) to form the serum group, while another 50 μL of the bacterial solution was mixed with 150 μL of PBS to form the control group. The mixtures were incubated at 37°C for 3 h. For colony enumeration, six rows of EP tubes were prepared, each containing six tubes with 900 μL of PBS. Samples from both the serum and control groups were taken, and 100 μL of each sample was serially diluted 10-fold in PBS. Diluted samples were plated onto LB agar and incubated at 37°C for 18 h. Colony-forming units (CFUs) were counted to assess bacterial survival.
Construction of a Mixed Infection Mouse Sepsis Model
BALB/c mice weighing 20–22 g (6–8 weeks old) were used and housed under clean-grade laboratory conditions. The experimental strains comprised AB WT, ABΔhns, ABhns+ and DH5α.33 The experimental groups were as follows: group 1 featured a mixed infection with AB WT and DH5α, group 2 involved a mixed infection with ABΔhns and DH5α, and group 3 contained a mixed infection with ABhns+ and DH5α. Both A. baumannii and E. coli were cultured to the logarithmic phase and adjusted to a bacterial concentration of 1.5 × 107 CFU/mL. After mixing in a 1:1 ratio, 0.5 mL of the mixed bacterial solution was intraperitoneally injected into the mice to construct the mouse sepsis model.34
Following a 7-day observation period, blood samples were collected from the mice and plated on both drug-free and drug-containing plates for colony counting. To separately screen the two bacterial strains, we selected a drug to which one strain (either A. baumannii or E. coli) is naturally resistant, while the other is sensitive. In this case, we chose ampicillin, as A. baumannii is naturally resistant to it, whereas E. coli is sensitive. By comparing the colony counts of A. baumannii in the blood samples, we assessed the changes in the invasive ability of the strains across the different experimental groups.35
Histopathological Analysis of Major Organs in Mice
Following a 7-day observation period, mice that either succumbed to the infection or were euthanized underwent histopathological examination of their lungs, liver, kidneys, and other organs. The tissues were harvested and processed through a series of steps: fixation in 4% paraformaldehyde, dehydration using a graded ethanol series, infiltrated, and embedded in paraffin. The paraffin-embedded samples were sectioned at a thickness of 2 μm, deparaffinized, rehydrated, and stained with hematoxylin and eosin (H & E). All H & E-stained sections were observed under an optical microscope (Eclipse Ci-L, Nikon, Japan).36
Detection of hns and hcp Gene Expression in Mouse Blood
The expression levels of the hns and hcp genes in mouse blood were assessed across different treatment groups. Total RNA was extracted from mouse blood samples by Trizol method adhering to the manufacturer’s instructions and reverse-transcribed to produce cDNA. Subsequently, RT-qPCR was performed as previously described, employing 16S rRNA as the reference gene.37 Utilizing the 2−ΔΔCt method, the relative expression levels of the hns and hcp genes were calculated, providing insights into their transcriptional activity in response to various treatments.
TMT-Labeled Quantitative Proteomics for Screening Differential Proteins
Single colonies of AB WT and ABΔhns were used for protein extraction using the SDT lysis method,35 followed by protein quantification using the BCA method. Each group contained three biological replicate samples. Appropriate amounts of protein from each sample were digested with trypsin by the Filter Aided Sample Preparation (FASP) method. The resulting peptides were desalted using a C18 cartridge to remove impurities, lyophilized to remove residual solvents, and then resuspended in 40 μL of Dissolution buffer. The concentration of the peptides was measured at OD280. Next, 100 μg of peptides from each sample were labeled according to the instructions of the TMT labeling kit from Thermo Fisher. The labeled peptides from each group were combined and fractionated using the AKTA Purifier 100. Each fraction was separated using the Easy nLC nano-HPLC system. The separated samples were then analyzed by mass spectrometry using the Q-Exactive mass spectrometer (Thermo Fisher). The raw files from the mass spectrometry analysis were processed using Proteome Discoverer 1.4 to search against the corresponding database, resulting in protein identification and quantitative analysis. Differentially expressed proteins were screened based on a fold change greater than 1.5 (up-regulated more than 1.5-fold or down-regulated less than 0.67-fold) and a p-value <0.05.
Electrophoretic Mobility Shift Assay (EMSA)
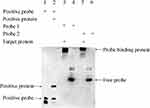
The binding interaction between H-NS and candidate genes was detected using an Electrophoretic Mobility Shift Assay (EMSA).38 Probes were synthesized by PCR. Utilizing the following primers: probe1F: 5′-TTACGCTGCGTAAGAAGCT-3′; probe1R: 5′-ACTAGCCCTAAATTATGGG-3′; probe2F: 5′-TCAAATGTATAACCAGCTG-3′; probe2R: 5′-TCAAGGTCAGCTAAAGAA-3′. The EMSA probe design is illustrated in Figure S1, with lane setting details in Figure 9. The labeled PCR products were analyzed using 1% agarose gel electrophoresis. After successful labeling, the molecular weight of the probe increased, resulting in a shifted electrophoretic band. The PCR products were purified using VAHTS DNA Clean Beads (Vazyme, Cat. No. N411-01). The labeled probe was then subjected to electrophoresis and membrane transfer. The membrane was washed, and signal detection was performed according to the LightShift® Chemiluminescent EMSA Kit, instructions (Thermo Fisher Scientific, US).
Statistical Analysis
The data were expressed as mean ± standard deviation. Statistical significance was assessed using Student’s s t-test. For all analyses, the following conventions were used: ns, not statistically significant, *P < 0.05, **P < 0.01, and ***P < 0.001. All statistical analyses were conducted using Prism 8.
Results
Construction of hns Knockout and Overexpression Strains
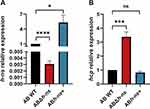
Both the ABΔhns and ABhns+ were successfully constructed, as illustrated in Figure 1A. In comparison to AB WT, the expression of the hns gene was significantly decreased in ABΔhns, whereas it was substantially elevated in ABhns+.
H-NS Negatively Regulates the Expression of T6SS Related Coding Gene hcp in A. baumannii
We analyzed the relative quantification of T6SS-related gene hcp before and after hns gene knockout and overexpression by RT-qPCR. The results revealed that the expression level of hcp, which encodes a secreted protein of the T6SS, was more than threefold higher in the ABΔhns strain compared to the AB WT strain. Conversely, there was no significant change in hcp expression in the ABhns+ strain (Figure 1B).
The Deletion of hns Confers ATCC19606 Competitive Advantage Against Commensal Flora
To assess the impact of hns gene knockout and overexpression on the competitive capacity of A. baumannii ATCC19606 against commensal flora, the survival of E. coli was evaluated after co-incubation with AB WT, ABΔhns, and ABhns+ strains for 4 h. The results indicated that compared with AB WT, E. coli survival significantly decreased in the ABΔhns group, while it increased in the ABhns+ group, indicating that the hns gene plays a crucial role in determining the competitive edge of A. baumannii ATCC19606 against commensal flora (Figure 2).
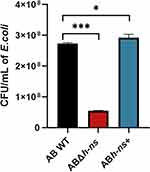 |
Figure 2 Changes of in vitro competitiveness of strains before and after hns gene knockout and overexpression. The *in the figure represents a statistical difference (*P < 0.05, ***P < 0.001). |
The Deletion of hns Increases Serum Resistance of ATCC19606
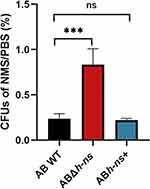
To investigate the impact of hns gene knockout and overexpression on the serum resistance of A. baumannii ATCC19606, colony counting was performed following serum treatment. The results demonstrated that the serum resistance of ABΔhns was enhanced compared with that of AB WT. A substantial difference in survival rates between ABΔhns and AB WT was observed in serum (Figure 3), suggesting that the hns gene may exert a negative regulatory effect on the serum resistance of A. baumannii.
The hns Gene Knockout Significantly Enhances the Invasiveness of A. baumannii to Blood
We constructed a co-infection septicemia mouse model with E. coli and ABΔhns, ABhns+ or AB WT, respectively. The colonies of A. baumannii in mouse blood were counted to evaluate the invasive ability of bacteria before and after hns gene knockout. The results showed that the number of ABΔhns in the blood of mice with mixed infection was significantly higher than that of AB WT (Figure 4), suggesting that hns gene deletion significantly enhances the invasive and competitive capacities of A. baumannii, making it more prone to cause bloodstream infections.
Effects of H-NS on Tissue Damage in Septicemic Mice
We observed the pathological changes in major tissues and organs of septicemia mice co-infected with E. coli and either ABΔhns, ABhns+ or AB WT (Figure 5). The results showed that the structure of endocardium, myocardium and epicardium was clear, with no obvious abnormality observed in the heart wall and heart cavity across all treatment groups. However, liver and spleen tissue were extensively necrotic. In the lung tissue, the structure of alveolar wall was loose and disordered, with numerous instances of alveolar necrosis. The renal tubules were irregularly arranged, with a large number of necrotic renal tubular epithelial cells and a small number of shed epithelial cells. There was no significant difference in the histopathology of different organs of mice in each treatment group, considering that the changes of secretion products, proteomics, metabolites, etc may occur at the molecular level, which are not intuitive in histology. Histopathological analysis revealed comparable tissue damage across groups, suggesting that H-NS may primarily influence molecular virulence factors rather than gross pathological outcomes.
 |
Figure 5 Pathology of main tissues and organs in mice co-infected with E. coli DH5α. |
Effect of H-NS on Expression of T6SS Related Coding Gene hcp in Mice
The expression levels of hns and hcp of AB WT, ABΔhns and ABhns+ in peripheral blood of mice mixed with E. coli were detected by RT-qPCR, respectively (Figure 6). The results indicated a negative correlation between the expression of hns and hcp in the peripheral blood of mice co-infected with E. coli, aligning with the in vitro findings (Figure 1), suggesting that hns can negatively regulate key genes related to T6SS.
Proteomic Analysis of AB WT and ABΔhns Strains
In order to understand the expression of related proteins following hns gene knockout, differential protein KEGG functional enrichment, volcano map and T6SS related differential protein expression analysis were performed comparing AB WT and ABΔhns. The results showed that 209 proteins were significantly upregulated and 33 proteins were significantly downregulated in ABΔhns compared to AB WT. These differentially expressed proteins were primarily involved in critical biological processes such as two-component system and biosynthesis of secondary metabolites (Figure 7A). With the difference ratio >1.5 or <0.67 and P <0.05 as the screening criteria, we found that the expression levels of T6SS-related proteins were consistently higher in ABΔhns group than in AB WT group (Figures 7B and 8), indicating that H-NS exerts an inhibitory effect on T6SS in A. baumannii.
 |
Figure 8 Heatmap of the expression of T6SS-related differential proteins. |
Regulation of H-NS on the Expression of T6SS Related Coding Gene hcp
To investigate whether H-NS regulates the hcp gene by direct binding, we purified the H-NS protein from A. baumanii ATCC19606 and performed gel migration experiments with the upstream hcp. Two specific DNA fragments, designated as C1 and C2 (depicted in Figure S1), were employed to assess H-NS binding to both the upstream and ORF regions of hcp, as illustrated in Figure 9. The results demonstrated that H-NS significantly retarded the migration of these two DNA fragments, confirming that the H-NS protein directly interacts with multiple sites in the upstream region of hcp.
Discussion
baumannii, a major pathogen in hospital-acquired infections, has garnered increasing attention due to the rise of multidrug-resistant strains.39 The World Health Organization has classified carbapenem-resistant A. baumannii as a critical priority pathogen for the development of new antibiotics.40 This bacterium is prevalent in hospital settings, yet the infections it causes often lead to severe clinical consequences, such as sepsis and other invasive infections, particularly in patients with compromised immunity. A. baumannii ranks 7th in blood specimens, accounting for 3.5%.41
T6SS is widely present in Gram-negative bacteria and significantly contributes to their pathogenicity.42 It endows bacteria with a competitive edge by transferring effector molecules, such as toxins, to neighboring pathogens or host cells, inducing cell lysis.43 During infection, T6SS causes damage to the “enemy” through a variety of mechanisms, such as destroying cell wall and membrane structure or inhibiting nucleic acid and protein formation, altering host cell signaling or regulating host immune response.44
The activation of T6SS in A. baumannii enables it to competitively inhibit and kill other bacteria, allowing it to emerge as the dominant pathogen.45 This can lead to severe invasive infections such as sepsis following mixed infections. Even with antibiotic treatment, the high pathogenicity and multidrug resistance of A. baumannii can result in severe outcomes. It can cause opportunistic nosocomial infections in immunocompromised patients, including wound infections, abdominal infections, urinary tract infections, central nervous system infections, and bacteremia, making clinical anti-infection treatment challenging.1 Therefore, understanding the specific regulatory mechanisms of T6SS activation/silencing is crucial for developing effective anti-infection therapies.46
H-NS is a globally distributed transcriptional regulator in Gram-negative bacteria that modulates bacterial biological characteristics by targeting and capturing horizontally transferred genetic material. The H-NS protein binds to specific DNA sequences within the bacterial genome, thereby “capturing” and regulating the expression of genes involved in important biological processes. T6SS-encoding genes are among them, which is a key virulence factor in many pathogenic bacteria.47 Studies have shown that H-NS regulates T6SS in various pathogens by sensing environmental conditions and controlling T6SS-related genes.48 In Vibrio parahemolyticus (V. parahemolyticus), H-NS regulates T6SS by sensing the salt concentration in the surrounding environment of the strain. As the salt concentration increases, the inhibition effect of H-NS on T6SS is relieved, enabling V. parahemolyticus to competitively kill E. coli.48 In Salmonella typhimurium (S. typhimurium), the H-NS protein causes the “silencing” of T6SS by targeting and binding T6SS-related genes. When macrophages are infected, the silencing effect is lifted, and the “sense-kill” mechanism of T6SS can enhance the pathogenicity of S. typhimurium.49 In enterohemorrhagic E. coli (EHEC), H-NS exerts total regulation on T6SS by inhibiting effect factor - katN.50 This study is the first to systematically explore the regulatory role of H-NS on T6SS in A. baumannii, revealing that H-NS inhibits T6SS expression by directly binding to the upstream region of the hcp gene. Previous studies have focused on the role of H-NS in other bacteria or on other regulatory mechanisms in A. baumannii, with little research systematically elucidating the regulatory role of H-NS on T6SS. Therefore, this study enriches the theoretical basis in this field and provides new perspectives for understanding the pathogenic mechanisms of A. baumannii and developing new anti-infection strategies.
The previous study conducted by our research group revealed that the hns gene in A. baumannii bloodstream infection isolates is highly conserved, which aligns with the findings reported elsewhere.48 According to the report, in A. baumannii, the insertion of an IS unit into the coding sequence of the hns gene results in enhanced virulence and motility, as well as a significant increase in its ability to adhere to eukaryotic cells. Transcriptomic analysis indicated that following the mutation of hns, the expression levels of various T6SS secreted proteins in the strain were significantly up-regulated, suggesting that H-NS may exert an inhibitory effect on A. baumannii T6SS, and the insertion of IS leads to H-NS mutation, thereby reducing its inhibitory effect on A. baumannii T6SS. The changes in virulence were verified through in vivo experiments using the Galleria mellonella.51 With a slight difference, in our study, through proteomic analysis, we found that the expression of T6SS-related proteins was increased in the knockout group compared with the wild type group. A mouse model of mixed infection sepsis was constructed to verify its effect on virulence, which further indicated that H-NS has an inhibitory effect on the T6SS of A. baumannii.
Based on prior research results, we hypothesized that H-NS plays an important regulatory role in the silencing or activation mechanism of A. baumannii T6SS. To validate this hypothesis, ATCC19606 hns gene knockout and over-expression strains were constructed, and a series of experiments were conducted to explore the potential regulatory targets and specific mechanisms by which H-NS regulates A. baumannii T6SS. Elucidating the regulatory pathways of H-NS on T6SS activation or silencing phenotypes in A. baumannii could provide a theoretical foundation for developing new anti-infection drugs targeting H-NS.
In this study, we constructed hns gene knockout and overexpression strains. We then conducted a series of in vitro and in vivo experiments, including RT-qPCR, bacterial competition assays, serum resistance tests, and a mouse septicemia model, to demonstrate the critical regulatory role of H-NS in A. baumannii’s competitive ability, serum resistance, and invasiveness. Furthermore, bioinformatics techniques were used to identify differentially expressed proteins regulated by H-NS, pinpointing potential targets of H-NS regulation of T6SS. Electrophoretic mobility shift assays revealed that H-NS might regulate T6SS by binding to multiple sites in the upstream region of hcp1. The H-NS-mediated “silencing switch” for T6SS activation provides a plausible explanation for A. baumannii outbreaks in hospital settings – where transient H-NS inhibition during antibiotic exposure or host stress could trigger hypervirulent phenotypes. These findings not only elucidate the multifaceted role of H-NS in A. baumannii pathogenicity but also provide strong support for developing innovative therapeutic strategies. However, unfortunately, Hcp is just a representative secreted protein of T6SS, not the entire type VI secretory system. Therefore, its more specific regulatory molecular mechanism is worthy of further exploration by researchers in the future. In future studies, researchers could explore the interaction of H-NS and other regulatory proteins in regulating T6SS or examine environmental factors that influence H-NS activity in clinical isolates. These insights could pave the way for tailored treatments against high-priority pathogens such as A. baumannii.
Conclusion
Overall, H-NS functions as a global regulatory factor in A. baumannii, modulating various biological characteristics by targeting and capturing horizontally transferred genetic material, including T6SS-encoding genes. Our study demonstrates that H-NS negatively regulates the expression of T6SS-related hcp gene in A. baumannii. The deletion of hns enhances the competitive ability and serum resistance of ATCC19606. Furthermore, the H-NS protein likely regulates T6SS by binding to multiple sites in the upstream region of hcp1. These findings highlight the multifaceted role of H-NS in the pathogenic mechanisms of A. baumannii, suggesting that H-NS could be a promising target for the developing novel clinical therapies.
Date Availability Statement
The datasets generated are available from the corresponding author on reasonable request.
Ethical Approval
This study was approved by the First Affiliated Hospital of Wenzhou Medical University Ethics Committee in Clinical Research (Acceptance Number: KY2024-R060). The mice were fed according to the Chinese National Standard for Laboratory Animals (GB14925-2010). All animal experiments were approved by Zhejiang Science and Technology Association SYXK [ID: SYXK (Zhejiang) 2018-0017] and conducted in accordance with Wenzhou Laboratory Animal Welfare and Ethics guidelines.
Acknowledgments
We thank professor Yunsong Yu for donating the plasmid pAT04 and pYMAb2 generously.28,52
Author Contributions
All authors significantly contributed to the work reported, whether through conception, study design, execution, data acquisition, analysis, or interpretation. They participated in drafting, revising, or critically reviewing the article, gave final approval of the version to be published, agreed on the chosen journal, and accepted responsibility for all aspects of the work.
Funding
This work was funded by Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022) and the Zhejiang Natural Science Fund (LY20H200004).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shi J, Cheng J, Liu S, Zhu Y, Zhu M. Acinetobacter baumannii: an evolving and cunning opponent. Front Microbiol. 2024;15:1332108. doi:10.3389/fmicb.2024.1332108
2. Tang A, Shi Y, Dong Q, et al. Prognostic differences in sepsis caused by gram-negative bacteria and gram-positive bacteria: a systematic review and meta-analysis. Crit Care. 2023;27(1):467. doi:10.1186/s13054-023-04750-w
3. Coulthurst S. The Type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165(5):503–515. doi:10.1099/mic.0.000789
4. Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15(1):9–21. doi:10.1016/j.chom.2013.11.008
5. Weber BS, Kinsella RL, Harding CM, Feldman MF. The secrets of Acinetobacter secretion. Trends Microbiol. 2017;25(7):532–545. doi:10.1016/j.tim.2017.01.005
6. Xiong C, Jiao H, Ran J, et al. A comprehensive understanding of the influence and molecular mechanism of exeA on the pathogenicity in Aeromonas hydrophila. Int J Biol Macromol. 2024;284(Pt 1):138080. doi:10.1016/j.ijbiomac
7. Colautti J, Kelly SD, Whitney JC. Specialized killing across the domains of life by the type VI secretion systems of Pseudomonas aeruginosa. Biochem J. 2025;482(1):1–15. doi:10.1042/BCJ20230240
8. Bray AS, Broberg CA, Hudson AW, et al. Klebsiella pneumoniae employs a type VI secretion system to overcome microbiota-mediated colonization resistance. Nat Commun. 2025;16(1):940. doi:10.1038/s41467-025-56309-8
9. Bezkorovayna V, Hayes BK, Gillett FN, et al. Delivery determinants of an Acinetobacter baumannii type VI secretion system bifunctional peptidoglycan hydrolase. mBio. 2025;16(2):e0262724. doi:10.1128/mbio.02627-24
10. Bongiovanni TR, Latario CJ, Le Cras Y, et al. Assembly of a unique membrane complex in type VI secretion systems of Bacteroidota. Nat Commun. 2024;15(1):429. doi:10.1038/s41467-023-44426-1
11. Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–353. doi:10.1038/nature12453
12. Durand E, Nguyen VS, Zoued A, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523(7562):555–560. doi:10.1038/nature14667
13. Jia F, Peng X, Yang X, et al. PqqF inhibits T6SS secretion by decreasing the pH in Serratia marcescens FS14. FEMS Microbiol Lett. 2024;371. doi:10.1093/femsle/fnae047
14. Wang X, Sun B, Xu M, et al. Crystal structure of the periplasmic domain of TssL, a key membrane component of Type VI secretion system. Int J Biol Macromol. 2018;120(Pt B):1474–1479. doi:10.1016/j.ijbiomac.2018.09.166
15. Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016;12(6):e1005735. doi:10.1371/journal.ppat.1005735
16. Pissaridou P, Allsopp LP, Wettstadt S, Howard SA, Mavridou DAI, Filloux A. The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc Natl Acad Sci U S A. 2018;115(49):12519–12524. doi:10.1073/pnas.1814181115
17. Hall CW, Zhang L, Mah TF. PA3225 is a transcriptional repressor of antibiotic resistance mechanisms in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61(8). doi:10.1128/aac.02114-16
18. Howard SA, Furniss RCD, Bonini D, et al. The breadth and molecular basis of Hcp-driven type VI secretion system effector delivery. mBio. 2021;12(3):e0026221. doi:10.1128/mBio.00262-21
19. Fei N, Ji W, Yang L, et al. Hcp of the type VI secretion system (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int J Mol Sci. 2022;23(17):9632. doi:10.3390/ijms23179632
20. Alves JA, Leal FC, Previato-Mello M, da Silva Neto JF. A quorum sensing-regulated type VI secretion system containing multiple nonredundant VgrG proteins is required for interbacterial competition in chromobacterium violaceum. Microbiol Spectr. 2022;10(4):e0157622. doi:10.1128/spectrum.01576-22
21. Proutière A, Drebes Dörr NC, Bader L, et al. Sporadic type VI secretion in seventh pandemic Vibrio cholerae. Microbiology. 2023;169(5). doi:10.1099/mic.0.001329
22. Dong JF, Liu CW, Wang P, Li L, Zou QH. The type VI secretion system in Acinetobacter baumannii clinical isolates and its roles in antimicrobial resistance acquisition. Microb Pathog. 2022;169:105668. doi:10.1016/j.micpath.2022.105668
23. Lin Y, Zhao D, Huang N, et al. Clinical impact of the type VI secretion system on clinical characteristics, virulence and prognosis of Acinetobacter baumannii during bloodstream infection. Microb Pathog. 2023;182:106252. doi:10.1016/j.micpath.2023.106252
24. Bai B, Eales BM, Huang W, et al. Clinical and genomic analysis of virulence-related genes in bloodstream infections caused by Acinetobacter baumannii. Virulence. 2022;13(1):1920–1927. doi:10.1080/21505594.2022.2132053
25. Kandolo O, Cherrak Y, Filella-Merce I, et al. Acinetobacter type VI secretion system comprises a non-canonical membrane complex. PLoS Pathog. 2023;19(9):e1011687. doi:10.1371/journal.ppat.1011687
26. Benomar S, Di Venanzio G, Feldman MF. Plasmid-encoded H-NS controls extracellular matrix composition in a modern Acinetobacter baumannii urinary isolate. J Bacteriol. 2021;203(21):e0027721. doi:10.1128/jb.00277-21
27. Qian C, Ma Z, Feng L, et al. Emergence of tet(X2) in Acinetobacter pittii confers clinical resistance to tigecycline. J Antimicrob Chemother. 2023;78(6):1543–1546. doi:10.1093/jac/dkad133
28. Zhang L, Fu Y, Han X, et al. Phenotypic variation and carbapenem resistance potential in OXA-499-producing Acinetobacter pittii. Front Microbiol. 2020;11:1134. doi:10.3389/fmicb.2020.01134
29. Singkham-In U, Higgins PG, Wannigama DL, Hongsing P, Chatsuwan T. Rescued chlorhexidine activity by resveratrol against carbapenem-resistant Acinetobacter baumannii via down-regulation of AdeB efflux pump. PLoS One. 2020;15(12):e0243082. doi:10.1371/journal.pone.0243082
30. Nguyen AT, Pham SC, Ly AK, Nguyen CVV, Vu TT, Ha TM. Overexpression of bla OXA-58 gene driven by ISAba3 is associated with imipenem resistance in a clinical Acinetobacter baumannii isolate from Vietnam. Biomed Res Int. 2020;2020:7213429. doi:10.1155/2020/7213429
31. Li L, Wang YN, Jia HB, et al. The type VI secretion system protein AsaA in Acinetobacter baumannii is a periplasmic protein physically interacting with TssM and required for T6SS assembly. Sci Rep. 2019;9(1):9438. doi:10.1038/s41598-019-45875-9
32. King LB, Swiatlo E, Swiatlo A, McDaniel LS. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol Med Microbiol. 2009;55(3):414–421. doi:10.1111/j.1574-695X.2009.00538.x
33. Li C, Xue H, Du X, Nyaruaba R, Yang H, Wei H. Outer membrane vesicles generated by an exogenous bacteriophage lysin and protection against Acinetobacter baumannii infection. J Nanobiotechnology. 2024;22(1):273. doi:10.1186/s12951-024-02553-x
34. Mansouri M, Sadeghpoor M, Abdollahi M, Vafaei AJ, Jalali Nadoushan M, Rasooli I. Synergistic immunoprotection by Oma87 and Bap against Acinetobacter baumannii sepsis model. Int Immunopharmacol. 2023;122:110650. doi:10.1016/j.intimp.2023.110650
35. Fereshteh S, Ajdary S, Sepehr A, et al. Immunization with recombinant DcaP-like protein and AbOmpA revealed protections against sepsis infection of multi-drug resistant Acinetobacter baumannii ST2(Pas) in a C57BL/6 mouse model. Microb Pathog. 2023;174:105882. doi:10.1016/j.micpath.2022.105882
36. Wu D, Zhou S, Hu S, Liu B. Inflammatory responses and histopathological changes in a mouse model of Staphylococcus aureus-induced bloodstream infections. J Infect Dev Ctries. 2017;11(4):294–305. doi:10.3855/jidc.7800
37. Zasada M, Madetko-Talowska A, Revhaug C, et al. Short- and long-term impact of hyperoxia on the blood and retinal cells’ transcriptome in a mouse model of oxygen-induced retinopathy. Pediatr Res. 2020;87(3):485–493. doi:10.1038/s41390-019-0598-y
38. Cui S, Xiao J, Wang Q, Zhang Y. H-NS binding to evpB and evpC and repressing T6SS expression in fish pathogen Edwardsiella piscicida. Arch Microbiol. 2016;198(7):653–661. doi:10.1007/s00203-016-1226-4
39. Ayoub Moubareck C, Hammoudi Halat D. Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics. 2020;9(3):119. doi:10.3390/antibiotics9030119
40. WHO bacterial priority pathogens list, 2024. Available from: https://www.who.int/publications/i/item/9789240093461.
41. CHINET 2023 data. Available from: https://www.chinets.com/Data/AntibioticDrugFast.
42. Chen Z, Mao Y, Song Y, et al. Refined egoist: the toxin-antitoxin immune system of T6SS. Microb Pathog. 2024;196:106991. doi:10.1016/j.micpath.2024.106991
43. Singh RP, Kumari K. Bacterial type VI secretion system (T6SS): an evolved molecular weapon with diverse functionality. Biotechnol Lett. 2023;45(3):309–331. doi:10.1007/s10529-023-03354-2
44. Allsopp LP, Bernal P. Killing in the name of: T6SS structure and effector diversity. Microbiology. 2023;169(7). doi:10.1099/mic.0.001367
45. Luo J, Chu X, Jie J, et al. Acinetobacter baumannii kills fungi via a type VI DNase effector. mBio. 2023;14(1):e0342022. doi:10.1128/mbio.03420-22
46. Repizo GD, Gagné S, Foucault-Grunenwald ML, et al. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One. 2015;10(9):e0138265. doi:10.1371/journal.pone.0138265
47. Liu Y, Zhou M, Bu Y, et al. Lysine acetylation regulates the AT-rich DNA possession ability of H-NS. Nucleic Acids Res. 2024;52(4):1645–1660. doi:10.1093/nar/gkad1172
48. Salomon D, Klimko JA, Orth K. H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology. 2014;160(Pt 9):1867–1873. doi:10.1099/mic.0.080028-0
49. Brunet YR, Khodr A, Logger L, et al. H-NS Silencing of the Salmonella pathogenicity island 6-encoded type VI secretion system limits Salmonella enterica serovar typhimurium interbacterial killing. Infect Immun. 2015;83(7):2738–2750. doi:10.1128/iai.00198-15
50. Wan B, Zhang Q, Ni J, et al. Type VI secretion system contributes to Enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog. 2017;13(3):e1006246. doi:10.1371/journal.ppat.1006246
51. Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LD, Paulsen IT, Brown MH. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun. 2013;81(7):2574–2583. doi:10.1128/iai.00065-13
52. Tucker AT, Nowicki EM, Boll JM, et al. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio. 2014;5(4):e01313–14. doi:10.1128/mBio.01313-14
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.