Back to Journals » International Journal of Nanomedicine » Volume 20
Umbilical Cord Mesenchymal Stromal Cell-Derived Small Extracellular Vesicles Modulate Skin Matrix Synthesis and Pigmentation
Authors Kee LT , Foo JB, How CW, Nur Azurah AG, Chan HH, Mohd Yunus MH, Ng SN, Ng MH , Law JX
Received 22 October 2024
Accepted for publication 24 December 2024
Published 4 February 2025 Volume 2025:20 Pages 1561—1578
DOI https://doi.org/10.2147/IJN.S497940
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Eng San Thian
Li Ting Kee,1 Jhi Biau Foo,2– 4 Chee Wun How,5 Abdul Ghani Nur Azurah,6 Hong Hao Chan,5 Mohd Heikal Mohd Yunus,7 See Nguan Ng,8 Min Hwei Ng,1 Jia Xian Law1
1Department of Tissue Engineering and Regenerative Medicine, Universiti Kebangsaan Malaysia, Cheras, Kuala Lumpur, Malaysia; 2School of Pharmacy, Taylor’s University, Subang Jaya, Selangor, Malaysia; 3Digital Health and Medical Advancements Impact Lab, Taylor’s University, Subang Jaya, Selangor, Malaysia; 4Non-Destructive Biomedical and Pharmaceutical Research Centre, Smart Manufacturing Research Institute, Universiti Teknologi MARA Selangor Campus, Puncak Alam, Selangor, Malaysia; 5School of Pharmacy, Monash University Malaysia, Bandar Sunway, Selangor, Malaysia; 6Department of Obstetrics and Gynaecology, Universiti Kebangsaan Malaysia Medical Centre, Kuala Lumpur, Malaysia; 7Department of Physiology, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia; 8Ming Medical Sdn Bhd, Petaling Jaya, Selangor, Malaysia
Correspondence: Jia Xian Law, Department of Tissue Engineering and Regenerative Medicine, Universiti Kebangsaan Malaysia, Cheras, Kuala Lumpur, Malaysia, Email [email protected]
Introduction: Research has unveiled the remarkable properties of extracellular vesicles derived from mesenchymal stromal cells (MSCs), particularly in promoting wound healing, aiding re-epithelialization, revitalizing aging skin, and inhibiting hyperpigmentation. However, investigations into the potential of small extracellular vesicles from umbilical cord-derived MSCs (UC-MSC-sEVs) in reducing scarring and preventing hyperpigmentation remain limited. Therefore, this study aims to evaluate the impact of UC-MSC-sEVs on the synthesis of the skin’s extracellular matrix (ECM) and pigmentation using in vitro models.
Methods: The study investigated the impact of characterized UC-MSC-sEVs on various aspects including the proliferation, migration, antioxidant activity, and ECM gene expression of human dermal fibroblasts (HDF). Additionally, the effects of UC-MSC-sEVs on the proliferation, melanin content, and tyrosinase (TYR) activity of human melanoma cells (MNT-1) were examined. Furthermore, ex vivo models were employed to evaluate the skin permeation of PKH26-labelled UC-MSC-sEVs.
Results: The findings indicated that a high concentration of UC-MSC-sEVs positively influenced the proliferation of HDF. However, no changes in cell migration rate were observed. While the expressions of collagen type 1 and type 3 remained unaffected by UC-MSC-sEVs treatment, there were dose-dependent increases in the gene expressions of fibronectin, matrix metallopeptidase (MMP) 1, and MMP 3. Furthermore, UC-MSC-sEVs treatment did not impact the antioxidative superoxide dismutase (SOD) expression in HDF. Although UC-MSC-sEVs did not alter the proliferation of MNT-1 cells, it did result in a dose-dependent reduction in melanin synthesis without affecting TYR activity. However, when it was applied topically, UC-MSC-sEVs failed to penetrate the skin barrier and remained localized within the stratum corneum layer even after 18 hours.
Conclusion: These results highlight the potential of UC-MSC-sEVs in stimulating HDF proliferation, regulating ECM synthesis, and reducing melanin production. This demonstrates the promising application of UC-MSC-sEVs in medical aesthetics for benefits such as scar reduction, skin rejuvenation, and skin lightening.
Keywords: mesenchymal stromal cell, extracellular vesicles, anti-scarring, pigmentation, medical aesthetic
Introduction
The quest to enhance or refine the human physique and rectify aesthetic imperfections has propelled the ascent of medical aesthetics. Within the domain of medical aesthetics, key priorities include advancing wound healing, anti-aging remedies, inhibiting hyperpigmentation, and combatting hair loss. In recent years, this discipline has garnered significant interest for its fusion with regenerative medicine to elevate one’s outward appearance. It encompasses a spectrum of interventions aimed at repairing, replacing, or regenerating human cells and tissues, employing cutting-edge techniques from regenerative medicine such as cellular therapies, natural or synthetic scaffold materials, and growth factors.
Among these strategies, extracellular vesicles (EVs) have emerged as promising therapeutic agents owing to their diverse biological functions and potential applications. EVs, composed of lipid bilayers, are released from nearly all cell types to regulate various biological processes.1–4 In the medical realm, they have been investigated as disease biomarkers and therapeutic carriers.5,6 These vesicles transport functional cargo such as proteins, lipids, and nucleic acids across cells, making them potential facilitators of tissue repair.1,2
EVs encompass a diverse family with distinct subclasses characterized by their subcellular origin, size, and composition.5 They are broadly classified into two main categories based on size: small extracellular vesicles (sEVs), typically under 200 nm, and large EVs (lEVs), typically exceeding 200 nm.3,4 sEVs, in particular, have gained prominence due to their compatibility with biological systems, presence protective lipid structure, small size, and capability to traverse barriers like the skin and blood-brain barrier.2 Notably, research has demonstrated the therapeutic potential of sEVs derived from various sources in skin conditions, including wound healing and regeneration.7–10 These findings suggest that sEVs hold promise in the realm of medical aesthetics.
Stem cell-derived EVs offer a multitude of beneficial effects in dermatology, including scar reduction, skin regeneration, and whitening effects.9,11–18 Of particular interest are sEVs derived from umbilical cord mesenchymal stromal cells (UC-MSCs), which offer distinct advantages over other MSC sources. The umbilical cord, housing a rich reserve of stem cells and supportive stromal tissue, provides a non-invasive and ethically uncontroversial MSC source. Furthermore, UC-MSCs demonstrate unique immunomodulatory properties and a higher proliferative capacity compared to their adult tissue counterparts, rendering them highly appealing for regenerative medicine.19–21
Previous findings showed that EVs from various stem cell sources effectively reduced scar formation by attenuating fibrosis through decreasing the excessive deposition of ECM components.22–24 Additionally, EVs obtained from human adipose tissue-derived stem cells and amniotic stem cells have been found to diminish intracellular melanin levels and regulate skin hyperpigmentation by inhibiting melanogenesis and facilitating melanosome degradation.18,25 However, research remains scarce on the distinctive therapeutic potential of small extracellular vesicles derived from umbilical cord mesenchymal stromal cells (UC-MSC-sEVs) in modulating scar formation and skin pigmentation. Hence, this study sought to assess the effectiveness of UC-MSC-sEVs in regulating the synthesis of extracellular matrix (ECM) and pigmentation of the skin using in vitro models.
Materials and Methods
Isolation, Culture and Characterization of Umbilical Cord Derived Mesenchymal Stromal Cells (UC-MSCs)
The umbilical cord samples were collected from mothers aged 18–44 who underwent natural or caesarean delivery at full term without pregnancy complications, infectious diseases, or a history of cancer at Hospital Canselor Tuanku Muhriz HCTM), Kuala Lumpur with informed consent forms and ethic approval (UKM PPI/111/8/JEP-2019-618). The isolation of UC-MSCs was done following a previously established protocol.26 In brief, the arteries and veins were removed from umbilical cord using sterile forceps, and the remaining tissue was minced. Subsequently, the minced tissue underwent enzymatic digestion using 0.6% collagenase type I (Worthington, USA) for 1 hour at 37°C in a shaking incubator. The isolated cells were then cultured in low glucose-Dulbecco’s Modified Eagle Medium (LG-DMEM; Sigma-Aldrich) supplemented with 1% GlutaMAX™ (Gibco, USA), 1% antibiotic-antimycotic (AA; Capricorn Scientific, Germany), and 10% fetal bovine serum (FBS; Capricorn Scientific). Once the cells attached, the FBS in the culture medium was replaced with 10% in-house prepared human platelet lysate (hPL) and 4 IU/mL of heparin (Heparinol®). hPL was prepared as described previously.27 Media changes were conducted every three days, and the cells were cryopreserved at passage two. After collecting three distinct biological samples (UC-MSCs), they were thawed from cryo-storage and pooled at passage four. The pooled UC-MSCs were characterized at passage five, and their conditioned medium was collected for the isolation of sEVs.
The pooled UC-MSCs underwent characterization following the guidelines recommended by the International Society for Cell and Gene Therapy (ISCT).28 Positive and negative markers for MSCs were identified using the Human MSC Analysis Kit (BD Biosciences, USA) as per the manufacturer’s instructions. In brief, 1×106 cells were resuspended in 100 µL of washing buffer and incubated with the antibody cocktails. The antibody cocktails included antibodies for positive surface markers (CD73, CD90, CD105) and negative surface markers (CD11b, CD19, CD34, CD45, Human leukocyte antigen-DR (HLA-DR)), and the cells were incubated in the dark at room temperature for 30 minutes. Subsequently, the cells were washed with washing buffer and analyzed using a flow cytometer (BD FACSVerse™).
The differentiation potential of UC-MSCs was evaluated using the StemPro® Adipogenesis Differentiation Kit and StemPro® Osteogenesis Differentiation Kit (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. In brief, UC-MSCs were seeded in 12-well plates and cultured in adipogenesis and osteogenesis differentiation media for 10 days, respectively. The differentiation media were refreshed every two to three days. After ten days of continuous induction, the cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) and stained with Oil Red O (Sigma-Aldrich) to visualize lipid droplets in adipogenic differentiated cells, and Alizarin Red (Sigma-Aldrich) to detect calcium deposition in osteogenic differentiated cells.
Collection of Conditioned Media for Small Extracellular Vesicles Isolation
When the UC-MSCs reached 70–80% confluence, the culture medium was aspirated, and the cells were rinsed three times with phosphate-buffered saline (PBS; Gibco). Subsequently, phenol red-free LG-DMEM basal medium (Sigma-Aldrich) was added. After incubating for 24 hours, the conditioned media (CM) was collected and centrifuged at 2,000 × g for 15 minutes at 4°C, followed by second centrifugation at 7,000 × g for 45 minutes at 4°C to remove dead cells and debris. Finally, the CM was filtered through a 0.22 μm filter to remove any particles larger than 220 nm.
sEVs Preparation Using Centricon® Plus-70 Filter Unit
Centricon® Plus-70 (100 kDa NMWL) (Merck Millipore, Germany) was used for isolating sEVs, following the method described in a previous study.29 In brief, the filter unit underwent pre-rinsing and was filled with filtered CM, then subjected to centrifugation at 3500 × g at 4°C. The duration of centrifugation was adjusted to avoid desiccation of the remaining solution. Subsequently, the filtrate collection cup was emptied, and the sample filter cup with the concentrate collection cup underwent reverse centrifugation at 1,000 × g for 2 minutes at 4°C to isolate the sEVs. Filtered PBS (Gibco) was added to the collected sEVs, followed by centrifugation at 3500 × g for 20 minutes at 4°C. After discarding the solution from the filtrate collection cup, the sample filter cup with the concentrate collection cup was subjected to reverse centrifugation at 1,000 × g for 2 minutes at 4°C to collect the isolated sEVs. These samples were then stored at −80°C for subsequent experiments. The sEVs samples obtained through this process were labeled as sEVs-C and utilized for experiments involving human dermal fibroblasts (HDF) and ex vivo skin samples.
sEVs Preparation Using Tangential Flow Filtration
sEVs isolation was also conducted via tangential flow filtration (TFF) using a 100-kDa MWCO ultrafiltration membrane filter capsule (Pall Corporation, USA). Initially, the filter capsule underwent washing with MiliQ water. Then, the normalized water permeability (NWP) was determined by applying three different pressures: 2.5 psi, 5 psi, and 10 psi to assess membrane performance. Buffer conditioning was performed using PBS (Gibco) to eliminate air bubbles from the Minimate™ TFF Capsule and system. Filtered CM was then added into the feed tank, with a magnetic stirrer continuously agitating to prevent gel formation. Simultaneously, diafiltration was carried out by passing two times the volume of PBS (Gibco) as the original conditioned media through the diafiltration tube immersed in the buffer solution. Once sEVs were concentrated to around 7 mL, the final volume was recirculated for 10 minutes at 60 rpm and collected from the reservoir. Subsequently, another 8 mL of PBS (Gibco) was introduced into the reservoir, recirculated through the filter for 10 minutes at 60 rpm, and collected. The obtained samples were stored at −80 °C for subsequent analysis. The collected EVs sample was designated as sEVs-TFF and was utilized for studies involving melanocytes.
Characterization of UC-MSC-sEVs
Particle Quantification and Size Distribution
To assess the size distribution and concentration of sEVs, nanoparticle tracking analysis (NTA) was conducted using the Nanosight NS300 (Malvern Panalytical, UK). UC-MSC-sEVs were diluted in filtered PBS (Gibco) before being introduced into the NTA chamber using disposable 1 mL syringes. The measurement settings were adjusted as follows: the camera level was set to level 14, the detection threshold was optimized to include as many particles as possible while restricting the count to 10–100 red crosses, and the number of blue crosses was maintained at five.
Protein Quantification
To quantify the protein content of sEVs, sEVs lysates were generated by incubating them with RIPA lysis buffer (Thermo Fisher Scientific) containing 1% Halt™ protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) for 30 minutes at 4°C, followed by centrifugation at 14,000 × g for 15 minutes at 4°C. The protein concentration of sEVs was subsequently quantified using the Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, the samples and prepared bovine serum albumin standards were added to a 96-well plate. Then, the working solution was added to each well and incubated for 30 minutes at 37°C. The optical density was measured at 562 nm.
sEVs Marker Expression
Total protein from cell lysate and sEVs were separated by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Immobilon®-P Polyvinylidene difluoride (PVDF) Membranes (0.45 μm pore size; Merck Millipore, Ireland). The membranes were blocked with 3% bovine serum albumin (BSA; Sigma-Aldrich) for 1 hour and incubated with primary antibodies: mouse anti-CD63 monoclonal antibody (Novus Biologicals, USA) (1: 1,000); mouse anti-TSG101 monoclonal antibody (Novus Biologicals) (1:1,000); mouse anti-HSP70 monoclonal antibody (R&D System, USA) (1:4,000); mouse anti-GP96 monoclonal antibody (R&D System) (1:6,000) overnight at 4°C, followed by incubation with Mouse IgG Horseradish Peroxidase (HRP)-conjugated Antibody (1:10,000) (R&D Systems, USA) for 1 hour at room temperature. Finally, the membranes were treated with Amersham ECL Prime Western blotting detection reagent (Cytiva, USA) and visualized using a gel imager (Amersham Imager 600).
Human Dermal Fibroblast Study
Isolation and Culture of HDF
Human dermal fibroblasts (HDF) were sourced from redundant tissue obtained with written informed consent from healthy patients aged 18–60 who underwent abdominoplasty at Hospital Canselor Tuanku Muhriz, Kuala Lumpur, without any infection, infectious diseases, chronic skin conditions, damaged skin in the abdominal area, or history of cancer. Skin samples measuring 3 cm² were meticulously cleaned and finely minced into small pieces. These tissue fragments underwent enzymatic digestion using 0.6% collagenase type I (Worthington) for 2–4 hours, followed by 0.05% trypsin-ethylenediamine tetraacetic acid (trypsin-EDTA; Gibco) for 10 minutes at 37°C in a shaking incubator. Subsequently, the cell suspension was centrifuged, and the pellet was resuspended in HDF culture medium (Nutrient Mixture F-12 and Dulbecco’s Modified Eagle Medium (FD medium; Sigma-Aldrich) supplemented with 10% FBS (Gibco) and 1% AA (Capricorn Scientific)) for in vitro expansion. The medium was refreshed every three days. Upon reaching 70–80% confluency, HDF were harvested using 0.05% trypsin-EDTA (Gibco) and utilized for subsequent experiments.
Cellular Uptake Assay
sEVs-C was labeled with PKH26 (Sigma Aldrich) following the manufacturer’s instructions. In brief, sEVs-C were incubated with 4 μM PKH26 Dye Solution in Diluent C with gentle mixing for 5 minutes. Excess labelling dye was eliminated using Amicon® Ultra-15 Centrifugal Filter Unit (100 kDa, Merck Milipore). HDF were seeded at a density of 5,000 cells/well in a 96-well plate for 24 hours. Then, the cells were incubated with PKH26-labeled sEVs at 37°C and 5% CO2 for 6 hours. After incubation, the cells were washed with PBS (Gibco), fixed with 4% paraformaldehyde (Sigma-Aldrich), and counterstained with Hoechst dye (Sigma-Aldrich) for 30 minutes at room temperature to visualize the nuclei. The cells were then observed using a fluorescence microscope (Nikon), and images were captured.
Cell Proliferation Assay
HDF were seeded at a density of 1,500 cells/well in a 96-well plate and treated with 0, 10, 25, and 50 μg/mL of sEVs-C in HDF culture medium for 24, 48, and 72 hours. Cell proliferation was assessed using a CCK8 kit (Elabscience, China) following the manufacturer’s protocol. Briefly, 10 μL CCK-8 buffer was added into each well and incubated at 37°C, 5% CO2 for 3 hours. Absorbance was measured at 450 nm using a microplate reader (BioTek, USA).
Wound Scratch Assay
HDF were seeded into 12-well plates at a density of 1.5×105 cells per well and incubated at 37°C and 5% CO2 to reach confluence. The cells were scratched using a sterilized 10 µL pipette tip. After washing with Dulbecco’s Phosphate Buffered Saline (DPBS; Sigma-Aldrich), different concentrations of sEVs-C (0, 10, 25, and 50 μg/mL) were added. Images of the scratched areas were captured at specific time points (0, 12, 24, 36 and 48 hours) using an inverted microscope (Nikon). At least three-microscope fields were determined for each condition. The wound area was measured using ImageJ software (version 1.53k). The wound closure rate (%) was calculated using the formula below:
where w(0) is wound area at 0 hour and w(t) is wound area at specific time.
Ribonucleic Acid (RNA) Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
HDF with seeding density of 3,000 cells per cm2 were exposed to varying concentrations of sEVs-C (0, 10, 25, and 50 μg/mL) until reaching 80–90% confluency. Then, the cells were washed thoroughly with PBS to remove any residual sEVs, and RNA was extracted directly from the cells using the NucleoSpin RNA kit (Macherey-Nagel, Germany). The LunaScript® RT SuperMix Kit (New England Biolabs, USA) was utilized for reverse transcription of total RNA to complementary DNA (cDNA) following the manufacturer’s instructions. Subsequently, polymerase chain reaction (PCR) was conducted using the generated cDNA as a template with the LunaScript® SYBR Green PCR Master Mix (New England Biolabs). The standard PCR cycle conditions comprised an initial denaturation at 95°C for 1 minute, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing-extension at 60°C for 30 seconds. Relative mRNA expression levels were analyzed using the 2(−∆∆Ct) method, normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Specific primer sequences employed in this study are detailed in Table 1.
 |
Table 1 Primers’ Sequence Targeting the ECM and ECM Modulation Genes |
Immunocytochemistry Analysis
HDF were plated at a density of 2×104 cells per well in a 48-well plate and maintained at 37°C in a 5% CO2 atmosphere. Upon reaching 80–90% confluence, the media was replaced with HDF culture media containing varying concentrations of sEVs-C (0, 10, 25, and 50 μg/mL) and incubated for 24 hours. Subsequently, the cells were rinsed with DPBS (Sigma-Aldrich) and fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 minutes at room temperature, followed by permeabilization with 0.5% Triton X-100 (Sigma-Aldrich) for 20 minutes. After blocking with 10% goat serum (Merck Millipore) for 1 hour at 37°C, the cells were incubated overnight at 4°C with primary antibodies, ie, rabbit anti-human collagen I antibody (COLI; Abcam, USA), and mouse anti-human alpha-smooth muscle actin antibody (αSMA; Abcam). Subsequently, the cells were treated with secondary antibodies, ie, goat anti-mouse IgG (Alexa Fluor® 488) (Abcam) and goat anti-rabbit IgG (Alexa Fluor® 594) (Abcam), for 2 hours at 37°C, and counterstained with Hoechst dye (Sigma-Alrich) for 30 minutes at room temperature. Images were acquired using a fluorescence microscope (Nikon). All primary and secondary antibodies were diluted at a ratio of 1:1,000. The fluorescent intensity of COLI and αSMA, as well as the number of nuclei, were quantified using ImageJ software (version 1.53k) to assess the relative protein expression levels.
Superoxide Dismutase (SOD) Assay
HDF were plated in 6-well plates at a density of 5×104 cells per well and incubated at 37°C with 5% CO2. Upon reaching 80–90% confluence, the medium was substituted with HDF culture medium containing various concentrations of sEVs-C (0, 10, 25, and 50 μg/mL) and incubated for 24 hours. Subsequently, HDF were lysed using radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) and centrifuged at 14,000 × g for 15 minutes at 4°C to collect the supernatant. Superoxide dismutase (SOD) activity in HDF was determined using the Superoxide Dismutase Activity Assay Kit (Sigma-Aldrich) following the manufacturer’s protocol. Briefly, 20 μL of sample, standard, and prepared dilution buffer were added to a 96-well plate. Then, 160 μL of WST working solution was added to each well, followed by the addition of 20 μL of xanthine oxidase working solution to initiate the SOD inhibition reaction. The plate was then incubated at room temperature for 30 minutes. Absorbance was measured at 450 nm using a microplate reader (BioTek), and SOD activity (units/mL) in each sample was calculated using the SOD standard curve.
Melanoma Cell (MNT-1) Study
MNT-1 Culture
MNT-1 cells were obtained from American Type Culture Collection (ATCC) (CRL-3450) and cultured in MNT-1 medium comprising DMEM-LG supplemented with 20% FBS, 10% AIM-V medium (Gibco), 1% sodium pyruvate (Sigma-Aldrich), 1% non-essential amino acids (NEAA; Sigma-Aldrich), and 1% AA at 37°C in a 5% CO2 incubator. Upon reaching 80–90% confluence, MNT-1 cells were detached using CTS™ TrypLE™ Select Enzyme (Gibco) and passage into cell culture plates for subsequent experiments.
Cellular Uptake of sEVs
sEVs-TFF was labeled with PKH26 (Sigma-Aldrich) following the manufacturer’s protocol. MNT-1 cells were seeded at a density of 7,500 cells/well in a 96-well plate for 48 hours. Then, the cells were incubated with PKH26-labeled sEVs for 6 hours. After incubation, the cells were washed with PBS, fixed with 4% paraformaldehyde (Sigma-Aldrich), and stained with Hoechst dye (Sigma-Aldrich). Imaging was conducted using a fluorescence microscope (Nikon).
Cell Proliferation Assay
MNT-1 cells were seeded at a density of 7500 cells/well in a 96-well plate and incubated for 48 hours at 37°C in a 5% CO2 incubator. Next, the cells were co-cultured with varying concentrations of sEVs-TFF (0, 10, 25, and 50 μg/mL) in MNT-1 medium for 24, 48, and 72 hours. Cell proliferation was assessed using a CCK8 kit (Elabscience) following the manufacturer’s instructions. Briefly, 10 μL CCK-8 Buffer was added into each well and incubate for 1 h at 37°C in a 5% CO2 incubator. Absorbance was measured at 450 nm using a microplate reader (BioTek) to measure cell proliferation rate.
Melanin Content and Tyrosinase Activity Assay
The melanin content and tyrosinase activity assays were conducted with adjustments based on the protocols described previously (Netcharoensirisuk et al 2021).30 MNT-1 cells were seeded at a density of 5×105 cells per well in a 6-well plate and cultured for 48 hours at 37°C in a 5% CO2 incubator. Upon reaching 80–90% confluence, the cells were treated with various concentrations of sEVs-TFF (0, 10, 25, and 50 μg/mL) in MNT-1 culture medium for 24 hours. Subsequently, the treated cells were harvested using CTS™ TrypLE™ Select Enzyme (Gibco). After centrifugation at 3000 rpm for 5 minutes at 37°C, the cells were resuspended in DPBS (Sigma-Aldrich) and divided into two portions for further analysis.
For tyrosinase activity assay, cell pellets were lysed with RIPA lysis buffer containing 1% Halt™ protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) for 45 minutes at 4°C. The lysed cells were then centrifuged at 12,000 rpm for 15 minutes at 4°C, and the protein concentration of the cell lysate from each treatment group was determined using the PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific). Next, 50 μg of protein from the supernatant was transferred into a 96-well plate and mixed with 50 μL of 15 mm levodopa (L-DOPA) (Sigma-Aldrich). The final volume was adjusted to 100 μL using PBS (Gibco) with pH 6.8. After incubation for 30 minutes at 37°C, dopachrome formation was measured by assessing the absorbance at 475 nm using a microplate reader (BioTek). The tyrosinase activity (%) was calculated using the formula below:
For the melanin content assay, cell pellets were dissolved in 250 μL of 1 N NaOH (EMSURE®, Germany) and incubated at 80°C for 2 hours. After centrifugation at 12,000 rpm for 10 minutes, supernatants were collected and added to a 96-well plate. The absorbance was measured at 405 nm using a microplate reader (BioTek). The melanin concentration was determined using a standard curve of synthetic melanin solutions with known concentrations. Subsequently, the melanin concentration was normalized to the total protein concentration determined using the PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific).
Ribonucleic Acid (RNA) Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
MNT-1 cells were seeded at a density of 2×105 cells per well in a 12-well plate and cultured for 48 hours at 37°C in a 5% CO2 incubator. Following this, the cells were treated with varying concentrations of sEVs-TFF (0, 10, 25, and 50 μg/mL) in MNT-1 culture medium for 24 hours. The cells were then thoroughly washed with PBS to remove any residual sEVs, and RNA was directly extracted from the cells using the Direct-zol RNA Miniprep Kits (Zymo Research, USA), following the manufacturer’s instructions. The total RNA was reverse transcribed into cDNA using the BlitzAmp cDNA synthesis kit (MiRXES, Singapore) according to the manufacturer’s protocol. PCR was conducted using the resulting cDNA as a template with the LunaScript® SYBR Green PCR Master Mix (New England Biolabs, USA). The standard PCR cycle conditions were 95°C for 1 minute, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing-extension at 60°C for 30 seconds. Melt curves were examined to confirm the specificity of the amplified product. Relative mRNA expression levels were analyzed using the 2(−∆∆Ct) method, with normalization to GAPDH. Specific primer sequences used in this study are detailed in Table 2.
 |
Table 2 Primers’ Sequences Targeting the Melanin Synthesis Gene |
Skin Permeation Analysis
sEVs-C was labeled with PKH26 (Sigma-Aldrich) following the manufacturer’s guidelines. Human skin samples, obtained with informed consent from patients and processed to remove the hypodermis, were preserved with intact dermis and epidermis and stored at −80°C until use. Upon thawing, the skin was rinsed with DPBS (Sigma-Aldrich) containing 1% AA (Capricorn Scientific), then cut into desired sizes. The labeled EVs was then applied to the skin surface. After an 18-hour incubation period, the skin strips were gently washed with PBS (Gibco) and dried. The skin strips were then embedded in Optimal cutting temperature (OCT) quick-freeze medium compound (Sakura Finetek, USA), snap-frozen in liquid nitrogen, and sectioned using a cryostat (HM525 NX Thermo Scientific, USA). Sections of 6 µm thickness were mounted on adhesion microscope slides (Citotest, China) and stained with 4′,6-diamidino-2-phenylindole (DAPI; Elabscience) for subsequent fluorescence imaging.
Statistical Analysis
Data analysis was performed using GraphPad Prism version 9.4.1 (GraphPad Software, Inc., USA), with results presented as the mean ± standard error of the mean. Statistical comparisons between the control and treatment groups were conducted using one-way and two-way analysis of variance (ANOVA). A p-value less than 0.05 was considered statistically significant.
Results
Human Dermal Fibroblast Study Characterization of UC-MSCs and UC-MSC-sEVs
UC-MSCs were isolated from umbilical cords and characterized based on their morphology, immunophenotype, and ability to differentiate into multiple lineages. At passage five, the pooled UC-MSCs exhibited a typical fibroblastic morphology, with elongated spindle-shaped cells adhering to plastic surfaces. Flow cytometry analysis revealed that over 99% of these cells expressed the surface markers CD73, CD90, and CD105, while the expression of negative markers such as HLA-DR, CD34, CD45, CD11b, and CD19 was less than 2%. Upon adipogenic induction, the cells transitioned to a polygonal shape with observable oil droplets stained by Oil Red O, indicative of adipogenesis. Similarly, osteogenic induction resulted in the deposition of calcium, confirmed by Alizarin Red staining, demonstrating their capacity for multilineage differentiation (Figure 1A-D).
UC-MSC-derived sEVs were isolated from conditioned medium using Centricon® Plus-70 (100 kDa NMWL) and TFF system (Figure 1E and F) and characterized following the guidelines outlined in MISEV 201830 to confirm their identity. The total protein concentrations of sEVs-C and sEVs-TFF were 741.9 ± 138.3 μg/mL and 775.4 ± 344.5 μg/mL, respectively, using the bicinchoninic acid assay. NanoSight analysis showed that the particle size of both sEVs-C and sEVs-TFF fell within the range of 50 nm to 200 nm, with mean sizes of 93.875 ± 5.866 nm and 127.250 ± 23.228 nm, respectively (Figure 1G and H). Concentration measurements indicated that each cell secreted approximately 3.226×103 ± 1.093×103 particles for sEVs-C and 2.446×103 ± 7.630×102 particles for sEVs-TFF. Immunoblotting analysis showed the expression of CD63, TSG101, and HSP70 in both sEVs-C and sEVs-TFF, while the negative marker (GP96) was absence in both sEVs (Figure 1I and J). These findings demonstrate the successful isolation of sEVs from UC-MSCs, meeting the accepted criteria for sEVs characterization.
UC-MSC-sEVs are Internalized by HDF, Promote HDF Proliferation but Do Not Affect HDF Migration and SOD Expression
To explore the uptake of sEVs by HDF, PKH26 dye was employed to label the sEVs. After a 6-hour incubation of HDF with UC-MSC-sEVs, the sEVs were observed in the perinuclear region of the HDF (Figure 2A).
To examine the impact of sEVs on HDF proliferation and migration, HDF were exposed to sEVs at varying concentrations (0, 10, 25, and 50 μg/mL) and assessed at different time intervals. As shown in Figure 2B, there were no significant differences in proliferation rates among all groups at 24 and 48 hours. However, by the 72-hour mark, sEVs notably boosted the cell proliferation at the concentrations of 25 and 50 μg/mL. Regarding cell migration, Figure 2C illustrates that HDF in all groups displayed migration over time, with no notable differences observed between the groups at any of the examined time points.
The level of SOD, an essential antioxidant enzyme for reducing oxidative stress, was measured in HDF treated with UC-MSC-sEVs. As shown in Figure 2D, the activity of SOD in HDF remained unchanged upon treatment with sEVs.
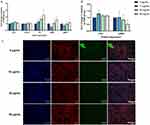
UC-MSC-sEVs Modulate HDF’s ECM Expression and Reduce the αSMA Expression
ECM plays a crucial role in maintaining tissue integrity, facilitating wound healing, and regulating tissue remodeling. As shown in Figure 3A, sEVs did not affect the gene expression of COLI and COLIII, while they dose-dependently upregulated the expressions of FN, MMP1, and MMP3. To confirm the gene expression of COLI, immunocytochemistry was performed, and the protein expression levels of the sEVs-treated groups were compared to the control group. As depicted in Figure 3B and C, the expressions of COLI and αSMA were not significantly altered by sEVs treatment. Specifically, the protein levels of COLI remained unchanged in the 10, 25, and 50 μg/mL groups. In contrast, the protein levels of αSMA showed a dose-dependent decrease.
Internalization of UC-MSC-sEVs Do Not Affect MNT-1 Proliferation, but Significantly Reduce Melanin Content by Modulating Melanogenesis-Related Gene Expression
PKH26-labeled sEVs were incubated with MNT-1 cells to observe their cellular internalization. After a 4-hour incubation, the sEVs were detected in the perinuclear region of the MNT-1 cells (Figure 4A).
Across the evaluated time points, MNT-1 cells in all groups exhibited proliferation, with no significant differences observed among the groups (Figure 4B). This suggests that sEVs-TFF did not exert any significant impact on MNT-1 proliferation.
UC-MSC-sEVs induced a concentration-dependent reduction in intracellular melanin content in MNT-1 cells (Figure 4C). A notable decrease in melanin content was observed when the cells were treated with 50 μg/mL of UC-MSC-sEVs. Additionally, slight reductions in tyrosinase activity were observed in MNT-1 cells exposed to UC-MSC-sEVs, although these changes were not statistically significant (Figure 4D). To gain further insight into the underlying mechanisms of melanin synthesis regulation, gene expression analysis was conducted. As depicted in Figure 4E, the results indicated that UC-MSC-sEVs did not affect the expression of TYRP1 but showed a reducing trend in the expression of MITF and TYRP2.
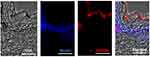
Penetration of UC-MSC-sEVs Is Limited to Stratum Corneum
To evaluate the skin permeation of UC-MSC-sEVs, PKH26 labeled sEVs were topically applied onto the surface of intact human skin. After an 18-hour incubation period, fluorescence was predominantly observed in the outer layer of the skin (stratum corneum), with no fluorescence detected in the deeper nucleated layer (stratum granulosum) (Figure 5).
Discussion
Previous research has demonstrated the promising benefits of EVs, including their role in combating scarring, promoting skin renewal and rejuvenation, and inhibiting hyperpigmentation. However, there is a notable gap in research focused on the distinct therapeutic potentials of sEVs obtained from UC-MSCs concerning scar reduction and skin pigmentation regulation, especially in comparison to EVs sourced from different types of stem cells. Therefore, this study aims to explore the effectiveness of UC-MSC-sEVs in influencing skin ECM production and pigmentation using in vitro models.
Before commencing in vitro analyses, we conducted an initial characterization of the proteome of MSC-sEVs (refer to Supplementary Method), aiming to gain preliminary insights into their protein composition. This step was crucial as proteins play pivotal roles in essential biological processes, including cell-to-cell communication.31 Through proteomic analysis, 148 proteins were identified in UC-MSC-sEVs after eliminating contaminated proteins (see Supplementary Result, Supplementary Figure S1 and Supplementary Table S1). Functional analysis (also depicted in Supplementary Result and Supplementary Figure S1) utilizing gene ontology indicated that MSC-sEVs protein predominantly found within extracellular exosomes and regions. They exhibit potential roles in facilitating skin healing and regeneration through multifaceted molecular and biological functions, as well as pathways. Furthermore, the results from STRING analysis highlighted a focus on downstream events and ECM degradation rather than upstream processes such as collagen formation and TGF-beta signaling, suggesting a potential mechanism of action for MSC-sEVs (refer to Supplementary Result, Supplementary Figure S2, Supplementary Table S2 and Table S3). Moreover, the absence of proteins associated with melanogenesis implies that the proteome of MSC-sEVs might not directly regulate melanin synthesis.
Apart from proteome profiling, we also performed an initial characterization of microRNA (miRNA) profiling in MSC-sEVs (see Supplementary Method), acknowledging the diverse cargo of EVs implicated in cellular processes. Small RNA-sequencing unveiled the existence of 245 known miRNAs and 34 novel miRNAs within MSC-sEVs, among which 26 miRNAs were exclusive to MSC-sEVs (depicted in Supplementary Result, Supplementary Figure S3 and Supplementary Table S4). Functional analysis indicated the participation of these miRNAs in modulating gene expression, cellular processes, and molecular functions (see Supplementary Result and Supplementary Figure S3).
Furthermore, an extensive literature review was conducted to explore the roles of miRNAs in skin biology, with specific emphasis on their participation in anti-scarring and hyperpigmentation processes. Numerous pathways have been identified to contribute to the wound healing process, with the TGF-β/SMAD signaling pathway being notably significant, particularly in the later stages of wound repair.32 Several miRNAs have been identified to target genes within the TGF-β and SMAD families associated with this pathway (Supplementary Table S5). These targeted genes, such as TGF-βR1, TGF-βR2, SMAD2, TGF-β1, SMAD3, SMAD4, SMURF2, IL-17RA, and EZH2, negatively regulate TGF signaling, thereby inhibiting myofibroblast differentiation and reducing collagen synthesis expression. Conversely, other targeted genes, including SIP1 and SMAD7, contribute to the elevation of COL1, COL3, and α-SMA levels.
Conversely, prior studies showed that certain miRNAs, such as miR-130a, miR-155, miR-181a, and miR-21, have been identified to target genes like CYLD, SHIP1, PTEN, and SPRY1, which activate the PI3K/AKT signaling pathway. These miRNAs exert positive effects on fibroblast functions such as proliferation and migration, while also enhancing α-SMA expression and increasing the expression of COL1A1 and COL3A1. Furthermore, a range of miRNAs, including miR-127-3p, miR-132, miR-145, miR-146b-5p, miR-21, miR-23b, and miR-99, target various pathways involved in regulating different aspects of the wound healing process. These pathways encompass proliferation, migration, angiogenesis, apoptosis, inflammation, ECM remodeling, and fibrosis in both fibroblasts and myofibroblasts (see Supplementary Table S5).
Melanogenesis is a complex process influenced by various external and internal factors, including UV rays, hormones, growth factors, and cytokines, which modulate cellular signaling pathways.33 Previous research has identified several miRNAs as key regulators of pigmentation. For instance, miR-125b-5p, miR-181a-5p, miR-25, miR-183, miR-218, and miR-340 have been shown to regulate MITF,18,34–38 leading to decreased expression of downstream genes such as TYR, TYRP1, and TYRP2/DCT, which ultimately affects melanin production. Other miRNAs target genes such as Myo5a, Wnt3a, IGF1R, SOX6, and receptor-interacting serine/threonine kinase 1 (riPK1),33,39–42 were also found to contribute to melanogenesis inhibition. Specific occurrences, like miR-199a targeting mTOR for melanosome degradation18 and miR-21 suppressing melanin synthesis through the EGFR/Akt pathway,43 further illustrate the complex regulatory network involved in melanogenesis (Supplementary Table S6).
To evaluate the potential of sEVs in anti-scarring interventions, we investigated various aspects of dermal fibroblast activities, including proliferation, migration, mRNA expression of ECM constituents, and protein expression of COL1 and αSMA, as well as antioxidant enzyme expression across different sEVs concentrations (0, 10, 25, and 50 μg/mL). HDF proliferation increased with higher sEVs concentrations, consistent with previous findings, indicating enhanced HDF proliferation with EVs treatment.44–46 However, sEVs did not affect HDF migration in our study, which contrary to some prior studies.47–50 This discrepancy suggests that HDF migration involves complex interactions influenced by multiple factors.
Considering the pivotal role of ECM remodeling in scarless wound healing, we examined the mRNA levels of ECM proteins (COLI, COLIII, FN, MMP1, and MMP3) to assess whether sEVs could influence ECM gene expression. Our results showed no significant impact of sEVs on the gene expression of COLI and COLIII, but we observed a dose-dependent increase in FN, MMP1, and MMP3 gene expressions. These findings were further supported by the protein expression of COLI, indicating that sEVs did not affect collagen production. Consistently, a previous study also reported minimal effects of exosomes on the gene expressions of COLI and COLIII.51 The unchanged collagen expression may be attributed to the diverse pathways identified in both Reactome and KEGG pathway analyses for MSC-sEVs proteome, which encompass processes related to ECM organization, degradation, collagen formation, and MMP activation, collectively regulating collagen synthesis, turnover, and maintenance. Furthermore, our comprehensive literature review on miRNA functions in skin biology unveiled various miRNAs capable of influencing collagen synthesis by targeting genes involved in the regulation of both the TGF-β/SMAD pathway and the PI3K/AKT pathway.
The presence of αSMA expression characterizes myofibroblasts, which transition from fibroblasts in response to skin injury. Myofibroblasts play crucial roles in matrix remodeling by promoting collagen synthesis to facilitate wound healing. However, prolonged persistence of myofibroblasts in the later stages of wound healing can result in scar formation.52,53 Our data indicate that sEVs reduced the protein expression levels of αSMA in a dose-dependent manner, suggesting that sEVs inhibited the phenotype transition of fibroblasts and prevented excessive collagen synthesis, thereby inhibiting the formation of granulation tissue. This finding aligns with the unchanged expression of collagen genes and proteins detected via RT-PCR and immunocytochemical staining, respectively.
The upregulation of FN, MMP1, and MMP3 by sEVs might be a part of the cellular response aimed at facilitating ECM remodeling. FN plays a crucial role in cell attachment and migration54 and contributes to tissue architecture and regulation of cellular processes,55 potentially improving skin texture and firmness. A slightly increase in FN expression in HDF following EVs treatment was also noted in a study by Kim et al.56 However, this slight increase in FN expression did not promote HDF migration, despite the importance of FN in cell attachment and migration. This could be attributed to insufficient levels of fibronectin necessary to initiate migration and the involvement of multiple factors in the complex process of cell migration. Additionally, MMPs typically inhibit collagen deposition during ECM remodeling and contribute to scar resolution. The observed increase in MMPs aligns with findings from STRING analysis, indicating MMP activation. Previous studies14,48,56 have also reported increased expression of MMP1 and MMP3, suggesting a potential role in preventing scar formation and promoting scarless wound healing. However, the biological activity of MMPs can be counteracted by TIMPs,57 and the ratio of MMPs to TIMPs has been proposed to contribute to scarless restoration.58 Therefore, further investigation into the MMPs-to-TIMPs ratio is warranted to confirm the potential of sEVs in promoting scarless wound healing.
sEVs did not affect the SOD activity of HDF, suggesting that they did not induce oxidative stress, which would prompt cells to increase SOD activity as a defense mechanism against oxidative damage. Similar findings were reported in previous studies involving EVs treatment.59,60 In the study by Lerner et al, it was observed that only EVs derived from oxidative-stressed cells induced an increase in CAT and SOD activity in recipient cells, indicating that EVs from stressed cells convey protective signals to recipient cells.59 Similarly, Yan et al found that UC-MSC-Exo did not induce SOD activity but did enhance the activity of another antioxidant enzyme, GPX.60 These results suggest that EVs cargo may have different affinities for activating or enhancing the activity of specific antioxidant enzymes.
Regulation of melanin synthesis is important in handling abnormal skin pigmentation. Thus, the impact of UC-MSC-sEVs on MNT-1 activity was evaluated, specifically proliferation, melanin synthesis, tyrosinase activity, and expression levels of melanogenesis-related genes, to ascertain their potential in inhibiting hyperpigmentation. Notably, sEVs did not influence MNT-1 proliferation rate, consistent with findings that EVs from human adipose tissue-derived stem cells (ADSCs) had no effect on B16F10 cell viability.25 As expected, sEVs exhibited a dose-dependent reduction in melanin synthesis, aligning with prior studies showing EVs from various sources (ADSCs, Korean seaweeds (Sargassum fusiforme and Codium fragile), and leaves and stems of Dendropanax morbifera) decreased intracellular melanin levels.12,25,61 TYR catalyzes the initial step of melanogenesis, converting L-tyrosine into L-DOPA.62 However, sEVs did not affect TYR activity although reducing melanin synthesis, suggesting EVs may modulate melanin production through mechanisms beyond direct tyrosinase inhibition.
Furthermore, key regulators of melanogenesis—TYRP1, MITF, and TYRP2—were analyzed to understand the underlying mechanism. The results showed that sEVs modulated melanogenesis-related gene expression by downregulating MITF and TYRP2. MITF is a major transcriptional regulator for melanogenesis-related enzymes and is crucial for melanocyte pigmentation, proliferation, and survival.63,64 TYRP2 plays a vital role in melanin synthesis.65 The observed downregulation of MITF and TYRP2 is expected to reduce melanin synthesis. However, the unexpected no changes in TYRP1 levels might counterbalance this effect by enhancing tyrosinase activation and stability, given TYRP1’s role in these processes.62 Therefore, the decrease in melanin content, alongside the downregulation of MITF and TYRP2, suggests a potential inhibitory role of sEVs-derived miRNAs in melanogenesis. Nonetheless, further research is needed to fully elucidate the effects of sEVs on melanogenesis mechanisms.
In addition, skin permeation analysis was conducted to assess the penetrability of MSC-sEVs into human skin. The results indicated that topically administered MSC-sEVs were limited to the stratum corneum, consistent with earlier findings,66 which showed that sEVs were restricted from further penetration into deeper epidermal layers. Importantly, despite limited penetration into deeper skin layers, the functionality of EVs remained intact, as they were still able to inhibit complement activation in the stratum corneum and subsequently suppress IL-17 secretion, reducing psoriasis-like inflammation in a mouse model of psoriasis.66
Limitation and Future Perspective
Throughout the study, there were a number of limitations. Although the proteomic and miRNA profiling results provide valuable insights into the composition and potential functions of UC-MSC-sEVs, it is important to note that our analyses were based on a single replicate of pooled samples. This small sample size limits the statistical power and generalizability of our findings. Therefore, caution interpretation of results should be considered, and further studies with larger sample sizes are warranted to validate our observations. Besides, due to limitations in sample availability, this study relied solely on frozen skin samples. Live skin samples are suggested to be used because they provide more accurate representation of the dynamic processes that occur within the skin and could confirm whether EVs can be uptake by other mechanisms. Besides, ex vivo models like MelanoDerm™ could provide additional validation of the effectiveness of EVs in inhibiting skin hyperpigmentation before proceeding to in vivo testing because this skin model mimics the in vivo conditions and can be used to predict the in vivo results.
Conclusion
In summary, sEVs displayed the ability to enhance proliferation and regulate ECM expression in HDF, while migration rate and antioxidant activity remained unchanged. On the other hand, UC-MSC-sEVs did not stimulate human melanoma cell growth but reduced pigmentation by decreasing melanin synthesis. These in vitro findings highlight the dose-dependent beneficial effects of UC-MSC-sEVs on skin cells, particularly fibroblasts and melanocytes, through different cellular mechanisms. This underscores the importance of considering sEV dose or charge in future studies and suggests a promising approach for addressing hypertrophic scars and hyperpigmentation, offering insights into the field of dermatology.
Ethics Approval and Statement
This study was approved by the National University of Malaysia (UKM) Research Ethics Committee (UKM PPI/111/8/JEP-2019-618), at UKM. Written informed consent was obtained from all participants prior to their involvement in this study at the Department of Obstetrics & Gynaecology at Hospital Canselor Tuanku Muhriz (HCTM), Kuala Lumpur. The study was performed in accordance with the ethical principles outlined in the Declaration of Helsinki.
Acknowledgments
We would like to specially thank the Faculty of Medicine, Universiti Kebangsaan Malaysia (Grant code: FF-2019-450/1) and Ming Medical Sdn. Bhd. (Grant code: FF-2019-450) for providing financial support in this work.
Disclosure
Ms Li Ting Kee reports a patent PI2024006022 pending to. Dr Min Hwei Ng reports a patent WJMSC exosomes for skin aesthetic use pending to None; and A company, Ming Medical Sdn Bhd, sponsors the study. However, academic researchers have developed the protocol for the derivation of the exosome entirely. Dr Jia Xian Law reports a patent An anti-scarring and anti-pigmentation composition pending to Universiti Kebangaan Malaysia and Ming Medical Sdn Bhd. The authors report no other conflicts of interest in this work.
References
1. Al-Masawa ME, Alshawsh MA, Ng CY, et al. Efficacy and safety of small extracellular vesicle interventions in wound healing and skin regeneration: a systematic review and meta-analysis of animal studies. Theranostics. 2022;12(15):6455–6508. doi:10.7150/thno.73436
2. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19(1):1–19. doi:10.1186/s12964-021-00730-1
3. Witwer KW, Van Balkom BWM, Bruno S, et al. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J Extracell Vesicles. 2019;8(1). doi:10.1080/20013078.2019.1609206
4. Welsh JA, Goberdhan DCI, O’Driscoll L, et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles. 2024;13(2). doi:10.1002/jev2.12404
5. Verweij FJ, Balaj L, Boulanger CM, et al. The power of imaging to understand extracellular vesicle biology in vivo. Nat Methods. 2021;18(9):1013–1026. doi:10.1038/s41592-021-01206-3
6. Ng CY, Kee LT, Al-Masawa ME, et al. Scalable production of extracellular vesicles and its therapeutic values: a review. Int J mol Sci. 2022;23(14):7986. doi:10.3390/ijms23147986
7. Quiñones-Vico MI, Sanabria-de la Torre R, Sánchez-Díaz M, et al. The role of exosomes derived from mesenchymal stromal cells in dermatology. Front Cell Dev Biol. 2021;9(April):647012. doi:10.3389/fcell.2021.647012
8. McBride JD, Rodriguez-Menocal L, Badiavas EV. Extracellular vesicles as biomarkers and therapeutics in dermatology: a focus on exosomes. J Invest Dermatol. 2017;137(8):1622–1629. doi:10.1016/j.jid.2017.04.021
9. Narauskaitė D, Vydmantaitė G, Rusteikaitė J, et al. Extracellular vesicles in skin wound healing. Pharmaceuticals. 2021;14(8). doi:10.3390/ph14080811
10. Ha DH, Kim HK, Lee J, et al. Mesenchymal stem/stromal cell-derived exosomes for immunomodulatory therapeutics and skin regeneration. Cells. 2020;9(1157):4–6.
11. Xu P, Xin Y, Zhang Z, et al. Extracellular vesicles from adipose-derived stem cells ameliorate ultraviolet B-induced skin photoagiAng by attenuating reactive oxygen species production and inflammation. Stem Cell Res Ther. 2020;11(1):1–14. doi:10.1186/s13287-020-01777-6
12. Jang B, Chung H, Jung H, et al. Extracellular vesicles from Korean Codium fragile and Sargassum fusiforme negatively regulate melanin synthesis. Mol Cells. 2021;44(10):736–745. doi:10.14348/molcells.2021.2167
13. Shen Y, Xu G, Huang H, et al. Sequential release of small extracellular vesicles from bilayered thiolated alginate/polyethylene glycol diacrylate hydrogels for scarless wound healing. ACS Nano. 2021;15(4):6352–6368. doi:10.1021/acsnano.0c07714
14. Wang L, Hu L, Zhou X, et al. Exosomes secreted by human adipose mesenchymal stem cells promote scarless cutaneous repair by regulating extracellular matrix remodelling. Sci Rep. 2017;7(1):1–12. doi:10.1038/s41598-017-12919-x
15. Oh M, Lee J, Kim YJ, Rhee WJ, Park JH. Exosomes derived from human induced pluripotent stem cells ameliorate the aging of skin fibroblasts. Int J mol Sci. 2018;19(6):1–18. doi:10.3390/ijms19061715
16. Zhang W, Bai X, Zhao B, et al. Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway. Exp Cell Res. 2018;370(2):333–342. doi:10.1016/j.yexcr.2018.06.035
17. Kim YJ, Mi Yoo S, Park HH, et al. Exosomes derived from human umbilical cord blood mesenchymal stem cells stimulates rejuvenation of human skin. Biochem Biophys Res Commun. 2017;493(2):1102–1108. doi:10.1016/j.bbrc.2017.09.056
18. Wang XY, Guan XH, Yu ZP, et al. Human amniotic stem cells-derived exosmal miR-181a-5p and miR-199a inhibit melanogenesis and promote melanosome degradation in skin hyperpigmentation, respectively. Stem Cell Res Ther. 2021;12(1):1–16. doi:10.1186/s13287-021-02570-9
19. Selich A, Zimmermann K, Tenspolde M, et al. Umbilical cord as a long-term source of activatable mesenchymal stromal cells for immunomodulation. Stem Cell Res Ther. 2019;10(1):1–14. doi:10.1186/s13287-019-1376-9
20. El Omar R, Beroud J, Stoltz JF, Menu P, Velot E, Decot V. Umbilical cord mesenchymal stem cells: the new gold standard for mesenchymal stem cell-based therapies? Tissue Eng - Part B Rev. 2014;20(5):523–544. doi:10.1089/ten.teb.2013.0664
21. Liu Q, Zhang J, Tang Y, Ma Y, Xue Z, Wang J. What is the impact of human umbilical cord mesenchymal stem cell transplantation on clinical treatment? Curr Gene Ther. 2021;22(4):319–330. doi:10.2174/1566523221666211014165341
22. Kee LT, Ng CY, Al‐masawa ME, et al. Extracellular vesicles in facial aesthetics: a review. Int J mol Sci. 2022;23(12):6742. doi:10.3390/ijms23126742
23. Zhang B, Gong J, He L, et al. Exosomes based advancements for application in medical aesthetics. Front Bioeng Biotechnol. 2022;10(December):1–24. doi:10.3389/fbioe.2022.1083640
24. Li Y, Xiao Q, Tang J, Xiong L, Li L. Extracellular vesicles: emerging therapeutics in cutaneous lesions. Int J Nanomed. 2021;16(August):6183–6202. doi:10.2147/IJN.S322356
25. Cho BS, Lee J, Won Y, et al. Skin brightening efficacy of exosomes derived from human adipose tissue-derived stem/stromal cells: a prospective, split-face, randomized placebo-controlled study. Cosmetics. 2020;7(4):1–12. doi:10.3390/cosmetics7040090
26. Chan AML, Amh N, Mohd Yunus MH, et al. Safety study of allogeneic mesenchymal stem cell therapy in animal model. Regen Ther. 2022;19:158–165. doi:10.1016/j.reth.2022.01.008
27. Bin Budi Harto PH, Bin Mahmud MH, Othman AHB, et al. Human platelet lysate promotes proliferation but fails to maintain chondrogenic markers of chondrocytes. Sains Malays. 2019;48(10):2169–2176. doi:10.17576/jsm-2019-4810-12
28. Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8(4):315–317. doi:10.1080/14653240600855905
29. Tan KL, Chia WC, How CW, et al. Benchtop isolation and characterisation of small extracellular vesicles from human mesenchymal stem cells. Mol Biotechnol. 2021;63(9):780–791. doi:10.1007/s12033-021-00339-2
30. Netcharoensirisuk P, Abrahamian C, Tang R, et al. Flavonoids increase melanin production and reduce proliferation, migration and invasion of melanoma cells by blocking endolysosomal/melanosomal TPC2. Sci Rep. 2021;11(1):1–14. doi:10.1038/s41598-021-88196-6
31. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi:10.1080/20013078.2018.1535750
32. Neupane YR, Handral HK, Alkaff SA, et al. Cell-derived nanovesicles from mesenchymal stem cells as extracellular vesicle-mimetics in wound healing. Acta Pharm Sin B. 2023;13(5):1887–1902. doi:10.1016/j.apsb.2022.10.022
33. Zhang T, Wang XF, Wang ZC, et al. Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed Pharmacother. 2020;129(3):110287. doi:10.1016/j.biopha.2020.110287
34. Dynoodt P, Mestdagh P, Van Peer G, et al. Identification of miR-145 as a key regulator of the pigmentary process. J Invest Dermatol. 2013;133(1):201–209. doi:10.1038/jid.2012.266
35. Wang X, Wu Y, Du P, et al. Study on the mechanism of miR-125b-5p affecting melanocyte biological behavior and melanogenesis in vitiligo through regulation of MITF. Dis Markers. 2022;2022:1–17. doi:10.1155/2022/6832680
36. Zhu Z, He J, Jia X, et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in alpaca (Lama pacos) skin melanocytes. Domest Anim Endocrinol. 2010;38(3):200–209. doi:10.1016/j.domaniend.2009.10.004
37. Du B, Liu X, Khan A, et al. miRNA-183∼96∼182 regulates melanogenesis, cell proliferation and migration in B16 cells. Acta Histochem. 2020;122(3):151508. doi:10.1016/j.acthis.2020.151508
38. Guo J, Zhang JF, Wang WM, et al. MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression. RNA Biol. 2014;11(6):732–741. doi:10.4161/rna.28865
39. Yang Y, Wei X, Bai J, et al. MicroRNA-340 is involved in ultraviolet B-induced pigmentation by regulating the MITF/TYRP1 axis. J Int Med Res. 2020;48(11). doi:10.1177/0300060520971510
40. Zhao Y, Wang P, Meng J, et al. MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting Wnt3a. Int J mol Sci. 2015;16(5):10921–10933. doi:10.3390/ijms160510921
41. Liu B, Zhang J, Hu S, et al. MicroRNA-379 mediates pigmentation, migration and proliferation of melanocytes by targeting the insulin-like growth factor 1 receptor. Exp Dermatol. 2020;29(5):467–476. doi:10.1111/exd.14095
42. Liu X, Du B, Zhang P, et al. MiR-380-3p regulates melanogenesis by targeting SOX6 in melanocytes from alpacas (Vicugna pacos). BMC Genomics. 2019;20(1):1–10. doi:10.1186/s12864-019-6343-4
43. Sun X, Wang T, Huang B, Ruan G, Xu A. MicroRNA-421 participates in vitiligo development through regulating human melanocyte survival by targeting receptor-interacting serine/threonine kinase 1. Mol Med Rep. 2020;21(2):858–866. doi:10.3892/mmr.2019.10878
44. Lin KY, Chen CM, Lu CY, Cheng CY, Wu YH. Regulation of miR-21 expression in human melanoma via UV-ray-induced melanin pigmentation. Environ Toxicol. 2017;32(8):2064–2069. doi:10.1002/tox.22421
45. Hu Y, Rao SS, Wang ZX, et al. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics. 2018;8(1):169–184. doi:10.7150/thno.21234
46. Zhang Y, Pan Y, Liu Y, et al. Exosomes derived from human umbilical cord blood mesenchymal stem cells stimulate regenerative wound healing via transforming growth factor-β receptor inhibition. Stem Cell Res Ther. 2021;12(1):1–14. doi:10.1186/s13287-021-02517-0
47. Deng M, Yu Z, Li D, et al. Human umbilical cord mesenchymal stem cell-derived and dermal fibroblast-derived extracellular vesicles protect dermal fibroblasts from ultraviolet radiation-induced photoaging: in vitro. Photochem Photobiol Sci. 2020;19(3):406–414. doi:10.1039/c9pp00421a
48. Ren S, Chen J, Duscher D, et al. Microvesicles from human adipose stem cells promote wound healing by optimizing cellular functions via AKT and ERK signaling pathways. Stem Cell Res Ther. 2019;10(1):1–14. doi:10.1186/s13287-019-1152-x
49. Zhao B, Zhang Y, Han S, et al. Exosomes derived from human amniotic epithelial cells accelerate wound healing and inhibit scar formation. J Mol Histol. 2017;48(2):121–132. doi:10.1007/s10735-017-9711-x
50. Go YY, Lee CM, Ju WM, Chae SW, Song JJ. Extracellular vesicles (Secretomes) from human trophoblasts promote the regeneration of skin fibroblasts. Int J mol Sci. 2021;22(13):6959. doi:10.3390/ijms22136959
51. Vu DM, Nguyen VT, Nguyen TH, et al. Effects of extracellular vesicles secreted by TGFβ-stimulated umbilical cord mesenchymal stem cells on skin fibroblasts by promoting fibroblast migration and ECM protein production. Biomedicines. 2022;10(8):1–12. doi:10.3390/biomedicines10081810
52. Tutuianu R, Rosca AM, Iacomi DM, Simionescu M, Titorencu I. Human mesenchymal stromal cell-derived exosomes promote in vitro wound healing by modulating the biological properties of skin keratinocytes and fibroblasts and stimulating angiogenesis. Int J mol Sci. 2021;22(12):6239. doi:10.3390/ijms22126239
53. Hinz B. Myofibroblasts. Exp Eye Res. 2015;142:56–70. doi:10.1016/j.exer.2015.07.009
54. Rippa AL, Kalabusheva EP, Vorotelyak EA. Regeneration of dermis: scarring and cells involved. Cells. 2019;8(6):607. doi:10.3390/cells8060607
55. Yue B. Biology of the extracellular matrix: an overview. J Glaucoma. 2014;23(8):S20–S23. doi:10.1097/IJG.0000000000000108
56. Xue M, Jackson CJ. Extracellular matrix reorganization during wound healing and its impact on abnormal scarring. Adv Wound Care. 2015;4(3):119–136. doi:10.1089/wound.2013.0485
57. Kim S, Lee SK, Kim H, Kim TM. Exosomes secreted from induced pluripotent stem cell-derived mesenchymal stem cells accelerate skin cell proliferation. Int J mol Sci. 2018;19(10):3119. doi:10.3390/ijms19103119
58. Yang C, Luo L, Bai X, et al. Highly-expressed micoRNA-21 in adipose derived stem cell exosomes can enhance the migration and proliferation of the HaCaT cells by increasing the MMP-9 expression through the PI3K/AKT pathway. Arch Biochem Biophys. 2020;681:1–31. doi:10.1016/j.abb.2020.108259
59. Dang CM, Beanes SR, Lee H, Zhang X, Soo C, Ting K. Scarless fetal wounds are associated with an increased matrix metalloproteinase-to-tissue-derived inhibitor of metalloproteinase ratio. Plast Reconstr Surg. 2003;111(7):2273–2285. doi:10.1097/01.PRS.0000060102.57809.DA
60. Lerner N, Chen I, Schreiber-Avissar S, Beit-Yannai E. Extracellular vesicles mediate anti-oxidative response—in vitro study in the ocular drainage system. Int J mol Sci. 2020;21(17):1–17. doi:10.3390/ijms21176105
61. Yan Y, Jiang W, Tan Y, et al. hucMSC exosome-derived GPX1 is required for the recovery of hepatic oxidant injury. Mol Ther. 2017;25(2):465–479. doi:10.1016/j.ymthe.2016.11.019
62. Lee R, Ko HJ, Kim K, et al. Anti-melanogenic effects of extracellular vesicles derived from plant leaves and stems in mouse melanoma cells and human healthy skin. J Extracell Vesicles. 2020;9(1):1703480. doi:10.1080/20013078.2019.1703480
63. Videira IF Dos S, Moura DFL, Magina S. Mechanisms regulating melanogenesis. An Bras Dermatol. 2014;88(1):76–83. doi:10.1007/978-3-642-45407-3_24
64. Pillaiyar T, Manickam M, Jung SH. Inhibitors of melanogenesis: a patent review (2009-2014). Expert Opin Ther Pat. 2015;25(7):775–788. doi:10.1517/13543776.2015.1039985
65. Xue L, Li Y, Zhao B, et al. TRP-2 mediates coat color pigmentation in sheep skin. Mol Med Rep. 2018;17(4):5869–5877. doi:10.3892/mmr.2018.8563
66. Zhang B, Lai RC, Sim WK, Choo ABH, Lane EB, Lim SK. Topical application of mesenchymal stem cell exosomes alleviates the imiquimod induced psoriasis-like inflammation. Int J mol Sci. 2021;22(2):1–13. doi:10.3390/ijms22020720
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.