Back to Journals » International Journal of Nanomedicine » Volume 20
Therapeutic Targeting in Ovarian Cancer: Nano-Enhanced CRISPR/Cas9 Gene Editing and Drug Combination Therapy
Authors Kim HK , Cheong H , Kim MY, Jin HE
Received 21 November 2024
Accepted for publication 26 February 2025
Published 30 March 2025 Volume 2025:20 Pages 3907—3931
DOI https://doi.org/10.2147/IJN.S507688
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Jie Huang
Hong-Kook Kim,1,2 Heedon Cheong,3 Moo-Yeon Kim,3 Hyo-Eon Jin2,3
1AI-Super Convergence KIURI Translational Research Center, Ajou University, Suwon, 16499, Republic of Korea; 2Research Institute of Pharmaceutical Science and Technology, Ajou University, Suwon, 16499, Republic of Korea; 3Department of Pharmacy, Ajou University, Suwon, 16499, Republic of Korea
Correspondence: Hyo-Eon Jin, Professor, Department of Pharmacy, Ajou University, Suwon, 16499, Republic of Korea, Tel +82-031-219-3466, Fax +82-0504-152-4017, Email [email protected]
Abstract: Ovarian cancer is the third most common gynecological cancer worldwide. Due to the high recurrence rate of advanced-stage ovarian cancer, often resulting from drug-resistant and refractory disease, various treatment strategies are under investigation. Genome editing of therapeutic target genes holds promise in enhancing cancer treatment efficacy by elucidating gene functions and mechanisms involved in cancer progression. The CRISPR/Cas9 system, in particular, shows great potential in ovarian cancer gene therapy and drug development. Targeting therapeutic genes such as BRCA1/2, P53, Snai1 etc, could improve the therapeutic strategy in ovarian cancer. CRISPR/Cas9 is a powerful gene-editing tool that there are many on-going clinical trials to treat various diseases including cancer. Nano-based delivery systems for CRISPR/Cas9 offer further therapeutic benefits, leveraging the unique properties of nanoparticles to improve delivery efficiency. Nano-based delivery systems could enhance the stability of CRISPR/Cas9 delivery formats (such as plasmid, mRNA, etc) and improve the delivery precision of delivery to target tumors. Additionally, combining CRISPR/Cas9 with targeted drug treatments, especially those aimed at genes associated with drug resistance, may significantly improve therapeutic outcomes in ovarian cancer. In this review, we discuss therapeutic target genes and their mechanisms in ovarian cancer, advances in nano-based CRISPR/Cas9 delivery, and the therapeutic potential of combining CRISPR/Cas9 with drug treatments for ovarian cancer.
Keywords: ovarian cancer, CRISRP/Cas9, therapeutic genes, nano-based delivery, anti-cancer drugs
Introduction
Ovarian cancer is a malignant tumor that originates in the ovary and can arise from the surface of ovary, the fallopian tubes, or the mesothelium-lined peritoneal cavity.1 Ovarian cancer is the third most common gynecologic malignancy, accounting for 313,959 new cases and 207,252 deaths worldwide in 2020, with a high mortality rate primarily due to late-stage diagnoses.2 Symptoms are often vague, including abdominal pain, dyspepsia, menstrual irregularities, and other mild digestive disturbances. Due to these nonspecific symptoms, early diagnosis is challenging, leading to a high mortality rate, as most cases are diagnosed at an advanced stage (stage III or IV).3
Ovarian cancer is generally classified into three types: epithelial ovarian cancer, germ cell tumors, and sex-cord-stromal tumors. Approximately 90% of ovarian cancers are epithelial, while germ cell tumors constitute about 3%, and sex-cord-stromal tumors account for less than 2%.4 Epithelial ovarian cancers develop in the ovarian epithelial tissue and are divided into two categories: type I and type II tumors. Type I tumors are slow-growing and include low-grade serous carcinoma (LGSC), mucinous carcinoma (MC), endometrioid carcinoma (EC), clear cell carcinoma (CCC), and transitional cell carcinoma (TCC). Type II tumors, by contrast, are aggressive, fast-growing neoplasms, including high-grade serous carcinoma (HGSC), carcinosarcoma, and undifferentiated carcinoma.5,6 HGSC comprises approximately 80% of epithelial ovarian cancers.4 Germ cell tumors, which arise from the reproductive cells that develop into eggs, typically occur in children, adolescents, and young women. Malignant ovarian germ cell tumors are often chemosensitive, allowing for a high probability of fertility preservation and cure through surgery and chemotherapy.7,8 Sex-cord-stromal tumors are a rare, heterogeneous group of neoplasms that develop from the sex cords or ovarian stromal cells. These tumors may be benign or malignant and can occur across a wide age range.9,10
The standard treatment for ovarian cancer involves cytoreductive surgery, followed by chemotherapy with platinum- and taxane-based regimens.11 In first-line treatment, the main chemotherapeutic agents are usually administered as cisplatin/paclitaxel or carboplatin/paclitaxel combinations.12 For some patients with advanced ovarian cancer (stage III or IV), treatment may begin with neoadjuvant (preoperative) chemotherapy to shrink the tumor before surgery.13 However, chemotherapy drugs have adverse events such as granulocytopenia, nausea, vomiting, and neurotoxicity.12 Additionally, 20–30% of patients experience chemoresistance during first-line treatment.14 Chemoresistance is often associated with genetic alterations such as mutations in TRP53, BRCA1/2, necessitating more targeted approaches to overcome these barriers.15
In maintenance therapy, two targeted agents are available: bevacizumab and poly (ADP-ribose) polymerase (PARP) inhibitors.14 Bevacizumab is an anti-angiogenesis agent that targets vascular endothelial growth factor (VEGF). It can be used in combination with chemotherapy in maintenance therapy. Also used with olaparib for platinum-sensitive recurrent ovarian cancer.16,17 PARP inhibitors work by blocking DNA repair pathways, inducing apoptosis particularly in homologous recombination (HR)-deficient cells, commonly seen in breast cancer gene (BRCA) 1/2 mutations. FDA-approved PARP inhibitors for ovarian cancer include olaparib, rucaparib, and niraparib.18 Niraparib is approved as frontline maintenance therapy for both HR-proficient and HR-deficient tumors. Olaparib is used for BRCA-mutated tumors or in combination with bevacizumab for HR-deficient tumors. Rucaparib is not approved for frontline maintenance but is used in maintenance therapy for recurrent platinum-sensitive disease.19 However, 80% of patients with advanced-stage ovarian cancer relapse within 18 months due to platinum-resistant or refractory cancer.14 While bevacizumab and PARP inhibitors have advanced ovarian cancer treatment, limitations remain, and some patients develop resistance to these therapies.20 Consequently, new treatment strategies for ovarian cancer are continually being explored.
The clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated 9 (Cas9) system represents a powerful tool for therapeutic gene editing.21 Using CRISPR/Cas9 gene editing to inhibit oncogene expression in ovarian cancer may reduce tumor growth and enhance drug sensitivity. However, there is a risk of off-target mutagenesis and limited specificity due to the instability of CRISPR/Cas9 components in body fluids. Nanoparticles offer a potential solution, enabling efficient and safe delivery of CRISPR/Cas9 to target sites.22 Nanoparticles are synthesized using various materials and formed into different shapes, resulting in a wide range of types, such as liposome, dendrimers, and viral nanoparticles, etc. These nanoparticles can enhance the stability of CRISPR/Cas9 delivery, improve cellular uptake in target cells, and reduce immunogenicity, ultimately leading to higher target gene editing efficiency.23,24
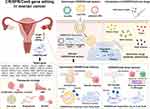
This review examines therapeutic target genes implicated in ovarian cancer progression, advancements in nanoparticle-based CRISPR/Cas9 delivery, and the synergistic effects of integrating gene editing with drug therapies for ovarian cancer treatment (Figure 1).
CRISPR/Cas9 Therapy
Structure and Editing Mechanisms of CRISPR/Cas9
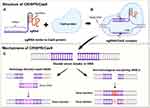
CRISPR/Cas system was discovered as an adaptive immune mechanism in bacteria and archaea allowing them to counteract invasive mobile genetic elements (MGEs).25,26 Cas9 is a class 2 endonuclease in the CRISPR-Cas system, playing a key role in cutting foreign DNA.27 This system stores sequence information of invading genetic material within CRISPR loci, creating a memory of previously encountered genetic elements.28
CRISPR loci are transcribed into CRISPR RNA (crRNA), which forms a complex with trans-activating CRISPR RNA (tracrRNA), binding the two RNA transcripts together.29 This crRNA-tracrRNA complex, also known as single guide RNA (sgRNA), binds to the Cas9 protein and directs it to the target DNA.30 A protospacer adjacent motif (PAM) sequence is required for the hybridization of crRNA to the DNA target, and this sequence is recognized by the Cas9 protein.31 Once the PAM sequence is identified, Cas9 cleaves the DNA three base pairs upstream of the PAM site.32
Following DNA cleavage, the damaged DNA can be repaired by two primary mechanisms: non-homologous end joining (NHEJ) and homology-directed repair (HDR).33 NHEJ is a straight forward repair process that may involve random insertions or deletions of nucleotides. In contrast, HDR uses single-stranded (ssDNA) or double-stranded DNA (dsDNA) as donor DNA, allowing for the insertion of desired genetic sequences34 (Figure 2).
Applications of CRISPR/Cas9
CRISPR/Cas9 offers significant potential in human gene therapy and drug development.35 CRISPR/Cas9 gene editing could be used to specific disrupt cancer-related genes thereby, inhibiting the expression of cancer-related proteins. This gene editing technique also enables the identification and study of cancer-associated genes as potential therapeutic targets.36
CRISPR/Cas9 offers several advantages over other gene therapy approaches, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and RNA interference (RNAi), and DNAzyme.
ZFNs are artificial endonucleases created by fusing a zinc finger DNA-binding domain to a non-specific DNA cleavage domain, allowing them to target specific genomic sequences. However, ZFNs have several disadvantages, including limited target site accessibility, low efficiency, high cost and time-consuming process.37,38 TALENs, on the other hand, are artificial restriction enzymes engineered by fusing a DNA-binding domain derived from transcription activator-like effectors (TALEs) with the catalytic domain of FokI endonucleases. TALENs can specifically target genomic sequences and are easier to design compared to ZFNs. However, TALENs larger size makes them more challenging to deliver into cells, and they exhibit relatively lower efficiency compared to CRISPR/Cas9.39,40 Unlike ZFNs and TALENs, which rely on large DNA segments (500–1500 bp) and can typically target only one gene at a time, CRISPR/Cas9 can efficiently target any gene sequence using a 20 bp protospacer within the guide RNA. This system allows for higher efficiency and the simultaneous targeting of multiple genes within the same cell.38
RNAi is a natural gene silencing process mediated by RNA molecules in cells. It can be achieved using chemically synthesized small interfering RNA (siRNA) or vector-based short hairpin RNA (shRNA). RNAi suppresses target gene expression by degrading specific mRNA through double-stranded RNA. Although RNAi enables rapid workflow and transient knockdown studies without causing permanent genetic changes. However, RNAi has high off-target effects and might result in incomplete knockdown of the target gene leading to ambiguous results. In contrast, CRISPR/Cas9 offers lower off-target effects and permanent gene knockout.41,42
Deoxyribozyme (DNAzyme) is a single-stranded DNA oligonucleotides with catalytic function that can silence specific genes through metal ions activity. DNAzyme exhibit high efficiency and can be constructed into multifunctional constructs. It can also be used in combination with CRISPR/Cas9 to enhance therapeutic efficiency. However DNAzyme has lower clinical applicability compared to CRIPSR/Cas9.43–46
Although CRISPR/Cas9 offers many advantages, it also has several limitations and risks. One major concern is its relatively high frequency (≥50%) of off-target effects, which could induce unintended gene in non-target genes.47 Additionally, CRISPR/Cas9 based gene therapy is expensive; for example, the world’s first approved CRISPR/Cas9 based therapy, “Casgevy”, costs approximately $2.2 million per patient. This high cost presents a significant barrier to accessibility for many patients.48 Furthermore, the long-term effects of CRISPR/Cas9 remain uncertain and insufficiently studied, raising concerns about potential unintended consequences that may arise in the future. These challenges have also led to ethical concerns regarding the widespread use of CRISPR/Cas9 therapy.49
Despite its limitations and associated risks, CRISRP/Cas9 based therapy holds great therapeutic potential, and many clinical trials are currently underway. Several clinical trials are exploring the applications of CRISPR/Cas9 in cancer treatment, including the inhibition of intracellular immune checkpoint CISH in gastrointestinal cancer (clinical trial ID NCT04426669), PD-1 and TCR knockout in mesothelin positive solid tumor (clinical trial ID NCT03545815), and PD-1 knockout in lung cancer (clinical trial ID NCT02793856).50
Recently, researchers have been exploring nanoparticle-mediated CRISPR/Cas9 delivery and the combination of the CRISPR/Cas9 system with anticancer drugs for cancer treatment. In this review, we discuss the use of CRISPR/Cas9 delivery systems combined with nanoparticles and anti-cancer drugs targeting ovarian cancer.
Therapeutic Target Genes in Ovarian Cancer
Therapeutic genome editing offers great potential in developing ovarian cancer treatments, by targeting specific genes associated with therapeutic strategies. Table 1 outlines therapeutic strategies and candidate target genes in ovarian cancer that are edited using the CRISPR/Cas9 system.
 |
Table 1 Therapeutic Target Genes in Ovarian Cancer |
Reduction of Proliferation, Migration, and Invasion
Proliferation, migration and invasion has an important role in cancer development and progression. Proliferation involves cancer cells copying their DNA and dividing, which increases the number of cancer cells.67 Migration is the directed movement of cancer cells within tissues or organs.68 Invasion allows cancer cells to penetrate extracellular matrices and infiltrate tissues, enabling their spread to different areas.69 To reduce proliferation, migration, and invasion in ovarian cancer, therapeutic gene editing has targeted genes such as B-cell-specific Moloney murine leukemia virus integration site 1 (BMI1), ATPase family AAA domain-containing protein 2 (ATAD2), protease-activated receptor 2 (PAR2), Egl-9 family hypoxia-inducible factor 1 (EGLN1), human endogenous retrovirus (HERV-K) envelope (env) gene, chemokine (C-C motif) ligand 2 (CCL2), and signal transducer and activator of transcription 3 participates (STAT3).
BMI1 is upregulated in a variety of cancers and regulates cell proliferation, cell cycle, and cell immortality.70 Using CRISPR/Cas9 to knockout BMI1 in SKOV3 cells significantly reduced ovarian cancer cell proliferation, migration, and invasion, promoting apoptosis and enhancing platinum sensitivity by modulating genes in the focal adhesion and PI3K/AKT pathways. Tumor size was dramatically reduced by BMI1 knockout in SKOV3 xenografted BALB/c nude mice.51
ATAD2 is a member of the ATPase family and is involved in promoting cell proliferation, migration, invasion, and metastasis while inhibiting apoptosis during cancer progression.71 Overexpression of ATAD2 is associated with low overall survival, and poor clinical outcomes.72 CRISPR/Cas9 knockdown of ATAD2 in SKOV3 and A2780 cells reduced proliferation and colony formation by downregulating the JNK-MAPK, P38, and ERK pathways.52
PAR2 is a member of the G-protein coupled receptor (GPCR) family that initiates signal transduction pathways.73 It is significantly overexpressed in patients with ovarian cancer based on The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx) and Gene Expression Omnibus (GEO) databases. Knockout of PAR2 in OV90 cells reduced migration and invasion, while knockout of downstream components (Gαq/11, Gα12/13, β-arrestin1/2) further inhibited PAR2-mediated migration and invasion.53
EGLN1 is an oxygen dependent, primary hypoxia-inducible factor 1 (HIF1) hydroxylase that generates a binding site for a ubiquitin ligase complex including the von Hippel-Lindau tumor suppressor gene (VHL), leading to HIF1 degradation.74 Genome-wide CRISPR/Cas9 studies identified EGLN1 as a dependency in clear cell ovarian cancer cells. EGLN1 knockout reduced proliferation by stabilizing and accumulating HIF1A, and VHL knockout also showed similar effects.54
HERVs are endogenous viral elements in the human genome that have lost the ability to be an active virus; however, HERV-derived element can be a pathological contributor to various diseases.75 The HERV-K envelope (env) protein is highly expressed in several cancers and may be associated with cancer progression.76 CRISPR/Cas9 knockout of HERV-K env in SKOV3 and OVCAR3 cells reduced proliferation, migration, and invasion, with distinct regulatory patterns of retinoblastoma (RB) and cyclin B1 proteins between ovarian cancer cell lines.55
CCL2 also known as monocyte chemoattractant protein-1, is a potent chemoattractant of immune cells and associated with tumor growth and progression.77 In Liu et al (2023), Knockout of CCL2 in A2780 cells reduced proliferation, migration, and invasion by downregulating the mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK)/mitogen-activated protein three kinase 19 (MAP3K19) signaling pathway. MAP3K19 is the key target of CCL2 regulation in ovarian cancer progression, MAP3K19-knockout in A2780 cells, reduced proliferation, migration and invasion.56
STAT3 is a member of the STAT family and a key regulator of tumorigenesis. STAT3 supports proliferation, migration, invasion, survival, and chemoresistance in ovarian cancer.78 In Lu et al (2019), STAT knockout in ovarian cancer cells reduced proliferation and migration, EMT, arrest cell cycle progression, altered stemness markers, and inhibited tumor growth in tumor xenograft mice. Knockout of STAT3 using CRISPR/Cas9 in four ovarian cancer cell lines, HEY, OVCAR3, OVCAR8 and SKOV3 reduced proliferation, migration, arrested cell cycle progression in the G2/M phase by downregulating key cell cycle mediators (CDKs, cyclins, cell division cycle proteins and checkpoint proteins), altered stem-like properties (ALDH1A1, ALDH1A3 and CD44) and inhibited tumor growth in xenograft NOG or athymic nude mice. STAT3 knockout in SKOV3 cells, the EMT-related key genes CDH1 (E-cadherin), CDH2 (N-cadherin, mesenchymal marker) were upregulated and SNAI2&3, ZEB1&2, KLF8, TWIST1&2, GSC, SIX1, and FOXC1 were downregulated.57
Inhibition of Epithelial-Mesenchymal Transition (EMT)
Epithelial-mesenchymal transition (EMT) is a biological program through which epithelial cells undergo multiple biochemical changes and acquire a mesenchymal phenotype. It plays a role in wound healing, embryonic development, and tissue fibrosis. However, it also occurs in cancer progression and is associated with cancer stem cell characteristics, tissue invasiveness and resistance to cancer treatment.79,80 EMT in ovarian cancer has been targeted by editing genes such as baculoviral IAP repeat containing 5 (BIRC5), snail family transcriptional repressor 1 (Snai1), microRNA-21 (miR-21), metal responsive transcription factor 1 (MTF1).
BIRC5, also known as survivin, is a member of the inhibitor of apoptosis (IAP) family and is known to be involved in apoptosis inhibition, metastasis and chemoresistance.81 In Zhao et al (2017), by using The Cancer Genome Atlas Program (TCGA) and Gene Expression Omnibus (GSE13876), BIRC5 was overexpressed in ovarian serous carcinoma and patients were associated with poor survival. BIRC5 knockout using lentiviral CRISPR/Cas9 nickase in SKOV3 and OVCAR3 cells inhibited EMT by modulating TGF-β pathway, enhancing epithelial markers (ie, cytokeratin 7) and reducing mesenchymal markers (ie, snail2, β-catenin, and vimentin). BIRC5 knockout resulted in inhibited cell proliferation, migration, invasion, cell survival and chemoresistance to paclitaxel.58
Snai1 (Snail or Snail1) is a transcription factor, Snai1 represses epithelial markers (E-cadherin, claudins and occludin) and promotes mesenchymal markers (vimentin, fibronectin, N-cadherin).82 CRISPR/Cas9n knockout of Snai1 in RMG-1 cells (ovarian adenocarcinoma) reduced migration, increased cell-cell adhesion, and upregulated epithelial markers.59
miRNA-21 (miR-21) is known to be highly expressed in cancer and promotes tumor growth, invasion, and metastasis.83 CRISPR/Cas9 disruption of miR-21 in SKOV3 and OVCAR3 cells inhibited EMT, reduced proliferation and migration, and increased epithelial markers. In addition, proliferation, migration, invasion and chemoresistance to paclitaxel were reduced in both miR-21 disrupted SKOV3 and OVCAR3 cells.60
MTF1, the zinc finger protein is associated with tumor growth and poor survival. CRISPR/Cas9 nickase knockout of MTF1 inhibited EMT in OVCAR3 and SKOV3 cells by upregulating epithelial cell markers (E-cadherin and cytokeratin 7) and tumor suppressor Kruppel-like factor 4 (KLF4), downregulating mesenchymal markers (Snai2 and β-catenin), and attenuating ERK1/2 and AKT pathway.61
Cell Cycle Regulation
Cell cycle dysregulation leads to uncontrolled cell proliferation and tumorigenesis.84 The Erythropoietin-producing hepatocellular (EPH) receptor A1 (EPHA1), part of the receptor tyrosine kinase family, was investigated as a target for ovarian cancer treatment. CRISPR/Cas9 knockdown of EPHA1 in SKOV3 and COV504 cells led to cell cycle arrest in the G0/G1 phase, reducing proliferation, migration, and invasion. Knockdown of EPHA1 also downregulated matrix metalloproteinase-2 (MMP2) and the proto-oncogene c-MYC, and upregulated ERK2.62
Reduction of Anoikis Resistance
Anoikis is a form of programmed apoptosis triggered when cells detach from the extracellular matrix (ECM) and attach to an inappropriate site.85 Cancer cells often acquire anoikis resistance, enhancing their aggressiveness. Therapeutic gene editing has targeted protein-L-isoaspartate (D-aspartate) O-methyltransferase (PCMT1) and Rho guanine nucleotide exchange factor (Rgnef) to reduce anoikis resistance in ovarian cancer.86
PCMT1 is an enzyme that recognizes and repairs the abnormal L-isoaspartyl residues in proteins.87 PCMT1 is associated with anoikis resistance in ovarian cancer and regulates the integrin‑FAK‑Src pathway by interacting with the ECM protein laminin subunit beta 3 (LAMB3), promoting cancer progression. CRISPR/Cas9 knockout of PCMT1 in SKOV3 cells induced detachment-induced apoptosis from the ECM and downregulated phosphorylation of FAK and Src.63
Rgnef is a Rho guanine nucleotide exchange factor (GEF) that acts as a downstream of integrin-FAK signaling and regulates Rho GTPase activity.88 In Kleinschmidt et al (2019), Rgnef was overexpressed in late-stage serous ovarian cancer which activates as a downstream of integrins and supports antioxidant genes, protecting ovarian cancer cells from oxidative stress resulting in anoikis resistance. CRISPR/Cas9 knockout of Rgnef in mouse ovarian carcinoma (MOVCAR) cells impaired anchorage-independent growth and triggered anoikis.64
Inhibition of Angiogenesis
Angiogenesis, the process of forming new blood vessels, is essential for tumor growth and metastasis, providing cancer cells with oxygen and nutrients to support rapid proliferation and spread.89,90 One of the key mediators of this process is vascular endothelial growth factor (VEGF), which induces abnormal structural and functional changes in tumor vasculature.91
Epidermal growth factor-like domain multiple 6 (EGFL6), a member of the epidermal growth factor (EGF) superfamily, has been studied as a target for angiogenesis inhibition. EGFL6 is abnormally expressed in various malignant tumors and contributes to tumor proliferation, invasion, migration, and angiogenesis.92 In a study by Zhu et al (2020), CRISPR/Cas9 knockout of EGFL6 in SKOV3 cells led to the downregulation of the fibroblast growth factor 2 (FGF-2)/platelet-derived growth factor subunit B (PDGFB) signaling pathway, indicating a reduction in angiogenesis, proliferation, and metastasis. Furthermore, EGFL6 knockout also downregulated N-cadherin, MMP2, MMP9, and vimentin, while upregulated E-cadherin, suggesting EMT inhibition. In a xenograft model using EGFL6-knockout SKOV3 cells in BALB/c nude mice, tumor growth and expression levels of EGFL6, VEGF-A, FGF-2, and PDGFβ were significantly reduced.65
Other Therapeutic Effect
In addition, other therapeutic strategies in ovarian cancer research focus on modulating the tumor microenvironment and enhancing immune responses. These strategies involve gene editing that enhances the activity of tumor-infiltrating lymphocytes (TILs), which is essential for overcoming immune suppression in the tumor microenvironment. Transforming growth factor-beta (TGF-β) is known to be expressed in most malignant tumors and plays a significant role in tumor-induced immunosuppression.93 In Fix et al (2022), CRISPR/Cas9 was used to knockout TGF-β receptor 2 (TGFBR2) in ovarian cancer TILs. This knockout rendered the TILs resistant to TGF-β-mediated immunosuppression, evidenced by a lack of phosphorylation of small signaling molecules against decapentaplegic homolog (SMAD), increased secretion of proinflammatory cytokines (IFN-γ and TNF-α), and improved cytotoxicity even in the presence of TGF-β. Similarly, TGFBR2 knockdown resulted in TILs that were resistant to TGF-β without affecting TIL expansion efficiency or T-cell receptor clonal diversity.66
Nano-Based CRISPR/Cas9 Delivery in Ovarian Cancer
Concept of Nanoparticle
Nano-based delivery systems are advanced technologies that package drugs, genes, or other therapeutic agents into particles at the nanoscale, allowing for precise delivery to targeted tissues or cells.94 These systems offer several advantages, including targeted delivery, enhanced solubility, sustained release, multifunctionality, and potential for personalized medicine. By attaching specific ligands or antibodies to the nanoparticle surface, these systems can achieve selective targeting, which enhances therapeutic efficacy and minimize the side effects.95
Nanoparticles are typically 1–100 nanometers in size and can be made from various materials and in different shapes. Surface modification with specific molecules increases selectivity for targeted tissues or cells. Nanoparticles can be engineered to deliver drugs, genes, or other therapeutic agents directly to specific sites, enhancing drug stability and preventing degradation in the bloodstream, which ultimately improves bioavailability.96–98
While the CRISPR/Cas9 system is a powerful tool for genome editing, delivering Cas9 (as plasmid DNA, mRNA, or protein) along with single guide RNA (sgRNA) to target sites is challenging. CRIPSR/Cas9 could be degraded by enzymes, acids, and other substances in body fluids. Other risks include cell damage, off-target mutagenesis, and poor specificity.99 Nanoparticle delivery provides a safer and more effective way to transport CRISPR/Cas9 components.22 Both viral and non-viral nanoparticles have been used in CRISPR/Cas9 delivery. Viral systems, including adenoviruses and lentiviruses, are efficient but can trigger immune responses and present manufacturing challenges. Non-viral nanoparticles, such as lipid, gold, magnetic, albumin, and polymeric nanoparticles, are easier to modify, cost-effective, and less immunogenic, though they may have lower transfection efficiency and uncertain long-term toxicity.
Nano-Based CRISPR/Cas9 Delivery in Ovarian Cancer
The effectiveness of CRISPR/Cas9 gene editing in ovarian cancer depends heavily on the chosen delivery strategy. Given the large size of Cas9 (160 kDa, 4300 bases) and sgRNA (~31 kDa, 130 bases), conventional viral and non-viral delivery methods can be limited by potential toxicity and inefficiency.100 Developing non-toxic, efficient delivery systems for Cas9 and sgRNA could improve therapeutic efficacy. Nanoparticles hold significant promise in this area, as summarized in Table 2, and various nanoparticles have been developed for CRISPR/Cas9 delivery in ovarian cancer, including amino-ionizable lipid nanoparticles, folate receptor-targeted cationic liposomes, reduction-sensitive fluorinated platinum (Pt) transfection nanoplatforms, multifunctional nucleus-targeting “core-shell” artificial viruses (RRPHC) and CRISPR-GPS nanocomplex.
 |
Table 2 Nano-Based CRISPR/Cas9 Delivery in Ovarian Cancer |
Amino – Ionizable Lipid Nanoparticles
Rosenblum et al (2020) designed an ionizable cationic lipid nanoparticle (LNP) to co-encapsulate Cas9 mRNA and sgRNA targeting polo-like kinase 1 (PLK1), a protein that plays a critical role in cell division (Figure 3). This amino-ionizable LNP effectively encapsulates negatively charged RNA through positively charged lipids.105 In this study, to reduce immunogenicity and enhance RNA stability, Cas9 mRNA was modified with 5-methoxyuridine, and IDT sgRNA XT was used. The encapsulation efficiency of Cas9 mRNA and sgRNA in CRISPR-LNPs (cLNPs) with different ionizable amino lipids was measured using a RiboGreen assay. Lipid 8 (L8) showed the highest efficiency, exceeding 90%. In vitro, cLNPs encapsulating sgPLK1 (sgPLK1-cLNPs) achieved 91% specific and efficient PLK1 gene editing in human serous ovarian adenocarcinoma Ovcar8 cells, resulting in G2-M phase cell cycle arrest and apoptosis. As a control, cLNPs encapsulating green fluorescent protein sgRNA (sgGFP-cLNPs) showed no reduction in cell viability, indicating low toxicity. In vivo, sgPLK1-cLNPs were evaluated in a glioblastoma model, where they achieved 68% PLK1 gene editing and significantly reduced tumor growth after a single dose. To overcome liver accumulation, epidermal growth factor receptor (EGFR)-coated sgPLK1-cLNPs were used, targeting ovarian tumors overexpressing EGFR. In an intraperitoneal Ovcar8 xenograft model, this treatment significantly reduced tumor growth and improved survival.100
 |
Figure 3 Design and therapeutic gene editing of amino-ionizable LNPs for PLK1-targeted CRISPR/Cas9 delivery. (A) Schematic illustration of CRISRP LNPs (cLNPs) encapsulating Cas9 mRNA and sgRNA. (B) Chemical structures of amino-ionizable lipids. (C) Encapsulation efficiency of Cas9 mRNA and sgRNA in different amino-ionizable lipids cLNPs. (D) In vitro gene editing (%) at the PLK locus in OV8 cells. (E) Schematic of injection into the mouse hippocampus. (F) In vivo gene editing (%) at the PLK locus in 005 GBM bearing mice. (G) Bioluminescence images of 005 BGM bearing mice after single dose treatment of cLNPs. (H) Tumor growth curve and survival (%) of 005 BGM bearing mice with a single dose of cLNPs. Abbreviations: Cas9, CRISPR associated protein 9; cLNPs, CRISPR lipid nanoparticles; CRISPR, clustered regularly interspaced short palindromic repeats; L1, lipid 1; L6, lipid 6; L8, lipid 8; L10, lipid 10; MC3, (6Z,9Z,28Z,31Z)-Heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino) butanoate; mRNA, messenger RNA; sgGFP, sgRNA green fluorescent protein; sgGFP-cLNPs, sgGFP encapsulated L8-cLNPs; sgPLK1, sgRNA polo-like kinase 1; sgPLK1, sgPLK1 encapsulated L8-cLNPs; sgRNA, single guide RNA. Note: Data from Rosenblum et al (2020).100. |
Folate Receptor-Targeted Cationic Liposome (F-LP)
Folate receptor-targeted cationic liposomes (F-LP) were constructed to deliver CRISPR plasmids which expresses Cas9 and sgRNA that targets DNA methyltransferase 1 (DNMT1) gene in ovarian cancer models. DNMT1 plays an important role in DNA methylation and is associated with tumorigenesis and chemoresistance. Aberrant overexpression of DNMT1 can lead to the inactivation of tumor suppressor genes and the maintenance of cancer stem cells.106 F-LP formulations incorporated folate-PEG-succinyl-Chol for receptor-specific targeting. Optimal binding between F-LP and gDNMT1 plasmids was achieved at a weight ratio of 12:1 or 16:1, with no free plasmid DNA observed in gel electrophoresis, suggesting a successful formulation of F-Lp/gDNMT1 complexes. In vitro, F-LP/gDNMT1 complexes demonstrated a transfection efficiency of 28.6% in SKOV3 cells, effectively disrupting DNMT1 expression and reducing DNA methylation. In vivo, these complexes inhibited both paclitaxel-sensitive and paclitaxel-resistant SKOV3 tumors in BALB/c mice, with minimal side effects compared to high-dose paclitaxel treatment. Tumor DNMT1 and HP1α expression levels were downregulated, further indicating reduced DNA methylation.101
Reduction-Sensitive Fluorinated-Platinum (Pt) Universal Transfection Nanoplatform (PtUTP-F)
A reduction-sensitive fluorinated platinum (Pt) nanoplatform (PtUTP-F) was developed for CRISPR/dCas9-mediated activation of cancer/testis antigen 45 (CT45), which enhances chemosensitivity in high-grade serous ovarian cancer (HGSOC).102 The CRISPR/dCas9 (dead Cas9) system is devoid of nuclease activity and is incapable of cleaving the target gene. However, it is still capable of binding to DNA strands and initiating transcription. In comparison to the CRISPR/Cas9 system, which has the potential to damage genes and cause off-target alteration, CRISPR/dCas9 is a safer and more controllable.107,108 Cancer/testis antigen 45 (CT45) is a protein phosphatase 4C (PP4C) inhibitor that can disrupt the DNA repair pathway and to act as a chemosensitivity modulator, with implications for long-term survival in HGSOC.109
PtUTP-F comprised Pt(IV), synthesized with hydrogen peroxide, and fluorinated polyethyleneimine (PEI1.8K-F). The platform effectively bound CRISPR constructs at a minimum mass ratio of 1:1. The pCT45 plasmid, which expresses sgRNA targeting the promoters of CT45, and the pCas9-VP64 plasmid, which transcribes the dCas9 activator, were used to generate the dCas9-CT45 construct. The dCas9-CT45 loaded PtUTP-F (PtUTP-F/dCas9-CT45) was prepared with an optimal ratio of 20:1:1 (WPtUTP-F/WpCT45/WdCas9). PtUTP-F/dCas9-CT45 achieved a transfection efficiency of 61.4% in A2780 cells, surpassing that of commercial Lipo600 (39.7%). In medium containing 10% serum, PtUTP-F exhibited a transfection efficiency of 40.1%, significantly higher than Lipo600 (13.4%). In vitro, PtUTP-F/dCas9-CT45 upregulated CT45 and downregulated PP4C, disrupted DNA repair pathways, and enhanced platinum drug sensitivity. In vivo, PtUTP-F/dCas9-CT45 inhibited tumor growth and increased CT45 expression in subcutaneous A2780 xenografts BALB/c nude mice.102 The design of the nanoparticle and in vitro transcription results in Lu et al (2021) are shown in Figure 4.
Multifunctional Nucleus Targeting “Core-Shell” Artificial Virus (RRPHC)
A multifunctional nucleus-targeting “core-shell” artificial virus (RRPHC) was constructed to deliver Cas9-hMTH1, targeting MutT homolog 1 (MTH1) (Figure 4).103 MTH1 plays a crucial role in maintaining the integrity of the genetic material by sanitizing oxidized deoxynucleoside triphosphate (dNTP) pools, preventing the incorporation of damaged bases into DNA replication, thereby avoiding genetic instability and cell death. MTH1 sanitizes oxidized nucleotides and prevents their incorporation into DNA. Its inhibition selectively induces apoptosis in cancer cells.110,111 RRPHC artificial virus (RRPHC/Cas9-hMTH1 nanoparticle) was constructed by combining the PF33/Cas9-hMTH1 nanoparticle with a versatile multifunctional shell (RGD-R8-PEG-HA, RRPH). The core nanoparticle (PF33/Cas9-hMTH1) was formed by binding Cas9-hMTH1 to fluorinated polymer PF33 at a mass ratio of 1:1. The shell (RGD-R8-PEG-HA) targeted CD44 receptors overexpressed in ovarian tumors, promoting cellular uptake and endosomal escape via hyaluronidase degradation. In vitro, RRPHC/pEGFP nanoparticles exhibited a transfection efficiency exceeding 90% in SKOV3 cells, outperforming Lipofectamine 3000. RRPHC/Cas9-hMTH1 effectively inhibited MTH1 expression, induced apoptosis, and reduced proliferation in ovarian cancer cells. In vivo, RRPHC/Cas9-hMTH1 significantly inhibited tumor growth and MTH1 expression in subcutaneous SKOV3 xenograft BALB/c nude mice.103
CRISPR-GPS Nanocomplex
A tandem peptide nanocomplexes was constructed to deliver sgRNA/Cas9 ribonucleoprotein (RNP) complex. The tandem peptide was composed of a cell-penetrating peptide (TP) and a αvβ3/αvβ5 integrin-targeting peptide (iRGD) conjugated with a lipid tail, forming palmitoyl-TP-iRGD (pTP-iRGD). The CRISPR-GPS (guiding peptide sequences) nanocomplex was assembled using pTP-iRGD to encapsulate sgRNA/Cas9 RNP at a mass ratio of sgRNA:Cas9:pTP-iRGD of 1:1:30. The CRISPR-GPS nanocomplex effectively targeted αvb3 integrin in OVCAR8 cells, facilitating co-localized sgRNA and Cas9 into the cell. Additionally, when the CRISPR-GPS nanocomplex carrying sgGFP1/Cas9 was introduced into OVCAR8 cells expressing destabilized GFP (OVCAR8/d2eGFP), it disrupted more than 10% of GFP expression. CRISPR-GPS nanocomplex efficiently delivered functional sgRNA/Cas9 complexes into the cells.104
Combination of CRISPR/Cas9 System and Other Drugs in Ovarian Cancer Treatment
Anticancer Drugs in Ovarian Cancer
Anticancer drugs commonly used in ovarian cancer fall into two main categories: chemotherapy drugs (eg, platinum and taxane-based agents) and targeted therapies, such as poly-ADP ribose polymerase inhibitors (PARPi) and vascular endothelial growth factor inhibitors (VEGFi).112,113 Platinum-based drugs bind specific regions of the double-stranded DNA structure in the nucleus of cancer cells, thereby inhibiting DNA replication and suppressing cancer cell proliferation.114 Taxanes inhibit the separation of microtubules during the mitotic process, thereby inhibiting cell division and proliferation.115 PARPi target enzymes involved in DNA repair, impeding the resolution of single-strand DNA breaks and leading to double-strand breaks.116 VEGFi inhibit angiogenesis, suppressing tumor growth by disrupting the blood supply.117 Figure 5 illustrates the mechanisms of action of platinum, taxane, PARPi, and VEGFi as well as their combination with the CRISP/Cas9 system.
Ovarian cancer remains a lethal malignancy, primarily due to its high recurrence rate and the development of resistance to anticancer drugs.118 Understanding the molecular mechanisms underlying drug resistance and identifying target genes through CRISPR/Cas9 can enhance drug response and therapeutic outcomes.119,120 Additionally, integrating nano-based delivery systems with anticancer drugs offers a promising approach for improving outcomes in drug-resistant cases. Table 3 summarizes the combination of CRISPR/Cas9 technology with chemotherapy for the treatment of ovarian cancer, highlighting therapeutic target genes and anticancer agents.
 |
Table 3 Combination of CRISPR/Cas9 System and Chemotherapy in Ovarian Cancer Treatment |
Combination of CRISPR/Cas9 Gene Editing and Platinum Drugs
The combination of CRISPR/Cas9 gene editing and platinum-based drugs offers a promising strategy to overcome drug resistance in cancer therapy by targeting specific genes associated with tumor survival and chemoresistance. By precisely knocking out genes such as eukaryotic translation elongation factor 1 delta (EEF1D), runt-related transcription factor 1 (RUNX1), fibrillin 1 (FBN1), adipocyte fatty acid binding protein (FABP4), salt-induced kinase 2 (SIK2), or carnitine palmitoyltransferase 1A (CPT1A), CRISPR/Cas9 enhances the sensitivity of cancer cells to platinum drugs like cisplatin and carboplatin, resulting in increased DNA damage and apoptosis.
The PI3K/Akt signaling pathway plays a crucial role in tumorigenesis, metastasis, and chemotherapy resistance.133 Eukaryotic translation elongation factor 1 delta (EEF1D) activates this pathway, promoting proliferation and drug resistance in cancer cells.134 In Xu et al (2022), EEF1D knockout in SKOV3 and cisplatin-resistant SKOV3 (SKOV/DDP) cells led to PI3K/Akt pathway inactivation, regulating Bax/Bcl-2 levels to enhance apoptosis and suppress DNA repair mechanisms. This increased sensitivity to cisplatin and reduced cancer cell survival.121
Runt-related transcription factor 1 (RUNX1), known for its role in leukemia, also promotes metastasis, angiogenesis, stemness, and chemoresistance in solid tumors.135 High RUNX1 expression in ovarian cancer correlates with poor survival in patients. RUNX1 suppresses cisplatin-induced apoptosis by upregulating BCL2 and suppressing the miR-17~92 cluster, which enhances apoptosis by targeting BCL2. Knockdown of RUNX1 using CRISPR/Cas9 increased apoptosis and cisplatin sensitivity in OVCAR3 and SKOV3 cells.122
Fibrillin 1 (FBN1), an extracellular matrix glycoprotein, influences proliferation, adhesion, and immune cell infiltration in the tumor microenvironment.136 FBN1 was highly expressed in cisplatin-resistant organoids and was associated with poor overall survival and progression-free survival. FBN1 knockout in cisplatin-resistant ovarian cancer organoids and OVCAR433 increased sensitivity to cisplatin by inhibiting glycolysis and angiogenesis. This effect was mediated through the downregulation of the vascular endothelial growth factor receptor 2 (VEGFR2)/signal transducer and activator of transcription 2 (STAT2) pathway and related genes, including vascular endothelial growth factor A (VEGFA), aldolase A (ALDOA), glucose transporter 1 (GLUT1), and angiopoietin-1 (Ang-1), as well as key genes in the FAK/AKT1 pathway. In subcutaneous cisplatin-resistant ovarian cancer organoid xenograft models using BALB/c nude mice, tumor size was significantly reduced with or without cisplatin treatment.123
Adipocyte fatty acid binding protein (FABP4), a member of the intracellular lipid-binding protein family, has been identified as a critical regulator of lipid-related metabolism in cancer cells. It has also been implicated in cancer cell aggressiveness and drug resistance.137 In the study by Mukherjee et al (2020), CRISPR knockout of FABP4 in OVCAR8 cells implanted in intraperitoneal xenograft immunocompromised athymic nude mice resulted in reduced metastatic tumors, including tumors in the omentum. This reduction appears to be associated with an increased ratio of 5-hydroxymethylcytosine (5-HMC) within the DNA. Furthermore, treatment with a FABP4 inhibitor in ID8 cells in intraperitoneal xenograft C57BL/6 mice reduced metastases. Combination treatment with carboplatin and the FABP4 inhibitor resulted in significantly smaller tumors than carboplatin treatment alone.124
In a study by Fan et al (2021), knockdown of SIK2 in OVCAR8 and SKOV3 cells increased sensitivity to carboplatin treatment. SIK2, a member of the AMPK family, plays a role in metabolic homeostasis and tumorigenesis.138 In SIK2-knockdown OVCAR8 cells, survivin—a protein that promotes tumor progression by deregulating apoptosis—was downregulated. This downregulation enhanced carboplatin-induced apoptosis.125
CPT1A is a key rate-limiting enzyme in fatty acid oxidation (FAO), that converts acyl-coenzyme As to acyl-carnitines. Recent studies have shown that FAO and CPT1A are associated with cancer growth and drug resistance.139 Huang et al (2021) demonstrated that FAO and oxidative phosphorylation (OXPHOS) metabolism are associated with platinum resistance in HGSOC. In the HGSOC intra-patient cell line PEO4, which overexpresses CPT1A, increased expression of the FAO/OXPHOS pathway was observed. Knockout of CPT1A in carboplatin-resistant PEO4 cells increased carboplatin sensitivity by upregulating carboplatin-induced reactive oxygen species, leading to increased DNA damage and apoptosis.126
Combination of CRISPR/Cas9 Gene Editing and Taxanes
Taxanes, such as paclitaxel, are widely used chemotherapeutic agents that target microtubules, disrupting cell division and inducing apoptosis in cancer cells.140 Combining CRISPR/Cas9 with taxanes allows researchers to identify and manipulate specific genes that influence microtubule dynamics and drug sensitivity, offering a promising approach to enhance therapeutic efficacy.
VASH2 accelerates angiogenesis and exhibits tubulin carboxypeptidase (TCP) activity related to microtubule polymerization.141 In Koyangi et al (2021), knockout of VASH2 in SKOV3 and SHIN-3 cells increased paclitaxel sensitivity by reducing TCP activity and detyrosinated tubulin levels, without affecting cisplatin sensitivity.127 Similarly, knockdown of B7 homolog 3 (B7-H3; CD276) in A2780 and OVCAR3 cells downregulated PI3K and AKT and reduced proliferation. Additionally, knockdown of BCL-2 in A2780 and OVCAR3 cells reduced B7-H3-induced chemoresistance to paclitaxel and cisplatin.128 B7-H3 was more highly expressed in ovarian tumors from patients with high malignancy compared to those with low malignancy.142 B7-H3 contributes to proliferation and drug resistance by activating the PI3K/AKT signaling pathway and upregulating downstream B-cell lymphoma 2 (BCL-2) in ovarian cancer cells.128
Combination of CRISPR/Cas9 Gene Editing and Other Drugs
In cancer therapy, inducing DNA damage to promote cell death is a common strategy.143 However, cancer cells employ multiple DNA repair mechanisms, with the non-homologous end joining (NHEJ) pathway being a critical mechanism for repairing DNA double-strand breaks (DSBs).144 Exonuclease 1 (Exo1), a 5′ to 3′ exonuclease, plays a key role in end processing during NHEJ.145 He et al (2020) observed increased expression of Exo1 in cisplatin- or doxorubicin-resistant SKOV3 cell lines. Knockout of Exo1 in these resistant cells increased sensitivity to ionizing radiation and reduced NHEJ efficiency. Similarly, knockdown of Exo1 using siRNA enhanced sensitivity to cisplatin and doxorubicin treatment.129
AT-rich interactive domain 1A (ARID1A), a subunit of the SWI-SNF complex, acts as a tumor suppressor in ovarian clear cell carcinoma (OCCC) and is mutated in approximately 50% of cases.146,147 ARID1A regulates proteins involved in the cell cycle and DNA repair. Loss of ARID1A in OCCC has been linked to resistance to platinum-based chemotherapy. ARID1A regulates proteins involved in the cell cycle and DNA repair, and its loss results in defective cell cycle control and an impaired G2/M DNA damage checkpoint.148 Gemcitabine, a pyrimidine antimetabolite, is sometimes used to treat OCCC after platinum-resistant recurrence.149 Kuroda et al (2019) demonstrated that ARID1A knockout in the RMG-1 OCCC cell line increased sensitivity to gemcitabine. Similarly, cytarabine, another pyrimidine antimetabolite, showed enhanced efficacy in ARID1A-knockout RMG-1 cells, unlike other drugs such as paclitaxel, carboplatin, or doxorubicin. This indicates that ARID1A loss is closely associated with the response to pyrimidine antimetabolites. Gemcitabine treatment in ARID1A-knockout RMG-1 cells increased the sub-G1 fraction of the cell cycle, indicating apoptosis induction, while no such effect was observed in ARID1A-proficient ES-2 cells. In vivo, gemcitabine suppressed tumor growth in ARID1A-deficient JHOC-9 xenografts in BALB/c-nu/nu mice but had no effect in ARID1A-proficient ES-2 xenografts. Additionally, a clinical case of an ARID1A-deficient OCCC patient who was resistant to paclitaxel and carboplatin showed a dramatic response to gemcitabine as a second-line treatment.130
In cancer stem cells, ATP-binding cassette (ABC) drug transporters such as breast cancer resistance protein (BCRP) and extracellular matrix (ECM) components like COL3A1 are linked to drug resistance.150,151 Aldehyde dehydrogenase isoform 1A1 (ALDH1A1) is a marker of ovarian cancer stem cells and is associated with chemoresistance.152 Nowacka et al (2022) reported that in the topotecan-resistant ovarian cancer cell line W1TR, ALDH1A1 expression was elevated compared to the drug-sensitive W1 line. Knockout of ALDH1A1 in W1TR cells increased sensitivity to topotecan treatment. However, in three-dimensional (3D) cell cultures conditions, ALDH1A1-knockout W1TR spheroids exhibited increased resistance to topotecan. Furthermore, BCRP and COL3A1 expression were upregulated in W1TR cells but were downregulated in ALDH1A1-knockout W1TR cells, and were nearly undetectable in drug-sensitive W1 cells. These findings suggests that factors such as spheroid density, ECM expression, and drug efflux capacity contribute to increased topotecan resistance in 3D spheroid cultures.131
Combination of CRISPR/Cas9 Gene Editing and PARP Inhibitors
The Cancer Genome Atlas Consortium reports that approximately 50% of ovarian high-grade serous carcinomas exhibit homologous recombination deficiency (HRD).153 HRD is an important biomarker for the efficacy of PARP inhibitor therapy in ovarian cancer.154 Mutations in transformation-related protein 53 (TRP53), BRCA1 and BRCA2 impair the repair of DNA double-strand breaks (DSBs) via homologous recombination (HR).155 In Walton et al (2016), knockout of TP53 and BRCA2 in ID8 ovarian cancer cells resulted in the loss of Rad51 foci formation, indicating HR deficiency. These cells showed increased sensitivity to the PARP inhibitor rucaparib compared to cells with only TRP53 knockout. Additionally, TRP53 and BRCA2 knockout in ID8 intraperitoneal xenografts in C57BL/6 mice led to slower orthotopic tumor growth compared to xenografts with only TP53 knockout.132
In a subsequent study by Walton et al (2017), BRCA1, Pten, and Nf1 were also knockout in TRP53-knockout ID8 cell clones. TRP53:BRCA1 and TRP53:BRCA2 double-knockout ID8 cells displayed homologous recombination deficiency and increased drug sensitivity to both rucaparib and the platinum drug cisplatin compared to TRP53-knockout cells. However, loss of Pten and Nf1 did not significantly affect sensitivity to either rucaparib or cisplatin.156
Combining the CRISPR/Cas9 System with Immunotherapy
Cancer immunotherapy enhances the immune system’s ability to fight cancer, often by targeting tumor-associated immune-suppressive mechanisms.157 Among these, immune checkpoint inhibitors have shown promising results in ovarian cancer. The most extensively studied immune checkpoint targets are programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1), which help reverse immunosuppressive signals in the tumor microenvironment.158
In Yahata et al (2019), PD-L1 was knockout in murine ovarian cancer ID8 cells and intraperitoneally inoculated into syngeneic mice. Mice with PD-L1 knockout cells exhibited prolonged survival and tumor shrinkage compared to controls. The therapeutic benefits were further enhanced with cisplatin treatment. Moreover, the anti-tumor effects of genetic PD-L1 knockout were stronger than pharmacological PD-L1 inhibition using anti-PD-L1 antibodies.159 In Paffenholz et al (2022), CRISPR/Cas9 was combined with transposon/transposase-based systems and in vivo organ electroporation (EPO-GEMM) to generate HGSOC model mice. CRISPR/Cas9 was used to target TP53, along with tumor suppressor genes (Pten, Rb) or an oncogene (Myc). Treatment of HGSOC model mice with a combination of cisplatin and anti–PD-1 antibodies resulted in greater tumor volume reduction and improved survival compared to treatment with cisplatin or anti–PD-1 alone.160
Conclusion
Ovarian cancer remains one of the most challenging malignancies due to its high relapse rate and persistent drug resistance. These challenges underscore the urgent need for innovative therapeutic strategies to improve patient outcomes. Key advancements include identifying candidate therapeutic target genes, developing nano-based CRISPR/Cas9 delivery systems, and combining CRISPR/Cas9 with conventional or novel drugs. CRISPR/Cas9 technology facilitates precise gene editing to explore the roles of genes in cancer proliferation, migration, invasion, EMT, cell cycle regulation, anoikis resistance, and angiogenesis. This versatile tool holds great potential for ovarian cancer gene therapy and drug development. Nano-based delivery systems further enhance therapeutic efficacy by enabling efficient, targeted delivery to tumor tissues or cells while minimizing side effects. The combination of CRISPR/Cas9 with nano-based systems offers a synergistic approach, enabling direct gene editing within tumors to amplify therapeutic outcomes. Addressing drug resistance remains a critical focus in ovarian cancer treatment. The integration of CRISPR/Cas9 with drug therapies has shown promise in overcoming resistance by targeting genes associated with chemoresistance, thereby increasing drug sensitivity in resistant cancer cells and improving treatment efficacy. The integration of CRISPR/Cas9 with nano-based systems and drug therapies could address existing research gaps and pave the way for future investigations in ovarian cancer treatment. Moreover, this approach has the potential to facilitate the clinical application of CRISPR/Cas9 mediated therapy in ovarian cancers by enhancing its efficacy and precision. In particular, the incorporation of CRISPR/Cas9 into precision medicine frameworks could enable personalized treatment strategies tailored to the genetic profile of individual patients, ultimately improving therapeutic outcomes and minimizing adverse effects. In summary, CRISPR/Cas9, in conjunction with nano-based delivery systems and drug therapies, represents a transformative approach in ovarian cancer treatment. Its application in identifying therapeutic target genes and overcoming drug resistance offers hope for improving patient outcomes and advancing cancer therapy. The combination of CRISPR/Cas9 with nano-based delivery systems and drug therapies holds significant promise for the future of ovarian cancer treatment. This strategy could enhance the safety and therapeutic efficiency of CRISPR/Cas9 gene therapy. Furthermore, the development of CRISPR/Cas9 as part of precision medicine approaches offers the potential for targeted, patient-specific treatments that maximize efficacy while reducing off-target effects. This promising therapeutic strategy may not only significantly advance the treatment of ovarian cancer, but also serve as a potential breakthrough for the treatment of other cancers in the future.
Acknowledgment
This research was supported by Korea Initiative for fostering University of Research and Innovation Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No. NRF2021M3H1A104892211), NRF grant funded by the Korea government (MSIT) (No. 2022R1A2C1004714), and the GRRC program of Gyeonggi province (GRRC Ajou2023-B04).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. doi:10.2353/ajpath.2010.100105
2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
3. Berek JS, Kehoe ST, Kumar L, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2018;143 Suppl 2:59–78. doi:10.1002/ijgo.12614
4. Stewart C, Ralyea C, Lockwood S. Ovarian Cancer: an Integrated Review. Semin Oncol Nurs. 2019;35(2):151–156. doi:10.1016/j.soncn.2019.02.001
5. Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int. 2014;2014:934261. doi:10.1155/2014/934261
6. Kurman RJ, Shih Ie M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186(4):733–747. doi:10.1016/j.ajpath.2015.11.011
7. Roth LM, Talerman A. Recent advances in the pathology and classification of ovarian germ cell tumors. Int J Gynecol Pathol. 2006;25(4):305–320. doi:10.1097/01.pgp.0000225844.59621.9d
8. Brown J, Friedlander M, Backes FJ, et al. Gynecologic Cancer Intergroup (GCIG) consensus review for ovarian germ cell tumors. Int J Gynecol Cancer. 2014;24(9 Suppl 3):S48–54. doi:10.1097/IGC.0000000000000223
9. Roth LM. Recent advances in the pathology and classification of ovarian sex cord-stromal tumors. Int J Gynecol Pathol. 2006;25(3):199–215. doi:10.1097/01.pgp.0000192271.22289.e6
10. Ray-Coquard I, Brown J, Harter P, et al. Gynecologic Cancer InterGroup (GCIG) consensus review for ovarian sex cord stromal tumors. Int J Gynecol Cancer. 2014;24(9 Suppl 3):S42–7. doi:10.1097/IGC.0000000000000249
11. Ledermann JA, Raja FA, Fotopoulou C, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24 Suppl 6:vi24–32. doi:10.1093/annonc/mdt333
12. McGuire WP 3rd, Markman M. Primary ovarian cancer chemotherapy: current standards of care. Br J Cancer. 2003;89 Suppl 3(Suppl 3):S3–8. doi:10.1038/sj.bjc.6601494
13. Sato S, Itamochi H. Neoadjuvant chemotherapy in advanced ovarian cancer: latest results and place in therapy. Ther Adv Med Oncol. 2014;6(6):293–304. doi:10.1177/1758834014544891
14. Lukanović D, Kobal B, Černe K. Ovarian cancer: treatment and resistance to pharmacotherapy. Reprod Med. 2022;3(2):127–140. doi:10.3390/reprodmed3020011
15. Wang L, Wang X, Zhu X, et al. Drug resistance in ovarian cancer: from mechanism to clinical trial. mol Cancer. 2024;23(1):66. doi:10.1186/s12943-024-01967-3
16. Eskander RN, Randall LM. Bevacizumab in the treatment of ovarian cancer. Biologics. 2011;5:1–5. doi:10.2147/BTT.S13071
17. Nakai H, Matsumura N. The roles and limitations of bevacizumab in the treatment of ovarian cancer. Int J Clin Oncol. 2022;27(7):1120–1126. doi:10.1007/s10147-022-02169-x
18. Zheng F, Zhang Y, Chen S, Weng X, Rao Y, Fang H. Mechanism and current progress of Poly ADP-ribose polymerase (PARP) inhibitors in the treatment of ovarian cancer. Biomed Pharmacother. 2020;123:109661. doi:10.1016/j.biopha.2019.109661
19. Smith M, Pothuri B. Appropriate Selection of PARP Inhibitors in Ovarian Cancer. Curr Treat Options Oncol. 2022;23(6):887–903. doi:10.1007/s11864-022-00938-4
20. Ortiz M, Wabel E, Mitchell K, Horibata S. Mechanisms of chemotherapy resistance in ovarian cancer. Cancer Drug Resistance. 2022;5(2):304. doi:10.20517/cdr.2021.147
21. Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi:10.1126/science.aat5011
22. Liu Q, Yang J, Xing Y, Zhao Y, Liu Y. Development of delivery strategies for CRISPR‐Cas9 genome editing. BMEMat. 2023;1(3). doi:10.1002/bmm2.12025
23. Aghamiri S, Talaei S, Ghavidel AA, et al. Nanoparticles-mediated CRISPR/Cas9 delivery: recent advances in cancer treatment. J Drug Delivery Sci Technol. 2020;56:101533.
24. Hii ARK, Qi X, Wu Z. Advanced strategies for CRISPR/Cas9 delivery and applications in gene editing, therapy, and cancer detection using nanoparticles and nanocarriers. J Mater Chem B. 2024;12:1467–1489. doi:10.1039/D3TB01850D
25. Newsom S, Parameshwaran HP, Martin L, Rajan R. The CRISPR-Cas mechanism for adaptive immunity and alternate bacterial functions fuels diverse biotechnologies. Front Cell Infect Microbiol. 2020;10:619763. doi:10.3389/fcimb.2020.619763
26. Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi:10.1126/science.1258096
27. Gostimskaya I. CRISPR-Cas9: a history of its discovery and ethical considerations of its use in genome editing. Biochemistry. 2022;87(8):777–788. doi:10.1134/S0006297922080090
28. Kick L, Kirchner M, Schneider S. CRISPR-Cas9: from a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered. 2017;8(3):280–286. doi:10.1080/21655979.2017.1299834
29. Arora L, Narula A. Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci. 2017;8:1932. doi:10.3389/fpls.2017.01932
30. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science. 2012;337(6096):816–821. doi:10.1126/science.1225829
31. Karvelis T, Gasiunas G, Siksnys V. Methods for decoding Cas9 protospacer adjacent motif (PAM) sequences: a brief overview. Methods. 2017;121-122:3–8. doi:10.1016/j.ymeth.2017.03.006
32. Thurtle-Schmidt DM, Lo TW. Molecular biology at the cutting edge: a review on CRISPR/CAS9 gene editing for undergraduates. Biochem mol Biol Educ. 2018;46(2):195–205. doi:10.1002/bmb.21108
33. Redman M, King A, Watson C, King D. What is CRISPR/Cas9?. Arch dis childhood-edu pract. 2016;101(4):213–215. doi:10.1136/archdischild-2016-310459
34. Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi:10.1146/annurev-biophys-062215-010822
35. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi:10.1016/j.cell.2014.05.010
36. Liu B, Saber A, Haisma HJ. CRISPR/Cas9: a powerful tool for identification of new targets for cancer treatment. Drug Discov Today. 2019;24(4):955–970. doi:10.1016/j.drudis.2019.02.011
37. Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16(4):707–717. doi:10.1038/mt.2008.20
38. Gupta RM, Musunuru K. Expanding the genetic editing tool kit: zFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124(10):4154–4161. doi:10.1172/JCI72992
39. Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi:10.1038/nbt.1755
40. Holkers M, Maggio I, Liu J, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41(5):e63. doi:10.1093/nar/gks1446
41. Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Delivery Rev. 2009;61(9):746–759. doi:10.1016/j.addr.2009.04.004
42. Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung V, Smith A. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 2015;43(7):3407–3419. doi:10.1093/nar/gkv226
43. Yan J, Bhadane R, Ran M, et al. Development of Aptamer-DNAzyme based metal-nucleic acid frameworks for gastric cancer therapy. Nat Commun. 2024;15(1):3684. doi:10.1038/s41467-024-48149-9
44. Yan J, Ma X, Liang D, et al. An autocatalytic multicomponent DNAzyme nanomachine for tumor-specific photothermal therapy sensitization in pancreatic cancer. Nat Commun. 2023;14(1):6905. doi:10.1038/s41467-023-42740-2
45. Yan J, Ran M, Shen X, Zhang H. Therapeutic DNAzymes: from structure design to clinical applications. Adv Mater. 2023;35(30):2300374. doi:10.1002/adma.202300374
46. Li F, Song N, Dong Y, et al. A proton-activatable DNA-based nanosystem enables co-delivery of CRISPR/Cas9 and DNAzyme for combined gene therapy. Angew Chem Int Ed Engl. 2022;61(9):e202116569. doi:10.1002/anie.202116569
47. Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. mol Ther Nucleic Acids. 2015;4:e264. doi:10.1038/mtna.2015.37
48. Rueda J, de Miguel Beriain I, Montoliu L. Affordable pricing of CRISPR treatments is a pressing ethical imperative. CRISPR J. 2024;7(5):220–226. doi:10.1089/crispr.2024.0042
49. Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387. doi:10.3389/fonc.2020.01387
50. Rafii S, Tashkandi E, Bukhari N, Al-Shamsi HO. Current status of CRISPR/Cas9 application in clinical cancer research: opportunities and challenges. Cancers. 2022;14(4):947. doi:10.3390/cancers14040947
51. Zhao Q, Qian Q, Cao D, Yang J, Gui T, Shen K. Role of BMI1 in epithelial ovarian cancer: investigated via the CRISPR/Cas9 system and RNA sequencing. J Ovarian Res. 2018;11(1):31. doi:10.1186/s13048-018-0406-z
52. Liu Q, Liu H, Li L, et al. ATAD2 predicts poor outcomes in patients with ovarian cancer and is a marker of proliferation. Int J Oncol. 2020;56(1):219–231. doi:10.3892/ijo.2019.4913
53. Jiang Y, Lim J, Wu KC, Xu W, Suen JY, Fairlie DP. PAR2 induces ovarian cancer cell motility by merging three signalling pathways to transactivate EGFR. Br J Pharmacol. 2021;178(4):913–932. doi:10.1111/bph.15332
54. Price C, Gill S, Ho ZV, et al. Genome-wide interrogation of human cancers identifies EGLN1 dependency in clear cell ovarian cancers. Cancer Res. 2019;79(10):2564–2579. doi:10.1158/0008-5472.CAN-18-2674
55. Ko EJ, Kim ET, Kim H, et al. Effect of human endogenous retrovirus-K env gene knockout on proliferation of ovarian cancer cells. Genes Genomics. 2022;44(9):1091–1097. doi:10.1007/s13258-022-01280-7
56. Liu W, Wang L, Zhang J, Cheng K, Zheng W, Ma Z. CC chemokine 2 promotes ovarian cancer progression through the MEK/ERK/MAP3K19 signaling pathway. Int J Mol Sci. 2023;24(13):10652.
57. Lu T, Bankhead A 3rd, Ljungman M, Neamati N. Multi-omics profiling reveals key signaling pathways in ovarian cancer controlled by STAT3. Theranostics. 2019;9(19):5478–5496. doi:10.7150/thno.33444
58. Zhao G, Wang Q, Gu Q, et al. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8(55):94666. doi:10.18632/oncotarget.21863
59. Haraguchi M, Sato M, Ozawa M. CRISPR/Cas9n-mediated deletion of the snail 1Gene (SNAI1) reveals its role in regulating cell morphology, cell-cell interactions, and gene expression in ovarian cancer (RMG-1) cells. PLoS One. 2015;10(7):e0132260. doi:10.1371/journal.pone.0132260
60. Huo W, Zhao G, Yin J, et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2017;8(1):57–64. doi:10.7150/jca.16723
61. Ji L, Zhao G, Zhang P, et al. Knockout of MTF1 inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2018;9(24):4578–4585. doi:10.7150/jca.28040
62. Cui Y, Wu BO, Flamini V, Evans BAJ, Zhou D, Jiang WG. Knockdown of EPHA1 using CRISPR/CAS9 suppresses aggressive properties of ovarian cancer cells. Anticancer Res. 2017;37(8):4415–4424. doi:10.21873/anticanres.11836
63. Zhang J, Li Y, Liu H, et al. Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis. J Exp Clin Cancer Res. 2022;41(1):24. doi:10.1186/s13046-022-02242-3
64. Kleinschmidt EG, Miller NLG, Ozmadenci D, et al. Rgnef promotes ovarian tumor progression and confers protection from oxidative stress. Oncogene. 2019;38(36):6323–6337. doi:10.1038/s41388-019-0881-8
65. Zhu W, Liu C, Lu T, et al. Knockout of EGFL6 by CRISPR/Cas9 mediated inhibition of tumor angiogenesis in ovarian cancer. Front Oncol. 2020;10:1451. doi:10.3389/fonc.2020.01451
66. Fix SM, Forget MA, Sakellariou-Thompson D, et al. CRISPR-mediated TGFBR2 knockout renders human ovarian cancer tumor-infiltrating lymphocytes resistant to TGF-beta signaling. J Immunother Cancer. 2022;10(7):e003750. doi:10.1136/jitc-2021-003750
67. Huff J. Mechanisms, chemical carcinogenesis, and risk assessment: cell proliferation and cancer. Am. J. Ind. Med. 1995;27(2):293–300. doi:10.1002/ajim.4700270213
68. Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3(5):362–374. doi:10.1038/nrc1075
69. Krakhmal NV, Zavyalova M, Denisov E, Vtorushin S, Perelmuter V. Cancer invasion: patterns and mechanisms. Acta Naturae. 2015;7(2 (25)):17–28. doi:10.32607/20758251-2015-7-2-17-28
70. Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30(3):372–378. doi:10.1002/stem.1035
71. Liu H, Wen Q, Yan S, et al. Tumor-promoting ATAD2 and its preclinical challenges. Biomolecules. 2022;12(8):1040. doi:10.3390/biom12081040
72. Han HJ, Huang QY, Huang LJ, Chang F, Diao QZ. Prognostic value of ATPase family, AAA+ domain containing 2 expression in human cancers: a systematic review and meta-analysis. Medicine. 2019;98(39):e17180. doi:10.1097/MD.0000000000017180
73. Adams MN, Ramachandran R, Yau MK, et al. Structure, function and pathophysiology of protease activated receptors. Pharmacol Ther. 2011;130(3):248–282. doi:10.1016/j.pharmthera.2011.01.003
74. Strocchi S, Reggiani F, Gobbi G, Ciarrocchi A, Sancisi V. The multifaceted role of EGLN family prolyl hydroxylases in cancer: going beyond HIF regulation. Oncogene. 2022;41(29):3665–3679. doi:10.1038/s41388-022-02378-8
75. Ko E-J, Cha H-J. The roles of human endogenous retroviruses (HERVs) in inflammation. Kosin Med J. 2021;36(2):69–78. doi:10.7180/kmj.2021.36.2.69
76. Ko EJ, Song KS, Ock MS, et al. Expression profiles of human endogenous retrovirus (HERV)-K and HERV-R Env proteins in various cancers. BMB Rep. 2021;54(7):368–373. doi:10.5483/BMBRep.2021.54.7.246
77. Hao Q, Vadgama JV, Wang P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal. 2020;18(1):82. doi:10.1186/s12964-020-00589-8
78. Liang R, Chen X, Chen L, et al. STAT3 signaling in ovarian cancer: a potential therapeutic target. J Cancer. 2020;11(4):837–848. doi:10.7150/jca.35011
79. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
80. Georgakopoulos-Soares I, Chartoumpekis DV, Kyriazopoulou V, Zaravinos A. EMT Factors and Metabolic Pathways in Cancer. Front Oncol. 2020;10:499. doi:10.3389/fonc.2020.00499
81. Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16:49. doi:10.1186/s12935-016-0326-1
82. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev mol Cell Biol. 2014;15(3):178–196. doi:10.1038/nrm3758
83. Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB, Cheng XC. MicroRNA-21 gene and cancer. Med Oncol. 2013;30(1):376. doi:10.1007/s12032-012-0376-8
84. Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther. 2013;138(2):255–271.
85. Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833(12):3481–3498. doi:10.1016/j.bbamcr.2013.06.026
86. Cai Q, Yan L, Xu Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene. 2015;34(25):3315–3324. doi:10.1038/onc.2014.264
87. Desrosiers R R, Fanélus I. Damaged proteins bearing L-isoaspartyl residues and aging: a dynamic equilibrium between generation of isomerized forms and repair by PIMT. Current aging sci. 2011;4(1):8–18. doi:10.2174/1874609811104010008
88. Miller NL, Lawson C, Kleinschmidt EG, Tancioni I, Uryu S, Schlaepfer DD. A non-canonical role for Rgnef in promoting integrin-stimulated focal adhesion kinase activation. J Cell Sci. 2013;126(Pt 21):5074–5085. doi:10.1242/jcs.135509
89. Folkman J, Merler E, Abernathy C, Williams G. Isolation of a tumor factor responsible for angiogenesis. J Exp Med. 1971;133(2):275. doi:10.1084/jem.133.2.275
90. Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2(3):213–219. doi:10.2147/vhrm.2006.2.3.213
91. Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69 Suppl 3:4–10. doi:10.1159/000088478
92. Kang J, Wang J, Tian J, Shi R, Jia H, Wang Y. The emerging role of EGFL6 in angiogenesis and tumor progression. Int J Med Sci. 2020;17(10):1320–1326. doi:10.7150/ijms.45129
93. Wojtowicz-Praga S. Reversal of tumor-induced immunosuppression by TGF-β inhibitors. Invest New Drugs. 2003;21:21–32. doi:10.1023/A:1022951824806
94. Wilczewska AZ, Niemirowicz K, Markiewicz KH, Car H. Nanoparticles as drug delivery systems. Pharmacol Rep. 2012;64(5):1020–1037. doi:10.1016/S1734-1140(12)70901-5
95. Sultana A, Zare M, Thomas V, Kumar TSS, Ramakrishna S. Nano-based drug delivery systems: conventional drug delivery routes, recent developments and future prospects. Med Drug Discovery. 2022;15:100134.
96. Yetisgin AA, Cetinel S, Zuvin M, Kosar A, Kutlu O. Therapeutic nanoparticles and their targeted delivery applications. Molecules. 2020;25(9):2193. doi:10.3390/molecules25092193
97. Basha M. Nanotechnology as a promising strategy for anticancer drug delivery. Curr Drug Deliv. 2018;15(4):497–509. doi:10.2174/1567201814666170516114411
98. Mudshinge SR, Deore AB, Patil S, Bhalgat CM. Nanoparticles: emerging carriers for drug delivery. Saudi Pharm J. 2011;19(3):129–141. doi:10.1016/j.jsps.2011.04.001
99. Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36(2):173–185. doi:10.1016/j.tibtech.2017.11.006
100. Rosenblum D, Gutkin A, Kedmi R, et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020;6(47):eabc9450. doi:10.1126/sciadv.abc9450
101. He ZY, Zhang YG, Yang YH, et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum Gene Ther. 2018;29(2):223–233. doi:10.1089/hum.2017.209
102. Lu H, Zhang Q, He S, et al. Reduction-sensitive fluorinated-pt(iv) universal transfection nanoplatform facilitating CT45-targeted CRISPR/dCas9 activation for synergistic and individualized treatment of ovarian cancer. Small. 2021;17(41):e2102494. doi:10.1002/smll.202102494
103. Li L, Song L, Liu X, et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11(1):95–111. doi:10.1021/acsnano.6b04261
104. Jain PK, Lo JH, Rananaware S, et al. Non-viral delivery of CRISPR/Cas9 complex using CRISPR-GPS nanocomplexes. Nanoscale. 2019;11(44):21317–21323. doi:10.1039/C9NR01786K
105. Cornebise M, Narayanan E, Xia Y, et al. Discovery of a novel amino lipid that improves lipid nanoparticle performance through specific interactions with mRNA. Adv Funct Mater. 2021;32(8):2106727.
106. Pathania R, Ramachandran S, Elangovan S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. doi:10.1038/ncomms7910
107. Liu Z, Liao Z, Chen Y, Han L, Yin Q, Xiao H. Application of various delivery methods for CRISPR/dCas9. Mol Biotechnol. 2020;62(8):355–363. doi:10.1007/s12033-020-00258-8
108. Baylis F. Counterpoint: the potential harms of human gene editing using CRISPR-Cas9. Clin Chem. 2018;64(3):489–491. doi:10.1373/clinchem.2017.278317
109. Coscia F, Lengyel E, Duraiswamy J, et al. Multi-level proteomics identifies CT45 as a chemosensitivity mediator and immunotherapy target in ovarian cancer. Cell. 2018;175(1):159–70e16. doi:10.1016/j.cell.2018.08.065
110. Gad H, Koolmeister T, Jemth AS, et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508(7495):215–21. doi:10.1038/nature13181
111. Gao W, Cao W, Sun Y, et al. AuNP flares-capped mesoporous silica nanoplatform for MTH1 detection and inhibition. Biomaterials. 2015;69:212–221. doi:10.1016/j.biomaterials.2015.08.021
112. Parmar MK, Ledermann JA, Colombo N, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099–2106.
113. Nero C, Ciccarone F, Pietragalla A, et al. Ovarian cancer treatments strategy: focus on PARP inhibitors and immune check point inhibitors. Cancers. 2021;13(6):1298. doi:10.3390/cancers13061298
114. Zhang C, Xu C, Gao X, Yao Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics. 2022;12(5):2115. doi:10.7150/thno.69424
115. Mosca L, Ilari A, Fazi F, Assaraf YG, Colotti G. Taxanes in cancer treatment: activity, chemoresistance and its overcoming. Drug Resist Updat. 2021;54:100742. doi:10.1016/j.drup.2020.100742
116. Curtin N. PARP inhibitors for anticancer therapy. Biochem Soc Trans. 2014;42(1):82–88. doi:10.1042/BST20130187
117. Soltau J, Drevs J. Mode of action and clinical impact of VEGF signaling inhibitors. Expert Rev Anticancer Ther. 2009;9(5):649–662. doi:10.1586/era.09.19
118. Cornelison R, Llaneza DC, Landen CN. Emerging therapeutics to overcome chemoresistance in epithelial ovarian cancer: a mini-review. Int J mol Sci. 2017;18(10):2171. doi:10.3390/ijms18102171
119. Norouzi-Barough L, Sarookhani MR, Sharifi M, Moghbelinejad S, Jangjoo S, Salehi R. Molecular mechanisms of drug resistance in ovarian cancer. J Cell Physiol. 2018;233(6):4546–4562. doi:10.1002/jcp.26289
120. Saber A, Liu B, Ebrahimi P, Haisma HJ. CRISPR/Cas9 for overcoming drug resistance in solid tumors. Daru. 2020;28(1):295–304. doi:10.1007/s40199-019-00240-z
121. Xu Q, Liu Y, Wang S, et al. Interfering with the expression of EEF1D gene enhances the sensitivity of ovarian cancer cells to cisplatin. BMC Cancer. 2022;22(1):1–15. doi:10.1186/s12885-022-09699-7
122. Xiao L, Peng Z, Zhu A, et al. Inhibition of RUNX1 promotes cisplatin-induced apoptosis in ovarian cancer cells. Biochem Pharmacol. 2020;180:114116. doi:10.1016/j.bcp.2020.114116
123. Wang Z, Chen W, Zuo L, et al. The Fibrillin-1/VEGFR2/STAT2 signaling axis promotes chemoresistance via modulating glycolysis and angiogenesis in ovarian cancer organoids and cells. Cancer Commun. 2022;42(3):245–265. doi:10.1002/cac2.12274
124. Mukherjee A, Chiang C-Y, Daifotis HA, et al. Adipocyte-induced FABP4 expression in ovarian cancer cells promotes metastasis and mediates carboplatin resistance. Cancer Res. 2020;80(8):1748–1761. doi:10.1158/0008-5472.CAN-19-1999
125. Fan D, Yang H, Mao W, et al. A novel salt inducible kinase 2 inhibitor, ARN-3261, sensitizes ovarian cancer cell lines and xenografts to carboplatin. Cancers. 2021;13(3):446. doi:10.3390/cancers13030446
126. Huang D, Chowdhury S, Wang H, et al. Multiomic analysis identifies CPT1A as a potential therapeutic target in platinum-refractory, high-grade serous ovarian cancer. Cell Rep Med. 2021;2(12):100471. doi:10.1016/j.xcrm.2021.100471
127. Koyanagi T, Saga Y, Takahashi Y, et al. Knockout of vasohibin-2 reduces tubulin carboxypeptidase activity and increases paclitaxel sensitivity in ovarian cancer. Cancer Med. 2021;10(8):2732–2739. doi:10.1002/cam4.3841
128. Zhou L, Zhao Y. B7-H3 induces ovarian cancer drugs resistance through an PI3K/AKT/BCL-2 signaling pathway. Cancer Manag Res. 2019;11:10205–10214. doi:10.2147/CMAR.S222224
129. He D, Li T, Sheng M, Yang B. Exonuclease 1 (Exo1) participates in mammalian non-homologous end joining and contributes to drug resistance in ovarian cancer. Med Sci Monit. 2020;26:e918751. doi:10.12659/MSM.918751
130. Kuroda T, Ogiwara H, Sasaki M, et al. Therapeutic preferability of gemcitabine for ARID1A-deficient ovarian clear cell carcinoma. Gynecologic Oncol. 2019;155(3):489–498. doi:10.1016/j.ygyno.2019.10.002
131. Nowacka M, Ginter-Matuszewska B, Świerczewska M, Sterzyńska K, Nowicki M, Januchowski R. Effect of ALDH1A1 gene knockout on drug resistance in paclitaxel and topotecan resistant human ovarian cancer cell lines in 2D and 3D model. Int J mol Sci. 2022;23(6):3036. doi:10.3390/ijms23063036
132. Walton J, Blagih J, Ennis D, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 2016;76(20):6118–6129. doi:10.1158/0008-5472.CAN-16-1272
133. Qiao M, Sheng S, Pardee AB. Metastasis and AKT activation. Cell Cycle. 2008;7(19):2991–2996. doi:10.4161/cc.7.19.6784
134. Xu H, Yu S, Peng K, et al. The role of EEF1D in disease pathogenesis: a narrative review. Ann Transl Med. 2021;9(20):1600. doi:10.21037/atm-21-5025
135. Lin TC. RUNX1 and cancer. Biochim Biophys Acta Rev Cancer. 2022;1877(3):188715. doi:10.1016/j.bbcan.2022.188715
136. Mahdizadehi M, Saghaeian Jazi M, Mir SM, Jafari SM. Role of fibrilins in human cancer: a narrative review. Health Sci Rep. 2023;6(7):e1434. doi:10.1002/hsr2.1434
137. Sun N, Zhao X. Therapeutic implications of FABP4 in cancer: an emerging target to tackle cancer. Front Pharmacol. 2022;13:948610. doi:10.3389/fphar.2022.948610
138. Sun Z, Jiang Q, Li J, Guo J. The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis. Signal Transduct Target Ther. 2020;5(1):150. doi:10.1038/s41392-020-00265-w
139. Schlaepfer IR, Joshi M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 2020;161(2). doi:10.1210/endocr/bqz046
140. Long BH, Fairchild CR. Paclitaxel inhibits progression of mitotic cells to G1 phase by interference with spindle formation without affecting other microtubule functions during anaphase and telephase. Cancer Res. 1994;54(16):4355–4361.
141. Nieuwenhuis J, Adamopoulos A, Bleijerveld OB, et al. Vasohibins encode tubulin detyrosinating activity. Science. 2017;358(6369):1453–1456. doi:10.1126/science.aao5676
142. Picarda E, Ohaegbulam KC, Zang X. Molecular Pathways: targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin Cancer Res. 2016;22(14):3425–3431. doi:10.1158/1078-0432.CCR-15-2428
143. Woods D, Turchi JJ. Chemotherapy induced DNA damage response: convergence of drugs and pathways. Cancer Biol Ther. 2013;14(5):379–389. doi:10.4161/cbt.23761
144. Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev mol Cell Biol. 2017;18(8):495–506. doi:10.1038/nrm.2017.48
145. Bahmed K, Seth A, Nitiss KC, Nitiss JL. End-processing during non-homologous end-joining: a role for exonuclease 1. Nucleic Acids Res. 2011;39(3):970–978. doi:10.1093/nar/gkq886
146. Jones S, Wang T-L, Shih I-M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. doi:10.1126/science.1196333
147. Katagiri A, Nakayama K, Rahman MT, et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25(2):282–288. doi:10.1038/modpathol.2011.161
148. Mandal J, Mandal P, Wang TL, Shih IM. Treating ARID1A mutated cancers by harnessing synthetic lethality and DNA damage response. J Biomed Sci. 2022;29(1):71. doi:10.1186/s12929-022-00856-5
149. Lorusso D, Di stefano A, Fanfani F, Scambia G. Role of gemcitabine in ovarian cancer treatment. Ann Oncol. 2006;17 Suppl 5:v188–94. doi:10.1093/annonc/mdj979
150. Moitra K, Lou H, Dean M. Multidrug efflux pumps and cancer stem cells: insights into multidrug resistance and therapeutic development. Clin Pharmacol Ther. 2011;89(4):491–502. doi:10.1038/clpt.2011.14
151. Kesh K, Gupta VK, Durden B, et al. Therapy resistance, cancer stem cells and ECM in cancer: the matrix reloaded. Cancers. 2020;12(10):3067. doi:10.3390/cancers12103067
152. Nwani NG, Condello S, Wang Y, et al. A novel ALDH1A1 inhibitor targets cells with stem cell characteristics in ovarian cancer. Cancers. 2019;11(4):502. doi:10.3390/cancers11040502
153. Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi:10.1038/nature10166
154. Jiang X, Li X, Li W, Bai H, Zhang Z. PARP inhibitors in ovarian cancer: sensitivity prediction and resistance mechanisms. J Cell Mol Med. 2019;23(4):2303–2313. doi:10.1111/jcmm.14133
155. Huo Y, Selenica P, Mahdi AH, et al. Genetic interactions among Brca1, Brca2, Palb2, and Trp53 in mammary tumor development. NPJ Breast Cancer. 2021;7(1):45. doi:10.1038/s41523-021-00253-5
156. Walton JB, Farquharson M, Mason S, et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci Rep. 2017;7(1):16827. doi:10.1038/s41598-017-17119-1
157. Dillman RO. Cancer immunotherapy. Cancer Biother Radiopharm. 2011;26(1):1–64. doi:10.1089/cbr.2010.0902
158. Yang C, Xia BR, Zhang ZC, Zhang YJ, Lou G, Jin WL. Immunotherapy for ovarian cancer: adjuvant, combination, and neoadjuvant. Front Immunol. 2020;11:577869. doi:10.3389/fimmu.2020.577869
159. Yahata T, Mizoguchi M, Kimura A, et al. Programmed cell death ligand 1 d isruption by clustered regularly interspaced short palindromic repeats/Cas9‐genome editing promotes antitumor immunity and suppresses ovarian cancer progression. Cancer Sci. 2019;110(4):1279–1292. doi:10.1111/cas.13958
160. Paffenholz SV, Salvagno C, Ho YJ, et al. Senescence induction dictates response to chemo- and immunotherapy in preclinical models of ovarian cancer. Proc Natl Acad Sci U S A. 2022;119(5). doi:10.1073/pnas.2117754119.
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.