Back to Journals » International Journal of Nanomedicine » Volume 19
Unexpected Inhibitory Role of Silica Nanoparticles on Lung Cancer Development by Promoting M1 Polarization of Macrophages
Authors Xiang M , Chen C , Chen Y, Zhang Y, Shi L, Chen Y , Li J , Li B , Zeng B, Xing HR , Wang J, Zou Z
Received 18 June 2024
Accepted for publication 19 October 2024
Published 1 November 2024 Volume 2024:19 Pages 11087—11104
DOI https://doi.org/10.2147/IJN.S472796
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xing Zhang
Meng Xiang,1,* Chengzhi Chen,2,* Yuting Chen,1,* Yuhan Zhang,1 Lei Shi,1 Yan Chen,1 Jie Li,2 Bowen Li,1 Bin Zeng,2 H Rosie Xing,1 Jianyu Wang,2 Zhen Zou2
1State Key Laboratory of Ultrasound in Medicine and Engineering, College of Biomedical Engineering, Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 2Molecular Biology Laboratory of Respiratory Disease, Key Laboratory of Clinical Laboratory Diagnostics (Ministry of Education), College of Laboratory Medicine, Chongqing Medical University, Chongqing, 400016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhen Zou; Jianyu Wang, Email [email protected]; [email protected]
Introduction: Inhalation exposure to silica nanoparticles (SiNPs) is frequently inevitable in modern times. Although the impact of SiNPs on the ecological niche of the lungs has been extensively explored, the role and mechanism of SiNPs in the microenvironment of lung tumors remain elusive.
Methods: In this investigation, Lewis lung carcinoma (LLC) was implanted into the left lung in situ after 28 days of intratracheal SiNPs injection into the lungs of mice. This study evaluates the effects of SiNPs on the tumor immune microenvironment both in vitro and in vivo. Our findings indicate that SiNPs can suppress lung cancer by modulating the immune microenvironment of tumors.
Results: SiNPs treatment promotes macrophage M1 polarization by activating both NF-κB pathway and glycolytic mechanisms. This phenomenon may be associated with lung inflammation and fluctuation in the pre-metastatic and metastatic microenvironments induced by SiNPs exposure in mice. Additionally, we have shown for the first time that SiNPs have an inhibitory effect on lung carcinogenesis and its progression.
Conclusion: This study uniquely demonstrates that SiNPs suppress lung cancer by promoting M1 polarization of macrophages in the immune microenvironment of lung tumors. Our findings are critical in exploring the interaction between SiNPs and lung cancer.
Keywords: silicon nanoparticles, lung cancer, macrophage polarization, NF-κB, glycolysis
Graphical Abstract:

Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with over 1.8 million deaths and approximately 2.2 million new cases each year.1 The risk factors associated with the development of lung cancer typically include smoking, a family history of cancer, lung diseases, and other harmful factors.2 In addition to these, air pollution is another major cause of lung cancer. Harmful substances in the air, primarily originating from modern industrial and domestic activities such as PM2.5, sulfur oxides, second-hand smoke, and cooking oil fumes, enter the lungs, causing inflammation, DNA damage, and epigenetic regulation, ultimately leading to the development of lung cancer.3,4
The production and distribution of engineered nanomaterials (ENMs) are rapidly expanding due to the significant advancements of nanotechnology. Silica nanoparticles (SiNPs) are among the most commonly utilized nanomaterials and have been extensively used in coatings, electronic packaging materials, plastic engineering, textile industry, biomedical engineering, and other fields. The global market for nano-silica was estimated at 4.6 billion dollars in 2021 (https://www.giiresearch.com).5 Therefore, occupational and accidental inhalation exposure to SiNPs is inevitable and the potential biological impacts of SiNPs on human health have attracted considerable concerns. Extensive studies have demonstrated that inhalation of SiNPs would directly result in lung injury and lung fibrosis in a dose-dependent manner.6,7 Although titanium dioxide, silica, and gold nanoparticles have been shown to induce endothelial cell leakage and promote breast cancer in vivo,8 the link between SiNP inhalation and lung cancer development is still an enigma.
SiNPs may enter into the distal lung tissues after inhalation, and then be recognized and internalized by professional phagocytes, such as macrophages.9 Macrophages are a heterogeneous population of immune cells, with distinct subtypes based on morphology, phenotype, and function. M1 macrophages play a crucial role in antitumor immunity by mediating pro-inflammatory processes in the tumor microenvironment (TME), while M2 macrophages promote tumor growth and metastasis, exhibiting primary tumor characteristics. The most diverse immune cells in the TME are M2 macrophages and a small subset of M1 macrophages, referred to as “tumor-associated macrophages” (TAMs), which are essential for tumor growth. Microbial endotoxin and cytokines such as interferon-γ, tumor necrosis factor-α, and granulocyte monocyte colony-stimulating factor (GM-CSF) often induce M1-type polarization, whereas tumors, viruses and parasites induce M2 polarization.10,11 In vitro and to a lesser extent in vivo studies have suggested that SiNPs activated macrophages and consequently caused inflammatory responses, which underpin of the biological impacts of SiNPs.12 As such, it is reasonable to speculate that SiNPs may affect the immune microenvironment of lung tumors by acting on macrophages.
The current study demonstrates that SiNPs can surprisingly exert an inhibitory effect on lung cancer. Mechanistically, on one hand, SiNPs participate in the regulation of macrophage M1 polarization by activating the NF-κB pathway through an inflammatory response; Additionally, SiNPs regulate glycolysis and sustain M1 phenotype of macrophages, which together contribute to the inhibition of lung cancer cell migration and invasion. Overall, our study offers new insights into the biological effects of SiNPs on lung cancer.
Materials and Methods
Cell Culture
The murine Lewis lung carcinoma (LLC) cell line was purchased from the CELL BANK and cultured in DMEM medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 ug/mL streptomycin. RAW264.7 cells were obtained from Cell Bank of pricella and cultured in RAW264.7 special medium (pricella).
Characterization of Silica Nanoparticle (SiNPs)
The silica nanoparticles used in this study were obtained from Sigma Aldrich Chemical Co. (MO, USA, Cat# 637246), with a surface area of 590–690 m²/g and a purity of 99.5%. The physicochemical characterization of SiNPs was conducted according to the procedures described in our previous study. In brief, the raw material was dispersed in sterile distilled water and cleaned with an ultrasonic cleaner (SB-5200DT, Ningbo Science Ltd., Ningbo, China) at 20% of the maximum amplitude for 20 minutes while being operated on ice. The nanoparticles were freshly prepared before each use. To determine the morphology of SiNPs, transmission electron microscopy (TEM) (Hitachi-7500, Hitachi, Ltd, Tokyo, Japan) was used to observe a total of 150 nanoparticles, and the average size was calculated. The chemical elemental composition of SiNPs was analyzed with field emission scanning electron microscopy (Hitachi-su8010, Hitachi, Ltd, Tokyo, Japan) and energy dispersive spectroscopy (Oxford X-MAN 50) (FE-SEM/EDS). The properties of the synthesized SiNPs (amorphous or crystalline) were characterized by X-ray diffraction (XRD) with a Rigaku Ultima IV (Japan). The scattering angle (2θ) and intensity were recorded from 5° to 100° at a scanning speed of 0.2°/min using Cu Kα radiation (1.54 Å). The zeta potential and particle size (dynamic light scattering) were measured at 25 ◦C using the Malvern Zeta sizer Nano ZS nanoparticle analyzer.
Bronchoalveolar Lavage Fluids (BALFs) and Cell Counts
All mice were sacrificed on the 28th day after SiNPs exposure. Following thoracotomy, the trachea was surgically exposed, facilitating the introduction of 1.5mL of phosphate-buffered saline (PBS) into the pulmonary parenchyma via syringe administration. Subsequently, collection of lavage fluid occurred after each iterative lavage, conducted thrice. A total volume of 1.8 mL of bronchoalveolar lavage fluid (BALF) was meticulously extracted from each murine specimen. The BALF samples were then subjected to centrifugation at 1000×g for a for 15 minutes at 4 degrees Celsius. Following centrifugation, the resultant supernatant was carefully retrieved and preserved at −20°C for subsequent analytical scrutiny. The cell pellet was suspended in 1mL of PBS and the total number of cells was counted using a hemocytometer.
Histological Examination
Two batches of mice were sacrificed at 28 days after SiNPs exposure and 30 days after LLC injection. After removing the lung tissues, they were fixed in 4% paraformaldehyde at 4°C for 48 h, paraffin embedded and sectioned at 5μm/slice. The lung tissue sections of the first batch of mice were dewaxed and stained with hematoxylin and eosin (H&E) staining and collagen fiber staining (Masson), while the lung tissue sections of the second batch of mice were stained using H&E only, and both sets of sections were observed under light microscopy to assess pulmonary histological changes.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total cellular or tissue RNA was extracted using miRNeasy Serum/Plasma Kit (QIAGEN, Germany, code: 1071073) according to the manufacturer’s instructions and reverse transcription was performed using PrimeScript RT Master Mix (Takara, Japan, code: RR036A) and MIR-X™ miRNA First-Strand Synthesis Kit (Takara, Japan, code: 638313). Quantitative PCR (qPCR) was performed using SYGreen Real-Time PCR Master Mix Kit (Japan, Takara, code: RR420A) for Fast Real-Time PCR System (Applied Biosystems, Thermo Fisher Science, USA). qPCR was performed using the 2-ΔΔCT method to calculate relative gene expression and normalized to TBP values. The primer sequences used are described in Supplementary Table 1.
Western Blot
Homogenized mice lung tissue and RAW264.7 cells were lysed in RIPA lysis buffer (Beyotime, Shanghai, China) containing 1 mm PMSF (Beyotime, Shanghai, China). And 1mM phosphatase inhibitors were added as needed and its concentration was determined. The supernatant was collected after centrifugation, and its protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China) and boiled at 100°C for 5 minutes. Fibronectin (FN) (ABMART) and Vinculin (protein loading control, Proteintech, Wuhan, China) were resolved on 8% SDS-PAGE gels, while β-actin (protein loading control, Proteintech, Wuhan, China), P65 (Proteintech, Wuhan, China) and phosphorylated P65 (P-P65) (Proteintech, Wuhan, China) were separated on 12% SDS-PAGE gels. Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, CA, USA) and incubated the membranes with appropriate primary and secondary antibodies according to the manufacturer’s instructions.
Transwell Migration and Invasion Assays
Transwell inserts (pore size 8 μm, BD Falcon) coated with Matrigel (100 μL, 1:10 dilution in serum-free medium, BD Biosciences) were used for invasion assays, while uncoated inserts were used for migration assays. Equal numbers of cells were seeded in the upper chamber with 300 μL of serum-free high-glucose DMEM, and 800 μL of high-glucose DMEM containing 20% fetal bovine serum (FBS) was added to the lower chamber. After 24 hours of incubation, cells on the upper surface of the porous membrane were removed with a cotton swab. The remaining cells were fixed with ice-cold methanol for 30 minutes, stained with crystal violet (Solarbio, China, code: G1062) for 10 minutes, and counted under a light microscope. For quantification, three random Transwell inserts were analyzed, and nine fields were counted per insert.
Raw264.7 and LLC Co-Culture Assay
In the experiments, RAW264.7 cells were pretreated with SiNPs, SiNPs+JSH23, or SiNPs+2-DG for 24 hours. Six-well Transwell inserts (pore size 8 μm, Corning) were used to separate LLC from the treated RAW264.7. After validation through concentration gradient experiments, RAW264.7 and LLC cells were respectively seeded into the lower and upper chambers of the Transwell system at a ratio of 5%, and subsequently co-cultured for a duration of 48 hours.
Flow Cytometry
Adherent macrophages were washed with PBS, collected, and resuspended in 100 μL of PBS containing 1% serum. The cells were then stained with antibodies specific to surface markers. The following antibodies were used: anti-Fc receptor (14-0161-82, eBioscience), PE-conjugated CD86 (159203, Proteintech, Wuhan, China), and Alexa Fluor 488-conjugated CD206 (Proteintech, Wuhan, China). Staining was performed at 4°C in the dark for 30 minutes. Flow cytometry analysis was subsequently conducted, and data were analyzed using FlowJo software.
Animals and Exposure
All the C57 mice used in the experiment were purchased from ENSIWEIER, Chongqing, China. Animal experiments were approved by the Ethics Committee of Chongqing Medical University (Approval No. SCXK2012-0001), and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (NIH publication no. 85–23, revised 1996).
RNA-Seq and Bioinformatics Analysis
For RNA-seq, lung tissues (PBS/SiNPs infusion models) were obtained from two groups of mice (n=5 per group) following the manufacturer’s protocol. RNA sequencing was performed using the Illumina platform. The sample sizes were NC: 5 and SiNPs: 5. The raw count matrix was normalized using DESeq2, and differentially expressed genes (DEGs) were identified based on a fold change > 1.5 and a p-value < 0.05. Subsequent analysis, including the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA), was conducted to elucidate the biological significance of the DEGs. Throughout the analysis process, we strictly adhered to mouse gene identifiers and annotation databases, without utilizing any human gene information. Therefore, all analysis results are related to mice. RNA-seq was performed by Majorbio, Shanghai, China.
Statistical Analysis
Statistical analysis of all experimental data was performed as mean ± standard deviation (S.D). of three independent replicates. Statistical comparison of the Student’s T-test was performed using GraphPad Prism 8.0. All data were expressed as mean ± S.D. *P<0.05 is considered statistically significant and is marked with an asterisk. **P<0.01 and ***P<0.001 were considered highly statistically significant and were marked with double and triple asterisks, respectively.
Results
Effects of SiNPS Exposure on Mice Pulmonary Function
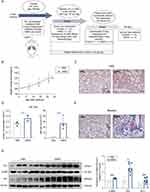
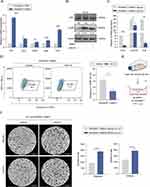
To provide a detailed characterization of SiNPs, we used transmission electron microscopy (TEM) to analyze the morphology and size of commercially available SiNPs. The average particle size of the SiNPs was 28 ± 13.416 nm. Additionally, the SiNPs were dispersed in distilled water using ultrasonication, and dynamic light scattering (DLS) was used to determine the hydrodynamic diameter of the SiNPs, which was found to be 255.84 nm. This size was larger than that observed with TEM, which may be due to the presence of a hydrated ion layer. The polydispersity index (PDI) and zeta potential of the SiNPs were also measured, with values of 0.48 and −26.86 mV, respectively (Figure 1A–C, Supplementary Figure 1A). The negative zeta potential contributes to the stability of the SiNPs.13 To determine the elemental composition of the SiNPs, field emission scanning electron microscopy with energy-dispersive spectroscopy (FE-SEM/EDS) was employed. The results indicated that the major elemental components of the SiNPs were Si and O (Figure 1D and E). To determine whether the SiNPs were crystalline or amorphous, X-ray diffraction (XRD) patterns were analyzed. A broad peak at 22° and the absence of other crystallization peaks confirmed the amorphous nature of the SiNPs (Supplementary Figure 1B).
In order to investigate the potential effects of SiNPs exposure on lung tissue, the establishment of allogeneic tumor grafts, and tumor metastasis, we designed an experimental procedure as shown in Figure 2A. Each group consisted of 25 mice, with 10 mice participating in the first 30 days of the experiment, 5 mice in the 60-day phase, and 10 mice reserved to observe the survival rate at day 88. In detail, female C57 mice were randomly assigned to receive either PBS or PBS-suspended SiNPs (5 mg/kg) via a single dose intratracheal instillation. After 28 days, a portion of the mice in each group (Control, n=10; SiNPs, n=10) were sacrificed to assess the extent of pulmonary inflammation and fibrosis. At the same time, 1×105 LLC-Luc cells were implanted into the left lung of each remaining mouse (Control, n=5; SiNPs, n=5). Lung tumor metastasis was observed on day 58, during which the body weight of all mice was recorded. All mice were sacrificed on day 88 and the survival rate during the experiment was statistically analyzed (Figure 2A).
Silica nanoparticles (SiNPs) have previously been shown to induce pulmonary inflammation and fibrosis. In this study, our objective was to investigate the impact of SiNPs on lung inflammation and fibrosis using a murine model. We assessed changes in body weight, lung tissue morphology, and bronchoalveolar lavage fluid (BALF) composition in mice exposed to SiNPs over a 28-day period. Initially, no significant difference in body weight was observed between the experimental and control groups (Figure 2B). Hematoxylin-eosin (H&E) staining revealed that exposure to SiNPs resulted in the disrupted alveolar structures thickened alveolar wall, and infiltration of inflammatory cells (Figure 2C). Quantification of cells in BAL fluid showed a significantly higher total number of cells and total protein levels in mice exposed to SiNPs compared to the control group (Figure 2D). Masson’s trichrome staining indicated the collagen deposition in the lung tissues of SiNP-treated mice, but not in those treated with PBS (Figure 2E). Furthermore, SiNP exposure led to increased expression of the fibrosis markers FN1 and α-SMA, as well as the oxidative stress marker HO-1 in protein levels in mouse lung tissue (Figure 2F). Our in vivo experiments demonstrate that although SiNP exposure does not affect body weight in mice, it can induce a moderate inflammatory response and increase pulmonary fibrosis, potentially impacting the overall health of the mice.
SiNPs Inhibit the Growth of Lung Tumors
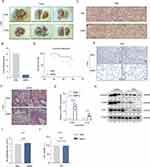
To investigate the potential effects of Silicon Nanoparticles (SiNPs) on lung tumor progression, we administered LLC-Luc cells in situ into the left lung of mice in both phosphate-buffered saline (PBS and SiNPs exposure cohorts). We observed the lung tumor formation in both groups of mice (Figure 3A), and quantified tumor fluorescence (Figure 3B), revealing that tumor growth was reduced in the SiNPs-exposed group compared to the control group. No significant differences have been detected in the distant organ metastasis (including heart, liver, spleen, and kidney) between the two groups of mice (Supplementary Figure 2A). Hematoxylin and eosin (H&E) staining of lung tissue revealed disrupted alveolar structures, hyperplastic and disorganized alveolar epithelium, and cellular heterogeneity in control mice, while the SiNPs group exhibited preserved alveolar structures with no apparent heterotypic proliferation (Figure 3C). Moreover, the survival rate of mice in the SiNPs-exposed group significantly surpassed that of the PBS-exposed group (Figure 3D). Immunohistochemical (IHC) staining for Ki67 further confirmed the tumor-inhibitory effect of SiNPs (Figure 3E).
We re-evaluated inflammation and fibrosis in the lungs of mice in both experimental groups. (SiNPs/PBS tracheal instillation for 28 days followed by in situ injection of LLC cells into the lung for 30 days). Masson’s trichrome staining confirmed increased collagen deposition in the lungs of SiNPs-exposed mice compared to the PBS group (Figure 3F), Furthermore, a consistent trend in fibrotic deposition within lung tumor nodules was observed in both groups (Supplementary Figure 2B). However, the disparity in collagen deposition between the two groups decreased compared to the 28-day period (Figure 1E). Quantitative real-time PCR analysis revealed elevated mRNA expressions of inflammatory cytokines Tnf-α and Il-1β in the lung tissues of SiNPs-exposed mice (Figure 3G). Additionally, compared to the PBS group, the expression levels of α-SMA, HO-1, and Occludin proteins were increased in lung tissues of SiNPs-exposed mice (Figure 3H). To determine whether SiNPs directly affect LLC cells, we exposed LLC cells to SiNPs in vitro and found no discernible effect on cell proliferation (Figure 3I) or toxicity (Figure 3J) compared to PBS, suggesting that SiNPs indirectly inhibit lung tumor progression.
SiNPs Inhibit Lung Cancer by Promoting M1 Polarization of Macrophages
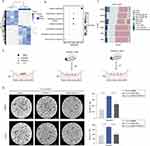
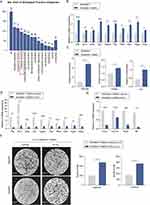
To further elucidate the mechanism by which SiNPs suppress lung cancer, we subjected lung tissues from both groups of mice to transcriptional analysis using high-throughput sequencing (RNA-seq), as shown in Figure 4A. A total of 780 genes were up-regulated, and 608 were down-regulated in the SiNPs-exposed group, with more than 2-fold differential expression compared to the control PBS-exposed group. GO analysis revealed significant enrichment of the immune response pathway (Figure 4B). Macrophages play a critical role in the immune microenvironment of the lung, and are one of the most prevalent cell populations involved in immune infiltration. Immune component analysis using EPIC-master showed a significant increase in M1-type macrophages in the SiNPs-exposed group compared to the control group (Figure 4C). To explore whether SiNPs could inhibit lung cancer by modulating macrophage phenotype, we established three co-culture systems (Figure 4D). In each system, Raw264.7 macrophages were co-cultured with PBS or SiNPs for 24 hours, followed by co-culture of either DMEM or pretreated Raw264.7 macrophages with LLC for 48 hours. We assessed the altered invasiveness of LLC in vitro and found that Raw264.7 pretreated with PBS enhanced the migratory and invasive ability of LLC compared to LLC co-cultured with DMEM. However, treatment with SiNPs (100 μg/mL) significantly reversed the stimulatory effect of Raw264.7 on the migration and invasion of LLC (Figure 4E). These findings suggest that SiNPs may inhibit lung cancer progression by modulating macrophage phenotype.
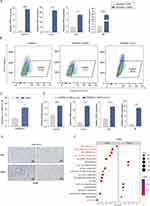
It is well-established that macrophages can respond to changes in the tumor immune microenvironment (TME) by undergoing reprogramming, which can alter their functional phenotype. M1-like macrophages exhibit tumor-killing properties, while M2-like macrophages are associated with promoting tumor growth and suppressing anti-tumor immunity. To investigate the effect of SiNPs on macrophage polarization, as shown in Supplementary Figure 2C, SiNPs treatment at effective concentrations had no significant cytotoxicity on RAW264.7 cells. We analyzed the expression levels of M1-like or M2-like biomarkers using quantitative polymerase chain reaction and flow cytometry. Our results demonstrated that SiNPs promoted M1-like activation of RAW264.7 macrophages, while M2-like biomarkers were not significantly altered. The LPS group was used as the positive control (Figures 5A–C, Supplementary Figure 2D and E). Similarly, this trend was observed in the polarization phenotype of RAW264.7 cells in the co-culture system (Figure 5D, Supplementary Figure 2F). To further evaluate whether SiNP-induced macrophage polarization is the primary factor leading to differences in tumorigenesis between the two groups of mice, we performed immunohistochemical (IHC) staining for CD86 on the lung tumors of both groups. We found that the number of M1 macrophages in the lung tumors of mice in the SiNPs group was significantly increased compared to those in the PBS group (Figure 5E). These data strongly suggest that SiNPs promote M1 polarization of macrophages, which could potentially exert tumor-inhibiting effects.
SiNPs-Mediated Activation of NF-κB Signaling Promotes the M1 Polarization of Macrophages
To elucidate further the mechanisms by which SiNPs induce macrophage M1 polarization and suppress tumors, we conducted downstream pathway analysis of differential expressed genes (logFC>1, p<0.05). GSEA was performed to assess the distribution of genes across various pathways, revealing the enrichment of several inflammation-related pathways. Notably, the TNF-α signaling via NF-κB pathway was significantly activated (Figure 5F). NF-κB signaling is a crucial for M1-like polarization of macrophages. To investigate whether SiNPs can activate the NF-κB pathway to promote macrophage polarization, we examined elevated inflammatory and apoptotic factors associated with NF-κB activation by qPCR (Figure 6A). Additionally, we assessed the levels of phosphorylated p65 (p-p65) protein in response to SiNPs exposure. Our results demonstrated that SiNPs exposure induced the activation of p-p65, which could be inhibited by the specific NF-κB inhibitor JSH-23, without significantly affecting total p65 levels (Figure 6B). In addition, qPCR and flow cytometry analyses revealed that SiNPs-induced M1 polarization was suppressed (Figure 6C and D). These findings suggest that SiNPs regulate M1 polarization in macrophages via the NF-κB pathway. To determine whether the activation status of NF-κB pathway affects the metastatic potential of LLC cells, we added JSH-23 to the co-culture system (Figure 6E). Our results showed that inactivation of NF-κB pathway significantly reversed the inhibitory effect of SiNPs on the migration and invasion of lung cancer cells (Figure 6F). These findings demonstrate that SiNPs promote macrophage M1 polarization and inhibit lung cancer cell metastasis by activating the NF-κB signaling pathway.
SiNPs Regulate Macrophage M1 Polarization by Promoting Macrophage Glycolysis
Metabolism regulates macrophage differentiation, polarization, mobilization and their antitumor response.14 To investigate the impact of SiNPs on macrophage metabolism, we performed GO analysis and found differential genes were enriched in metabolic processes (Figure 7A). Among these processes, glycolysis is crucial for maintaining macrophage M1 polarization. To confirm the association between SiNPs exposure and glycolysis, we conducted qPCR analysis, which revealed higher mRNA expression levels of key glycolytic enzymes in the SiNPs-exposed group compared to the PBS-exposed group, including Pfkl, Eno1,Ldha, Gpi, Gapdh, Hk2,Pfkm,Pgk1,Pgam1,Aldoa (Figure 7B). Furthermore, SiNPs exposure increased glucose, ATP, and lactate levels in macrophages (Figure 7C). Treatment with 2-deoxy-D-glucose (2-DG), a glycolysis inhibitor, reduced the expression of key glycolytic enzymes and the M1 polarization marker in macrophage (Figure 7D and E). In a co-culture system, the addition of 2-DG enhanced LLC cells migration and invasion compared to the SiNPs group. These findings suggest that SiNPs promote macrophage polarization by regulating glycolysis and possess the ability to inhibit lung cancer.
Discussion
The widespread use of nanomaterials requires a detailed assessment of their effects on biological systems. Recent studies investigating the impact of nanomaterials on these systems, even in non-clinical applications, have revealed that prolonged exposure to nanomaterials can alter the tumor microenvironment, leading to the formation of new tumors or an increase in metastatic sites in cancer patients.8,15 Furthermore, due to frequent inhalation the respiratory tract, nanomaterials may affect the pre-tumor microenvironment in lung cancer, making them a potentially significant factor in its development.16,17 Nickel oxide (NiO) and titanium dioxide (TiO2) are two types of nanomaterials recognized as carcinogenic s by the International Agency for Research on Cancer and the European Chemical Agency Committee for Risk Assessment,18–20 Furthermore, high aspect ratio nanoparticles (HARNs) and multi-walled carbon nanotubes (MWCNTs), both classified as nanomaterials, have raised concerns due to their potential to induce lung cancer through inhalation.21 Intriguingly, our study is the first to identify an inhibitory effect of SiNPs on lung cancer development, primarily manifested by the suppression of tumor growth, an increase in survival rates in vivo, and the inhibition of migration and invasion in vitro.
How to explain the unexpected phenomenon? Several factors need to be considered. Firstly, in the experimental group, the infiltrating inflammatory cells, total cell count and protein content in the lungs of mice exposed to SiNPs increased, indicating the initiation of local inflammatory responses. These inflammatory responses altered the immune microenvironment of the lung and, as a result, affected both the pre-metastatic niche and the metastatic niche, leading to the inhibition of lung cancer development and progression in vivo. These inflammatory responses altered the immune microenvironment in the lungs, subsequently affecting both the pre-metastatic and metastatic niches, which led to the inhibition of lung cancer development and progression in vivo. Secondly, RNA-seq data revealed significant enrichment of immune response pathways, particularly the accumulation of M1-type macrophages, which are known to inhibit cancer cell development in the SiNP-exposed group compared to the control group. These findings suggest that SiNP exposure creates a unique lung microenvironment that hinders the colonization of lung cancer cells. Thirdly, comparison with other current studies on the effects of nanomaterials on tumors, although Zhu et al also used tracheal drips to cause mice to inhale nanomaterials, its tumor model is implanting breast cancer cells into the mammary fat pad of mice. The animal model that David et al used was to implant breast cancer cells under the skin and then inject nanomaterials into the tail vein.8,21 These models primarily examined the effects of nanomaterials on tumor invasion and metastasis by focusing on the vascular endothelial system and blood circulation. In contrast, our study involved tracheal instillation of SiNPs into mouse lungs for 28 days, followed by in situ implantation of LLC. This tumor model offers the advantage of directly observing the effects of SiNP-induced alterations in the tumor microenvironment on lung cancer development, providing more accurate and relevant results.
Tumor immune infiltration is a complex ecosystem in which tumor-associated macrophages play a crucial role in this process.22 Our study observed an increase in M1 macrophages in the SiNPs exposure group, confirmed by PCR and FACS. To further elucidate the mechanism by which SiNPs promote M1 polarization in macrophages. We conducted a bioinformatic analysis of RNA-seq data. The results suggest that SiNPs exposure influences M1 polarization through biological processes primarily associated with oxidative stress, mitochondrial damage, apoptosis, and acute inflammatory responses. Firstly, our findings demonstrate that SiNPs exposure induces the polarization of RAW264.7 macrophages from M0 to M1 by activating the NF-κB signaling pathway, a critical event for promoting M1 polarization. Previous studies suggests that when SiNPs enter the lungs, they are phagocytosed by macrophages.9 The deposition of SiNPs in macrophages can impair lysosomal function and autophagic flux, leading to the activation of NF-κB signaling.23,24 Subsequently, macrophages secreted more cytokines such as TNF-α and IL-6, resulting in M1 phenotype activation. Additionally, SiNPs can induce oxidative stress in pulmonary epithelial cells, leading to the production of inflammatory factors such as reactive oxygen species (ROS), C-X-C motif chemokine ligand 1 (CXCL-1), CXCL-8, and interleukin-1 (IL1).5,7 These factors have been suggested as potential mechanisms for activating NF-κB signaling and promoting M1 polarization in macrophages.25,26 Secondly, we discovered that SiNPs sustain macrophage M1 polarization by promoting glycolytic metabolism. Inhalation of SiNPs can damage mitochondrial structure and function in macrophages, impairing oxidative phosphorylation.27,28 Moreover, SiNPs exposure increases ROS production and enhances the expression and stability of inducible nitric oxide synthase (iNOS), leading to glycolysis and promoting M1 macrophage polarization.14,29 Collectively, these mechanisms contribute to the suppression of lung cancer by SiNPs through macrophage polarization. This study raises new questions, such as how SiNPs might influence other metabolic changes in macrophages and affect development of lung cancer. On the one hand, alveolar macrophages, the first line of defense for lung phagocytosis, are the primary cells to come into contact with inhaled SiNPs. Upon contact, macrophages are attracted to and phagocytose silica particles. On the other hand, tumor-associated macrophages, the predominant immune cell population within tumors,30 may explain the sensitivity of macrophages to SiNPs. Previous studies indicate that nanomaterial effects on macrophages vary depending on particle size and surface area, and that these effects are concentration-dependent.5,31 In cellular experiments, the doses of SiNPs we used inhibited the migration and invasion of tumor cells without significantly affecting tumor cell proliferation and viability. This largely avoided the direct toxicity of the material, ensuring the accuracy of experimental results. This approach is consistent with our animal model, in which the SiNP dosage was selected to minimize the overall impact on the health of the mice. In contrast to the findings at 28 days, observations at 58 days reveal a narrowing discrepancy in fibrotic deposition within the pulmonary tissues of both mouse cohorts. This trend suggests a potential reduction in the detrimental impact of SiNPs on pulmonary health with prolonged exposure duration. However, there remains no precise method for converting in vitro doses to in vivo conditions, and further research is needed to explore the effects of SiNPs with varying characteristics (eg, size, surface area, shape)32 on lung tumors and the differences in dosing. Additionally, these questions require further investigation. The current study suggests that SiNPs inhibit lung cancer by inducing an inflammatory environment in the lungs, thereby affecting tumor colonization. However, it remains unclear whether SiNPs would exert similar inhibitory effects in established lung cancer models, such as those induced by carcinogens or genetically engineered mouse (GEM) models. Existing research indicates that once lung tumors are established, the tumor microenvironment (TME) undergoes complex changes. Unlike the acute inflammatory response induced by SiNPs, chronic inflammation associated with tumor persistence generates cytokines that drive tumorigenesis by inducing genetic mutations, inhibiting apoptosis, and promoting angiogenesis, leading to dysregulated inflammatory signaling pathways. This chronic inflammation also facilitates the formation of an immunosuppressive TME by recruiting immunosuppressive cells such as M2 tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs), further advancing tumor progression.33,34 In this context, the impact of SiNPs on macrophage polarization and tumor metabolic reprogramming within the TME may become more complex. Under chronic inflammatory conditions, SiNPs may promote the transition of macrophages from the M1 to M2 phenotype, potentially accelerating tumor progression in conjunction with the Warburg effect. Alternatively, SiNPs may reprogram M2 macrophages, similar to other nanomaterials such as iron oxides, or interact with apoptotic and autophagic pathways to inhibit tumor growth. These possibilities warrant further investigation.
Conclusion
This study is the first to demonstrate that SiNPs can inhibit lung cancer development by promoting the M1 polarization of macrophages within the tumor microenvironment (Graphical abstract created in Biorender.com). These findings extend beyond the traditional understanding of SiNPs’ effects on biological systems, offering novel insights into the role of nanomaterials in lung cancer progression. By revealing how SiNPs influence immune modulation within the tumor microenvironment, this research deepens our understanding of the broader implications of nanomaterials in the context of lung cancer biology.
Abbreviation
ENMs, engineered nanomaterials; SiNPs, silica nanoparticles; TME, tumor microenvironment; TAMs, tumor-associated macrophages; GM-CSF, granulocyte monocyte colony-stimulating; TEM, transmission electron microscopy; PDI, polydispersity index; LLC, lung carcinoma; NiO, Nickel oxide; TiO2, Titanium dioxide; HARN, High aspect ratio nanoparticles; MWCNTs, Multi-walled carbon nanotubes.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
All animal experiments were approved by the Ethics Committee of Chongqing Medical University (#SCXK2012-0001), and animal care and experimentation with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH publication no. 85–23, revised 1996).
Funding
This work is supported by the National Natural Science Foundation of China (82073277 and 82173247), Chongqing Talents: Exceptional Young Talents Project (CQYC2020058650), the Natural Science Foundation of Chongqing (cstc2021ycjh-bgzxm0105), Program for Youth Innovation in Future Medicine, Chongqing Medical University (W0038), the Science and Technology Project Affiliated to the Education Department of Chongqing (KJQN202100404), Science and Technology Project of Chongqing Yuzhong District (20200110) and Project of Chongqing Natural Science Foundation Innovation and Development Fund (Municipal Education Commission) (2022NSCQ-LZX0020). Z.Z. and C.C. are granted by Chongqing Bayu Young. Scholar program. Research and Innovation Projects for Graduate Students of Chongqing (CYS22376).
Disclosure
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1. Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–263. doi:10.3322/caac.21834
2. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535–554.
3. Xue Y, Wang L, Zhang Y, Zhao Y, Liu Y. Air pollution: a culprit of lung cancer. J Hazard Mater. 2022;434:128937. doi:10.1016/j.jhazmat.2022.128937
4. Berg CD, Schiller JH, Boffetta P, et al. Air pollution and lung cancer: a review by International Association for the Study of Lung Cancer Early Detection and Screening Committee. J Thorac Oncol. 2023;18(10):1277–1289.
5. Guo C, Zhao X, Ma R, et al. Silica nanoparticles promoted pro-inflammatory macrophage and foam cell transformation via ROS/PPARgamma/NF-kappaB signaling. Sci Total Environ. 2023;881:163430. doi:10.1016/j.scitotenv.2023.163430
6. Lo Re S, Lecocq M, Uwambayinema F, et al. Platelet-derived growth factor-producing CD4+ Foxp3+ regulatory T lymphocytes promote lung fibrosis. Am J Respir Crit Care Med. 2011;184(11):1270–1281. doi:10.1164/rccm.201103-0516OC
7. Wang M, Li J, Dong S, et al. Silica nanoparticles induce lung inflammation in mice via ROS/PARP/TRPM2 signaling-mediated lysosome impairment and autophagy dysfunction. Part Fibre Toxicol. 2020;17(1):23. doi:10.1186/s12989-020-00353-3
8. Peng F, Setyawati MI, Tee JK, et al. Nanoparticles promote in vivo breast cancer cell intravasation and extravasation by inducing endothelial leakiness. Nat Nanotechnol. 2019;14(3):279–286. doi:10.1038/s41565-018-0356-z
9. Hamilton RF, Thakur SA, Holian A. Silica binding and toxicity in alveolar macrophages. Free Radic Biol Med. 2008;44(7):1246–1258. doi:10.1016/j.freeradbiomed.2007.12.027
10. Xia Y, Rao L, Yao H, Wang Z, Ning P, Chen X. Engineering macrophages for cancer immunotherapy and drug delivery. Adv Mater. 2020;32(40):e2002054. doi:10.1002/adma.202002054
11. Zhu S, Yi M, Wu Y, Dong B, Wu K. Roles of tumor-associated macrophages in tumor progression: implications on therapeutic strategies. Exp Hematol Oncol. 2021;10(1):60. doi:10.1186/s40164-021-00252-z
12. Benmerzoug S, Rose S, Bounab B, et al. STING-dependent sensing of self-DNA drives silica-induced lung inflammation. Nat Commun. 2018;9(1):5226. doi:10.1038/s41467-018-07425-1
13. Singh G, Arora H, Hariprasad P, Sharma S. Development of clove oil based nanoencapsulated biopesticide employing mesoporous nanosilica synthesized from paddy straw via bioinspired sol-gel route. Environ Res. 2023;220:115208. doi:10.1016/j.envres.2022.115208
14. Mehla K, Singh PK. Metabolic regulation of macrophage polarization in cancer. Trends Cancer. 2019;5(12):822–834. doi:10.1016/j.trecan.2019.10.007
15. Lu X, Zhu Y, Bai R, et al. Long-term pulmonary exposure to multi-walled carbon nanotubes promotes breast cancer metastatic cascades. Nat Nanotechnol. 2019;14(7):719–727. doi:10.1038/s41565-019-0472-4
16. Wang Z, Zhai Z, Chen C, et al. Air pollution particles hijack peroxidasin to disrupt immunosurveillance and promote lung cancer. Elife. 2022;11:e75345.
17. Saber AT, Poulsen SS, Hadrup N, Jacobsen NR, Vogel U. Commentary: the chronic inhalation study in rats for assessing lung cancer risk may be better than its reputation. Part Fibre Toxicol. 2019;16(1):44. doi:10.1186/s12989-019-0330-4
18. Braakhuis HM, Gosens I, Heringa MB, et al. Mechanism of action of TiO(2): recommendations to reduce uncertainties related to carcinogenic potential. Annu Rev Pharmacol Toxicol. 2021;61:203–223. doi:10.1146/annurev-pharmtox-101419-100049
19. Sezer S, Yucel A, Ozhan Turhan D, Emre FB, Sarikaya M. Comparison of ZnO doped different phases TiO(2) nanoparticles in terms of toxicity using zebrafish (Danio rerio). Chemosphere. 2023;325:138342. doi:10.1016/j.chemosphere.2023.138342
20. Borm PJ, Schins RP, Albrecht C. Inhaled particles and lung cancer, part B: paradigms and risk assessment. Int J Cancer. 2004;110(1):3–14. doi:10.1002/ijc.20064
21. Kuempel ED, Jaurand MC, Moller P, et al. Evaluating the mechanistic evidence and key data gaps in assessing the potential carcinogenicity of carbon nanotubes and nanofibers in humans. Crit Rev Toxicol. 2017;47(1):1–58.
22. Liu Y, Xu R, Gu H, et al. Metabolic reprogramming in macrophage responses. Biomark Res. 2021;9(1):1. doi:10.1186/s40364-020-00251-y
23. Wu Y, Jin Y, Sun T, et al. p62/SQSTM1 accumulation due to degradation inhibition and transcriptional activation plays a critical role in silica nanoparticle-induced airway inflammation via NF-kappaB activation. J Nanobiotechnology. 2020;18(1):77.
24. Wang J, Yu Y, Lu K, et al. Silica nanoparticles induce autophagy dysfunction via lysosomal impairment and inhibition of autophagosome degradation in hepatocytes. Int J Nanomed. 2017;12:809–825. doi:10.2147/IJN.S123596
25. Chen X, Tang J, Shuai W, Meng J, Feng J, Han Z. Macrophage polarization and its role in the pathogenesis of acute lung injury/acute respiratory distress syndrome. Inflamm Res. 2020;69(9):883–895. doi:10.1007/s00011-020-01378-2
26. Lawrence T. Macrophages and NF-kappaB in cancer. Curr Top Microbiol Immunol. 2011;349:171–184.
27. Lozano O, Silva-Platas C, Chapoy-Villanueva H, et al. Amorphous SiO2 nanoparticles promote cardiac dysfunction via the opening of the mitochondrial permeability transition pore in rat heart and human cardiomyocytes. Part Fibre Toxicol. 2020;17(1):15. doi:10.1186/s12989-020-00346-2
28. Zhao X, Xu H, Li Y, et al. Proteomic profiling reveals dysregulated mitochondrial complex subunits responsible for myocardial toxicity induced by SiNPs. Sci Total Environ. 2023;857(Pt 1):159206. doi:10.1016/j.scitotenv.2022.159206
29. Mills EL, Kelly B, Logan A, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167(2):457–70e13. doi:10.1016/j.cell.2016.08.064
30. Reddon H, Patel Y, Turcotte M, Pigeyre M, Meyre D. Revisiting the evolutionary origins of obesity: lazy versus peppy-thrifty genotype hypothesis. Obes Rev. 2018;19(11):1525–1543. doi:10.1111/obr.12742
31. Ruan C, Wang C, Gong X, et al. An integrative multi-omics approach uncovers the regulatory role of CDK7 and CDK4 in autophagy activation induced by silica nanoparticles. Autophagy. 2021;17(6):1426–1447. doi:10.1080/15548627.2020.1763019
32. Inoue M, Sakamoto K, Suzuki A, et al. Size and surface modification of silica nanoparticles affect the severity of lung toxicity by modulating endosomal ROS generation in macrophages. Part Fibre Toxicol. 2021;18(1):21. doi:10.1186/s12989-021-00415-0
33. Denk D, Greten FR. Inflammation: the incubator of the tumor microenvironment. Trends Cancer. 2022;8(11):901–914.
34. Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. doi:10.1186/s13045-019-0760-3
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.