Back to Journals » Cancer Management and Research » Volume 16
Tiaogan Bushen Xiaoji Formula Enhances the Sensitivity of Estrogen Receptor- Positive Breast Cancer to Tamoxifen by Inhibiting the TGF-β/SMAD Pathway
Authors Lu J, Li Z, Liu X, Xu B, Zhang W
Received 28 May 2024
Accepted for publication 29 August 2024
Published 9 September 2024 Volume 2024:16 Pages 1189—1204
DOI https://doi.org/10.2147/CMAR.S477399
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Jiafeng Lu,1,* Zhaoyan Li,2,* Xingjing Liu,2 Bin Xu,1 Weiyu Zhang2
1Department of Pharmacy, Ruijin Hospital, Shanghai Jiaotong University, Shanghai, People’s Republic of China; 2Department of Traditional Chinese Medicine, Ruijin Hospital, Shanghai Jiaotong University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Weiyu Zhang, Department of Traditional Chinese Medicine, Ruijin Hospital, Shanghai Jiaotong University, Shanghai, 200025, People’s Republic of China, Tel +86-021-67888394 ; Tel +86-13818103558, Email [email protected] Bin Xu, Department of Pharmacy, Ruijin Hospital, Shanghai Jiaotong University, Shanghai, 200025, People’s Republic of China, Tel +86-021-67888176 ; Tel +86-18917762607, Email [email protected]
Background: The resistance to endocrine therapy can lead to recurrence and metastasis of breast cancer (BC), affecting the survival period. Tiaogan Bushen Xiaoji (TGBSXJ) Formula, a traditional Chinese medicine (TCM) decoction, has been widely used in the treatment of estrogen receptor-positive (ER+) BC. However, the underlying mechanism of TGBSXJ Formula in ER+BC treatment has not been totally elucidated.
Methods: Network pharmacology (NP) and RNA sequencing were used to predict the candidate ingredients and explore the potential targets of TGBSXJ Formula. Then, the results of NP and RNA sequencing were investigated by in vitro experiments.
Results: Active ingredients of TGBSXJ Formula mainly included Mangiferin, Rutin, Anemarrhena asphodeloides saponin BII, Ganoderic acid A and Acacetin, etc. A protein-protein interaction (PPI) network was created based on the active ingredients of TGBSXJ Formula and target genes of ER+ BC, in which TGF-β, MMP2 and SMAD3 were defined as the hub genes. In vitro experiments showed that TGBSXJ Formula significantly inhibited the viability, colony ability and migration of ER+ BC cells, and significantly increased the sensitivity to TAM. Western blot analysis showed that TGBSXJ Formula significantly downregulated TGF-β, E-cadherin, MMP2, MMP9, N-cadherin, p-Smad2 and p-Smad3 in ER+ BC cells.
Conclusion: TGBSXJ Formula increases the sensitivity of ER+ BC cells to TAM by inhibiting the TGF-β/Smad signaling pathway.
Keywords: estrogen receptor-positive breast cancer, network pharmacology, traditional Chinese medicine, endocrine resistance
Background
Breast cancer (BC), a predominant malignancy in women, accounts for approximately 30% of newly diagnosed cancers worldwide,1 The five-year survival of metastatic BC is lower than 30%.2 Pathologically, nearly 70% of BCs are estrogen receptor positive (ER+). Because of their strong dependency on the estrogen-estrogen receptor axis, estrogen inhibitors and estrogen receptor antagonists have been the main treatments of ER+BC for decades.3 The National Comprehensive Cancer Network (NCCN) recommends a long-term endocrine therapy for 5–10 years after surgery to ER+BC patients, aiming to prevent metastases, local recurrences and contralateral tumors.4
Tamoxifen (TAM), a synthetic non-steroidal anti-estrogenic agent, has been a common hormonal therapy for both premenopausal and postmenopausal women with either early stage or advanced estrogen receptor alpha-positive (ERα+) BC.5 Regardless of menopausal status, a 5-year TAM-based endocrine therapy significantly reduces the risk of distant and local recurrences of BCs by 10%-30%.6
Despite its significant contributions in reducing recurrence and mortality risks, over 40% of TAM-treated BC patients eventually develop acquired resistance, severely compromising oncologic outcomes and contributing to 90% of cancer-related deaths.7 This resistance is often attributed to ER deficiency, structural and functional abnormalities, tumor heterogeneity, and complex interactions between estrogen signaling networks and other cellular pathways.8 Thus, effective approaches to reduce the dosage of TAM and its resistance in ER+BC patients are urgently needed.
Traditional Chinese Medicine (TCM), renowned for its anti-cancer and chemoprotective properties with low toxicities, have been traditionally utilized as adjuvant therapies for cancers.9 TCM offer synergistic anti-cancer effects, enhance chemosensitivity, alleviate drug resistance and adverse effects, reduce toxicity, provide pain relief, and improve patients’ quality of life.10 The therapy of “Tiaogan Bushen Xiaoji (Regulating Liver, Nourishing Kidney, and Eliminating Accumulation)” is a TCM treatment for breast cancer, summarized by Prof. Shen Xiaoheng, a renowned TCM practitioner in Shanghai, and it has achieved good clinical efficacy, especially in endocrine therapy resistance.
Tiaogan Bushen Xiaoji (TGBSXJ) Formula is a type of TCM decoction composed of Astragalus membranaceus (Fisch). Bge., Ganoderma lucidum (Leyss. Ex Fr). Karst., Atractylodes macrocephala Koidz., Anemarrhena asphodeloides Bge., Citrus reticulata Blanco cv., Buthus martensii Karsch, Prunella vulgaris L. and Curcuma phaeocaulis Val., formulated with the principles of “soothing liver, nourishing kidney and eliminating pathogenic factors”.11
Due to the multi-targeted nature of human diseases and the complex components of TCM herbal products, network pharmacology (NP) has emerged to predict the possible pharmacological mechanisms of TCM herbs. It links drug actions and disease targets, and quantitatively interprets the overall regulatory mechanisms of drugs.12
In the present study, we constructed a “TGBSXJ Formula-active ingredients-targets” network based on the active ingredients of TGBSXJ Formula and targets of ER+ BC screened from online databases. A protein-protein interaction (PPI) network was created to obtain the hub genes by calculating the degree of network. Then, enrichment analyses of interested targets were performed to screen the potential signaling pathways of involved in the efficacy of TGBSXJ Formula in ER+ BC. In vitro experiments were finally conducted to validate the predicted molecular mechanism of TGBSXJ Formula. A schematic diagram of this working is depicted in Figure 1.
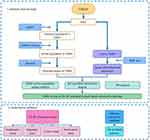 |
Figure 1 The workflow of network pharmacology analysis and validation of TGBSXJ Formula on ER+ BC. |
Methods
Components and Preparation of TGBSXJ Formula
The TCM decoction pieces were provided by Shanghai Wanshi cheng Pharmaceutical Co., Ltd (China) and identified by InnoiHealth Co., Ltd (China). Morphological, microscopic, and phytochemical identifications were conducted in accordance with the Pharmacopoeia of the People’s Republic of China (2020 edition). Astragalus membranaceus (Fisch). Bge. (12 g), Ganoderma lucidum (Leyss. Ex Fr). Karst. (9 g), Atractylodes macrocephala Koidz. (9 g), Anemarrhena asphodeloides Bge. (9 g), Citrus reticulata Blanco cv. (15 g), Buthus martensii Karsch (3 g), Prunella vulgaris L. (15 g) and Curcuma phaeocaulis Val. (15 g) were weighed and processed in a herbal decocting machine pot (YJD20L, Beijing Donghuayuan Medicine Equipment). After adding 1 L of distilled water and boiling for 1 h, herbal liquid was collected and filtered using an 80-mesh sieve. The liquid was concentrated in a vacuum concentrator (Rotavapor R-200, Buchiglas China Co., Ltd.) at 60~80°C and pressure ranging from −0.03 to −0.09 MPa. Using vacuum Freeze-dryer (LGJ-18T, Movel Scientific Instrument Co., Ltd) with a series of pre-set parameters (cold trap temperature, −81.1°C, compressor temperature, −13.0°C, vacuum pressure 56 Pa, drying time, 72 hours), the freeze-dried concentrated extract was obtained at a yield of 32.1% (w/w, dried extracts/crude herbs).
Screening of Active Ingredients in TGBSXJ Formula and Their Targets
The active ingredients of Astragalus membranaceus (Fisch). Bge., Ganoderma lucidum (Leyss. Ex Fr). Karst., Atractylodes macrocephala Koidz., Anemarrhena asphodeloides Bge., Citrus reticulata Blanco cv., Buthus martensii Karsch, Prunella vulgaris L. and Curcuma phaeocaulis Val. in TGBSXJ Formula were searched through the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://old.tcmsp-e.com/tcmsp.php) (TCMSP Version 2.3),13 and those with an oral bioavailability (OB) ≥30% and drug likeness (DL) ≥ 0.10 were selected. The Gene symbols were obtained from the UniProt database (https://www.uniprot.org) (UniProt consortium 2023).14
An Intersection Dataset of Targets of TGBSXJ Formula and ER+ BC
With “Estrogen receptor-positive breast cancer” as the search term, the potential targets of ER+ BC were selected by the DisGeNET database (https://www.disgenet.org) (version 7.0).15 A Venn diagram was drawn to clarify the intersection dataset between the ER+ BC-related targets and the potential targets of TGBSXJ Formula.
Construction of a “TGBSXJ Formula-Active Ingredients-Targets” Network and a PPI Network
A “TGBSXJ Formula-active-ingredients-targets” network was constructed by Cytoscape 3.7.1, and its built-in tool ‘Network Analyzer’ was used to calculate the degree of each node and screen major active ingredients. A PPI network was constructed using the STRING database (https://cn.string-db.org/) (version 12.0).16 Modular clustering of the protein network was conducted to obtain the core proteins with the highest degrees by the MCODE plugin in Cytoscape (version 7.2).
Enrichment Analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov) (version 2021).17 GO terms in biological process (BP), molecular function (MF) and cellular component (CC), and enriched signaling pathways were depicted in bubble plots. All the algorithm of the database are shown in Table 1.
 |
Table 1 The Algorithm of the Database |
RNA Sequencing
MCF-7 cells induced with blank control or Dulbecco’s Modified Eagle Medium (DMEM) containing TGBSXJ Formula (1200μg/mL) in five biological replicates were cultured for 48 hours. Total RNA extracted by TRIzol was examined for RNA integrity using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, US). RNA concentration and purity were measured using the Qubit® 3.0 Fluorometer (Life Technologies, CA, USA) and Nanodrop One spectrophotometer (Thermo Fisher Scientific Inc, USA), respectively. RNA sequencing was performed in Shanghai Sinotech Genomics Co., Ltd.
Quantitative Analysis of Major Components
Chromatographic separations were performed using an Agilent 1290 UPLC (Agilent Technologies, Santa Clara, CA, US) and a Thermo Orbitrap Fusion (ThermoFisher Scientific, USA). The samples were collected on a Waters/ACQUITY UPLC HSS T3 Column, 100*2.1 mm, 1.8 μm column with a temperature of 40°C, and a flow rate of 0.4 mL/min. In positive (POS) ion mode, the mobile phase consisted of solvent A (water+0.1% formic acid) and solvent B (acetonitrile), and the gradient elution conditions were set as follows: 0–1 min, 5% phase B; 1–18 min, 95% phase B; 18–20 min, 95% phase B; 20–20.5 min, 5% phase B, 20.5–25 min, 5% phase B; 25 min, stop. One primary mass spectrometry scan (100 ms) triggered 10 secondary mass spectrometry scans (500 ms), and both the first and secondary scanning ranges were 150 m/z - 1000 m/z. To evaluate the stability of the HPLC-MS during acquisition, TGBSXJ Formula sample was acquired after the standard samples.
Cell Culture and Treatment
Human ER+ BC cell lines MCF-7 and T47D were provided by the Chinese Academy of Sciences. MCF-7 cells were cultured in DMEM, and T47D cells were cultured in Roswell Park Memorial Institute (RPMI) 1640, both types of them containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were placed in an incubator with 5% CO2 at 37 °C.
MCF-7 cells were treated with blank control, 10 μM TAM, 1200 μmol/L TGBSXJ Formula, 10 μM TAM + 1200 μmol/L TGBSXJ Formula or 5 μM TAM + 1200 μmol/L TGBSXJ Formula for indicated time points. T47D cells were treated with blank control, 13 μM TAM, 780 μmol/L TGBSXJ Formula, 13 μM TAM + 780 μmol/L TGBSXJ Formula or 6.5 μM TAM + 780 μmol/L TGBSXJ Formula for indicated time points.
Cell Viability Assay
The viability of the ER+ BC cell lines MCF-7 and T47D was detected by CCK-8 assays. Cells were seeded in a 96-well plate with 3000 cells per well. After a 24-hour culture, the cells were induced with TGBSXJ Formula (0, 125, 250, 500, 1000, 2000, 4000 and 8000 μg/mL) or TAM (234–118-0, Sigma-Aldrich; 0, 1, 2, 4, 8, 16 and 64 μmol/L) at varying concentrations for 48 hours. Fresh medium containing 10% of Cell Counting Kit-8 (CCK-8) solution was added per well and cultured for 2 hours, followed by the measurement of optical density (OD) at 490 nm using a Biotek Synergy microplate reader.
Wound Healing Assay
Wound healing assays were performed to evaluate the capacity of cell migration and repair processes in damaged tissues. Cells were seeded in a 6-well plate and allowed to grow to confluence. A straight line was plotted per well using the tip of a pipette. After the induction of TGBSXJ Formula and/or TAM for 0 hours and 48 hours, cell migration to the wound space was captured and wound healing rate was calculated using Image J. Wound healing rate (%) = (wound width at 0 hours – wound width at 24 hours) / wound width at 0 hours × 100%.
Colony Formation Assay
Colony formation assays were performed to further determine the inhibitory effect of TGBSXJ Formula on the tumorigenicity of ER+ BC cells. Every 3×102 MCF-7 and T47D cells were seeded in a 6-well plate and incubated overnight. After induction of TGBSXJ Formula and/or TAM for seven days, cell colonies were fixed in 4% paraformaldehyde and stained with crystal violet. Colonies containing 30 single cells or more were captured under a stereomicroscope and counted.
Migration Assays
Transwell assays were performed to evaluate inhibitory effect of TGBSXJ Formula on the penetrating ability of ER+ BC cells. To For the migration assay, every 5×103 cells were seeded on the upper chamber (8μm) in24-well plates (Corning) with 300μL of serum-free medium containing TGBSXJ Formula and/or TAM. 600 μL of fresh medium containing 1% FBS was added in the bottom chamber. After a 24-h culture, the cells having migrated to the bottom were stained in 0.1% crystal violet for 30 minutes and captured under a microscope. Similarly, cells were seeded on the upper chamber of the Transwell insert with 50 μL of pre-coated Matrigel and 500 μL of fresh medium containing 10% FBS was added in the bottom chamber. Cells having invaded to the bottom at 24 hours were stained and captured.
Western Blot
Western blotting was performed to analyze the protein expression levels in both MCF-7 and T47D cells following different treatment regimens. Total proteins were extracted from cells using RIPA buffer containing protease and phosphatase inhibitors. Protein lysates, obtained following a 30-minute incubation on ice, a 15-second sonication and a 20-minute centrifugation at 4°C and 12,000 rpm, were quantified by BCA assay (Beyotime, Shanghai, China), loaded on 10% SDS-PAGE gels, and transferred to polyvinylidene fluoride membranes (Invitrogen, MA, USA). They were immersed in blocking buffer (EpiZyme, Shanghai, China) and incubated with the following primary antibodies overnight at 4°C and in a 1:1000 ratio: anti-TGF-β (Proteintech, IL, USA), anti-E-cadherin (CST, MA, USA), anti-MMP2 (CST, MA, USA), anti-MMP9 (CST, MA, USA), anti-N-cadherin (CST, MA, USA), anti-p-Smad2 (CST, MA, USA) and anti-p-Smad3 (CST, MA, USA). Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000, Abcam, Cambridge, UK) at room temperature for 1 hour. Protein bands were visualized using Immobilon Western Chemiluminescent HRP Substrate (Millipore, MA, USA) and quantified using Image J.
Statistical Analysis
SPSS 26.0 and GraphPad Prism 6.01 were used for statistical analysis and figure formatting, respectively. Measurement data with a normal distribution were compared using the independent sample t-test between groups, and one-way ANOVA among three and more groups. A non-parametric rank sum test was performed to compare non-normal measurement data. P<0.05 was considered as statistically significant.
Results
Active Ingredients in TGBSXJ Formula and Potential Targets of ER+ BC
To predict the candidate active ingredients and potential targets in TGBSXJ Formula. A total of 176 active ingredients from 9 TCM herbs constituting TGBSXJ Formula were screened in the TCMSP database, including 56 ingredients from the Astragalus membranaceus (Fisch). Bge., 89 from Ganoderma lucidum (Leyss. Ex Fr). Karst., 51 from Atractylodes macrocephala Koidz., 51 from Anemarrhena asphodeloides Bge., 11 from Citrus reticulata Blanco cv., 29 from Hedyotis diffusa Willd., 49 from Prunella vulgaris L., and 63 from Curcuma phaeocaulis Val. Moreover, 450 target genes of active ingredients in TGBSXJ Formula were screened using the TCMSP and UniProt.
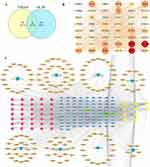
A total of 510 potential targets of ER+ BC were screened through the DisGeNET database. Then, a set of intersection data containing 60 targets of TGBSXJ Formula and ER+ BC was obtained (Figure 2A).
PPI Network
PPI networks were constructed by the STRING database. Through topological analyses, 60 nodes and 737 edges with an average degree of 24.32 and betweenness of 2.93 were obtained. TGF-β1, MMP2 and PSMAD3 were selected as the candidate targets for further analyses because of their higher degrees (Figure 2B).
A network of “TGBSXJ Formula-active ingredients-ER+ BC ” was drawn by Cytoscape 3.7.1, yielding 51 nodes and 1912 edges with an average degree of 25.32.
Sorted by the degree value, the core candidate active ingredients of TGBSXJ Formula were identified as follows: Mangiferin, Hesperidin, Rutin, Anemarrhena asphodeloides saponin BII, Ganoderic acid A, Acacetin, Anemarrhena saponin A-III and Desmethylpinosylvin, and TGF-β1, MMP2 and PSMAD3 were also captured as potential targets (Figure 2C).
GO and KEGG Pathway Enrichment Analyses
GO and KEGG pathway enrichment analyses of the intersection dataset were performed using the DAVID database. A total of 586 GO entries were obtained, including 463, 42 and 81 terms of BP, CC and MF, respectively. The major GO entries included protein binding, enzyme binding, positive regulation of transcription, apoptotic process, RNA polymerase II promoter transcription, DNA binding, etc (Figure 3A).
A total of 143 KEGG terms were significantly enriched in the intersected targets, including pathways in cancer, receptor activation, proteoglycans in cancer, breast cancer, etc. The major pathways included PI3K/Akt signaling pathway, MAPK signaling pathway, FoXO signaling pathway, HIF-1 signaling pathway, TGF-βsignaling pathway, etc (Figure 3B).
Transcriptome of Differentially Expressed Genes (DEGs) in ER+ BC Cells Treated with TGBSXJ Formula
To Compensate for the limitations of the NP, RNA-seq was performed to determine the changes in gene expression induced by TGBSXJ Formula. DEGs with p-value<0.05 and log fold change (FC)>2 in MCF-7 cells either treated with and without TGBSXJ Formula were screened. Among the 203 DEGs, 75 were upregulated and 128 downregulated (Figure 3C). The Fragments Per Kilobase of transcript per Million mapped reads (FPKM) corresponding to DEGs were visualized by clustering heatmap, showing a different hierarchical clustering between groups (Figure 3D). A total of 940 GO entries were obtained, including 767, 55 and 118 terms of BP, CC and MF, respectively. The major GO entries included response to type I interferon, NADP+, cadmium ion, retinal metabolic process, etc (Figure 3E). A total of 196 KEGG terms were significantly enriched in the intersected targets, including Chemical carcinogenesis, Metabolism of xenobiotics by cytochrome P450, Tryptophan metabolism, etc. The major pathways included RIG-I-like receptor signaling pathway, ErbB signaling pathway, P53 signaling pathway, HIF-1 signaling pathway, GnRH signaling pathway and TGF-βsignaling pathway, etc (Figure 3F).
HPLC Profile of TGBSXJ Formula
According to the results of NP, eight potential components (Mangiferin, Hesperidin, Rutin, Anemarrhena asphodeloides saponin BII, Ganoderic acid A, Acacetin, Anemarrhena saponin A-III and Desmethylpinosylvin) of TGBSXJ Formula were determined by high-performance liquid chromatography (HPLC). Five marker components were identified by comparing their retention times (RT) with those of standard samples (Figure 4). The RT of multistands (RTM) and samples (RTS) are shown in Table 2.
 |
Table 2 Retention Time of the Compounds |
TGBSXJ Formula Inhibited the Survival of ER+ BC Cells in vitro
The CCK-8 results showed that TGBSXJ Formula dose-dependently inhibited the viability of MCF-7 (IC50 values of 1302.5 μg/mL, 1118.9 μg/mL and 679.4 μg/mL at 24, 48 and 72 hours, respectively) and T47D cells (IC50 values of 1099 μg/mL, 776.9 μg/mL and 1000 μg/mL at 24, 48 and 72 hours, respectively) (Figure 5A and B). The effects of TAM (Figure 5C) on cell viability were investigated that TAM induction at varying concentrations for 48 hours inhibited the survival of MCF-7 (IC50 value=10.52 μM) and T47D cells (IC50 value=13.54 μM) in a dose-dependent manner (Figure 5D). In the following study, MCF-7 and T47D cells were induced with 10μM and 13μM TAM, respectively.
TGBSXJ Formula Reversed TAM-Induced Inhibitions on the Proliferation and Migration of ER+ BC Cells in vitro
The synergistic effects of TGBSXJ Formula combined with TAM on inhibiting proliferation and migration of the ER+BC (MCF-7 and T47D) cells were evaluated using three experimental assays: the wound healing assay, colony formation assay, and Transwell migration assay.
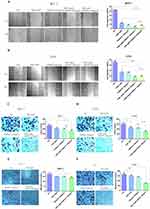
The wound healing assay was conducted to assess the migratory ability of the ER+BC cells treated with TGBSXJ Formula, TAM, and their combination. After creating standardized wounds in the cell monolayer, images were captured at 0 and 48 hours. Cells not treated with any drugs exhibited significant wound closure, approximately 41.94% in MCF-7 cells and 71.10% in T47D cells at 48 hours (p < 0.01). In contrast, treatment with TAM alone reduced wound closure to 12.74% in MCF-7 cells and 31.80% in T47D cells. Remarkably, the combination treatment of TGBSXJ Formula and TAM resulted in only 4.13% wound closure for MCF-7 cells and 15.45% for T47D cells, indicating a significant reduction in cell migration (p < 0.01), and demonstrating the enhanced anti-migratory effect when TGBSXJ Formula is included in the treatment regimen (Figure 6A and B).
The colony formation assay was performed to assess the survival and proliferation of the ER+BC cells. The results indicated that MCF-7 cells treated with TAM alone had no significant reduction in colony formation compared to the control group (p > 0.05), while T47D cells exhibited a slight but significant reduction in colony formation, with a decrease of approximately 23.2% in the number of colonies compared to the control (p < 0.05). However, the combination of TGBSXJ Formula and TAM led to an even more pronounced inhibition, with an additional reduction of approximately 24.7% in the number of colonies formed by MCF-7 cells and 33.7% by T47D cells compared to TAM alone (p < 0.01). These results suggest that TGBSXJ Formula not only augments the anti-proliferative effects of TAM but also promotes cell death in cancer cell lines over time (Figure 6C and D).
The Transwell migration assays further evaluated the invasive capability of ER+BC cells following treatment. In this assay, the combined treatment of TGBSXJ Formula and TAM significantly reduced the number of cells that migrated through the transwell membrane. Specifically, there was a 19.9% decrease in the number of MCF-7 cells and an 54.6% decrease in T47D cells that migrated compared to the untreated control group (MCF-7: p < 0.05, T47D: p < 0.01). For cells treated with TAM alone, migration decreased by 13.2% and 9.8%, respectively (p < 0.05). These findings indicate that TGBSXJ Formula markedly enhances the anti-invasive properties of TAM (Figure 6E and F).
TGBSXJ Formula Protected Against ER+ BC by Inhibiting the TGF-β /Smad Signaling Pathway
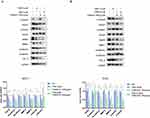
The effects of the TGBSXJ Formula in combination with TAM on the expression levels of key proteins involved in ER+BC progression were evaluated using Western blot analysis (Figure 7A and B).
TGF-β
Treatment with TAM alone resulted in a moderate reduction in TGF-β levels in both MCF-7 and T47D cells, demonstrating a decrease of 26.8% in MCF-7 cells and an increase of 13.6% relative to the control group (MCF-7: p < 0.01, T47D: p < 0.05). In contrast, the combination of TGBSXJ Formula and TAM led to a reduction of TGF-β levels with 80.8% in MCF-7 cells and 75.3% in T47D cells compared to the TAM alone group (p < 0.01).
E-Cadherin and N-Cadherin
Treatment with TAM alone resulted in a reduction in E-cadherin levels of 20.5% in MCF-7 and 14.5% in T47D cells relative to the control group. However, this decrease did not achieve statistical significance (p > 0.05). In contrast, the combination of TGBSXJ Formula and TAM led to a reduction of E-cadherin levels with 31.3% in MCF-7 cells and an increase with 71.9% in T47D cells compared to the TAM alone group (p < 0.05). Conversely, N-cadherin, a marker associated with epithelial-to-mesenchymal transition (EMT), was downregulated. The combination treatment significantly decreased N-cadherin expression by about 25.0% in MCF-7 cells and 36.7% in T47D relative to the TAM alone group (p < 0.01).
MMP-2 and MMP-9
The matrix metalloproteinases MMP-2 and MMP-9 are critical for tumor invasion and metastasis. TAM alone reduced MMP-2 levels by 10.4% in MCF-7 cells and increased 35% in T47D cells compared to the control group (MCF-7: p < 0.05, T47D: p > 0.05). The TGBSXJ and TAM combination reduced these levels even further, with MMP-2 by 37.7% in MCF-7 cells and 46.8% in T47D cells (MCF-7: p < 0.05, T47D: p < 0.01) compared to the TAM alone group. The combination treatment significantly decreased MMP-9 expression by about 4.4% in MCF-7 cells and 36.7% in T47D relative to the TAM alone group (p < 0.05).
p-Smad2 and p-Smad3
Both phosphorylated Smad proteins, p-Smad2 and p-Smad3, serve as downstream mediators of TGF-β signaling. Treatment with TAM alone reduced p-Smad2 levels by 12.0% in MCF-7 cells and increased by 5.2% in T47D cells relative to the control group. However, this decrease did not achieve statistical significance (p > 0.05). In contrast, combination treatment resulted in a striking inhibition of p-Smad2 and levels, with reductions exceeding 57.0% in MCF-7 cells and 33.3% in T47D cells compared to TAM alone group (p < 0.01). The combination treatment significantly decreased p-Smad3 expression by about 15.6% in MCF-7 cells and 54.0% in T47D relative to the control group (p < 0.01).
Discussion
Tamoxifen, an essential treatment of ER+BC, has been used for more than three decades. Currently, it is utilized as a chemo-preventive agent for patients.18 However, the acquired resistance to TAM is a leading cause of treatment failure.19 The mechanisms underlying endocrine resistance are complex and multifaceted, encompassing both intrinsic factors present at diagnosis and acquired changes during therapy. Intrinsic resistance may arise from pre-existing tumor characteristics, such as mutations in the ER or dysregulation of key signaling pathways.20 Acquired resistance, on the other hand, often results from adaptive responses of the tumor to the selective pressure exerted by long-term hormone treatment. These adaptive changes can include alterations in receptor expression, activation of growth factor signaling pathways, and changes in the tumor microenvironment.21
Clinical studies and preclinical investigations have suggested that some TCM possess anti-estrogenic properties,22 which can modulate estrogen signaling pathways and potentially restore sensitivity to endocrine therapies.23 Additionally, TCM may help regulate the tumor microenvironment,24 reduce inflammation,24 and inhibit signaling pathways associated with tumor growth and metastasis.25
TCM emphasizes a holistic view towards BC, that the occurrence of breast cancer is related to multiple factors, including emotional factors, diet, lifestyle, and environmental impacts. TGBSXJ Formula is a TCM decoction composed of 9 herbal medicines, exerting an anti-BC property through “soothing the liver and tonifying the kidney, supporting the energy and strengthening the essence”. In the present study, in vitro evidences have supported the role of TGBSXJ Formula in inhibiting the viability, colony formation ability and migration of ER+BC cells, which synergizes with TAM.
The NP analysis of the TGBSXJ Formula has revealed significant insights into its multi-targeted mechanisms of action in the treatment of ER+BC. By employing systems biology approaches, Mangiferin, Rutin, Anemarrhena asphodeloides saponin BII, Ganoderic acid A and Acacetin were screened as the main active ingredients of TGBSXJ Formula, which have been verified anti-tumor properties.26–30
Our studies demonstrate that TGBSXJ Formula enhances the sensitivity of ER+BC cells to Tamoxifen (TAM), as evidenced by several assays, including wound healing assays, colony formation, and Transwell migration tests. These experiments reveal that the combination of TGBSXJ Formula and TAM significantly inhibits the migratory, proliferative, and invasive capabilities of ER+BC cells compared to the use of TAM alone. This suggests that TGBSXJ Formula not only plays a crucial role in mitigating the aggressive behavior of these cancer cells but also works synergistically with TAM.
In our study, the integration of NP analysis with RNA sequencing offers a more comprehensive approach to elucidate the underlying mechanisms of the TGBSXJ Formula in ER+BC treatment. While NP provides valuable insights into the potential molecular targets and pathways influenced by the compounds within TGBSXJ Formula, RNA sequencing offers a more granular view of gene expression changes that really occur in response to treatment. The number of common GO entries between the NP and RNA sequencing is 196, and the matching rate accounts for 33.45%. The entries with high rich factor are positive regulation of interferon-alpha production, positive regulation of fibroblast proliferation, estrogen metabolic process, oxidoreductase activity, regulation of gene expression, etc. The number of common KEGG entries between the NP and RNA sequencing is 119, and the matching rate accounts for 82.07%. The pathways with high rich factor are Steroid hormone biosynthesis, Chemical carcinogenesis - DNA adducts, Metabolism of xenobiotics by cytochrome P450, ErbB signaling pathway, TGF-beta signaling pathway, etc. According to the results of NP analysis and RNA sequencing, TGF-β, SMAD3 and MMP2 were selected as the potential targets and TGF-β/SMAD pathway as the mechanism of TGBSXJ Formula on ER+BC (Figure 8).
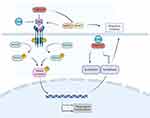 |
Figure 8 The overview of the regulatory mechanism involved in the anti-cancer effect of TGBSXJ Formula on ER+ BC. |
TGF-βis a prototypical ligand of the TGF-β superfamily that governs signaling transduction through receptor serine/threonine kinases.31 It is also a proinflammatory cytokine chiefly involved in tumorigenesis, exerting a dichotomous role in human cancers genesis either as a promoter or a suppressor.32,33 TGF-β activation leads to the resistance to anti-estrogens.33 Smad2 and Smad3, two receptor-regulated effector proteins (R-Smads), are involved in the TGF-β signaling pathway. Their phosphorylation activated by TβRI at a C-terminal SSXS motif, results in nuclear accumulation of R-Smad.34 Phosphorylation is a key mechanism to trigger nuclear translocation and activation of Smads, thereby regulating the transcription of target genes. Phosphorylated Smad proteins are typically overexpressed in BC tissues and closely related to the poor prognosis.35 MMP-2 and MMP-9, two members of the family of matrix metalloproteinases (MMPs), are responsible for tissue remodeling and repair by degrading various proteins in the extracellular matrix (ECM).36 Through cleaving latent TGF-β binding protein (LTBP), MMP-2 and MMP-9 stimulates the activation of TGF-β and regulates the activity and bioavailability in the ECM.37 Additionally, TGF-β can also regulate the expressions and activities of MMP-2 and MMP-9 in a reciprocal manner. Sun et al reported that TGF-β mediates gene transcriptions and protein syntheses of MMP-2 and MMP-9 through the Smad signaling pathway.38 E-cadherin belongs to the family of calcium-dependent adhesion molecules of epithelial cells, exerting an important role in tissue growth and development.39 It was associated with upregulation of genes involved in transforming growth factor-β (TGFβ), reactive oxygen species and apoptosis signalling pathways.40 N-cadherin, also known as neural cadherin, serves as a calcium-dependent single-chain transmembrane glycoprotein that mediates homotypic and heterotypic cell adhesion. Abnormal expressed N-cadherin is closely related to the progression of human malignanciest by mediating tumor cells’ transformation, adhesion, apoptosis, angiogenesis, invasion and metastasis.41 Both E-cadherin and N-cadherin are closely associated with TGF-β in the processes of epithelial-mesenchymal transition (EMT) and angiogenesis.42
Western blot analysis further elucidated the underlying mechanisms by showing that TGBSXJ Formula suppresses the above proteins related to TGF-β/SMAD signaling pathway, with pronounced effects observed in conjunction with TAM treatment. TGBSXJ Formula appears to enhance the efficacy of TAM, reinforcing the hypothesis that combination therapy can yield better clinical outcomes for patients facing endocrine resistance.
Conclusions
In summary, our study screened the active ingredients of TGBSXJ Formula and the targets of ER+ BC using online databases. Through NP approaches and RNA sequencing, we identified the hub gene and enriched signaling pathways underlying the pharmacological mechanism of TGBSXJ Formula for the treatment of ER+ BC. Our in vitro experiments further demonstrated that TGBSXJ Formula increased the sensitivity of ER+ BC to TAM by inhibiting cell proliferation and migration via inhibiting the TGF-β/Smad signaling pathway. Our data indicate that TGBSXJ Formula could be new strategy to enhance efficacy and therapeutic duration of tamoxifen, which holds significant promise in addressing issues related to TAM resistance in ER+BC. TGBSXJ Formula into treatment regimens could improve therapeutic responses and, ultimately, patient prognosis. Further investigations into the molecular mechanisms and the clinical implications of this combination could be one of the effective strategies in managing ER+BC.
Acknowledgments
The authors are grateful to the Pharmacy Department and the Traditional Chinese Medicine Department of Ruijin Hospital, School of Medicine, Shanghai Jiaotong University for providing us with a superior research platform and learning environment.
Funding
This work was supported by (1) the National Natural Science Foundation of China (No. 8230150159), (2) Special research project of traditional Chinese medicine of Shanghai Municipal Commission of Health and Family Planning (No. 2016LP017), (3) Scientific research project of traditional Chinese medicine of Jiading District Health Commission of Shanghai Municipality (No. 2023-KY-ZYY-02) and (4) Ruijin Hospital Youth Training Program (No. KY20240295).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Kashyap D, Pal D, Sharma R. Global increase in breast cancer incidence: risk factors and preventive measures. Biomed Res Int. 2022;2022:9605439. doi:10.1155/2022/9605439
3. Davies C, Godwin J, Gray R, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378(9793):771–784. doi:10.1016/S0140-6736(11)60993-8
4. Gradishar WJ, Moran MS, Abraham J, et al. NCCN guidelines(r) insights: breast cancer, version 4.2021. J Natl Compr Canc Netw. 2021;19(5):484–493. doi:10.6004/jnccn.2021.0023
5. Ojo D, Wei F, Liu Y, et al. Factors promoting tamoxifen resistance in breast cancer via stimulating breast cancer stem cell expansion. Curr. Med. Chem. 2015;22(19):2360–2374. doi:10.2174/0929867322666150416095744
6. Chan CWH, Miaskowski C, McCarthy A, et al. Tamoxifen-related endocrine symptoms in Chinese patients with breast cancer: study protocol clinical trial (SPIRIT Compliant). Medicine. 2020;99(8):e19083. doi:10.1097/MD.0000000000019083
7. Wilson TR, Longley DB, Johnston PG. Chemoresistance in solid tumours. Ann Oncol. 2006;17(Suppl 10):x315–324. doi:10.1093/annonc/mdl280
8. Mills JN, Rutkovsky AC, Giordano A. Mechanisms of resistance in estrogen receptor positive breast cancer: overcoming resistance to tamoxifen/aromatase inhibitors. Curr. Opin. Pharmacol. 2018;41:59–65. doi:10.1016/j.coph.2018.04.009
9. Gezici S, Şekeroğlu N. Current perspectives in the application of medicinal plants against cancer: novel therapeutic agents. Anti-Cancer Agents Med Chemi. 2019;19(1):101–111.
10. Su XL, Wang JW, Che H, et al. Clinical application and mechanism of traditional Chinese medicine in treatment of lung cancer. Chinese Med J. 2020;133(24):2987–2997. doi:10.1097/CM9.0000000000001141
11. Sun Xueran YK, Lingling LV. Evaluation on clinical efficacy of liver-regulating, kidney-tonifying and mass-eliminating therapy by stage for advanced breast cancer. World Chin Med. 2019;1(14):115–121. (in Chinese).
12. Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt H. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci. 2022;43(2):136–150. doi:10.1016/j.tips.2021.11.004
13. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi:10.1186/1758-2946-6-13
14. Bateman A, Martin M-J, Orchard S, UniProt. The universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–d531. doi:10.1093/nar/gkac1052
15. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–d839. doi:10.1093/nar/gkw943
16. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–d646. doi:10.1093/nar/gkac1000
17. Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi:10.1186/gb-2003-4-5-p3
18. Merikhian P, Ghadirian R, Farahmand L, Mansouri S, Majidzadeh AK. MUC1 induces tamoxifen resistance in estrogen receptor-positive breast cancer. Expert Rev Anticancer Ther. 2017;17(7):607–613. doi:10.1080/14737140.2017.1340837
19. Wiebe VJ, Osborne CK, Fuqua SA, DeGregorio MW. Tamoxifen resistance in breast cancer. Crit Rev Oncol Hematol. 1993;14(3):173–188. doi:10.1016/1040-8428(93)90008-R
20. Saatci O, Huynh-Dam KT, Sahin O. Sahin O: endocrine resistance in breast cancer: from molecular mechanisms to therapeutic strategies. J Mol Med. 2021;99(12):1691–1710. doi:10.1007/s00109-021-02136-5
21. Raheem F, Karikalan SA, Batalini F, El Masry A, Mina L. Metastatic ER+ breast cancer: mechanisms of resistance and future therapeutic approaches. Int J Mol Sci. 2023;24(22):16198. doi:10.3390/ijms242216198
22. Ni W, Fan H, Zheng X, et al. Cryptotanshinone inhibits ERα-dependent and -independent BCRP oligomer formation to reverse multidrug resistance in breast cancer. Front Oncol. 2021;11:624811. doi:10.3389/fonc.2021.624811
23. Wang Y, Yue W, Lang H, Ding X, Chen X, Chen H. Resuming sensitivity of tamoxifen-resistant breast cancer cells to tamoxifen by tetrandrine. Integr Cancer Ther. 2021;20:1534735421996822. doi:10.1177/1534735421996822
24. Liu J, Wang Y, Qiu Z, et al. Impact of TCM on tumor-infiltrating myeloid precursors in the tumor microenvironment. Front Cell Dev Biol. 2021;9:635122. doi:10.3389/fcell.2021.635122
25. Wang Y, Zhang Q, Chen Y, et al. Antitumor effects of immunity-enhancing traditional Chinese medicine. Biomed Pharmacothe. 2020;121:109570. doi:10.1016/j.biopha.2019.109570
26. Tan HY, Wang N, Li S, et al. Repression of WT1-mediated LEF1 transcription by mangiferin governs β-catenin-independent wnt signalling inactivation in hepatocellular carcinoma. Cell Physiol Biochem. 2018;47(5):1819–1834. doi:10.1159/000491063
27. Zhang B, Zhao J, Li S, Zeng L, Chen Y, Fang J. Mangiferin activates the Nrf2-ARE pathway and reduces etoposide-induced DNA damage in human umbilical cord mononuclear blood cells. Pharm Biol. 2015;53(4):503–511. doi:10.3109/13880209.2014.927890
28. Huo M, Xia A, Cheng W, et al. Rutin promotes pancreatic cancer cell apoptosis by upregulating miRNA-877-3p expression. Molecules. 2022;27(7):2293. doi:10.3390/molecules27072293
29. Zhou M, Zhang G, Hu J, et al. Rutin attenuates sorafenib-induced chemoresistance and autophagy in hepatocellular carcinoma by regulating BANCR/miRNA-590-5P/OLR1 axis. Int J Biol Sci. 2021;17(13):3595–3607. doi:10.7150/ijbs.62471
30. Zhang G, Dong J, Lu L, et al. Acacetin exerts antitumor effects on gastric cancer by targeting EGFR. Front Pharmacol. 2023;14:1121643. doi:10.3389/fphar.2023.1121643
31. Zi Z, Chapnick DA, Liu X. Dynamics of TGF-β/Smad signaling. FEBS Lett. 2012;586(14):1921–1928. doi:10.1016/j.febslet.2012.03.063
32. Aashaq S, Batool A, Mir SA, Beigh MA, Andrabi KI, Shah ZA. TGF-β signaling: a recap of SMAD-independent and SMAD-dependent pathways. J Cell Physiol. 2022;237(1):59–85. doi:10.1002/jcp.30529
33. Babyshkina N, Dronova T, Erdyneeva D, Gervas P, Cherdyntseva N. Role of TGF-β signaling in the mechanisms of tamoxifen resistance. Cytokine Growth Factor Rev. 2021;62:62–69. doi:10.1016/j.cytogfr.2021.09.005
34. Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi:10.1016/S0092-8674(03)00432-X
35. Yu L, Di Y, Xin L, et al. SND1 acts as a novel gene transcription activator recognizing the conserved Motif domains of Smad promoters, inducing TGFβ1 response and breast cancer metastasis. Oncogene. 2017;36(27):3903–3914. doi:10.1038/onc.2017.30
36. Zhao X, Chen J, Sun H, Zhang Y, Zou D. New insights into fibrosis from the ECM degradation perspective: the macrophage-MMP-ECM interaction. Cell Biosci. 2022;12(1):117. doi:10.1186/s13578-022-00856-w
37. Tatti O, Vehviläinen P, Lehti K, Keski-Oja J. MT1-MMP releases latent TGF-β1 from endothelial cell extracellular matrix via proteolytic processing of LTBP-1. Exp. Cell. Res. 2008;314(13):2501–2514. doi:10.1016/j.yexcr.2008.05.018
38. Sun Y, Zhou QM, Lu YY, et al. Resveratrol inhibits the migration and metastasis of MDA-MB-231 human breast cancer by reversing TGF-β1-induced epithelial-mesenchymal transition. Molecules. 2019;24(6): 1131.
39. Giannotta M, Trani M, Dejana E. VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev Cell. 2013;26(5):441–454. doi:10.1016/j.devcel.2013.08.020
40. Padmanaban V, Krol I, Suhail Y, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573(7774):439–444. doi:10.1038/s41586-019-1526-3
41. Cao ZQ, Wang Z, Leng P. Aberrant N-cadherin expression in cancer. Biomed Pharmacothe. 2019;118:109320. doi:10.1016/j.biopha.2019.109320
42. Wei J, Wu L, Yang S, et al. E-cadherin to N-cadherin switching in the TGF-β1 mediated retinal pigment epithelial to mesenchymal transition. Exp Eye Res. 2022;220:109085. doi:10.1016/j.exer.2022.109085
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.