Back to Journals » Drug Design, Development and Therapy » Volume 19
Deciphering the Pharmacological Potential of Kouqiangjie Formula for the Treatment of Diabetic Periodontitis Based on Network Pharmacology, Machine Learning, Molecular Dynamics, and Animal Experiments
Authors Wu Y , Li J, Liu M, Gao R, Li H, Xie Y, Hu Q, Wei J, Zhao L, Li L
Received 1 October 2024
Accepted for publication 13 March 2025
Published 20 March 2025 Volume 2025:19 Pages 2103—2129
DOI https://doi.org/10.2147/DDDT.S494066
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Muzammal Hussain
Yeke Wu,1 Jiawei Li,2 Min Liu,3 Ranran Gao,4 Huijing Li,5 Yunfei Xie,6 Qiongying Hu,7 Jing Wei,5 Lixing Zhao,8 Li Li9
1Department of Stomatology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China; 2School of Basic Medical Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China; 3Department of Gynaecology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China; 4Department of Gynaecology, Henan Provincial People’s Hospital, Zhengzhou, 450000, People’s Republic of China; 5College of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China; 6Department of Nuclear Medicine, Sichuan Provincial People’s Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, 610072, People’s Republic of China; 7Department of Laboratory Medicine, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China; 8State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Department of Orthodontics, West China Hospital of Stomatology, Sichuan University, Chengdu, 610072, People’s Republic of China; 9Department of Radiology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China
Correspondence: Li Li, Department of Radiology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072, People’s Republic of China, Tel\Fax +86 028 87783457, Email [email protected]
Background: Periodontitis (PD) and type 2 diabetes mellitus (T2DM) represent interlinked global health burdens, commonly causing significant clinical complications when coincident. Therefore, managing both conditions (T2DM with periodontitis, DP) simultaneously poses considerable challenges, necessitating novel therapeutic strategies. KQJF has been clinically proven to treat DP with good efficacy, but its pharmacological substances and targets are not clear and urgently need to be clarified.
Aim: To define the potential active components and targets of KQJF for the treatment of DP.
Materials and Methods: The investigation commenced with the application of UPLC-Q-TOF/MS analysis to delineate the active constituents of KQJF and their associated targets in addressing DP. Additionally, the research incorporated subsequent methodologies such as machine learning, network pharmacology, molecular docking, molecular dynamics simulations, and a DP rat model was established and validated by in vivo experiments using H&E staining, immunohistochemistry, quantitative real-time PCR, and Western blot.
Results: KQJF was found to contain 49 prototype compounds and 121 metabolites with potential activity against PD and T2DM. Network pharmacology revealed 66 overlapping genes between the pharmacological targets of KQJF and known targets of PD and T2DM. Further exploration through PPI network and enrichment analyses illuminated the involvement of multi-target and multi-pathway mechanisms. Molecular docking and dynamics simulations confirmed the robust interactions between key compounds within KQJF and proteins associated with the diseases. In vivo validation demonstrated that KQJF treatment ameliorated DP-associated histopathological changes and modulated the expression of crucial proteins (including ABCG2, CCND1, CDKN1B, HIF1A, and PIK3R1) in a DP rat model.
Conclusion: In summary, KQJF exhibits potential therapeutic benefits for DP through a multi-component and multi-target approach, potentially offering a novel integrative treatment strategy. This study underscores the importance of integrating traditional medicine with modern molecular techniques to explore novel therapeutic avenues for complex comorbid conditions, providing a blueprint for future pharmacological explorations.
Keywords: Kouqiangjie Formula, T2DM with periodontitis, network pharmacology, machine learning, molecular dynamics
Graphical Abstract:

Introduction
Diabetes mellitus (DM) has long been acknowledged not only for its primary metabolic impairments but also for its role in developing secondary complications among which periodontal disease stands prominent. It’s increasingly clear that diabetes exacerbates the incidence and progression of periodontitis (PD), impacting significantly on oral health and, hence, overall quality of life.1,2 Conversely, severe PD can adversely affect glycemic control and contribute to the progression of diabetes,3 demonstrating an intricate bi-directional relationship. T2DM with periodontitis (DP), as a common oral complication in patients with DM, it has become one of the hot issues in dentistry and diabetes research worldwide.4 With changes in human lifestyle and diet, the number and age of diabetic patients are on the rise, further fueling the prevalence of DP. This growing co-morbidity calls for innovative approaches in both diagnostics and therapeutics.
Chinese medicine posits that dampness-heat and toxicity, stagnation of blood and blood stasis, and depletion of qi and yin are the central pathomechanisms of DP. The key to treating this condition lies in clearing heat and toxins, relieving dampness, harmonizing blood, and nourishing yin and qi. Kouqiangjie Formula (KQJF), a clinical formulation developed collaboratively by our group and the hospital’s pharmacy department, consists of twelve medicinal ingredients, including Rhizoma Smilacis Glabrae (20 g, the dried rhizome of Smilax glabra Roxb)., Taraxaci Herba (20 g, the dried whole herb of Taraxacum mongolicum Hand.-Mazz., T. borealisinense Kitam. Or plants of the same genus)., Herba Portulacae (20 g, the above ground parts of Portulaca oleracea L)., Polygonati Rhizoma (20 g, the dried rhizomes of Polygonatum kingianum Collett & Hemsl., P. sibiricum F. Delaroche. or P. cyrtonema Hua)., Plantaginis Herba (20 g, the dried whole herb of Plantago asiatica L. or P. depressa Willd)., Agrimoniae Herba (15 g, the dried above-ground parts of Agrimonia pilosa Ledeb)., Semen Coicis (20 g, the tried mature seed kernels of Coix lacryma-jobi var. ma-yuen (Rom.Caill). Stapf), Chuanxiong Rhizoma (10 g, the dried rhizome of Ligusticum chuanxiong Hort)., Sepiae Endoconcha (20 g, the dried inner shell of Sepiella maindroni de Rochebrune or Sepia esculenta Hoyle)., Cyathulae Radix (10 g, the dried root of Cyathula officinalis K.C.Kuan), Radix Glycyrrhizae (10 g, the dried roots and rhizomes of Glycyrrhiza uralensis Fisch)., and Fructus Mume (10 g, the dried nearly mature fruit of Prunus mume Siebold & Zucc).5 As well as the names of the botanical sources of the constituent herbs of the formula have been checked on World Flora Online (www.worldfloraonline.org). This formulation aims to clear heat, detoxify toxins, relieve dampness, and harmonize blood to support qi and yin. Primarily used for addressing symptoms such as soreness and distension of teeth, mastication pain, dry mouth, halitosis, gingival swelling, epistaxis, and pus overflow due to dampness and heat, as well as chronic PD with similar manifestations, KQJF has demonstrated significant therapeutic efficacy during its clinical application over several years.6 Notably, no apparent adverse reactions have been reported, and the formulation has secured a Chinese invention patent (No. ZL202211294394.7). However, its composition is complex, and the main active ingredients and mechanisms that exert the efficacy have not been fully revealed.
Advances in the field of biopharmaceuticals have led to the adoption of cutting-edge techniques such as serum pharmacochemistry analysis, which is considered a practical means of discovering the effective substances for the treatment of disease in herbal medicines by understanding the chemical composition of serum following drug administration.7,8 Furthermore, the development of predictive models using network pharmacology and machine learning canvases an integral strategy allowing for the identification of novel biomarkers and therapeutic targets.9–11 Additionally, molecular docking and molecular dynamics simulations serve pivotal roles in drug development by predicting how small molecules interact with their protein targets, thus providing insights into the molecular basis of drug action.12,13 Complementing these in silico approaches, animal models have been indispensable in simulating the human conditions of DP, thus providing critical insights into the disease process, and evaluating the efficacy of potential pharmacological interventions prior to human trials. In conclusion, the amalgam of traditional and novel research methods such as network pharmacology, molecular docking, molecular dynamics simulations, and strategic animal experiments creates a robust platform for the holistic understanding and innovative treatment of the nexus between diabetes and periodontal disease.
Hence, the objective of this investigation is to utilize a diverse array of technical methodologies, including serum pharmacochemistry analysis, machine learning, network pharmacology, molecular dynamics, molecular docking, and animal experimentation, to thoroughly explore the active constituents and underlying mechanisms of KQJF in the treatment of DP. This interdisciplinary approach not only aids in deciphering the mechanisms by which KQJF addresses DP but also paves the way for innovative therapies that could significantly alleviate the burden of these interconnected diseases. Furthermore, it serves as a valuable methodological reference for the study of other compound agents.
Materials and Methods
Serum Pharmacochemistry Analysis
Sample Preparation
To investigate the pharmacokinetic properties of KQJF, we utilized six Sprague-Dawley rats. These were allocated into two groups: a control group and a KQJF-treated group. The administered dose for the KQJF-treated group was set at 78 g/kg/d, informed by the dosage reported in high-dose treatments from previous literature.5 We administered the treatment via oral gavage over a period of three days. Blood samples were collected at 0.5, 1, and 2 hours following the final dose on the third day. Serum was isolated from these samples, pooled in equal volumes, and then subjected to UPLC-Q-TOF/MS analysis to study the drug’s systemic presence and characteristics.
UPLC-Q-TOF/MS Analysis Conditions
The KQJF sample underwent analysis utilizing a Waters H-Class ultra-high performance liquid chromatography (UHPLC) system in conjunction with an AB Sciex Triple TOF® 4600 high-resolution mass spectrometer. Chromatographic separation was achieved utilizing a Waters ACQUITY® UPLC® HSS T3 column (2.1 × 150 mm, 1.8 µm) maintained at 30°C. The mobile phase comprised 0.1% formic acid in water and acetonitrile, delivered at a flow rate of 0.3 mL/min. An injection volume of 5 µL was utilized, and detection occurred over a wavelength range spanning 190–400 nm. Mass spectrometric analysis was conducted in both positive and negative ion modes.
Network Pharmacology and Bioinformatics Analysis
Target Acquisition for KQJF
To identify bioactive compounds entering the bloodstream from the KQJF, the UPLC-Q-TOF-MS method was employed, successfully detecting 49 blood-borne components. Subsequently, typical SMILES structures corresponding to these active components were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). These SMILES formats of the screened compounds were then input into the Swiss Target Prediction database (http://www.swisstargetprediction.ch/) to forecast potential targets, using a selection threshold where the Probability of target prediction is greater than 0. This process yielded UniProt IDs for the target proteins of the selected compounds. Using the ID mapping tool on UniProt (https://www.uniprot.org/), these UniProt IDs were converted into Gene symbols, with duplicates being removed to finalize the list of active component targets. Further target prediction was performed through the Comparative Toxicogenomics Database (CTD, https://ctdbase.org/) for the 49 bloodborne components, enhancing the robustness of target identification. Finally, the predicted targets from both the Swiss Target Prediction and CTD databases were consolidated and deduplicated, culminating in a comprehensive list of KQJF-related targets for subsequent analyses.
Acquisition of Disease-Related Targets
To explore the gene expression patterns linked to PD, we conducted a search in the Gene Expression Omnibus (GEO) database using “periodontitis” as the query term. Our investigation led us to the dataset GSE16134, which is built upon the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. This dataset comprises samples from 241 PD patients and 69 healthy individuals, serving as the primary dataset for our analysis. Subsequently, our focus shifted to identifying genes associated with T2DM. Employing “type 2 diabetes mellitus” as the search query, we retrieved pertinent gene information from two distinct disease gene repositories: GeneCards (https://www.genecards.org/) and the CTD (https://ctdbase.org/). The retrieved data from both databases were systematically compiled to identify potential T2DM disease targets, forming the basis for our subsequent analyses.
PD Disease Differential Analysis and Weighted Gene Co-Expression Network Analysis (WGCNA)
In our investigation, we utilized R software to preprocess the chosen dataset (GSE16134) for normalization and differential analysis. Employing a cutoff of |log2 fold change (FC)| ≥ 0.585 with an adjusted P value < 0.05, we identified differentially expressed genes (DEGs). Subsequently, we applied the “WGCNA” package in R to conduct weighted gene co-expression network analysis (WGCNA) for module identification. To ensure result accuracy, we included the top 25% of genes exhibiting the most significant expression variations in the WGCNA. An optimal soft-thresholding power was determined to construct a weighted adjacency matrix, which was then transformed into a topological overlap matrix (TOM). Using hierarchical clustering, modules were delineated based on TOM dissimilarity, with a minimum module size of 100. Each module was assigned a distinct color for ease of identification. The module eigengene, representing the principal component of gene expression patterns within each module, was utilized to summarize its expression profile. To quantify the relationship between these modules and disease status, we calculated module significance (MS), reflecting the correlation between the module eigengene and clinical traits of interest. Additionally, gene significance (GS) was evaluated as the correlation between individual gene expression and clinical phenotypes, providing insights into potential gene-disease associations.
Target Screening for DP Treatment With KQJF
To delineate the potential interactions among targets associated with PD identified through the WGCNA analysis, main components of KQJF, and T2DM, we intersected the sets of relevant targets from each category. We utilized the “Venn” package in R software to perform a visual analysis and generate a Venn diagram that illustrates the overlapping targets among these conditions.
Constructing Protein-Protein Interaction (PPI) Networks
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, available at https://cn.string-db.org/) is a renowned bioinformatics resource designed to aggregate and disseminate data concerning protein-protein and gene-protein interactions. For the current study, intersecting targets were uploaded to the STRING database, applying a minimum interaction score threshold of 0.4 to refine the results. After this filtering process, both the PPI network diagram and the corresponding TSV (Tab-separated Values) file were downloaded and preserved for further analysis. Subsequently, the visualization and multidimensional network construction of the PPI, under the study designation “KQJF-PD-T2DM”, were carried out using Cytoscape software (version 3.9.0). This software facilitates a detailed visual representation of the interaction networks, providing a platform for in-depth biological interpretation and analysis.
Functional Enrichment Analysis of GO and KEGG Pathways
Using the R software, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses to investigate the potential biological functions and principal signaling pathways involved in the treatment of DP with KQJF. The analyses were based on a significance threshold set at a q-value of <0.05. The results were ordered in descending importance based on their p-values, allowing for the identification of statistically significant enriched terms. Specifically, the top 10 GO terms across three categories, including biological process (BP), cellular component (CC), and molecular function (MF). And the top 20 KEGG pathways were systematically extracted and are presented herein.
Core Gene Identification
We employed three distinct machine learning algorithms, namely SVM-RFE (Support Vector Machine-Recursive Feature Elimination), LASSO (Least Absolute Shrinkage and Selection Operator), and Degree analysis, to pinpoint key genes among intersected targets. The LASSO method and the SVM classifier were utilized to categorize biomarkers for PD diagnosis. Ten-fold cross-validation using the glmnet package was conducted to distinguish between PD patients and healthy controls, minimizing overfitting and enhancing model reliability. To refine the selection of high-potential genes, the SVM-RFE algorithm was executed with the e1071 and svmRadial packages. Subsequently, topological analysis using the Cytoscape software was applied to the intersecting targets to identify the top 20 targets in the PPI network. Genes that intersected across the three selection algorithms were considered crucial for the diagnostic analysis of DP. To gauge the predictive efficacy of the DP diagnostic model, we employed the pROC package. The accuracy of the prediction model was evaluated by computing the Area Under the Receiver Operating Characteristic (ROC) Curve (AUC), where higher AUC values signify enhanced predictive accuracy. Furthermore, line plots were generated to project the diagnostic efficacy for DP.
Gene Set Enrichment Analysis (GSEA)
To investigate the associations between core target genes and signaling pathways, we divided the PD cohort into two subgroups based on the median expression levels of core target genes. Subsequently, we conducted GSEA analysis for each subgroup, setting the threshold for statistical significance at adjusted p-value less than 0.05.
Immunocyte Infiltration Analysis
Immunocyte infiltration analysis was conducted utilizing the CIBERSORT algorithm, accessible at http://cibersortx.stanford.edu, to ascertain the relative proportions of 22 types of infiltrating immune cells within each tissue sample. The immune score for each sample was computed utilizing the “ESTIMATE” algorithm. Additionally, the relationship between signature genes and quantities of infiltrating immune cells was evaluated through Spearman’s rank correlation analysis, implemented in R software.
Molecular Docking
Firstly, key protein structures related to the identified core targets from the intersectional target screening are retrieved from the RCSB Protein Data Bank (PDB format). Concurrently, structural data of key active compounds are extracted from the PubChem database. Before docking, these proteins and small molecule compounds are preprocessed using the AutoDock software (version 1.1.2), which also facilitates the conversion of formats and the analysis of the binding sites on the proteins, aiming to locate active docking pockets. Following the preparation, both target proteins and small molecule compounds are imported into the AutoDock Vina software for docking. This involves setting the coordinates for the docking sites and performing docking validation. The outcomes of the docking trials are represented in a tripartite table, detailing the binding affinities between the small molecules and the target proteins. For a subset of the results, visualizations are rendered using the PyMOL software to provide a clear depiction of the molecular interactions and bindings observed.
Molecular Dynamics Analysis
Molecular dynamics simulations were performed using GROMACS 2022 software. Small molecules were parameterized with the General Amber Force Field (GAFF), while proteins used the AMBER14SB force field and TIP3P water model. Simulations ran under constant temperature and pressure with periodic boundary conditions. Constraints on hydrogen bonding were enforced with LINCS, and an integration timestep of 2 fs was used. Electrostatic interactions were calculated via the Particle-Mesh Ewald (PME) with a cutoff of 1.2 nm. Non-bonded interactions were truncated at 10 Å, and the neighbor list updated every 10 steps. Temperature was controlled with a V-rescale thermostat at 298 K, and pressure was maintained at 1 bar with the Berendsen barostat. Equilibration involved 100 ps of both NVT and NPT simulations at 298 K. A 100 ns production MD simulation was conducted, with snapshots saved every 10 ps.
Experimental Verification
Establishment of a T2DM Rat Model
Eighteen healthy male Sprague-Dawley rats from Sichuan Viton Lihua Laboratory Animal Technology Co. (Chengdu, China) were used to develop a T2DM model. Initially, twelve rats were fed a high-fat, high-sugar diet for two weeks, followed by fasting and intraperitoneal injections of streptozotocin (STZ) at 30 mg/kg for three consecutive days. Two weeks post-STZ, the rats underwent glucose and insulin tolerance tests (IGTT and ITT) with glucose measurements at specific intervals during the tests, and repeated blood glucose level assessments over four weeks. A T2DM model was confirmed in rats showing three consecutive fasting blood glucose levels exceeding 11.1 mmol/L and random levels above 16.7 mmol/L.
Establishment of a PD Model in T2DM Rats
In an experiment spanning seven weeks, 12 T2DM model rats developed periodontitis via a combined anesthesia approach using intraperitoneal sodium pentobarbital and inhaled isoflurane. They had orthodontic ligatures soaked in Porphyromonas gingivalis W83 and tied around their first molars, supplemented by bi-daily injections of P. gingivalis lipopolysaccharide (LPS) into the gingival sulcus. Meanwhile, six control rats received saline injections similarly. After four weeks, periodontal health was assessed by criteria including pocket probing depth, attachment loss, x-ray confirmed alveolar bone resorption, tooth mobility, and furcation involvement, indicating significant periodontal degradation in the experimental group compared to controls.
Experimental Grouping and Administration
Following the successful induction of a DP rat model in 12 specimens, they were promptly administered KQJF at a concentration of 0.78 g/mL, with a dosage of 19.5 g/kg/day, twice daily. Concurrently, rats in the control group received an equivalent volume of 0.9% (w/v) normal saline (NS) twice daily for two weeks before euthanasia. Eighteen SD rats were randomly divided into three cohorts (6 per group): a control group (NC), a DP model group, and a DP+KQJF treatment group. The entire experimental duration spanned 7 weeks.
H&E Staining
To analyze tissue and cellular structure, maxillae were first decalcified in a 15% EDTA solution at room temperature, with solution changes every three days, for a duration of six weeks. Subsequently, the samples underwent dehydration using a graded series of alcohols and were then paraffin-embedded. Sagittal sections, 4 μm thick, were prepared, deparaffinized, rehydrated, and stained using hematoxylin and eosin (H&E). Morphological examination was conducted using a microscope (Eclip80i; Nikon, Tokyo, Japan), and images were captured with a digital scanner (Panoramic 250; 3DHISTECH, Budapest, Hungary).
Immunohistochemistry Analysis
Sections were first deparaffinized, hydrated, and then subjected to antigen retrieval using 0.1% trypsin at 37°C for 30 minutes. This was followed by a 3% hydrogen peroxide treatment to inhibit peroxidase activity and a blocking step involving 5% bovine serum albumin (BSA) for 60 minutes at ambient temperature. Primary antibodies from various sources, including rabbit anti-HIF-1α (Affinity, Cincinnati, OH, USA) and CCND1 (Affinity), PI3KR1 (Abcam, Cambridge, MA, USA), CDKN1B (Proteintech, Rosemont, IL, USA), and ABCG2 (Proteintech) were prepared in 1% BSA and applied to the sections, which were then incubated overnight at 4°C. Detection was achieved using a diaminobenzidine (DAB) kit (Servicebio, Wuhan, China) and nuclei were stained with haematoxylin. The resultant images were analyzed using Image-Pro Plus software (version 7.0, Media Cybernetics, Inc., Rockville, MD, USA).
Quantitative Real-Time PCR
Total RNA was extracted from mouse periodontal tissues using TRIzol reagent, and RNA concentration was quantified. cDNA was synthesized with the BeyoRT™ II cDNA First-strand Synthesis kit (Beyotime, Shanghai, China) under the following conditions: 25 °C for 10 min, 55 °C for 15 min, and 85 °C for 5 min. Amplification was performed on an ABI Prism® 7300 Real-time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) using the BeyoFast™ SYBR Green qPCR Mix kit (Beyotime) with the following cycling parameters: initial denaturation at 95 °C for 60s, followed by 40 cycles of denaturation at 95 °C for 15s, annealing at 60 °C for 15s, and extension at 72 °C for 45s. Primer sequences were designed using Primer 5.0 software (Supplementary Table S1) and synthesized by Shanghai Bioengineering Co., Ltd., with GAPDH as the internal control. Threshold cycle (Ct) values were recorded, and mRNA expression levels were calculated using the 2−ΔΔCt method. Each sample was prepared in triplicate, and experiments were conducted at least three times.
Western Blot Analysis
Proteins were extracted from periodontal tissues in the NC, DP, and DP+KQJF groups using RIPA buffer supplemented with protease and phosphatase inhibitors (Solarbio, Beijing, China). Quantification of proteins was conducted using the BCA method (Beyotime, Shanghai, China). Equal amounts of protein were separated via SDS-PAGE and subsequently transferred to PVDF membranes (Merck Millipore, Billerica, MA, USA) at 100 V for 1 hour. Membranes were then subjected to overnight incubation at 4°C with primary antibodies targeting HIF-1α, CCND1 (both from Affinity), PI3KR1 (Abcam), CDKN1B, ABCG2 (both from Proteintech), and GAPDH (Affinity). Following this, the blots were probed with HRP-linked secondary antibodies (MultiSciences, Hangzhou, China) and visualized using an ECL system (BL523B; Biosharp, Beijing, China). Band densities were quantified using Image-Lab software (Bio-Rad, version 6.1; Hercules, CA, USA), with GAPDH serving as the normalization control.
Statistical Analysis
Statistical analyses were conducted utilizing IBM SPSS Statistics (version 23.0; IBM, Armonk, NY, USA). Data are expressed as mean ± standard deviation (SD). For multiple group comparisons, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was applied. Statistical significance was defined as a p-value < 0.05.
Results
Identification of Components Absorbed Into the Blood by KQJF
To delineate the pharmacologically active compounds in KQJF, we employed UPLC-Q-TOF/MS for the analysis of serum samples from the blank and KQJF groups. The total ion chromatograms pre- and post-KQJF administration are depicted in Figure S1A-D, respectively. Leveraging multistage mass spectrometry data and integrating results from the characterization of the components present in KQJF (Supplementary Table S2 and Figure S2), as well as relevant literature concerning the herbs contained in KQJF, we identified a repertoire of 49 prototype compounds including Citric acid, Macrozamin, Neochlorogenic acid, Protocatechuic aldehyde, Cryptochlorogenic acid among others, in the medicated serum samples (Supplementary Table S3). Additionally, 121 metabolites were detected (Supplementary Table S4). This comprehensive profiling not only enhances our understanding of the active constituents of KQJF but also underscores the complex transformation those components undergo post-administration.
Acquisition of KQJF Drug-Related Targets and DP Disease-Related Targets
To identify molecular targets related to KQJF, our initial queries utilizing the Swiss Target Prediction database and the CTD database revealed 448 and 570 potential targets, respectively. Upon merging and deduplicating these datasets, a consolidated list of 873 unique targets associated with KQJF was generated. Subsequent analysis was directed towards PD datasets (GSE16134). To mitigate batch effects among the samples, normalization procedures were implemented prior to differential analysis (Figure S3A-B). Differential expression analysis was performed utilizing the “limma” package in R software, identifying 156 DEGs. These findings were visually depicted in a volcano plot (Figure 1A) and a heatmap (Figure 1B). Subsequent exploration involved retrieving targets associated with T2DM, utilizing the GeneCards database (with a relevance score threshold of ≥10) and the CTD database (with an inference score of ≥30). This search yielded 6333 and 6376 potential targets from each respective database. Merging and deduplicating these lists resulted in a total of 9551 unique T2DM-associated targets.
Identification of Key Gene Modules Associated With PD Through WGCNA Analysis
To pinpoint key gene modules linked to PD, we utilized WGCNA analysis to establish co-expression networks and modules in both healthy controls and PD patients. Gene expression variance was computed across all genes within the GSE16134 dataset, with the top 25% of genes exhibiting the highest variance selected for subsequent analysis. Setting a soft-threshold power of 13 yielded a scale-free topology fit index (R²) of 0.9, determining the formation of co-expressed gene modules (Figure 2A). Using the dynamic tree cutting method, we identified nine distinct co-expressed modules, color-coded, and presented as a heatmap of the topological overlap matrix (TOM), showcasing their connectivity (Figure 2B–C). These gene modules were subsequently correlated with clinical traits, comparing similarities and adjacencies in expressions between the control group and PD patients. Among these, the blue module showed the strongest association with PD, containing 1713 genes (Figure 2D). Furthermore, genes within the blue module demonstrated significant positive correlation across diverse PD samples (correlation coefficient = 0.76, P-value = 8.9E-192) (Figure 2E). This evidence underscores the relevance of the blue module to the pathophysiology of PD.
Intersecting Targets Screening and “KQJF-DP Disease” Network Construction
To unveil the potential mechanisms driving the therapeutic efficacy of KQJF against DP, we intersected the pharmacological targets of KQJF with the established targets implicated in both PD and T2DM. This analysis identified 66 overlapping genes (Figure 3A). To further investigate the core components of KQJF, their targets, and their potential association with DP, we employed the Cytoscape software (version 3.9.0) to construct the “KQJF-DP” network. This network was generated by importing a specifically prepared “Network.xlxs” file into Cytoscape, resulting in a network comprising 98 nodes, including 30 active component nodes, 66 target nodes, and 2 disease nodes, and 263 edges (Figure 3B). Using the Network analyzer plugin, we analyzed the network’s topological parameters, finding an average degree of 5.367, network heterogeneity of 1.762, network density of 0.055, and centrality of 0.638. In this network, active components are represented by diamond shapes, diseases by inverted triangles, and corresponding drug-target interactions and disease interactions by circles. Each edge indicates the interaction between KQJF and its target nodes, and also between DP and its target nodes. Nodes with higher degrees within the network are considered central or “hub” nodes. Noteworthy central nodes among the active components include Luteolin (Degree = 28), Ferulic acid (Degree = 15), Chlorogenic acid (Degree = 13), Ligustilide (Degree = 9), and Glycyrrhizic acid (Degree = 6) (Table 1). These findings suggest that KQJF may exert its therapeutic effects through multiple, strongly interactive components that serve as central hubs in the network. Moreover, the network reveals multiple components acting simultaneously on multiple targets, as well as instances where multiple components interact with a single target, highlighting the multi-component, multi-target modulatory nature of KQJF in the treatment of DP.
 |
Table 1 Ranking List of Core Compounds of KQJF for DP Treatment |
PPI Network Construction for Intersecting Targets
In cellular systems, the functionality of proteins largely hinges on their ability to form macromolecular complexes through interactions, as isolated proteins seldom perform cellular functions independently. Consequently, the study of PPI networks is crucial for dissecting cellular organization, biological processes, and functions, highlighting the need for enhanced research in this domain. Understanding these networks is particularly vital for unraveling the mechanisms behind the therapeutic effects of KQJF on DP diseases. To elucidate the interaction dynamics among targets implicated in DP, we analyzed the intersection targets within a PPI network. Specifically, 66 proteins associated with DP intersection targets were uploaded into the STRING database to clarify the relationships between these component targets. This analysis yielded a network comprising 66 proteins with 231 interactions. In this network representation, darker colors denote higher degree values indicating a protein’s prominence in the network, with larger node sizes reflecting greater significance. The drug-disease intersection interactions and the PPI network are depicted in Figure 3C. The derived PPI data were then visualized using the Cytoscape software, with results presented in Figure 3D. To pinpoint the core targets potentially crucial in the treatment of DP by the KQJF, the top 20 proteins ranked by degree values were deemed central targets, as listed in Table 2. These 10 core targets are posited to play significant roles in the onset and progression of DP under treatment with KQJF.
 |
Table 2 Top 20 Core Targets for Degree Value |
Enrichment Analysis
Through the application of the “Cluster Profiler” package in R software, a comprehensive GO functional enrichment and KEGG pathway enrichment analysis was conducted for 66 intersecting targets involved in DP. The GO analysis resulted in 1375 entries, distributed across biological processes (1223 entries), molecular functions (92 entries), and cellular components (60 entries). Key biological processes included positive regulation of cytokine production, response to toxic substances, chemokine production, response to drugs, and response to steroid hormones. In terms of molecular function, the analysis highlighted activities such as cyclin-dependent protein serine/threonine kinase regulator activity and cytokine receptor binding. The cellular components analysis predominantly involved entities like the serine/threonine protein kinase complex and the cyclin-dependent protein kinase holoenzyme complex (Figure 4A). These results suggest potential molecular targets involved in inflammation and cell cycle processes, which are closely associated with the pathogenesis of DP. This indicates that the treatment efficacy of KQJF in DP may involve several biological processes.
 |
Figure 4 Enrichment analysis and construction of “active ingredient-target-pathway” network diagram. (A) GO analysis; (B) KEGG analysis; (C) The “active ingredient-target-pathway” network diagram. |
Further KEGG pathway analysis utilizing the same “Cluster Profiler” package revealed 105 significantly enriched pathways (q-value <0.05), including critical signaling pathways such as the FoxO, HIF-1, JAK-STAT, AMPK, AGE-RAGE in diabetic complications, TNF, PI3K-Akt, p53, B cell receptor, and NOD-like receptor pathways (Figure 4B). In the “active ingredient-target-pathway” (Figure 4C), from the outside to the inside, the signaling pathway is represented by pink nodes, the purple nodes represent the active ingredient, and the target is represented by green circular nodes. The network, consisting of 72 nodes connected by 177 edges, was analyzed using the Network Analyzer plugin, revealing an average of 4.917 neighbors per node, network heterogeneity of 0.795, network density of 0.069, and centrality of 0.219. This multi-component, multi-target, and multi-pathway characteristic of the KQJF’s active components underscores its therapeutic potential against DP through multiple layers of interaction and regulatory mechanisms.
Identification and Diagnostic Potential of Key Genes for PD Using Machine Learning Algorithms
To assess the diagnostic potential of intersection targets in PD compared to a healthy population, we employed three machine learning algorithms (SVM-RFE, LASSO, and Degree) on a PD dataset to isolate meaningful core targets capable of distinguishing PD patients. Using the SVM-RFE algorithm, we identified 37 out of 66 intersection targets as core targets (Figure 5A). The LASSO algorithm selected 23 genes from the same set of intersection targets (Figure 5B), and the top 20 targets were determined based on the Degree values from the PPI network (Figure 5C). By combining the results of these three algorithms, five genes (Including CCND1, PIK3R1, CDKN1B, ABCG2, and HIF1A) were identified as key genes (Figure 5D). To evaluate the diagnostic capability of these key genes in differentiating PD samples from healthy controls, expression profiling and specificity analysis were conducted. Results, depicted in a line graph (Figure 5E), demonstrated that these key genes have significant accuracy and specificity in distinguishing between PD and control samples. This evidence underscores the high diagnostic accuracy and specificity of these core targets in differentiating PD patients from healthy controls. Furthermore, expression levels and ROC curve analyses of these core targets were performed (Figure 5F–G). The results indicated significantly elevated expression levels of CCND1, PIK3R1, CDKN1B, and HIF1A in PD patients, with all corresponding AUC values exceeding 0.5. This underscores the robust diagnostic potential of these markers for PD.
GSEA Analysis of the Five Core Targets
To further elucidate the interplay between key molecular targets and PD, we employed GSEA analysis to investigate the associations between five pivotal targets and various signaling pathways implicated in PD. The outcomes demonstrated distinct pathway affiliations for each target, offering valuable insights into the potential mechanisms underlying PD. ABCG2 was significantly associated with pathways including DNA replication, fatty acid elongation, lipoic acid metabolism, mismatch repair, and nucleotide excision repair, suggesting a multifaceted role in cellular maintenance and metabolic regulation (Figure 6A). CCND1 exhibited strong links with pathways such as beta-alanine metabolism, biosynthesis of unsaturated fatty acids, fatty acid elongation, terpenoid backbone biosynthesis, and tyrosine metabolism, highlighting its involvement in lipid synthesis and amino acid processing (Figure 6B). CDKN1B showed significant correlations with N-glycan biosynthesis, other glycan degradation, protein export, selenocompound metabolism, and various types of N-glycan biosynthesis, indicating its critical role in protein glycosylation and degradation processes (Figure 6C). Similarly, HIF1A was notably associated with N-glycan biosynthesis, other glycan degradation, protein export, selenocompound metabolism, and viral protein interactions with cytokine and cytokine receptors, demonstrating its participation in immune response modulation and metabolic adaptation in hypoxic conditions (Figure 6D). Lastly, PIK3R1 revealed substantial associations with circadian rhythm, DNA replication, the Fanconi anemia pathway, mismatch repair, and nucleocytoplasmic transport, underscoring its involvement in fundamental cellular functions such as DNA maintenance and intracellular transport (Figure 6E). These findings, underscore the intricate network of signaling pathways engaged by the identified hub genes in PD, thus providing a richer understanding of the disease’s etiology and potential therapeutic targets.
 |
Figure 6 GSEA analysis of the five core targets. (A) ABCG2; (B) CCND1; (C) CDKN1B; (D) HIF1A; (E) PIK3R1. |
Immune Cell Infiltration Analysis
The etiology of PD is notably intricate, with the immune system playing a pivotal role in its pathogenesis. To explore the variances in the immune microenvironment between PD patients and healthy controls, we employed the CIBERSORT algorithm. As presented in Figures 7A–B, notable positive correlations were observed in PD samples with various immune cell types, including naive B cells, plasma cells, naive CD4 T cells, gamma delta T cells, activated memory CD4 T cells, M0 macrophages, and neutrophils. Conversely, memory B cells, follicular helper T cells, CD8 T cells, regulatory T cells (Tregs), resting NK cells, activated NK cells, M1 macrophages, resting dendritic cells, and resting mast cells were significantly negatively correlated. Furthermore, we explored the relationship between the expression levels of core target genes and the immune microenvironment. Our findings indicated significant positive correlations in resting dendritic cells with ABCG2, CCND1, and PIK3R1, while showing a negative correlation with CDKN1B. CD8 T cells exhibited significant negative correlations with ABCG2, CDKN1B, and HIF1A, but a positive correlation with CCND1 (Figure 7C). These results imply that alterations in the immune microenvironment in PD patients could be closely linked to key genes such as ABCG2, CCND1, CDKN1B, PIK3R1, and HIF1A. This underscores the complex interplay between the immune system and molecular targets in the pathophysiology of PD and possibly extends to broader oncological contexts.
Molecular Docking
Molecular docking studies have provided deeper insights into the interaction between the components of KQJF and their potential therapeutic targets for DP. Core targets were identified based on results from biomarker prediction, leading to the retrieval of several PDB files for key proteins, specifically ABCG2 (PDB ID: 8BHT), CCND1 (PDB ID: 2W96), CDKN1B (PDB ID: 7ORG), HIF1A (PDB ID: 1H2K), and PIK3R1 (PDB ID: 5FOR). Extensive analysis of the “active component-target” network’s Degree results identified the top five active compounds: Luteolin, Ferulic acid, Chlorogenic acid, Ligustilide, and Glycyrrhizic acid. The strength of interaction between target proteins and active components is quantified using docking scores. A more favorable interaction is indicated by a more negative docking score, suggesting stronger binding affinity. Notably, a binding energy less than −5 kcal/mol indicates significant receptor-ligand activity, with exceptionally strong interactions below −7 kcal/mol. Figure 8A displays a heatmap detailing the binding energies between these 14 compounds and their respective targets. Results showed that KQJF’s small molecules interact effectively with their targets. Particularly, the five compounds with optimal binding were further analyzed to elucidate the interactions between the small molecule active components and protein receptors. Utilizing AutoDock Vina and PyMOL software, interaction diagrams were generated, highlighting the surrounding amino acid residues and their binding modes. Chlorogenic acid’s interaction with CCND1 exhibited a binding energy of −5.2032 kcal/mol, mainly forming hydrogen bonds with ARG B:139, ARG B:181, LYS B:142, GLN B:168, and LEU B:171 (Figure 8B). Glycyrrhizic acid demonstrated a highly favorable interaction with ABCG2, with a binding energy of −7.8345 kcal/mol, engaging in hydrogen bonds with LYS A:86, SER A:87, SER A:88, GLY A:85, ILE A:63, LYS A:61, LYS A:97, SER A:655, and ASP A:98 (Figure 8C). Ferulic acid showed a binding energy of −5.1499 kcal/mol with CCND1, mainly interacting through a hydrogen bond with GLU B:263 (Figure S4A). Ligustilide with HIF1A recorded a binding energy of −5.1743 kcal/mol, primarily forming hydrogen bonds with GLN A:204 and ALA A:300 (Figure S4B). Luteolin with HIF1A demonstrated a binding energy of −6.0907 kcal/mol, primarily forming hydrogen bonds with THR S:796, GLN A:314, ALA A:300, GLN A:204, and TRP A:179 (Figure S4C). These findings underscore the therapeutic potential of KQJF in targeting PD_T2DM through effective molecular interactions between its components and specific disease-related targets.
 |
Figure 8 Molecular docking results of active ingredients in KQJF with five core targets. (A) Heat map of molecular docking binding energy; (B) Chlorogenic acid-CCND1; (C) Glycyrrhizic acid-ABCG2. |
Molecular Dynamics Simulations
Based on the molecular docking results, we found that Glycyrrhizic acid had the lowest binding energy to ABCG2, CDKN1B and HIF1A. Therefore, we further performed molecular dynamics simulation analysis. The root mean square deviation (RMSD) serves as a critical indicator of system stability by summing the deviations of all atoms in a conformation from their target positions at a given moment. As depicted in Figure 9A, there is a gradual stabilization of the complex (ABCG2-Glycyrrhizic acid) structure as indicated by the RMSD, suggesting increasing structural stability over time. The radius of gyration (Rg) provides insights into overall structural changes and can be employed to assess the compactness of protein structures. Significant changes in Rg indicate system expansion. According to Figure 9B, the Rg of the complex remains relatively stable, underscoring a consistently stable configuration. The root mean square fluctuation (RMSF) quantifies the flexibility of amino acid residues within the protein. This metric is graphically represented in Figure 9C. For understanding the interaction of Glycyrrhizic acid on the ABCG2 surface, one approach involves evaluating the initial docking sites by measuring the distance between the centroid of the residue at the docking site and the centroid of Glycyrrhizic acid. Furthermore, the proximity between Glycyrrhizic acid and ABCG2 centroid is analyzed. Figure 9D illustrates minimal fluctuations in these distances, not exceeding 0.5 nm and demonstrating gradual stabilization, which confirms the stability of the distance between the Glycyrrhizic acid and ABCG2. The buried solvent-accessible surface area (SASA) enveloping the Glycyrrhizic acid provides a measure of the interface size between Glycyrrhizic acid and ABCG2, offering insights into their binding state. As shown in Figure 9E, fluctuations in the buried SASA diminish progressively, suggesting a gradual stabilization in the contact area and, consequently, a stabilization in the complex formation. Hydrogen bonding, a pivotal force in protein-ligand interactions that reflects the strength of electrostatic interactions, is discussed lastly. Figure 9F indicates that the number of hydrogen bonds between Glycyrrhizic acid and ABCG2 remains relatively consistent, fluctuating mainly between one to five bonds. This observation underscores the stability of electrostatic interactions within the complex. In addition, the molecular dynamics simulation results also showed that Glycyrrhizic acid binds to CDKN1B (Figure 9G–L) and HIF1A (Figure 9M–R) with equally good stability.
In Vivo Animal Experimental Validation
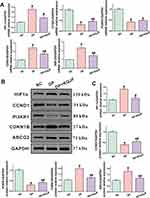
In order to validate the results of network pharmacology, molecular docking, molecular dynamics and machine learning experiments, we established a DP rat model, and firstly, we used H&E staining to carefully observe the morphological changes of the maxilla in different groups, and the results showed that the periodontal tissues of rats in the NC group had normal morphology and structure, and no obvious histopathological damage changes were seen; compared with the blank group, the DP group saw widening of the periodontal membrane gap, morphological and structural destruction and defects of the interdental papillae, degeneration and necrosis of the combined epithelium, the gingival sulcus epithelium and gingival epithelial cells (black arrows), a large number of neutrophils infiltration (red arrows), fibrous tissue proliferation (blue arrows), neonatal capillary formation (yellow arrowheads), and exposed dentin; Compared with the DP model group, the DP+KQJF treatment group displayed significant preservation of tissue architecture with reduced inflammatory cell infiltration (red arrowheads) (Figure 10A). This suggests that KQJF treatment ameliorates the histopathological changes associated with DP. Further, we analyzed the protein expression of five core targets screened by network pharmacology and machine learning. Immunohistochemical analysis demonstrated a notable increase in HIF-1α, CCND1, and PI3KR1 expression in the DP+KQJF group compared to the DP model group. Furthermore, KQJF treatment significantly downregulated the expression of CDKN1B and ABCG2, indicating a potential reversal of pathological processes in periodontal tissues (Figure 10B–C). These molecular changes point towards an anti-inflammatory and tissue-protective role of KQJF in the context of diabetic periodontitis. The qRT-PCR and Western blot analyses were consistent with the immunohistochemical findings, affirming the modulation of pivotal signaling proteins involved in cell cycle regulation, apoptosis, and metabolic stress responses. The increase in HIF-1α, CCND1, and PI3KR1 mRNA and protein levels alongside the decrease in CDKN1B and ABCG2 was significantly evident in the DP+KQJF group compared to the DP model group (Figure 11A–B). The protein quantitative analysis reaffirmed the significant upregulation and downregulation of these proteins, suggesting that KQJF activates a protective cellular program against the pathological backdrop of DP (Figure 11C). These results indicate that KQJF not only mitigates histological and molecular disturbances induced by PD in the presence of T2DM but also suggests the modulation of distinct biochemical pathways critical for disease progression and tissue integrity. Thus, KQJF emerges as a promising therapeutic candidate for managing DP patients.
Discussion
The present study elucidates the multifaceted pharmacological impact of KQJF on DP, providing a comprehensive molecular and cellular mechanistic insight. Our integration of advanced UPLC-Q-TOF/MS analytical methods, network pharmacology, machine learning, molecular simulations, and in vivo validations delineated the bioactive compounds in KQJF, their target interactions, and the subsequent modulation of disease pathways, underlining its potential as a therapeutic intervention.
Initially, our analyses revealed a complex blend of 49 prototype compounds and 121 metabolites derived from KQJF, underscoring the broad spectrum of bioactive molecules potentially instrumental in modulating pathological processes in DP. Notably, components such as Citric acid, Cryptochlorogenic acid, and Neochlorogenic acid, among others, exhibited significant transformations post-administration, suggesting metabolic modifications pivotal in eliciting therapeutic outcomes. It has been found that the concentration of citric acid used as a root conditioner affects the behavior of human periodontal ligament fibroblasts, with 10% citric acid for 90 seconds resulting in a more rapid proliferation of fibroblasts from human periodontal ligament compared to that with 50% citric acid.14 Cryptochlorogenic acid exhibits potent anti-inflammatory and antioxidant effects by inhibiting NF-κB/MAPK signaling pathways and promoting Nrf2 nuclear translocation, suggesting its therapeutic potential against LPS-induced inflammation and oxidative stress.15 The water extract from Lycium barbarum L. has been found to contain neochlorogenic acid and so on (up to 6.06%). This extract effectively mitigates symptoms of T2DM in rats by modulating blood glucose and lipid levels, repairing tissue injuries, reversing metabolic alterations, and restoring gut microbiota dysbiosis induced by T2DM.16 Luteolin protects pancreatic cells by reducing oxidative stress and improving insulin resistance, leading to lower blood sugar levels.17,18 It also inhibits cytokines associated with PD, thus mitigating its progression.19 Ferulic acid helps alleviate oxidative damage caused by T2DM, suppresses inflammation, and manages blood sugar levels by enhancing insulin sensitivity.20 Chlorogenic acid reduces oxidative stress in diabetes, preserves pancreatic function, and lowers blood sugar levels by inhibiting glucose absorption.21 It also suppresses inflammation in PD, aiding in tissue healing.22 In conclusion, there are studies that have demonstrated the potential of these ingredients to have a better treatment for periodontitis or T2DM. However, there are no studies demonstrating the relationship between these ingredients and DP treatment.
The elucidation of drug-related targets using databases such as Swiss Target Prediction and CTD, combined with differential expression analyses in gingival samples from DP individuals, allowed us to construct a robust list of targets which intersect with the molecular signatures of PD and T2DM. These insights provided a foundation to explore key gene modules via WGCNA analysis, identifying the blue module as particularly correlated with DP pathophysiology, thus prioritizing potential therapeutic targets. Furthermore, the construction of intersecting targets screening and the “KQJF-DP disease” network facilitated the identification of central compounds like Luteolin and Ferulic acid, which likely play critical roles in modulating disease mechanisms through their interactions with multiple targets and pathways.23 This network-centric approach not only highlighted the multi-target interactions but also reinforced the concept of network pharmacology where a holistic view is adopted rather than a single-target focus.
Recent studies have underscored the interplay between metabolic diseases such as T2DM and inflammatory conditions like PD, highlighting a bidirectional exacerbation influenced by common pathological processes such as inflammation, immune dysregulation, and altered cellular signaling.24,25 Correspondingly, our findings reveal that KQJF targets several pathways pivotal to both PD and T2DM, including JAK-STAT, PI3K-Akt, and TNF signaling pathways, corroborating with the literature suggesting these pathways are critical nodal points in the pathogenesis of both conditions.26–30 The intersection of KQJF’s active components with these pathways demonstrates the formula’s capability to modulate key biological processes such as cytokine production, immune response, and cell cycle regulation, which are significantly dysregulated in DP. The molecular docking and dynamics simulations offered a microscale glimpse into the interaction dynamics at the molecular level, providing evidence for the potential binding efficiency and stability of interactions between key proteins and KQJF-derived compounds. Notably, these molecular interactions, particularly those exhibiting high binding affinity and stability such as ABCG2 and HIF1A, which have been implicated in recent studies to play substantial roles in cellular protection against oxidative stress and apoptosis in diabetic environments.31,32 In evaluating immune cell contributions, our use of the CIBERSORT algorithm highlighted significant shifts in the immune cell landscape, suggesting KQJF may exert substantial immunomodulatory effects. This is particularly relevant given recent reports which indicate that modulation of specific immune cells and cytokines can substantially impact PD progression and T2DM complications.33–35 Such as, PD is strongly associated with M1 and M2 macrophage polarisation, and studies have shown increased M1 macrophage activity and a relative decrease in M2 macrophages during periodontitis, leading to inflammatory progression and impeding tissue repair. Therapeutic strategies focus on modulating macrophage subtype balance to promote inflammatory relief and tissue healing.36,37
Further, the validation of our findings through in vivo experimental approaches not only corroborated the molecular and computational predictions but also showcased the real-world efficacy of KQJF in mitigating the histopathological manifestations of DP. The modulation of key protein expressions involved in cell cycle regulation, apoptosis, and metabolic stress responses evident in treated groups emphasized the potential of KQJF as a multi-target therapeutic agent in managing DP, particularly in the milieu of T2DM, provides compelling evidence of KQJF’s therapeutic potential.
Critically, while our findings align with the emerging trends in metabolic and inflammatory disease research, certain limitations exist. Our study primarily focuses on biochemical and cellular pathways without extensive exploration into the long term clinical outcomes and potential side effects, which are essential for translational relevance. Further longitudinal and clinical studies will be required to translate these findings into therapeutic protocols.
Conclusion
In conclusion, KQJF emerges as a promising integrative treatment strategy for managing the complex interplay of DP, targeting key molecular pathways, and altering the immune profile to potentially mitigate disease progression. Excitingly, this study epitomizes the synergy between traditional herbal medicine and modern scientific methods, shedding light on the intricate molecular networks in disease and offering promising avenues for therapeutic interventions with broad-spectrum efficacy. Future research should aim to further validate these findings in larger cohorts and diverse population samples while also exploring the prospective adverse effects and optimal dosing strategies to maximize therapeutic gains.
Abbreviations
BSA, Bovine serum albumin; BP, Biological Process; CTD, Comparative Toxicogenomics Database; CC, Cellular Component; DP, T2DM with periodontitis; DEGs, Differentially expressed genes; DAB, Diaminobenzidine; GO, Gene Ontology;
GEO, Gene Expression Omnibus; GSEA, Gene Set Enrichment Analysis; GS, Gene significance; GAFF, General Amber Force Field; H&E, Hematoxylin and eosin; KEGG, Kyoto Encyclopedia of Genes and Genomes; KQJF, Kouqiangjie Formula; LPS, lipopolysaccharide; MS, Module significance; MF, Molecular Function; NS, Normal saline; NC, Blank control group; PME, Particle-Mesh Ewald; PPI, protein-protein interaction; PD, Periodontitis; ROC, Receiver Operating Characteristic; STZ, Streptozotocin; SD, Standard deviation; TOM, Topological overlap matrix; T2DM, Type 2 diabetes mellitus; UHPLC, Ultra-high performance liquid chromatography; WGCNA, Weighted gene co-expression network analysis.
Data Sharing Statement
Data will be made available on request from the corresponding author.
Ethics Approval and Consent to Participate
All animal studies in this research were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and received approval from the Laboratory Animal Management Committee at Chengdu University of Traditional Chinese Medicine (2023KL-115).
The GEO, GeneCards, and CTD are public databases, and the patients included in these databases have provided ethical approval for their data usage. Users can freely download relevant data for research purposes and publish related articles. Consequently, the Human Research Ethics Committee at Chengdu University of Traditional Chinese Medicine has reviewed and waived the requirement for ethical approval for this study.
Author Contributions
All authors significantly contributed to the reported work, whether in the conception, study design, execution, data acquisition, analysis, interpretation, or in all of these areas. They participated in drafting, revising, or critically reviewing the article; provided final approval for the version to be published; agreed on the journal to which the article has been submitted; and are accountable for all aspects of the work.
Funding
This research received financial support from several sources, including the National Natural Science Foundation of China (82074303, 81973684), the Natural Science Foundation of Sichuan Province (2023NSFSC1760).
Disclosure
The authors declare no competing interests in this work.
References
1. Simpson TC, Clarkson JE, Worthington HV, et al. Treatment of periodontitis for glycaemic control in people with diabetes mellitus. Cochrane Database Syst Rev. 2022;4(4):Cd004714. doi:10.1002/14651858.CD004714.pub4
2. Li Y, Huang Z, Pan S, et al. Resveratrol alleviates diabetic periodontitis-induced alveolar osteocyte ferroptosis possibly via regulation of SLC7A11/GPX4. Nutrients. 2023;15(9). doi:10.3390/nu15092115
3. C-z W, Y-h Y, H-h L, et al. Epidemiologic relationship between periodontitis and type 2 diabetes mellitus. BMC Oral Health. 2020;20(1):204. doi:10.1186/s12903-020-01180-w
4. Li B, Xin Z, Gao S, et al. SIRT6-regulated macrophage efferocytosis epigenetically controls inflammation resolution of diabetic periodontitis. Theranostics. 2023;13(1):231–249. doi:10.7150/thno.78878
5. Wu Y, Liu M, He X, et al. A breakthrough in periodontitis treatment: revealing the pharmacodynamic substances and mechanisms of Kouqiangjie formula. J Ethnopharmacol. 2024;323:117738. doi:10.1016/j.jep.2024.117738
6. He X, Zhou H, Wei J, et al. Effect of Kouqiangjie gargle combined with periodontal basic therapy on periodontal status and gingival crevicular fluid inflammatory cytokines in patients with chronic periodontitis complicated with type 2 diabetes mellitus. Tradi Chin Drug Res Clin Pharmacol. 2023;34(5):691–696.
7. Zhao X, Su H, Chen H, et al. Integrated serum pharmacochemistry and network pharmacology to explore the mechanism of Yi-Shan-Hong formula in alleviating chronic liver injury. Phytomed. 2024;128:155439. doi:10.1016/j.phymed.2024.155439
8. Qin J, Xiang G, Gao H, Meng X, Wang S, Zhang Y. Determination of the pharmacodynamic substances and mechanism of Shiwuwei Saierdou Pills against cholestatic hepatitis through chemical profile identification and network pharmacology analysis. Arabian J. Chem. 2024;17(2):105504. doi:10.1016/j.arabjc.2023.105504
9. Xiang G, Yang L, Qin J, Wang S, Zhang Y, Yang S. Revealing the potential bioactive components and mechanism of Qianhua Gout Capsules in the treatment of gouty arthritis through network pharmacology, molecular docking and pharmacodynamic study strategies. Heliyon. 2024;10(10):e30983. doi:10.1016/j.heliyon.2024.e30983
10. Feng M, Luo F, Wu H, et al. Network pharmacology analysis and machine-learning models confirmed the ability of YiShen HuoXue decoction to alleviate renal fibrosis by inhibiting pyroptosis. Drug Des Devel Ther. 2023;17:3169–3192. doi:10.2147/dddt.s420135
11. Noor F, Asif M, Ashfaq UA, Qasim M, Tahir Ul Qamar M. Machine learning for synergistic network pharmacology: a comprehensive overview. Briefings Bioinf. 2023;24(3). doi:10.1093/bib/bbad120
12. Santos LHS, Ferreira RS, Caffarena ER. Integrating molecular docking and molecular dynamics simulations. Meth mol Biol. 2019;2053:13–34. doi:10.1007/978-1-4939-9752-7_2
13. Sun Z, Wang Y, Pang X, Wang X, Zeng H. Mechanisms of polydatin against spinal cord ischemia-reperfusion injury based on network pharmacology, molecular docking and molecular dynamics simulation. Bioorg Chem. 2023;140:106840. doi:10.1016/j.bioorg.2023.106840
14. Caba-Paulino CE, Manfredi GGP, Zangrando MSR, et al. The concentration of citric acid as dental root conditioner influences the behavior of fibroblasts from human periodontal ligament. Arch Oral Biol. 2020;118:104839. doi:10.1016/j.archoralbio.2020.104839
15. Zhao XL, Yu L, Zhang SD, et al. Cryptochlorogenic acid attenuates LPS-induced inflammatory response and oxidative stress via upregulation of the Nrf2/HO-1 signaling pathway in RAW 264.7 macrophages. Int Immunopharmacol. 2020;83:106436. doi:10.1016/j.intimp.2020.106436
16. Zhao XQ, Guo S, Lu YY, et al. Lycium barbarum L. leaves ameliorate type 2 diabetes in rats by modulating metabolic profiles and gut microbiota composition. Biomed Pharmacothe. 2020;121:109559. doi:10.1016/j.biopha.2019.109559
17. Pradhan G, Kulkarni YA. Diabetes and its complications: role of luteolin, a wonder chemical from the natural source. Current Diabetes Rev. 2024. doi:10.2174/0115733998285798240217084632
18. Wang Z, Zeng M, Wang Z, Qin F, Chen J, He Z. Dietary luteolin: a narrative review focusing on its pharmacokinetic properties and effects on glycolipid metabolism. J Agri Food Chem. 2021;69(5):1441–1454. doi:10.1021/acs.jafc.0c08085
19. Balci Yuce H, Toker H, Yildirim A, Tekin MB, Gevrek F, Altunbas N. The effect of luteolin in prevention of periodontal disease in Wistar rats. J Periodontol. 2019;90(12):1481–1489. doi:10.1002/jper.18-0584
20. Li X, Wu J, Xu F, et al. Use of ferulic acid in the management of diabetes mellitus and its complications. Molecules. 2022;27(18). doi:10.3390/molecules27186010
21. Williamson G. Protection against developing type 2 diabetes by coffee consumption: assessment of the role of chlorogenic acid and metabolites on glycaemic responses. Food Funct. 2020;11(6):4826–4833. doi:10.1039/d0fo01168a
22. Huang X, Liu Y, Shen H, Fu T, Guo Y, Qiu S. Chlorogenic acid attenuates inflammation in LPS-induced Human gingival fibroblasts via CysLT1R/Nrf2/NLRP3 signaling. Int Immunopharmacol. 2022;107:108706. doi:10.1016/j.intimp.2022.108706
23. Guo C, Huang Q, Wang Y, et al. Therapeutic application of natural products: NAD+ metabolism as potential target. Phytomed. 2023;114:154768. doi:10.1016/j.phymed.2023.154768
24. Almubarak A, Tanagala KKK, Papapanou PN, Lalla E, Momen-Heravi F. Disruption of monocyte and macrophage homeostasis in periodontitis. Front Immunol. 2020;11:330. doi:10.3389/fimmu.2020.00330
25. Miller CS, Ding X, Dawson DR 3rd, Ebersole JL. Salivary biomarkers for discriminating periodontitis in the presence of diabetes. J clin periodontol. 2021;48(2):216–225. doi:10.1111/jcpe.13393
26. Chen J, Meng X. Aronia melanocarpa anthocyanin extracts improve hepatic structure and function in high-fat diet-/streptozotocin-induced T2DM mice. J Agri Food Chem. 2022;70(37):11531–11543. doi:10.1021/acs.jafc.2c03286
27. Wu F, Shao Q, Xia Q, et al. A bioinformatics and transcriptomics based investigation reveals an inhibitory role of Huanglian-Renshen-Decoction on hepatic glucose production of T2DM mice via PI3K/Akt/FoxO1 signaling pathway. Phytomed. 2021;83:153487. doi:10.1016/j.phymed.2021.153487
28. Yang X, Liu Y, Tang Z, Song Z, Liu C, Wang C. Total flavonoids of Hippophae rhamnoides L. improves type 2 diabetes symptoms in rats through down-regulating of the DAG/PRKCA/MAPK10/p65/TNF-α signalling pathway. J Ethnopharmacol. 2024;318(Pt A):116962. doi:10.1016/j.jep.2023.116962
29. Gao Z, Weng X, Yu D, et al. Porphyromonas gingivalis-derived lipopolysaccharide promotes glioma cell proliferation and migration via activating akt signaling pathways. Cells. 2022;11(24):4088. doi:10.3390/cells11244088
30. Shen Z, Zhang R, Huang Y, et al. The spatial transcriptomic landscape of human gingiva in health and periodontitis. Sci China Life Sci. 2024;67(4):720–732. doi:10.1007/s11427-023-2467-1
31. Wang LY, Cheng KC, Li Y, Niu CS, Cheng JT, Niu HS. Glycyrrhizic acid increases glucagon like peptide-1 secretion via TGR5 activation in type 1-like diabetic rats. Biomed Pharmacothe. 2017;95:599–604. doi:10.1016/j.biopha.2017.08.087
32. Tan D, Tseng HHL, Zhong Z, Wang S, Vong CT, Wang Y. Glycyrrhizic acid and its derivatives: promising candidates for the management of type 2 diabetes mellitus and its complications. Int J mol Sci. 2022;23(19):10988. doi:10.3390/ijms231910988
33. Zhang S, Gang X, Yang S, et al. The alterations in and the role of the Th17/Treg balance in metabolic diseases. Front Immunol. 2021;12:678355. doi:10.3389/fimmu.2021.678355
34. Cinkajzlová A, Mráz M, Haluzík M. Adipose tissue immune cells in obesity, type 2 diabetes mellitus and cardiovascular diseases. J Endocrinol. 2021;252(1):R1–r22. doi:10.1530/joe-21-0159
35. Yang B, Pang X, Li Z, Chen Z, Wang Y. Immunomodulation in the treatment of periodontitis: progress and perspectives. Front Immunol. 2021;12:781378. doi:10.3389/fimmu.2021.781378
36. Sun X, Gao J, Meng X, Lu X, Zhang L, Chen R. Polarized macrophages in periodontitis: characteristics, function, and molecular signaling. Front Immunol. 2021;12:763334. doi:10.3389/fimmu.2021.763334
37. Garaicoa-Pazmino C, Fretwurst T, Squarize CH, et al. Characterization of macrophage polarization in periodontal disease. J clin periodontol. 2019;46(8):830–839. doi:10.1111/jcpe.13156
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.