Back to Journals » Drug Design, Development and Therapy » Volume 19
Tubeimoside I Inhibits the Proliferation of Liver Cancer Through Inactivating NF-κB Pathway by Regulating TNFAIP3 Expression
Authors Zhang Y , Zhou M, Zhu L , Chen L , Zhang H , Huang Z , Zhou H
Received 2 December 2024
Accepted for publication 28 February 2025
Published 13 March 2025 Volume 2025:19 Pages 1895—1908
DOI https://doi.org/10.2147/DDDT.S507656
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Yajun Zhang,1,* Mingqin Zhou,2,* Liwen Zhu,3,* Lichan Chen,4 Haohua Zhang,3 Zhen Huang,5 Hongzhong Zhou3
1Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China; 2Department of Critical Care Medicine, Cancer Hospital of Shantou University Medical College, Shantou, People’s Republic of China; 3Department of Laboratory Medicine, Shenzhen Institute of Translational Medicine, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Medical Innovation Technology Transformation Center of Shenzhen Second People’s Hospital, School of Pharmaceutical Sciences, Health Science Center, Shenzhen University, Shenzhen, People’s Republic of China; 4Shenzhen Third People’s Hospital, Southern University of Science and Technology, Shenzhen, People’s Republic of China; 5Department of Laboratory Medicine of Pingshan District Maternal and Child Healthcare Hospital, Shenzhen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhen Huang, Email [email protected] Hongzhong Zhou, Email [email protected]
Purpose: This study aims to evaluate the therapeutic potential of tubeimoside I (TBMS1), a monomer compound extracted from the tubers of Chinese herb Bolbostemma paniculatum (Maxim). Franquet (Cucurbitaceae), in the treatment of liver cancer. Specifically, we sought to elucidate the underlying mechanisms through which TBMS1 exerts its anticancer effects.
Methods: The effects of TBMS1 on the viability, proliferation, and apoptosis of two liver cancer cell lines, MHCC97-H and SNU-449, were comprehensively assessed using Cell Counting Kit-8 (CCK-8), colony formation, 5-ethynyl-2′-deoxyuridine (EDU) assay, and flow cytometry assays. To uncover the molecular mechanisms, RNA sequencing was performed to identify the downstream targets of TBMS1. Additionally, we utilized network pharmacology to predict TBMS1 targets in liver cancer and employed Venn diagram analysis to integrate these predictions with our experimental findings. Pathway enrichment analysis was conducted using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases to elucidate the biological processes involved. Furthermore, a subcutaneous xenograft tumor model was established to investigate the in vivo antitumor efficacy of TBMS1.
Results: In vitro experiments demonstrated that TBMS1 significantly enhanced cell apoptosis and inhibited the growth of liver cancer cells. Both network pharmacology predictions and RNA-seq analyses revealed that the downstream target genes of TBMS1 were highly enriched in the NF-κB signaling pathway. Notably, we observed a significant upregulation of TNFα-induced protein 3 (TNFAIP3) expression with increasing concentrations of TBMS1. In vivo studies further confirmed that TBMS1 treatment dramatically reduced the volume and weight of liver cancer tumors compared to controls.
Conclusion: Our study provides compelling evidence that TBMS1 suppresses liver cancer progression by inactivating the NF-κB pathway and regulating TNFAIP3 expression. These findings offer novel insights and a theoretical basis for the development of targeted therapies for liver cancer.
Keywords: tubeimoside-I, hepatocellular carcinoma, TNFAIP3, NF-κB, proliferation
Introduction
Liver cancer ranks third globally in terms of cancer-related mortality and is the sixth most common type of cancer,1 with significant geographic heterogeneity.2 The most prevalent type of liver tumor is hepatocellular carcinoma (HCC), accounts for about 90% of reported cases.3 Chronic hepatitis caused by metabolic liver diseases such as hepatitis B and C virus infection and alcohol are potential pathophysiological factors that lead to HCC.4 Patients who are usually diagnosed with HCC are mostly in the middle to late stages, yielding a poor prognosis. However, owing to the absence of effective early diagnostic methods and the limitations of local treatment, systemic therapies are the only viable option for those with unresectable or metastatic HCC.5 Systemic treatments, such as immune checkpoint inhibitors and molecularly targeted drugs, have advanced significantly.6,7 At the same time, with the progress of technology, the technology of using artificial intelligence computing to predict the potential new targets of old drugs for new use is also being explored.8 However, due to drug resistance, metastasis, and frequent recurrence, HCC has one of the poorest prognoses, requiring new therapeutic approaches.
Traditional antitumor therapy combination with traditional Chinese medicine (TCM) to inhibit tumor progression as an adjuvant and alternative therapy have recently attracted attention.9,10 In practice, TCM can significantly reduce drug resistance, improve tumor inhibition, and improve side effects such as cancer-related bone marrow inhibition.11 Furthermore, Liu et al found that TCM adjuvant therapy could improve the survival prognosis of HCC patients by establishing a Cox multivariate regression model and using Log rank test analysis.12 The tuber of Bolbostemma paniculatum (Maxim). Franquet (Cucurbitaceae), commonly referred to as tubeimoside (TBMS), is used as a Chinese medicinal herb. It has been used most frequently to treat snake venom and irritation. Subsequently, the main component TBMS1 was isolated and identified as a triterpene saponin with a unique macrocyclic structure.13 It exhibited anti-inflammatory and antioxidant properties.14 TBMS1 has been confirmed to have significant anticancer activity against lung cancer, breast cancer, melanoma, and glioblastoma. The related mechanisms include inhibiting cell proliferation, inducing apoptosis, promoting autophagy and inducing cell cycle arrest.15–18 TBMS1 may be a new anti-tumor agent according to these findings. However, the exact effects of TBMS1 and the specific mechanism of action on HCC remain lacking.
The NF-κB signaling typically undergoes canonical or noncanonical activation through pattern recognition receptor (PRRS) control the course of cancer by promoting cell proliferation and differentiation, for example.19 As one of the pathways of inflammation-mediated HCC development, cytokine signaling is usually regulated by TNF-α, IL-6, NF-κB, JNK and STAT3.20,21 Moreover, the NF-kB pathway’s activation causes chronic liver inflammation and advances HCC.22,23 TNFAIP3 is a typical anti-inflammatory molecule linked to cancer and other human disorders. Studies suggested that TNFAIP3 plays a dual role in promoting and inhibiting solid tumors. For example, TNFAIP3 has anti-tumor effect in hepatocellular carcinoma and colorectal cancer, but it is a potential oncogene in breast cancer and gastric cancer.24–27 Moreover, as an ubiquitin-editing enzyme, TNFAIP3 can effectively downregulate NF-κB signaling.28,29 According to the above studies, TNFAIP3 may function as a tumor suppressor in HCC and its expression may affect the progression of HCC.
In this study, TBMS1 weakened liver cancer proliferation and promoted apoptosis both in vitro and in vivo. The RNA-Seq study and network pharmacology predictions revealed that the downstream target genes of TBMS1 were significantly enriched in the NF-κB pathway. Moreover, TNFAIP3 expression was notably elevated, concomitant with increased TMBS1 concentration. This study offers novel insights and a theoretical foundation for medication-target-based liver cancer treatment.
Materials and Methods
Target Prediction of TBMS1 in HCC
The SMILEs number of TBMS1 was searched from the PubChem chemical information database.30 Then, the related targets of TBMS1 were screened by Swiss TargetPrediction database,31 Pharmmapper database,32 and Super-PRED database.33 In the GeneCards,34 Therapeutic Target Database (TTD)35 and OMIM database (https://omim.org/), searching with “liver cancer” or “hepatocellular carcinoma” as the keyword to screened HCC-related targets. All targets collected in the above databases were standardized by the UniProt and NCBI. After removing the repeat targets, 1009 disease targets and 407 TBMS1-related targets were screened.
PPI Construction of TBMS1-HCC Targets
By taking the intersection of HCC-related and TBMS1-related targets, 132 overlapping targets were obtained, which were presented by Venn diagram. Then, the protein interaction network was constructed by importing the intersection gene into the STRING database.36 Network analysis and calculation of target proteins with high correlation were performed by Cytoscape 3.10.1 software.37
GO and KEGG Pathway Enrichment Analysis
GO and KEGG enrichment analysis of crossing targets were performed by Metascape.38 P < 0.05 for the KEGG and GO pathways was deemed significant. The results were visualized and plotted by an online platform for data analysis and visualization.39
Cell Lines and Cell Culture
Human hepatoma cell lines MHCC97-H and SNU-449 were procured from Jennio Biotech (GuangZhou, China). DMEM (Cytiva, Shanghai, China) supplemented with 10% FBS (ExCell Bio, Suzhou, China) was used to cultivate cells at 37 °C in a humidified incubator with 5% CO2. After observing the growth of cells, 70–80% density was used in the studies that followed.
Drug Treatment
Tubeimoside I (TBMS1) was purchased from Target Mol (T2715, Boston, MA, USA). TBMS1 powder was processed into a 22.5 mg/mL (15 mm) stock solution with DMSO (Solarbio, Beijing, China) and stored at –80 °C. TBMS1 was added to the medium and diluted to different concentrations with cell culture medium containing 10% serum in different proportions.
Cell Viability Assay
The CCK-8 reagent (HY-K0301, MCE, New Jersey, USA) was used to measure cell viability in accordance with instructions. Cells were seeded in 96-well plates at 1×104 cells/well. The group included the control group (TBMS1-free DMSO) and groups treated with different amounts of TBMS1 (0, 0.5, 1, 5, 10, 20, 25, and 50µM) for 24h, and six parallel replicates for each group. Then, 10 μL of CCK-8 solution was added and incubated for two hours at 37 °C. At 450 nm, absorbance was finally measured by Microplate Readers (Biotek Epoch, California, USA).
Colony Formation Assay
MHCC97-H and SNU-449 cells were planted in 6-well plates at a count of 2000 cells/well. Cells were grown in the medium containing different concentrations of TBMS1 (0μM, 5μM, 10μM, 15μM) for 14 days. After the number of cells in a single clone exceeded 50 under observation, the cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet, rinsing with cold PBS and dried. Subsequently, positive clones were counted by Image J software (National Institutes of health, Bethesda, Maryland, USA) and each group contained three independent parallel replicate wells.
EdU Assay
Cell proliferation was identified in accordance with the Cell-Light EdU Apollo488 In Vitro Kit’s instructions (C10310-3, RIB-BIO, Guangzhou, China). MHCC97-H and SNU-449 cells were treated with 0, 10, 20, and 25 μM TBMS1 for 24h after being plated in 24-well plates with 1×104 cells/well. EDU solution (50uM) diluted in culture medium was added to the treated cells and incubated with cells for 2h at 37 °C. After fixation and staining, the cells were permeabilized with PBS containing 0.5% tritonX-100 for 10 minutes, and then incubated with glycine (2 mg/mL). Lastly, positive cells were labeled by staining with 1 × Apollo solution and nuclear staining was performed with Hoechst 33342 for 30 minutes. A fluorescence microscope (Zeiss, Oberkochen, Germany) was used to collect images and a minimum of 5 fields/well were randomly selected for positive cell counting.
Flow Cytometry Analysis of Apoptosis
In accordance with the manufacturer’s recommendations, cell apoptosis of MHCC97-H and SNU-449 cells was measured using the Annexin V-PE/7-AAD Apoptosis Detection Kit (Vazyme, Nanjing, China). 5 μL of Annexin V-PE and 7-AAD reagent were mixed in the cell suspension with 400 μL 1× binding buffer, and the cells were stained for 15 minutes. Each sample contained 1×104 cells, and flow cytometry (NOVOCYTE 3130, ACEA Biosciences, California, USA) was used to quantify the apoptotic cells and further analyze them.
RNA-Seq Analysis of MHCC97-H Cells Induced by TBMS1
After being seeded at 1.5×106 cells/well in 6-well plates, MHCC97-H cells were exposed to TBMS1 at an IC50 concentration for 24h. Cell samples were treated with TRIzol and transferred to centrifuge tubes. Samples were treated with TRIzol and transferred to centrifuge tubes for total RNA extraction. The library was constructed by mRNA enrichment, fragmentation, reverse transcription, cDNA synthesis, PCR amplification and quantification, then subjected to Illumina sequencing. Three separate experiments provided samples for the collection. RNA-seq analysis was performed by Novogene Bio (Beijing, China).
Quantitative Reverse-Transcriptase Polymerase Chain Reaction (qRT-PCR) Assay
RNA was extracted from MHCC97-H and SNU-449 cells or tissues using TRIzol and reverse transcribed into cDNA using the M-MLV reverse transcriptase kit (Accurate Biology, Hunan, China). Quantitative qRT-PCR reactions were performed using SYBR Premix Ex Taq kit (Accurate Biology, Hunan, China). GAPDH served as the housekeeping gene and all experiments were repeated at least three times. Relative gene expression was assessed by calculating the results using the 2−ΔΔCT approach. Table 1 lists the primer sequences.
 |
Table 1 The Primer Sequences of qRT-PCR |
Western Blot Assay
Proteins of MHCC97-H and SNU-449 cells and tissues were extracted by cleavage with RIPA (Beyotime, Shanghai, China) mixed with protease inhibitors (HY-K0011, MCE, New Jersey, USA). A BCA standard curve was made according to the BCA Protein Assay kit (23227, Thermo scientific, Massachusetts, USA) instructions to determine the total protein concentration. SDS-PAGE electrophoresis was carried out according to the loading volume of 30μg. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, Bedford, MA, USA) and then blocked with 1xTBST (Epizyme BIO, Shanghai, China) dissolved in 5% skim milk powder for 1h 30min at room temperature on a shaker. The primary antibody was diluted in proportion and incubated at 4°C overnight. After washing with 1xTBST, secondary antibodies of the corresponding species were incubated for 1 h at room temperature on a shaker. The images were visualized and collected by the ChemiDoc TMMP Imaging System (Bio-Rad, California, USA) after infiltration of the bands with the ECL hypersensitive luminescent solution (WBKLS0500, Millipore, Massachusetts, USA), and the gray value was analyzed by ImageJ software. These dilution concentrations and primary antibodies were employed: Dilution ratios for anti-TNFAIP3 (Affinity, California, USA) and anti-GAPDH (Proteintech, Wuhan, China) were 1:500 and 1:10000. Goat anti-mouse secondary antibody (ZENBIO, Chengdu, China) and goat anti-rabbit secondary antibody (Proteintech, Wuhan, China) had dilution ratios of 1:8000 and 1:3000, respectively.
Xenograft Tumor Model
Male BALB/c nude mice (4–6 weeks old) were kept in an SPF level sterile environment after being purchased from HFK Bioscience in Beijing. After gathering MHCC97-H cells and modifying the concentration to 2×106/150 μL DMEM, the cells were implanted into the nude mices’ right hind leg. The nude mice were randomly split into two groups at random: the treated group and the negative control group (n = 5 in each group). The treated group received an intraperitoneal injection of TBMS1 (4 mg/kg), while the control group’s mice received an intraperitoneal injection of the same amount of sterile PBS. The length (L) and width (W) of the tumor were measured every two days, and the volume (V = L × W2/2) was computed. Tumors were excised and weighed, and the tissues were immediately isolated and fixed in 4% PFA immediately after 21 days of treatment with the injection. Tumor samples from five mice from each sample were collected for immunohistochemical staining, the residual tumor samples were used for qRT-PCR or Western blot assays. All operations were carried out in compliance with the institutional rules, and all animal experiments were authorized by the Shenzhen Second People’s Hospital (protocol code: 2023–262-01, data of approval: 14 November 2023).
Immunohistochemistry (IHC)
Tissue sections of transplanted tumors were stained with H&E and IHC staining with Ki67 (1:8000, Proteintech, Wuhan, China), TNFAIP3 (1:200, Affinity, Melbourne, Australia), Caspase-7 (1:400, Proteintech, Wuhan, China) and RARP (1:300, Proteintech, Wuhan, China), which was using the DAB chromogenic solution (ZSGB-BIO, Beijing, China).
Statistical Analysis
All data analysis was carried out by GraphPad Prism 9.0 software (GraphPad Software, San Diego, CA, USA), and results were presented as mean ±SD. Every experiment was performed in triplicate, each replicate was performed as an independent experiment or observation. Independent sample test (t-test) was used for comparison between two groups. One-way analysis of variance (ANOVA) was used for comparison between multiple groups.*P < 0.05 was deemed statistically significant.
Results
TBSM1 Inhibits Proliferation of HCC Cells
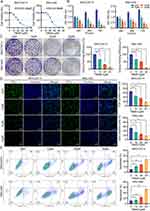
The effect of TBMS1 on the growth and proliferation of HCC cells was examined using the CCK-8, colony formation, and EDU assays. First, TBMS1 was administered at doses ranging from 0 to 50 μM to treat MHCC97-H and SNU-449 cells to ascertain the half maximal inhibitory concentration (IC50). As illustrated in Figure 1A, the IC50 values were 20.28μM and 22.98μM, respectively. In addition, we found that the inhibition of cell viability in both HCC cell lines increased with TBMS1 treatment concentration and time (Figure 1B). The colony formation assay’s results demonstrated that the quantity of positive colonies decreased with increasing drug concentration compared with the TBMS1 0μM group, indicating that TBMS1 can inhibit MHCC97-H and SNU-449 cells’ colony formation (Figure 1C). Additionally, the EDU experimental results showed that TBMS1 hindered DNA replication activity, which in turn inhibited the proliferation of MHCC97-H and SNU-449 cells. Following TBMS1 treatment, the number of edu-labeled green fluorescent cells was much lower (Figure 1D). The flow cytometric measurements indicated that cell apoptosis was significantly increased in MHCC97-H and SNU-449 cells treated with TBMS1 at doses of 10μM, 20μM, and 25μM (Figure 1E). According to these findings, TBMS1 may, in a concentration-dependent way, dramatically reduce the proliferative activity and encourage death of HCC cells.
Prediction of Anti-HCC Targets and Pathway of TBMS1 by Network Pharmacology
To explore and determine the mechanism of TBMS1 inhibition of liver cancer proliferation, we screened 407 TBMS1-related targets from the SwissTargetPrediction, Pharmmapper, and SuperPRED databases through target prediction and database search. Moreover, 1089 targets of liver cancer were collected from the GeneCards, TTD, OMIM databases. Subsequently, 132 potential targets of TBMS1 in HCC are shown by Venn diagram (Figure 2A). In addition, the protein interaction network (PPI) of intersection target genes was constructed by String database, and the network diagram was drawn and analyzed by Cytoscape software 3.10.1 (Figure 2B). This includes 132 nodes and 2201 edges, with an average node degree of 33.3. The CytoNCA plugin was used to calculate and screen the clusters with high correlation by comparing the Degree value. Genes located at intermediate nodes may serve as significant targets of TBMS1 in HCC treatment. To further analyze the biological functions of the target genes, we performed GO and KEGG enrichment analyses by Metascape. GO ontology enrichment analysis with p < 0.05 as the screening criterion. The top 15 enrichment results of biological process (BP), molecular function (MF) and cellular component (CC) for the three GO categories are shown in Figure 2C. The results showed that the target biological functions of TBMS1 in liver cancer were mainly enriched in response to hormone, enzyme-linked receptor protein signaling pathway, cell population proliferation and response to heterotypic biomass stimulation. In addition, the top 15 results of KEGG enrichment analysis (p < 0.05) suggested that the potential pathways of target genes included pathways in cancer, MAPK signaling pathway, Th17 cell differentiation, and transcriptional misregulation in cancer (Figure 2D). Also, to investigate the connection between target genes and pathways in more detail, we constructed the “drug-target-pathway” network by Cytoscape software 3.10.1 and calculated the correlation degree. The results showed that genes and functional pathways colored dark purple have higher correlations and are more closely related to the anti-HCC effect of TBMS1 (Figure 2E). Finally, synthesizing the above results, we propose that TBMS1 may participate in the regulation of cancer-related pathways through enzyme-linked receptor signaling to mediate HCC progression.
TBMS1 Exerts Anti-HCC Effect by Inactivating the NF-κB Pathway
To further validate the predictions of network pharmacology, RNA sequencing was performed on MHCC97-H cells treated with TBMS1 at half inhibitory concentration for 24h. Four hundred and forty-one up-regulated and 134 down-regulated genes were among the 575 differentially expressed genes (DEGs, p<0.05 and |log2 FC|≥ 1) that were evaluated, which were displayed by volcano plot (Figure 3A). The top 20 results of the GO and KEGG enrichment analysis were displayed after functional enrichment analysis of the differentially expressed genes was completed. The associated signaling pathways of the DEGs were mostly focused on cytokine–cytokine receptor interaction and NF-kappa B signaling pathway (Figure 3B–E). Interestingly, network pharmacological prediction and RNA-seq analysis were co-enriched in the NF-κB pathway. Next, we used qRT-PCR experiment to measure the NF-κB pathway gene’s mRNA expression level, including DUSP5, TNFAIP3, PLAU, IL1α, IL1β, TRAF1, etc. The mRNA levels of TNFAIP3 significantly increased with increasing concentrations of TBMS1 (Figure 3F). Concurrently, we observed a significant increase in TNFAIP3 protein levels with increasing TMBS1 concentration (Figure 3G). Thus, we hypothesized that TBMS1 regulates NF-κB pathway by regulating TNFAIP3 protein expression level.
TBMS1 Inhibits HCC Proliferation in vivo
To look at how TBMS1 affects HCC cells in vivo, human MHCC97-H cells were transplanted subcutaneously in nude mice. With compared to the control group, the tumor weight and volume of the nude mice in the TBMS1-treated group were noticeably lower (Figure 4A–C). This suggested that TBMS1 was able to inhibit tumor growth in vivo. Furthermore, we conducted immunohistochemical analysis and HE staining on tumor tissue slices and discovered that the focal necrosis was found in the center and edge of tumor tissue in TBMS1-treated group. The TBMS1-treated group had decreased levels of Ki67 (a marker of proliferation) and increased levels of Caspase 7 and RARP (markers of apoptosis) compared to the control group. Meanwhile, TNFAIP3 levels in the TBMS1-treated tumor tissues significantly increased (Figure 4D and E). Similarly, the TBMS1-treated group had higher TNFAIP3 mRNA and protein levels (Figure 4F and G). The above results suggest that TBMS1 inhibits the growth of HCC by regulating TNFAIP3 expression to inactivate the NF-κB pathway. Flow chart of TBMS1 therapy for liver cancer is presented in Figure 5.
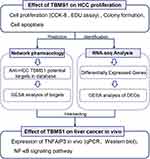 |
Figure 5 Flow chart of TBMS1 therapy for liver cancer. |
Discussion
Liver resection and liver transplantation are the primary curative treatment options for liver cancer. However, these therapies are limited in improving the outcomes due to a high morbidity and rate of death.40 The poor prognosis of HCC has necessitated novel treatment options to improve patient survival. In recent years, the use of natural compounds as potential antineoplastic agents has provided new strategies to improve the poor prognosis caused by malignant tumors with lower systemic toxicity and fewer adverse reactions. Chinese patented medicines and extracts have shown advantages for treating liver cancer.41 Yang et al found that the TCM compound FZJDXJ can inhibit the growth and migration of HCC by mediating AKT/CyclinD1/p21/p27 pathway.42 It has been reported that polysaccharide components in various Chinese herbal medicines can inhibit tumor progression by inhibiting abnormal proliferation and cell cycle-induced cancer cell death.43 For example, berberine (the active ingredient of aqueous extract of Rhizoma coptidis) and Curcumin (a component of turmeric) have been demonstrated to have some anti-liver cancer properties.44,45 Tubeimoside-I (TBMS1) is a major natural ingredient isolated and extracted from the tuber of Chinese medicinal herb Bolbostemma paniculatum (Maxim). Franquet (Cucurbitaceae). Although previous studies reported a related effect of TBMS1 on hepatoma cell HepG2,46 the specific mechanism of TBMS1’s anti-HCC action is yet unknown.
In our study, TBMS1 primarily exhibited anti-HCC activity by inhibiting viability and proliferation of tumor cells such as colony formation and DNA replication and promoting apoptosis. Furthermore, the volume and weight of nude mouse tumor tissues significantly decreased 21 days following the intraperitoneal injection of TBMS1 in vivo. These results indicate that TBMS1 exerts a key inhibitory effect on HCC proliferation. Network pharmacology analysis can systematically elucidate drug–organism interactions by predicting target profiles and pharmacological mechanisms. Firstly, we used network pharmacology to explore the underlying mechanisms by which TBMS1 affects HCC proliferation, then we determined 407 TBMS1-related target and 15 associated pathways in HCC. In addition, we applied RNA sequencing to furthermore identify differentially expressed genes after TBMS1 treatment. Interestingly, we found the results of network pharmacological prediction and RNA-seq analysis were co-enriched in the NF-κB pathway. NF-κB is involved in various pathological processes and is a transcription factor related to inflammation. Additionally, it has been discovered that NF-κB has a significant role in controlling inflammatory mediators linked to liver cancer.47 Therefore, inhibition of NF-κB signaling by targeting may help to intervene in HCC progression.
Next, we targeted TNFα-induced protein 3 (TNFAIP3), a differential gene that was significantly altered in the NF-κB pathway after TBMS1 treatment. Studies have shown that TNFAIP3 is a central modulator of cellular immunity and inflammation. As a ubiquitin-editing protein, TNFAIP3 can induce deubiquitination of related proteins and down-regulate NF-κB signaling, thereby playing an anti-inflammatory role.48–50 Our experimental results found that treatment with TBMS1 could increase TNFAIP3’s mRNA and protein expression in liver cancer cells. Also, TNFAIP3 expression was also significantly increased in tumor tissues of TBMS1-treated group. These results imply that TBMS1 may prevent the growth and trigger apoptosis of liver cancer by controlling TNFAIP3 expression and deactivating NF-κB. However, the underlying mechanism of the association between TNFAIP3 and TBMS1 anti-HCC activity remains unclear. For example, overexpression or knockdown of TNFAIP3 in HCC cells will affect the inhibitory effect of TBMS1 on HCC activity in vitro and in vivo, and to explore whether TNFAIP3 is involved in mediating TBMS1’s effect on NF-κB pathway. A comprehensive mechanism merits further investigation. To sum up, these present results suggest that TNFAIP3 plays a key role in TBMS1-mediated anti-HCC activity.
Conclusion
This study proved that TBMS1 suppressed HCC cells from proliferating both in vitro and in vivo. The findings also suggested that TBMS1 inactivated NF-κB pathway activity by regulating TNFAIP3 expression, thereby inhibiting the proliferation of HCC. Our research offers a novel perspective on the therapeutic application of TBMS1 as a potential antitumor agent for treating HCC.
Abbreviations
HCC, hepatocellular carcinoma; TBMS1, Tubeimoside I; TNFAIP3, TNFα-induced protein 3; CCK-8, Cell Counting Kit-8; EDU, 5-ethynyl-2′-deoxyuridine; IC50, half maximal inhibitory concentration; PVDF, polyvinylidene fluoride; ANOVA, analysis of variance; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; MF, molecular function; CC, cellular component; DEGs, differentially expressed genes.
Acknowledgment
We would like to thank Editage (www.editage.cn) for English language editing.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Guangdong Science and Technology Foundation (2023B0101200003); Shenzhen Medical Research Funds (A2303053); National Natural Science Foundation of China (82203870); Shenzhen Science and Technology Foundation (JCYJ20240813141015020; KCXFZ20230731094904008; KJZD20230923115359001; JSGG20220301090003004); National Key R&D Programmes (Grant No. 2022YFC2302700); Guangdong Basic and Applied Basic Research Foundation (2024A1515013237); Medical-Engineering Interdisciplinary Research Foundation of Shenzhen University(2023YG009). Shenzhen Portion of Shenzhen-Hong Kong Science and Technology Innovation Cooperation Zone (HTHZQSWS- KCCYB-2023060). Team-based Medical Science Research Program (2024YZZ09); Shenzhen Third People’s Hospital (23260G1001).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J Clin. 2024;74(3):229–263. doi:10.3322/caac.21834
2. Li Q, Ding C, Cao M, et al. Global epidemiology of liver cancer 2022: an emphasis on geographic disparities. Chin Med J. 2024;137(19):2334. doi:10.1097/CM9.0000000000003264
3. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
4. Llovet JM, Pinyol R, Kelley RK, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. doi:10.1038/s43018-022-00357-2
5. Liu Z, Lin Y, Zhang J, et al. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J Exp Clin Canc Res. 2019;38(1):447. doi:10.1186/s13046-019-1412-8
6. Tang W, Chen Z, Zhang W, et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5(1):87. doi:10.1038/s41392-020-0187-x
7. Donne R, Lujambio A. The liver cancer immune microenvironment: therapeutic implications for hepatocellular carcinoma. Hepatology. 2023;77(5):1773–1796. doi:10.1002/hep.32740
8. Zhang Z, Huang D, Feng J, et al. PDE5 inhibitors against cancer via mediating immune cells in tumor microenvironment: AI-based approach for future drug repurposing exploration. Interdiscip Med. 2024;2(3):e20230062. doi:10.1002/INMD.20230062
9. Ling CQ, Fan J, Lin HS, et al. Clinical practice guidelines for the treatment of primary liver cancer with integrative traditional Chinese and Western medicine. J Integr Med. 2018;16(4):236–248. doi:10.1016/j.joim.2018.05.002
10. Zhang Y, Lou Y, Wang J, Yu C, Shen W. Research status and molecular mechanism of the traditional Chinese medicine and antitumor therapy combined strategy based on tumor microenvironment. Front Immunol. 2020;11:609705. doi:10.3389/fimmu.2020.609705
11. Xu W, Li B, Xu M, Yang T, Hao X. Traditional Chinese medicine for precancerous lesions of gastric cancer: a review. Biomed Pharmacother. 2022;146:112542. doi:10.1016/j.biopha.2021.112542
12. Liu X, Li M, Wang X, et al. Effects of adjuvant traditional Chinese medicine therapy on long-term survival in patients with hepatocellular carcinoma. Phytomedicine. 2019;62:152930. doi:10.1016/j.phymed.2019.152930
13. Kong F-H, Zhu D-Y, Xu R-S. Structural study of tubeimoside i, a constituent of tu-bei-mu. Tetrahedron Lett. 1986;27(47):5765–5768. doi:10.1016/S0040-4039(00)85321-6
14. Wang CL, Gao MZ, Gao DM, et al. Tubeimoside-1: a review of its antitumor effects, pharmacokinetics, toxicity, and targeting preparations. Front Pharmacol. 2022;13:941270. doi:10.3389/fphar.2022.941270
15. Shi H, Bi H, Sun X, et al. Tubeimoside-1 inhibits the proliferation and metastasis by promoting miR-126-5p expression in non-small cell lung cancer cells. Oncol Lett. 2018;16(3):3126–3134. doi:10.3892/ol.2018.9051
16. Jiang S-L, Guan Y-D, Chen X-S. Tubeimoside-1, a triterpenoid saponin, induces cytoprotective autophagy in human breast cancer cells in vitro via Akt-mediated pathway. Acta Pharmacol Sin. 2019;40(7):919–928. doi:10.1038/s41401-018-0165-9
17. Cao J, Zhao E, Zhu Q, et al. Tubeimoside-1 inhibits glioblastoma growth, migration, and invasion via inducing ubiquitylation of MET. Cells. 2019;8(8):774. doi:10.3390/cells8080774
18. Du J, Dong Z, Tan L, et al. Tubeimoside I inhibits cell proliferation and induces a partly disrupted and cytoprotective autophagy through rapidly hyperactivation of MEK1/2-ERK1/2 cascade via promoting PTP1B in melanoma. Front Cell Dev Biol. 2020;8:607757. doi:10.3389/fcell.2020.607757
19. Ibrahim S, Zhu X, Luo X, Feng Y, Wang J. PIK3R3 regulates ZO-1 expression through the NF-kB pathway in inflammatory bowel disease. Int Immunopharmacol. 2020;85:106610. doi:10.1016/j.intimp.2020.106610
20. Yang YM, Kim SY, Seki E. Inflammation and liver cancer: molecular mechanisms and therapeutic targets. Semin Liver Dis. 2019;39(1):26–42. doi:10.1055/s-0038-1676806
21. Pua KH, Chew CL, Lane DP, Tergaonkar V. Inflammation-associated genomic instability in cancer. Genome Instabil Dis. 2020;1(1):1–9. doi:10.1007/s42764-019-00006-6
22. Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8(2):108–118. doi:10.1038/nrgastro.2010.213
23. Karin M. NF-kappaB as a critical link between inflammation and cancer. Csh Perspect Biol. 2009;1(5):a000141. doi:10.1101/cshperspect.a000141
24. Shi Y, Wang X, Wang J, Wang X, Zhou H, Zhang L. The dual roles of A20 in cancer. Cancer Lett. 2021;511:26–35. doi:10.1016/j.canlet.2021.04.017
25. Wang T, Xu X, Xu Q, et al. miR-19a promotes colitis-associated colorectal cancer by regulating tumor necrosis factor alpha-induced protein 3-NF-κB feedback loops. Oncogene. 2017;36(23):3240–3251. doi:10.1038/onc.2016.468
26. Chen H, Hu L, Luo Z, et al. A20 suppresses hepatocellular carcinoma proliferation and metastasis through inhibition of Twist1 expression. mol Cancer. 2015;14:186. doi:10.1186/s12943-015-0454-6
27. High A20 expression negatively impacts survival in patients with breast cancer - PubMed. Available from: https://pubmed.ncbi.nlm.nih.gov/31449546.
28. Boone DL, Turer EE, Lee EG, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5(10):1052–1060. doi:10.1038/ni1110
29. Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Sig Transduct Target Ther. 2020;5(1):1–23. doi:10.1038/s41392-020-00312-6
30. PubChem 2023 update - PubMed. Available from: https://pubmed.ncbi.nlm.nih.gov/36305812/.
31. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
32. Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–W360. doi:10.1093/nar/gkx374
33. Nickel J, Gohlke BO, Erehman J, et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 2014;42:W26–31. doi:10.1093/nar/gku477
34. The genecards suite | springerLink. Available from: https://link.springer.com/chapter/10.1007/978-981-16-5812-9_2.
35. Zhou Y, Zhang Y, Zhao D, et al. TTD: therapeutic target database describing target druggability information. Nucleic Acids Res. 2024;52(D1):D1465–D1477. doi:10.1093/nar/gkad751
36. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi:10.1093/nar/gkac1000
37. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
38. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
39. Tang D, Chen M, Huang X, et al. SRplot: a free online platform for data visualization and graphing. PLoS One. 2023;18(11):e0294236. doi:10.1371/journal.pone.0294236
40. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188314. doi:10.1016/j.bbcan.2019.188314
41. Liao K, Gong L, Yang Y, et al. A comprehensive review of research progress in Chinese medicines for primary liver cancer treatment. Tradit Med Res. 2022;7:10–18. doi:10.53388/TMR20220207263
42. Yang X, Feng Y, Liu Y, et al. Fuzheng Jiedu Xiaoji formulation inhibits hepatocellular carcinoma progression in patients by targeting the AKT/CyclinD1/p21/p27 pathway. Phytomedicine. 2021;87:153575. doi:10.1016/j.phymed.2021.153575
43. Wang N, Tan HY, Li L, Yuen MF, Feng Y. Berberine and coptidis rhizoma as potential anticancer agents: recent updates and future perspectives. J Ethnopharmacol. 2015;176:35–48. doi:10.1016/j.jep.2015.10.028
44. Tian WT, Zhang XW, Liu HP, Wen YH, Li HR, Gao J. Structural characterization of an acid polysaccharide from Pinellia ternata and its induction effect on apoptosis of Hep G2 cells. Int J Biol Macromol. 2020;153:451–460. doi:10.1016/j.ijbiomac.2020.02.219
45. Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 2013;15(1):195–218. doi:10.1208/s12248-012-9432-8
46. Yin Y, Chen W, Tang C, et al. NF-κB, JNK and p53 pathways are involved in tubeimoside-1-induced apoptosis in HepG2 cells with oxidative stress and G2/M cell cycle arrest. Food Chem Toxicol. 2011;49(12):3046–3054. doi:10.1016/j.fct.2011.10.001
47. Monkkonen T, Debnath J. Inflammatory signaling cascades and autophagy in cancer. Autophagy. 2018;14(2):190–198. doi:10.1080/15548627.2017.1345412
48. Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10(5):332–341. doi:10.1038/nrc2775
49. Lin SC, Chung JY, Lamothe B, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J mol Biol. 2008;376(2):526–540. doi:10.1016/j.jmb.2007.11.092
50. Wertz IE, O’Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi:10.1038/nature02794
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.