Back to Journals » International Journal of General Medicine » Volume 18
The Prognostic Significance of TMSB4X in Glioma Patients
Authors Li S, Fan T, Hao Z, Zhang Z, Han X, Li J, Wang Y, Zhou H, Xue X
Received 8 February 2025
Accepted for publication 23 May 2025
Published 6 June 2025 Volume 2025:18 Pages 2923—2940
DOI https://doi.org/10.2147/IJGM.S521390
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Dana Kristjansson
Sijie Li,1 Tianyi Fan,1 Zengyao Hao,1 Zizhou Zhang,1 Xuetao Han,1 Jiayuan Li,1 Yu Wang,1 Huandi Zhou,1– 3 Xiaoying Xue1,3
1Department of Radiotherapy, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China; 2Department of Central Laboratory, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 3Hebei Key Laboratory of Etiology Tracing and Individualized Diagnosis and Treatment for Digestive System Carcinoma, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China
Correspondence: Xiaoying Xue, Department of Radiotherapy, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China, Email [email protected]
Objective: The goal of this study was to analyze in depth the importance of the thymosin beta 4 X-linked gene (TMSB4X) in the disease process of gliomas for the prediction of patient prognosis.
Methods: We explored the expression of TMSB4X by analyzing datasets from The Cancer Genome Atlas (TCGA) datasets and the Chinese Glioma Genome Atlas (CGGA). Tumor sample tissues and corresponding paracancerous tissues of glioma patients were collected, and TMSB4X expression in glioma tissues was analyzed using Real-Time PCR to investigate its prognostic significance in glioma patients. In addition, we have plotted the ROC curves for the survival rate, performed univariate and multivariate Cox analyses and constructed nomogram. Differential expression analyses were performed on the basis of median expression levels of the TMSB4X gene, and Intuitive graphical presentation and visualisation of the 403 proteins screened were carried out using the Cytoscape software platform. (version: 3.9.1). The top 50 core genes were then screened using the cyto Hubba algorithm, these genes were analyzed using The Gene Ontology Database (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). TCGA datasets was utilized for Gene Set Enrichment Analysis (GSEA) to obtain potential NF-κB-related pathways in gliomas.
Results: Independent prognostic analyses show that the TMSB4X gene was identified as an independent prognostic indicator for gliomas. In GOKEGG and GSEA analyses, the analyses indicate that TMSB4X may play a crucial role in glioma patients by up-regulating I-kappaB kinase/NF-kappaB signaling. In glioma patients, upregulation of the I-kappaB kinase and NF-kappaB signalling pathways plays a crucial role, which may be linked to CASP1.
Conclusion: The findings showed that TMSB4X was identified as an potential independent risk factor in the prognosis of glioma patients, a finding that implies that TMSB4X has the potential to serve as a key biomarker for predicting the prognostic status of glioma patients.
Keywords: thymosin beta 4 X-linked, glioma, prognosis
Introduction
Gliomas, the most common primary malignant tumors in the central nervous system, account for approximately 81% of all malignant brain tumors. Their invasive growth patterns and marked heterogeneity contribute to clinical challenges such as poor therapeutic response, high recurrence rates, and short patient survival.1,2 These cancers develop from astrocytes, oligodendrocytes, or ependymal cells and show an infiltrative growth pattern in which the tumor boundaries are not clearly recognizable. World Health Organization (WHO) classifies gliomas as Grade II–IV, depending on the pathophysiological properties of gliomas, survival times span a wide range, from patients with WHO grade II gliomas, which may have a survival of more than 10 years, to patients with WHO grade IV gliomas, which may have a survival of only a few months.3,4 The most aggressive subtype, glioblastoma, accounts for 45% of all gliomas, and the overall 5-year survival rate for patients is 5%.5 Although the treatments for gliomas cover a wide range of methods such as surgery, chemotherapy, radiotherapy and the emerging immunotherapy, the overall survival of patients is relatively short. This phenomenon likely stems from two major characteristics of gliomas themselves: first, their high degree of heterogeneity, and second, their complex epigenetic mechanisms. Together, these two major factors may contribute to the difficulties in the treatment of gliomas and the limited survival of patients.6 It is therefore important to find effective prognostic markers in order to develop more effective treatments, which is also the key to prolong the survival of glioma patients and improve their quality of life. The clinical heterogeneity of gliomas is intricately linked to their complex molecular characteristics. Molecular biomarkers such as isocitrate dehydrogenase (IDH) mutations, 1p/19q codeletion, and O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation have been incorporated into diagnostic and prognostic evaluation frameworks.7 For instance, significant survival disparities persist among IDH-wildtype glioma patients, suggesting that additional molecular drivers may contribute to disease progression.8 However, existing biomarkers still exhibit limitations in guiding personalized therapeutic decision-making. Moreover, the dynamic regulatory networks of intercellular communication within the tumor microenvironment (TME)—particularly interactions between tumor cells and stromal components (eg, fibroblasts, immune cells)—are increasingly recognized as critical determinants of therapeutic resistance.9 Therefore, exploring novel molecular mechanisms underlying the malignant progression and microenvironment remodeling of gliomas is critical for overcoming current therapeutic bottlenecks.
Thymosin beta 4 is a member of the thymosin family, a highly conserved, hormone-like small 44 amino acid polypeptide.10 Thymosin beta 4 promotes angiogenesis, increases the viability of endothelial progenitor cells (EPCs), triggers cell proliferation and migration as well as the formation of intracellular capillary-like structures, inhibits apoptosis and inflammation.11 TMSB4X not only plays a role as a coregulator of fibroblast phenotype, but also plays a key role in promoting intercellular communication between fibroblasts and PC cells.12 There are many studies on the link between TMSB4X and cancer. TMSB4X expression was abnormally high in all types of thyroid cancer, and high TMSB4X expression correlated with advanced features of the disease.13 In prostate cancer, fibroblast TMSB4X opposes fibroblast reprogramming and can critically regulate intercellular communication.12 TMSB4X enhances metastasis of diffuse-type gastric cancer and growth of stomach organoid-generated tumor and promotes tumor clonality and antitumorigenicity.14 In ovarian cancer, increased TMSB4X expression accelerates ADSC-mediated proliferation, invasion and migration.15 The study also revealed that TMSB4X may be an effective target for the treatment of head and neck squamous cell carcinoma (HNSCC) and cervical cancer.16,17 Nevertheless, our current understanding of how TMSB4X is involved in the developmental process of gliomas and the molecular mechanisms behind it is still relatively scarce. The present study aims to meticulously investigate the specific molecular mechanisms underlying the prognostic impact of TMSB4X on gliomas.
Materials and Methods
Human Pan-Cancer Analysis and Sample Information
Acquisition of integrated and normalized pan-cancer dataset: We acquired a comprehensive unified and standardized broad cancer dataset, TCGA TARGET GTEx (PANCAN, N = 19131, G = 60,499), from the UCSC (https://xenabrowser.net/) database. Subsequently, we specifically extracted the expression information of the TMSB4X gene in these samples. On this basis, we further refined the sample classification, including Solid Tissue Normal, Primary Solid Tumor, Primary Tumor, Normal Tissue, Primary Blood Derived Cancer - Bone Marrow, Primary Blood Derived Cancer - Peripheral Blood. For the convenience of data processing, we performed a log2(x+0.001) logarithmic transformation for all expression values, and finally excluded cancer samples with less than 3 samples in single cancer. Finally, the expression data of 34 cancer types (GBM, GBMLGG, LGG, UCEC, BRCA, CESC, LUAD, ESCA, STES, KIRP, KIPAN, COAD, COADREAD, PRAD, STAD, HNSC, KIRC, LUSC, LIHC, WT, SKCM, BLCA, THCA, READ, OV, PAAD, TGCT, UCS, ALL, LAML, PCPG, ACC, KICH, and CHOL) were obtained to study the expression of TMSB4X in pan-cancer tumors and normal samples.
In addition to screening sample sources such as peripheral blood-derived cancer (TCGA-LAML), primary tumors, metastases from TCGA-SKCM, primary bone marrow-derived cancer, primary solid tumors, and recurrent bone marrow-derived cancer samples, we also acquired data from TCGA prognostic studies previously published in Cell, which are renowned for their high-quality prognosis datasets. We further obtained TARGET follow-up data from the UCSC cancer browser (https://xenabrowser.net/datapages/).18 We additionally applied a log2(x+0.001) transformation to each expression value and omitted cancer samples where the count was fewer than 10 within a particular cancer type. Finally, we obtained expression data and overall survival data of 38 cancer types (GBM, GBMLGG, LGG, UCEC, BRCA, CESC, LUAD, ESCA, STES, SARC, KIRP, KIPAN, COAD, COADREAD, PRAD, STAD, HNSC, KIRC, LUSC, THYM, LIHC, SKCM, BLCA, SKCM-M, THCA, MESO, READ, OV, UVM, PAAD, TGCT, UCS, LAML, PCPG, ACC, DLBC, KICH, CHOL), expression data, disease-specific survival data and progression-free interval data of 37 cancer types (GBM, GBMLGG, LGG, UCEC, BRCA, CESC, LUAD, ESCA, STES, SARC, KIRP, KIPAN, COAD, COADREAD, PRAD, STAD, HNSC, KIRC, LUSC, THYM, LIHC, SKCM, BLCA, THCA, SKCM-M, MESO, READ, OV, UVM, PAAD, TGCT, UCS, PCPG, ACC, DLBC, KICH, CHOL). After downloading the harmonized pan-cancer dataset from the UCSC database (TCGA Pan-Cancer, PANCAN, N = 10535, G = 60,499), we extracted the expression data for the TMSB4X gene from each sample, focusing specifically on those derived from primary blood-related cancers (Peripheral Blood) and primary tumors. We then applied a log2(x+0.001) transformation to normalize each expression value. Subsequently, we filtered out any tumor types that had fewer than 3 samples. Ultimately, we obtained expression data for 13 cancer types (GBMLGG, LGG, CESC, ESCA, STES, KIPAN, STAD, UCEC, HNSC, KIRC, LIHC, PAAD, CHOL) to explore the differences in TMSB4X expression across various grades of pan-cancer.
Specimens
Human tumor samples were continuously recruited at the Second Hospital of Hebei Medical University from November 2019 to March 2025. Collect tumor and normal tissue samples from 6 patients with gliomas who underwent their first surgery and have not previously received radiotherapy or chemotherapy, 32 tumor tissue specimens from glioma patients in the same condition were collected, then store them at −80 °C for later use. Human glioma tissues were deemed as exempt tissues by the Ethics Committee for Human Investigation of the Second Hospital of Hebei Medical University.
RNA Isolation and qPCR
Corresponding cancer and paracancerous tissues of glioma patients were completely dissolved in TRIzol reagent (Invitrogen) in a homogenizer, and total RNA was extracted.
The extracted total RNA concentration was measured with a NanoDrop 2000 instrument. cDNA was synthesized utilizing a reverse transcription kit sourced from Thermo Fisher Scientific, located in Waltham, MA, USA. The qPCR system (BioRad’s CFX96TM Option Module, United States) was used to conduct Quantitative PCR, employing SYBR Green from YEASEN, Shanghai, China (Hieff qPCR SYBR Green Master Mix). The fold change was calculated using the formula 2−[(Ct of target gene) − (Ct of GAPDH)] method. The sequences of the primers are as follows: TMSB4X forward: CCAGACTTCGCTCGTACTCG;TMSB4X reverse: AAAGGGGCAGCACAGTCATT;GAPDHforward:CATGAGAAGTATGACAACAGCCT; GAPDH reverse: AGTCCTTCCACGATACCAAAGT.
The Expression Analysis of TMSB4X
We downloaded the TCGA datasets to analyze the expression of TMSB4X in tumor and normal tissues, then verified it by Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) and Human Protein Atlas online analysis (https://www.proteinatlas.org/).
Validation of Prognostic Signal of TMSB4X and Correlation Analysis of Clinicopathological Features
We based our prognostic analyses on two important databases, The Cancer Genome Atlas (TCGA at https://portal.gdc.cancer.gov), which covers 703 patients, and The Chinese Glioma Genome Atlas (CGGA at http://www.cgga.org.cn/), which includes 693 patients. We divided all the patients into two groups, the TMSB4X high expression group and the TMSB4X low expression group, based on the median TMSB4X expression level. To validate the association between TMSB4X expression and glioma prognosis, we used the Kaplan-Meier (KM) survival analysis method, which was further validated in conjunction with the receiver operating characteristic curve (ROC). We performed a detailed analysis of TMSB4X expression levels in 703 patients from The Cancer Genome Atlas (TCGA) database based on several clinicopathological features, such as WHO grading, patient age, IDH mutation status, 1p19q co-deletion status, and primary treatment outcomes. Similarly, we also explored the TMSB4X expression levels of 693 patients based on the Chinese Glioma Genome Atlas (CGGA) dataset, taking into account several clinicopathological features such as WHO classification, IDH mutation status, patient age, and 1p19q co-deletion status.
Differentially Expressed Genes Analysis
The mRNA sequencing data for glioma was obtained and organized from the TCGA datasets. These data were then categorized into high- and low-expression groups, based on the median TMSB4X expression level, and subjected to differential expression analysis. By applying the criteria of adj. p value < 0.05 and |logFC| > 2, we identified significantly up-regulated and down-regulated Differentially Expressed Genes (DEGs). To visualize the results of this analysis, a volcano plot was created using the “ggplot2” software package.
Core Genes Screening and Pathway Mechanism Analysis
Input the selected DEGs in STRING (https://string-db.org/), set the minimum required interaction score to medium confidence (0.4), and hide disconnected nodes in the network, the 403 selected proteins were visualized using Cytoscape software (version: 3.9.1). Then we used the cyto Hubba algorithm to select the top 50 core genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to analyze the potential mechanism of TMSB4X function with the “cluster Profiler” software package. In addition, we also adopted the method of GSEA (gene set enrichment analysis) to dig deeper into the situation related to functional enrichment. Specifically, we consider a genome to be significantly enriched when NES > 1, p < 0.05, FDR < 0.05.
Statistical Analysis
Data was analysed using R software (version: 4.2.1, http://www.r-project.org/). To assess whether there is a significant difference in the expression level of the TMSB4X gene between glioma tissues and normal tissues, Wilcoxon rank sum test was used for the analysis. In order to determine whether the expression levels of TMSB4X genes were significantly different between glioma tissues and normal tissues, we used the Wilcoxon rank sum test as an analytical tool. To further confirm the specific expression status of the TMSB4X gene in the 6 pairs of samples, we implemented qPCR experiments for verification and set the statistical significance level at p < 0.05 as the basis for judging that the expression differences were statistically significant. To explore the potential association between TMSB4X expression levels and patients’ clinical characteristics, we applied the Kruskal–Wallis test as well as the Wilcoxon rank sum test for data analysis. Further, to assess the prognostic impact of TMSB4X, we performed prognostic analyses using the Kaplan-Meier survival curve method and the Cox proportional risk regression model for overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI), p < 0.05 indicated that the observed differences were statistically significant.
TMSB4X expression in 6 pairs of samples was verified by qPCR, and p < 0.05 was considered a statistically significant difference. The relationship between TMSB4X expression and clinical characteristics was evaluated using both the Kruskal–Wallis test and the Wilcoxon rank-sum test. For prognostic analysis, which encompassed overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI), the Kaplan-Meier method and Cox regression were employed, p < 0.05 indicates a statistically significant difference. Using qPCR technology, we validated TMSB4X gene expression levels in 32 samples. For progression-free survival (PFS) analysis, the Kaplan-Meier method and Cox regression were employed, p < 0.05 indicates a statistically significant difference.
In the process of mechanism analysis, we used GSEA software. We determined the results from the analysis to be statistically significant if they simultaneously met the three conditions, NES > 1, p < 0.05 and FDR < 0.05.
Ethical Statement
Our research has been approved by the Ethics Committee of the Second Hospital of Hebei Medical University. Our research strictly adheres to the principles and guidelines of the Helsinki Declaration, ensuring that the rights and interests of all human subjects involved in the study are fully respected and protected. In the process of research design and implementation, we adhere to the voluntary principle of informed consent, ensuring that each participant fully understands the research objectives, methods, potential risks, and potential benefits, and makes participation decisions without undue external influence. At the same time, we are committed to maintaining the privacy and anonymity of our subjects, ensuring that the collection, processing, and storage of research data comply with ethical standards and legal requirements. In summary, our research is conducted under ethical review and supervision, fully complying with the ethical principles advocated by the Helsinki Declaration, committed to promoting the integrity and progress of scientific research, while safeguarding the dignity and rights of human subjects.
Results
TMSB4X Expression in Multiple Cancer Types and Its Impact on Prognosis
Figure 1 is the workflow for our study. We obtained a standardized pan-cancer dataset from the UCSC database and went through a screening process to finalize the expression data for 34 cancers. On this basis, we further compared the differences in the expression levels of TMSB4X between tumor tissues and normal tissues. As shown in Figure 2A, TMSB4X expression levels were significantly higher in GBM, GBMLGG, LGG, STES, KIRP, KIPAN, COAD, COADREAD, STAD, HNSC, KIRC, LIHC, WT, BLCA, THCA, OV, PAAD, TGCT, ALL, and CHOL tissues compared to normal tissues. However, compared to normal tissues, it showed a trend of down-regulated expression in LUAD, PRAD, LUSC, SKCM, LAML, and PCPG. We performed differential analysis of TMSB4X expression in different tumour types within each clinical staging sample and showed that in 6 tumours, namely GBMLGG, LGG, STES, STAD, HNSC, and PAAD, the TMSB4X expression was statistically significantly different in the 6 tumours. We analyzed the differences in TMSB4X expression in samples from a wide range of tumours at various clinical stages, and in particular, significant differences in expression levels were observed in 6 tumours: GBMLGG, LGG, STES, STAD, HNSC, and PAAD (Figure 2B).
 |
Figure 1 The workflow chart of the study. |
Further, to investigated the specific role and impact of TMSB4X expression levels on the survival prognosis of pan-cancer patients, we performed several analyses including OS, DSS, and PFI. Specifically, in the OS analysis, we found that patients with high TMSB4X expression in 9 tumour types, namely GBMLGG, LGG, ESCA, STES, KIPAN, STAD, LIHC, UVM, and PAAD, had a significantly worse prognosis; on the contrary, in 3 tumours, namely SKCM, SKCM-M, and OV, patients with low TMSB4X expression showed a poorer prognosis (Figure 2C). In DSS analysis, high expression of TMSB4X is related to poor prognosis in 7 tumor types (GBMLGG, LGG, ESCA, STES, KIPAN, UVM, PAAD), additionally, in SKCM, THCA, SKCM-M, and OV, low expression of TMSB4X is related to poor prognosis (Figure 2D). As for PFI analysis, among the 10 types of tumor (GBMLGG, LGG, ESCA, STES, KIPAN, PRAD, HNSC, GBM, UVM, PAAD), the high expression had a poor prognosis. Moreover, a poor prognosis for low expression was observed in SKCM and SKCM-M (Figure 2E).
In Gliomas, A Significant Increase in the Expression Level of TMSB4X was Observed
Our study, utilizing the TCGA datasets, showed that compared with normal samples, TMSB4X expression was significantly increased in glioma samples (p < 0.001, Figure 3A). Apart from this, we used the website Gene Expression Profiling Interactive Analysis (GEPIA) as a platform to conduct the corresponding online data analysis and obtained similar results, and we saw that TMSB4X had significant high expression in GBM (Figure 3B). We extracted cancer and paracancerous tissues of 6 glioma patients, and extracted RNA for qPCR experiments. It was observed that TMSB4X expression in tumor tissues was greatly higher than that in adjacent cancer tissues (p < 0.05, Figure 3C), further verifying the high expression of TMSB4X in gliomas. In addition, we conducted an online analysis on The Human Protein Atlas website to verify the high expression of TMSB4X at the protein level in glioma (Figure 3D and E).
For Glioma Patients with High Level Expression of TMSB4X, They Tend to Have a Poorer Prognosis
In the survival prognostic analysis using the TCGA dataset, we comprehensively considered the 3 important indicators of OS, DSS to PFI. Through in-depth analyses, we found that among glioma patients, those who exhibited high TMSB4X expression characteristics generally had a poorer prognostic status (p < 0.001, Figure 4A–C). The OS analysis described above was verified by the analysis of CGGA dataset (p < 0.001, Figure 4D). We selected tumour tissue samples from 32 glioma patients and analysed the expression level of TMSB4X in these samples by RNA extraction and qPCR experimental methods, relevant clinical information of all patients is shown in Table 1. Kaplan-Meier survival analysis demonstrated that the median PFS was significantly shorter in the high-expression group compared to the low-expression group (Figure 4E, p = 0.007).
 |
Table 1 Relevant Clinical Information on the Patient Samples We Collected |
To further validate the potential of TMSB4X as a prognostic marker, we constructed a ROC curve analysis using the TCGA dataset. The results showed that TMSB4X performed well in predicting 1, 3 and 5 year survival rates with AUC values as high as 0.854, 0.870 and 0.809, respectively (Figure 4F), which indicated that TMSB4X had high predictive accuracy. To enhance the reliability of the conclusions, we also used the CGGA dataset for independent validation. In the ROC curve analysis of the CGGA dataset, TMSB4X also demonstrated its potential as a prognostic marker, with AUC values of 0.774, 0.764, and 0.699 in predicting 1, 3, and 5 year survival, respectively (Figure 4G), which was slightly lower than the results of the TCGA dataset but still maintained a relatively high predictive efficacy.
Exploring Potential Correlations Between TMSB4X Expression Levels and Clinicopathological Features
To further understand the impact of TMSB4X on the prognosis of gliomas, we analysed 7 clinicopathological features based on the TCGA dataset (p < 0.001, Table 2). As shown in Figure 5A, the expression level of TMSB4X showed an increasing trend as the WHO classification level increased (p < 0.001), and in addition, it was higher in patients over 60 years of age (p < 0.001, Figure 5B). The expression of TMSB4X was lower in patients with IDH mutation and co-expression of 1p19q (p < 0.001, Figure 5C and D). In terms of primary therapy outcomes, TMSB4X expression was higher in patients in the progressive disease (PD) group than in the other three groups (SD, PR, CR) (p < 0.001, Figure 5E). Furthermore, we verified the above conclusions by analyzing the CGGA datasets (p < 0.001, Figure 5F–I), the specific data analysis is shown in Table 3.
 |
Table 2 Relationship Between TMSB4X Expression and Clinicopathological Features Based on the TCGA Datasets |
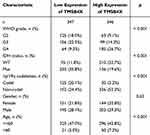 |
Table 3 Relationship Between TMSB4X Expression and Clinicopathological Features Based on the CGGA Datasets |
Univariate and Multivariate Regression Analysis and Establishment of Nomogram
We conducted univariate and multivariate regression analysis based on TCGA datasets. Univariate regression analysis showed that IDH mutation status, WHO grade, 1p19q expression status, age, and the level of TMSB4X expression is notably associated with survival prognosis (HR = 2.438; 95% CI [2.191–2.712]; p < 0.001); multivariate regression analysis showed that IDH mutation status, WHO grade, age, and TMSB4X expression level (HR =1.225; 95% CI [1.014–1.481]; p = 0.035), were significantly correlated with survival prognosis (Table 4). The study showed that TMSB4X was established as an independent and prognostic marker of predictive value in gliomas. To reinforce this finding, we validated the results of univariate and multivariate regression analyses with the help of the CGGA dataset (Figure 6A and B). Univariate and multivariable Cox regression analyses (adjusted for age and WHO grade) based on qPCR expression profiles and clinical data from our single-center cohort of 32 glioma patients demonstrated that elevated TMSB4X expression independently predicted worse PFS (Table 5, HR =1.345; 95% CI [1.064–1.700]; p = 0.013). Subsequently, based on the rich clinical features contained in the TCGA dataset, such as the mutation status of IDH, WHO grade, the expression pattern of 1p19q, patient age, gender, and the expression of TMSB4X, we constructed a quantitative prediction model, which was visually presented in graphical form (Figure 6C). Ultimately, in the group of patients evaluated for 3-year survival, we observed that the survival times predicted by the model showed a high degree of agreement with the actual survival times, which was further confirmed and validated by the excellent agreement of the calibration curves (Figure 6D–F).
 |
Table 4 Univariate and Multivariate Regression Analysis Based on the TCGA Datasets |
 |
Table 5 Univariate and Multivariate Regression Analysis Based on Patient Data from Our Center |
Screening of Differentially Expressed Genes and Enrichment Analysis
We performed a single gene differential analysis of TMSB4X and identified a total of 470 DEGs (Figure 7A). Then we used STRING online analysis to analyze DEGs and obtained a protein-protein interaction (PPI) network of 403 proteins, which was visualized using Cytoscape software (Figure 7B). We selected the top 50 core genes via the cyto Hubba algorithm (Figure 7C). GO KEGG analysis of these core genes showed that biological process (BP) of these core genes were enriched in antigen processing and presentation, viral life cycle, and positive regulation of I-kappaB kinase/NF-kappaB signaling. It was enriched in endopeptidase complex in the process of cellular component (CC); enriched KEGG pathways include pathetic Escherichia coli infection, bladder cancer, p53 signaling pathway, and viral mycocarditis (Figure 7D). Our GSEA analysis of up-regulated and down-regulated genes was similarly enriched in these processes (Figure 7E–K). By visualizing the intersection of genes corresponding to the positive regulation of I-kappaB kinase/NF kappaB signaling biological process (BP) enriched by the above two analysis methods, we can obtain two core genes, CASP1 and FASLG (Figure 7L).
Spearman Correlation Analysis of Core Genes, Expression in Glioma, and their Relationship with Survival and Prognosis
There was a significant correlation between the expression of the core gene CASP1 and the expression of TMSB4X (R = 0.855, p < 0.001, Figure 8A). Through in-depth analysis of the TCGA dataset, we observed significantly higher CASP1 expression levels in glioma samples compared to normal brain tissue samples (p < 0.01, Figure 8B). To further validate this finding, we performed an online analysis using the GEPIA website, which showed that CASP1 expression in tumour samples was significantly higher than that in normal samples in both GBM and LGG (Figure 8C). Overall survival (OS) analysis based on the TCGA dataset revealed that the group of patients with high CASP1 expression exhibited a poorer prognostic profile (p < 0.001, Figure 8D), a finding that was similarly validated in analyses of DSS and PFI (p < 0.001, Figure 8E and F). On the other hand, there is a notable correlation between the expression of the core gene FASLG and the expression of TMSB4X (R = 0.552, p < 0.001, Figure 8G). However, the expression levels of FASLG did not show significant differences between glioma and normal samples according to the TCGA dataset and the results of the GEPIA online analysis (Figure 8H and I). High expression of FASLG was equally predictive of a poorer patient prognosis in OS, DSS and PFI analyses assessing survival outcomes (p < 0.001, Figure 8J–L).
Discussion
In recent years, numerous studies have focused on an approach called pan-cancer analysis, which reveals commonalities and differences between tumours through comprehensive analysis across multiple cancer types. This broad perspective not only greatly contributes to the accuracy and efficiency of early cancer diagnosis, but also opens up new avenues for exploring and identifying potential therapeutic targets, revolutionising the possibilities for cancer treatment.19,20 The breakthrough progress in this methodology has primarily benefited from the implementation of large-scale projects such as the International Cancer Genome Consortium (ICGC) and TCGA, whose generated multi-omics data has provided a foundational resource for discovering “pan-cancer driver genes”21 In recent years, exploring meaningful molecular targets has been the research focus in the field of tumor medicine. TMSB4X is an actin chelating peptide, binds to actin monomers until cell signals trigger filament formation.16 Overall, TMSB4X plays a role in actin based processes such as cell migration and extracellular matrix (ECM) remodeling.22 Previous studies have shown that TMSB4X is associated with various cancers such as thyroid cancer, prostate cancer, and gastric cancer, however, no studies have yet analysed the specific role of TMSB4X in multiple cancers (ie pan-cancer) in depth. In our study, we can see that TMSB4X is abnormally expressed in 26 cancer species, with significant upregulation in 20 cancer species, especially in GBM, LGG, and GBMLGG patients. In addition, we also observed significant differences in the expression of TMSB4X in different clinical stages of 6 cancer species, including GBMLGG patients. Moreover, the level of TMSB4X expression is closely associated with poor prognostic indicators (including OS, DSS, and PFI) in patients, particularly those with LGG and GBMLGG. Thymosin beta 4 demonstrates therapeutic potential across neurological disorders by enhancing plasticity in both the central and peripheral nervous systems (CNS/PNS) and facilitating neurovascular remodeling, which collectively contribute to neurological restoration.23
Thymosin beta 4 enhances the proliferation of oligodendrocyte progenitor cells (OPCs) and promotes their maturation into myelinating oligodendrocytes, processes that collectively likely mediate its therapeutic benefits in experimental autoimmune encephalomyelitis (EAE).24 When administered 24 hours or later following neural injury or neurodegenerative disease initiation, Thymosin beta 4 stimulates multiple regenerative processes including neovascularization, neural progenitor differentiation, axonal/neurite extension, and oligodendrocyte production. These synergistic mechanisms ultimately lead to measurable improvements in functional performance and behavioral parameters. Previous studies have shown that Thymosin beta 4-mediated oligodendrocyte regeneration serves as a critical convergent mechanism underlying functional recovery in both acute neural trauma and chronic neurodegenerative conditions.25 Given the highly heterogeneous nature of gliomas, individual prognostic factors, specific clinical symptoms, possible side effects of treatment, and tumour progression must be thoroughly considered when developing a personalised treatment plan for patients with gliomas to ensure relevance and efficacy of the treatment plan.6 Therefore, we further investigated the functional role of TMSB4X in glioma.
In our research, TMSB4X was found to be highly expressed in glioma patients through analysis of TCGA datasets data. This result was validated using GEPIA and Human Protein Atlas online analysis, and further validation was performed using qPCR of tumor and normal tissue samples paired with 6 glioma patients. By performing Kaplan-Meier survival analysis as well as ROC curve assessment of data from the TGGA and CGGA databases, the research demonstrated a noteworthy statistical link between the level of TMSB4X expression and the survival outlook for glioma patients, highlighting TMSB4X’s potential as a useful indicator for predicting glioma prognosis. This study enrolled 32 glioma patients, from whom fresh-frozen tumor tissue samples were collected. Total RNA was extracted, reverse-transcribed into cDNA, and subjected to qPCR. The relative expression of TMSB4X was normalized to GAPDH and calculated. Based on the median TMSB4X expression level, patients were stratified into high-expression and low-expression groups. Kaplan-Meier survival analysis revealed a significantly shorter median PFS in the high-expression group compared to the low-expression group. Multivariate Cox proportional hazards regression analysis adjusted for age and WHO grade confirmed that high TMSB4X expression was an independent adverse prognostic factor for PFS. To further explore the clinical significance of TMSB4X in gliomas, we deeply analysed the expression patterns of TMSB4X under different clinicopathological features based on the TGGA and CGGA datasets. By implementing univariate and multivariate Cox regression analyses, we systematically evaluated the association between TMSB4X and the prognosis of glioma patients. In addition, we constructed a clinical correlation nomogram that visualises the evidence for TMSB4X as an independent prognostic factor for glioma, highlighting its importance in clinical decision-making. Finally, the calibration curve verified the matching between actual survival and predicted survival. To further explore the relevant mechanisms of TMSB4X affecting glioma, we analyzed the DEGs identified by single gene differential analysis of TMSB4X using string online analysis to obtain protein interaction networks and visualized them using Cytoscape. Subsequently, the top 50 core genes were screened using the Cyto Hubba algorithm, and GOKEGG analysis was performed on these core genes. In addition, we performed GSEA analysis of up- and down-regulated genes. We visualized the intersection of the genes corresponding to the positive regulation of I-kappaB kinase/NF-kappaB signaling biological process (BP) enriched by the above two analysis methods, respectively, and we could obtain two core genes, namely CASP1 and FASLG. Using in-depth profiling of the TCGA dataset, we identified a core gene, CASP1, the expression level of which showed an extremely strong positive correlation with TMSB4X (R = 0.855, p < 0.001). Further, validation by the GEPIA online platform confirmed that the expression of CASP1 in glioma tissues was significantly higher than that in normal brain tissues (p < 0.01). Not only that, CASP1 expression was also associated with multiple poor prognostic indicators in glioma patients, including OS, DSS and PFI.
Apoptosis, a form of programmed cell death, constitutes a crucial defense mechanism for the host against pathogen invaders. CASP1 is strongly associated with pyroptosis.26 In pyroptosis, innate immune sensors respond to pathogen -related molecular patterns PAMPs and damage-related molecular patterns DAMPs, mediating the formation of inflammasomes, subsequently, CASP1 recruitment and self activation occurred.22 Activated caspases have the ability to fragment gasdermin-D into its N-terminal fragment, known as GSDMD-N. GSDMD-N migrates to the membrane and triggers lytic cell death by forming pores with several subunits.27,28 Taiyu Shen et al found that activation of NF-κB signaling and NLRP3 inflammasome by 0.6 and 1.2 mm FFA was observed with elevated p-NF-κB/NF-κB ratios, increased protein abundance of NLRP3 and CASP1, CASP1 activity, and increased mRNA abundance of IL1B and IL18.29 Many studies have shown that NF-κB promotes glioma progression.30,31 Research finds that TRIM21 via NF-κB/NLRP3 inflammasome pathway inhibits M1 phenotype polarization of microglia and neuroinflammation-mediated neuronal damage.32 Previous studies have demonstrated that Resveratrol (RSV) exerts anti-tumor effects against glioblastoma by suppressing the overactivation of the NLRP3 inflammasome through the JAK2/STAT3 signaling pathway.33 Research has indicated that the activation of NF-κB is associated with poor radiation response and shortened survival in patients with GBM.34 Rui-Chao Chai et al found that YTHDF2 accelerated UBXN1 mRNA degradation via METTL3-mediated m6A, which, in turn, promoted NF-κB activation, promoting malignant progression of gliomas.35 Based on these findings, we propose a hypothesis that TMSB4X may have a significant impact on the survival of glioma patients via the CASP1/NF-κB pathway.
Limitations
This study also has some limitations. First, it would be desirable to validate the role of TMSB4X with a larger dataset. Secondly, the expression of TMSB4X at the protein level needs to be further validated by collecting glioma samples. Thirdly, our hypothesized TMSB4X/CASP1/NF-κB pro-oncogenic pathway in gliomas needs to be confirmed by more experimental studies.
Conclusion
In conclusion, our study examined the link between TMSB4X expression and the progression and prognosis of gliomas. The results revealed TMSB4X as a significant potential prognostic biomarker for glioma patients.
Abbreviations
ADSC, adipose-derived mesenchymal stem cell; ACC, adrenocortical carcinoma; ALL, acute lymphoblastic leukemia; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; COADREAD, colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; GBMLGG, glioma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIPAN, pan-kidney cohort (KICH+KIRC+KIRP); KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; SKCM-M, skin cutaneous melanoma metastasis; STAD, stomach adenocarcinoma; STES, stomach and esophageal carcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; WT, high-risk Wilms tumor.
Data Sharing Statement
The data used to support the results of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
The study was approved by the Human Investigation Ethical Committee of the Second Hospital of Hebei Medical University, and the written informed consent was obtained from all patients.
Acknowledgments
Thanks to all participants in the Radiotherapy Department and Central Laboratory of the Second Hospital of Hebei Medical University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Hebei Natural Science Foundation (Grant Number H2023206913; J230003; H2022206572) and the S&T Program of Hebei (Grant Number 236Z7718G).
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12.
2. Ostrom QT, Price M, Neff C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 2023;25(12 Suppl 2):iv1–iv99. doi:10.1093/neuonc/noad149
3. Cordier D, Krolicki L, Morgenstern A, Merlo A. Targeted radiolabeled compounds in glioma therapy. Sem Nucl Med. 2016;46(3):243–249. doi:10.1053/j.semnuclmed.2016.01.009
4. Weller M, Wen PY, Chang SM, et al. Glioma. Nat Rev Dis Prim. 2024;10(1):33. doi:10.1038/s41572-024-00516-y
5. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi:10.1093/neuonc/nou087
6. Xu H, Zhang A, Han X, et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas. Cancer Immunol Immunother. 2022;71(3):645–660. doi:10.1007/s00262-021-03022-2
7. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. doi:10.1093/neuonc/noab106
8. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi:10.1016/j.cell.2015.12.028
9. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. doi:10.1016/j.ccell.2017.02.009
10. Makowiecka A, Mazurkiewicz E, Mrówczyńska E, et al. Changes in biomechanical properties of A375 cells due to the silencing of TMSB4X expression are not directly correlated with alterations in their stemness features. Cells. 2021;10(4):769. doi:10.3390/cells10040769
11. Xing Y, Ye Y, Zuo H, Li Y. Progress on the function and application of thymosin β4. Front Endocrinol. 2021;12:767785. doi:10.3389/fendo.2021.767785
12. Wu Y, Clark KC, Nguyen EV, et al. Proteomic characterisation of prostate cancer intercellular communication reveals cell type-selective signalling and TMSB4X-dependent fibroblast reprogramming. Cell Oncol. 2022;45(6):1311–1328. doi:10.1007/s13402-022-00719-z
13. Kuo CY, Jhuang JY, Huang WC, Cheng SP. Aberrant expression of thymosin beta-4 correlates with advanced disease and BRAF V600E mutation in thyroid cancer. J Histochem Cytochem. 2022;70(10):707–716. doi:10.1369/00221554221138370
14. An HW, Kim SY, Kwon JW, et al. In vivo CRISPR-Cas9 knockout screening using quantitative PCR identifies thymosin beta-4 X-linked that promotes diffuse-type gastric cancer metastasis. Mol Carcinog. 2021;60(9):597–606. doi:10.1002/mc.23326
15. Chu Y, You M, Zhang J, et al. Adipose-derived mesenchymal stem cells enhance ovarian cancer growth and metastasis by increasing thymosin beta 4X-linked expression. Stem Cell Int. 2019;2019:9037197. doi:10.1155/2019/9037197
16. Chi LH, Chang WM, Chang YC, et al. Global proteomics-based identification and validation of thymosin beta-4 X-linked as a prognostic marker for head and neck squamous cell carcinoma. Sci Rep. 2017;7(1):9031. doi:10.1038/s41598-017-09539-w
17. Xia C, He Z, Cai Y. Quantitative proteomics analysis of differentially expressed proteins induced by astragaloside IV in cervical cancer cell invasion. Cell Mol Biol Lett. 2020;25(1):25. doi:10.1186/s11658-020-00218-9
18. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e11. doi:10.1016/j.cell.2018.02.052
19. Ju Q, Li X, Zhang H, Yan S, Li Y, Zhao Y. NFE2L2 is a potential prognostic biomarker and is correlated with immune infiltration in brain lower grade glioma: a pan-cancer analysis. Oxid Med Cell Longev. 2020;2020:3580719. doi:10.1155/2020/3580719
20. Cen X, Lan Y, Zou J, et al. Pan-cancer analysis shapes the understanding of cancer biology and medicine. Cancer Commun. 2025. doi:10.1002/cac2.70008
21. Underwood T. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi:10.1038/s41586-020-1969-6
22. Padmanabhan K, Grobe H, Cohen J, et al. Thymosin β4 is essential for adherens junction stability and epidermal planar cell polarity. Development. 2020;147(23). doi:10.1242/dev.193425
23. Shomali N, Baradaran B, Deljavanghodrati M, et al. A new insight into thymosin β4, a promising therapeutic approach for neurodegenerative disorders. J Cell Physiol. 2020;235(4):3270–3279. doi:10.1002/jcp.29293
24. Zhang J, Zhang ZG, Li Y, et al. Thymosin beta4 promotes oligodendrogenesis in the demyelinating central nervous system. Neurobiol Dis. 2016;88:85–95. doi:10.1016/j.nbd.2016.01.010
25. Chopp M, Zhang ZG. Thymosin β4 as a restorative/regenerative therapy for neurological injury and neurodegenerative diseases. Expert Opin Biol Ther. 2015;15 Suppl 1(sup1):S9–12. doi:10.1517/14712598.2015.1005596
26. Johnson DE, Cui Z. Triggering pyroptosis in cancer. Biomolecules. 2025;15(3):348.
27. Yang Z, Chen Z, Wang Y, et al. A novel defined pyroptosis-related gene signature for predicting prognosis and treatment of glioma. Front Oncol. 2022;12:717926. doi:10.3389/fonc.2022.717926
28. Kaczor S, Szewczyk-Roszczenko O, Pawlak D, Hermanowicz A, Hermanowicz JM. GSDM family and glioma. Biochim Biophys Acta Rev Cancer. 2025;1880(2):189283. doi:10.1016/j.bbcan.2025.189283
29. Shen T, Li X, Jin B, et al. Free fatty acids impair autophagic activity and activate nuclear factor kappa B signaling and NLR family pyrin domain containing 3 inflammasome in calf hepatocytes. J Dairy Sci. 2021;104:11973–11982.
30. Zhang J, Guo Z, Xie Q, Zhong C, Gao X, Yang Q. Tryptophan hydroxylase 1 drives glioma progression by modulating the serotonin/L1CAM/NF-κB signaling pathway. BMC Cancer. 2022;22(1):457. doi:10.1186/s12885-022-09569-2
31. Sim N, Li Y. NF-κB/p52 augments ETS1 binding genome-wide to promote glioma progression. Commun Biol. 2023;6(1):445. doi:10.1038/s42003-023-04821-2
32. Xiao T, Wan J, Qu H, Li Y. Tripartite-motif protein 21 knockdown extenuates LPS-triggered neurotoxicity by inhibiting microglial M1 polarization via suppressing NF-κB-mediated NLRP3 inflammasome activation. Arch Biochem Biophys. 2021;706:108918. doi:10.1016/j.abb.2021.108918
33. Zhang C, Peng Q, Tang Y, et al. Resveratrol ameliorates glioblastoma inflammatory response by reducing NLRP3 inflammasome activation through inhibition of the JAK2/STAT3 pathway. J Cancer Res Clin Oncol. 2024;150(3):168. doi:10.1007/s00432-024-05625-5
34. Bhat K, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi:10.1016/j.ccr.2013.08.001
35. Chai RC, Chang YZ, Chang X, et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14(1):109. doi:10.1186/s13045-021-01124-z
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Development and Validation of a Prognostic Molecular Phenotype and Clinical Characterization in Grade III Diffuse Gliomas Treatment with Radio-Chemotherapy
Gu W, Tang J, Liu P, Gan J, Lai J, Xu J, Deng J, Liu C, Wang Y, Zhang G, Yu F, Shi C, Fang K, Qiu F
Therapeutics and Clinical Risk Management 2025, 21:35-53
Published Date: 6 January 2025








