Back to Journals » Cancer Management and Research » Volume 17
Habitat Analysis in Tumor Imaging: Advancing Precision Medicine Through Radiomic Subregion Segmentation
Authors Wu LX, Ding N, Ji YD, Zhang YC, Li MJ, Shen JC, Hu HT , Jin L, Yin SN
Received 14 December 2024
Accepted for publication 19 March 2025
Published 1 April 2025 Volume 2025:17 Pages 731—741
DOI https://doi.org/10.2147/CMAR.S511796
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Ling Xiao Wu, Ning Ding, Yi Ding Ji, Yi Chi Zhang, Meng Juan Li, Jia Cheng Shen, Hai Tao Hu, Long Jin, Sheng Nan Yin
Department of Medical Imaging, Suzhou Ninth People’s Hospital, Wujiang, Suzhou, Jiangsu, People’s Republic of China
Correspondence: Long Jin; Sheng Nan Yin, Department of Medical Imaging, Suzhou Ninth People’s Hospital, Wujiang, Suzhou, Jiangsu, People’s Republic of China, Tel +8618550232113; +8618862301079, Email [email protected]
Abstract: Radiomics received a lot of attention because of its potential to provide personalized medicine in a non-invasive manner, usually focusing on the analysis of the entire lesion. A new method called habitat can identify subregional phenotypic changes within the lesion, thereby improving the ability to distinguish heterogeneity. The clustering method can be applied to multiple measurement parameters to separate different tumor habitats by segmentation. A data-driven repeatable voxel clustering method to identify subregions reflecting live tumors will be valuable for clinical diagnosis and further treatment. In this review, we aim to briefly summarize the widely used cluster analysis algorithms in subregion segmentation and the application of habitat analysis in tumor imaging. By analyzing many literatures, the commonly used K-means algorithm and other algorithms such as hierarchical clustering and consensus clustering are summarized. By identifying intratumoral heterogeneity, the key findings of habitat analysis in oncology are described, such as tumor differentiation, grading, and gene expression status. The latest progress and innovations in predicting tumor therapeutic effects and prognosis using habitat analysis are reviewed, including multimodal imaging data fusion, integration with artificial intelligence technologies, and non-invasive diagnostic methods. The limitations and challenges of habitat analysis in tumor imaging are also discussed, such as dependence on image quality and imaging techniques, insufficient automation and standardization, difficulties in biological interpretation, and lack of clinical validation. Finally, future directions for increasing the level of automation and standardization of habitat analysis to improve its accuracy and efficiency and reduce reliance on expert intervention are proposed. Habitat analysis represents a significant advancement in radiomics, offering a nuanced understanding of tumor heterogeneity. By leveraging sophisticated clustering algorithms and integrating multimodal imaging data, habitat analysis has the potential to transform clinical decision-making, enabling more precise diagnostics and personalized treatment strategies, ultimately advancing the field of precision medicine.
Keywords: habitat, cluster analysis, tumor imaging, K-means, radiomics
Introduction
Radiomics refers to the use of advanced imaging technologies and computational methods to extract and analyze a large number of quantitative features from medical images (such as CT, MRI, PET, etc), in order to reveal the heterogeneity and underlying biological information of the entire lesion (especially tumors), thereby providing support for disease diagnosis, prognosis assessment, and monitoring of treatment response. As the importance of tumor microenvironment and its impact on cancer progression and treatment response have been confirmed,1,2 an emerging method that can better quantify intra-tumor heterogeneity, namely habitat, has received increasing attention.3–5 Habitat analysis can classify tumors into subregions containing voxel clusters with similar characteristics, and personalize subregions.6,7 It usually involves a variety of imaging techniques, including magnetic resonance imaging (MRI), positron emission tomography (PET), computed tomography (CT), etc., and their combination use, such as PET/CT and MRI/CT.8–10 In recent years, habitat analysis has been widely used in the medical field, especially in the field of tumor imaging, and there have been a lot of research work on tumor identification, grading, expression, tumor prognosis assessment, therapeutic effect monitoring, and therapeutic resistance analysis.11,12 Jiaqi Li et al developed ITH score, which quantifies intra-tumor heterogeneity (ITH) of non-small cell lung cancer (NSCLC) by integrating local radiomic features and global pixel distribution patterns, which is helpful for tumor classification.13 Wu, Hao et al conducted a habitat analysis of PET/CT images to predict the Ki-67 status in high-grade serous ovarian cancer and to explore the stratification effect of this model on progression-free survival (PFS) in ovarian cancer patients. These studies demonstrate the application of habitat analysis in the study of tumor heterogeneity, where both the inter-tumor heterogeneity between different lesions and the intra-tumor heterogeneity within the same lesion are crucial for patient prognosis. This represents a significant challenge in the field of individualized and precise cancer treatment, providing important scientific evidence for the diagnosis and treatment of tumors. As a non-invasive analytical method, habitat analysis may provide an alternative to traditional biopsy to reduce the risk and discomfort of patients.14 In terms of new drug development and evaluation, habitat analysis can help to identify specific tumor regions where drugs act, accelerate the drug evaluation process, and show great application prospects.15 Integrate habitat analysis with genomic data to explore the genetic and molecular characteristics of tumors, providing a basis for personalized treatment. The most commonly used method for regional segmentation in habitat analysis is the K-means clustering algorithm.16,17 In addition, other algorithms such as Consensus Clustering,18 Hierarchical Clustering,19 Fuzzy-C-Means (FCM) clustering algorithm,20 Simple Linear Iterative Clustering (SLIC) algorithm,21 and Otsu thresholding method22 have also been applied in scientific research. Each method has its specific application scenarios and advantages, and researchers select the appropriate clustering technique based on the characteristics of the data and the research objectives. In the rapid development of habitat analysis research, there are also many challenges: the first is the standardization of technology; the second is model validation and generalization;23 in addition, the reproducibility and repeatability of radiomics features are the main challenges in the field, affecting the reliability and effectiveness of the models;24 the transformation of habitat analysis from research to clinical practice needs to overcome multiple barriers, including technical, ethical, and legal ones. In this brief review, our goal is to summarize the research progress, application prospects, and challenges of habitat analysis in the field of tumor imaging in recent years.
Habitat Imaging Model Algorithm
Subregion segmentation is an image analysis technique that reflects the internal heterogeneity of tumors by partitioning tumors into multiple subregions.25 Each subregion may have different biological characteristics and therapeutic responses.8 Subregion segmentation can be based on different image features (such as density, signal strength, texture, count or SUV, etc), which can be achieved by manual segmentation26,27 or by various algorithms (such as cluster analysis).28 In tumor habitat analysis, cluster analysis is used to group similar voxels together, which helps to identify subregions with similar biological characteristics within tumors and quantify significant regional changes in tissues after treatment.29 Ruili Wei et al used the K-means clustering algorithm to cluster four habitats (H1, H2, H3, and H4) to identify subregions with similar biological characteristics within tumors.30 In the study to evaluate the early response and prediction results of OPSCC in oropharyngeal squamous cell carcinoma, a robust consensus clustering method was used to divide the primary tumor and the involved lymph nodes into subregions based on 18F-FDG PET and comparative CT imaging.21 Tumor habitats may be characterized by different cell density, vascular distribution and metabolic activity. This research method can help to improve the description of tumor heterogeneity and provide a new perspective for the precise treatment of tumors. In this section, we illustrate some emerging applications of cluster analysis methods in tumor habitat analysis.
K-means Clustering Algorithm
In the field of habitat analysis, K-means clustering algorithm plays a key role. The general process includes initialization, that is, K data points are randomly selected as the initial clustering center; Each data point is assigned to the nearest cluster center to form K clusters; Calculate the average of all points in each cluster; Repeat the assignment and update steps until the cluster center no longer changes or a predetermined number of iterations is reached; Indicators such as contour coefficient and Calinski-Harabasz index were used to evaluate the clustering effect.31 K-means algorithm is simple and efficient, suitable for large-scale data sets, and provides an effective automated method for habitat analysis. It has been widely used in various medical image analysis, especially in tumor imaging and radiomics research, showing many characteristics and advantages.32 First, the K-means algorithm can automatically divide tumors into multiple subregions according to their image features (such as signal intensity, texture, etc). The imaging features of these subregions help to reveal the heterogeneity within tumors, which is very important for understanding the biological characteristics of tumors and predicting clinical outcomes. Second, K-means clustering can improve the performance of the model by extracting the features of different subregions of the tumor. Third, K-means clustering provides a more objective and repeatable way to define tumor subregions without relying on manual segmentation by subjective expert judgment; Fourth, K-means clustering can achieve multi-parameter and multi-sequence fusion, which helps to capture tumor features from different angles and enhance the generalization ability of the model. Fifth, K-means clustering can be compatible with advanced imaging techniques to extract deeper tumor features through combined use.33 At the same time, K-means clustering algorithm also faces many challenges: for very large data sets, it may require a lot of computing resources and time; It is sensitive to noise and outliers, which may affect the quality of clustering results. The interpretability of clustering results may be limited, especially if the clustering lacks a clear physical meaning.34 To sum up, K-means clustering algorithm has been widely used in many fields due to its high efficiency, and its advantages and characteristics make it a valuable tool in radiomics research. However, there are also some challenges, especially when dealing with complex data structures and high-dimensional data. Future research may focus on improving the algorithm’s stability, adaptability, and interpretability.
Otsu Threshold Method
Otsu method is a technique based on image threshold segmentation, which is used to automatically divide an image or data set into background and foreground. This technology determines the optimal threshold by maximizing inter-class variance or minimizing intra-class variance, so that the difference in grayscale values between the foreground and background after segmentation is maximized, while the grayscale differences within each part are minimized. Otsu threshold method is a dynamic threshold segmentation algorithm, it divides the image into two parts according to the gray level of the image, and finds a suitable gray level as the threshold by calculating the variance, and then divides the image. This method is simple, efficient and insensitive to parameter selection. It is especially suitable for threshold segmentation and simple binary classification in image processing. Many scholars used the Otsu sectionalized method to divide the region of interest (ROI) of glioma.35,36 In this way, the high and low expression clusters of each map are obtained. By defining each possible cluster combination as a distinct habitat, spatial habitat maps are then drawn in each patient’s image. However, threshold-based approaches have limitations.
Other Cluster Analysis Methods
In cluster analysis methods, in addition to K-means clustering algorithm, Beig, Niha et al. In this study, hierarchical clustering algorithm was used to perform cluster analysis on Differentially Expressing Genes (DEGs).37 Hierarchical clustering does not require the number of clusters to be specified in advance; it organizes the data by building a cluster hierarchy. It can be condensed (starting with a single data point and gradually merging into clusters) or split (starting with all data points as a cluster and gradually splitting into a single data point). This algorithm can provide a hierarchical view of the data; For large data sets, the computational complexity is high, and it is suitable for scenarios where small and medium-sized data sets and data hierarchy are required. Some studies have mentioned the use of consensus clustering to identify tumor subregions.38 Consensus clustering is a robust clustering method that determines the final clustering result by randomly resampling the data several times and performing the clustering, calculating the consistency between different clustering results. Consensus clustering is insensitive to initial conditions. Can identify the inherent cluster structure in the data; Due to the need to repeat the clustering process many times, the calculation cost is high, and it is suitable for scenarios requiring high stability and reliability of clustering results, such as gene expression data analysis. In addition, Parra, N Andres et al mentioned in their research the use of Fuzzy-C-Means clustering algorithm to process DCE-MRI data.20 Tabassum, Mehnaz et al mentioned NMF (Non-negative Matrix Factorization) as a clustering method in their research.39 Kazerouni, Anum et al mentioned the clustering algorithm for multi-parameter voxel data from DW-MRI and DCE-MRI.40 Wu, Jia et al mentioned the use of Simple Linear Iterative Clustering (SLIC) algorithm to conduct super pixel segmentation of CT scan and 18F-FDG PET imaging data.21 These clustering methods have their specific application scenarios, each has its own advantages and limitations, which method to choose depends on the specific data characteristics and analysis objectives.
Future Feasible Directions
In response to the limitations of the K-means clustering algorithm, future research should focus on improving the stability and adaptability of the algorithm. For instance, by introducing noise handling mechanisms and optimizing initialization methods, the algorithm’s capability to process complex data structures and high-dimensional data can be enhanced. Concurrently, exploring new clustering effectiveness evaluation indicators to enhance the interpretability of clustering results is essential. The integration of multimodal data from various imaging techniques (such as MRI, PET, CT, etc) is also a significant direction for future development. By developing more advanced fusion algorithms to capture tumor features from different perspectives, a more comprehensive revelation of the biological characteristics of tumors can be achieved. For example, leveraging deep learning technology to realize automatic feature extraction and fusion of multimodal data can improve the accuracy and generalization ability of models.
Habitat Analysis
Tumor heterogeneity refers to the differences in genotype and phenotype of cells within the tumor, which has an important impact on the prognosis and treatment response of the tumor. Tumor heterogeneity exists not only between different lesions (inter tumor heterogeneity), but also between different cells within the same lesion (intra tumor heterogeneity),41 which is a major challenge in the field of precision cancer therapy and brings great difficulties to the individualized treatment of tumors. We are concerned about the application of habitat analysis in tumor imaging.42 These applications demonstrate the potential of habitat analysis in oncology research and clinical practice, including tumor diagnosis, classification, prognostic assessment, treatment response monitoring, and the development of personalized treatment strategies. In scientific research, after completing subregional segmentation with habitat analysis, it is necessary to carry out feature extraction, selection, model establishment and evaluation. Radiomics and machine learning methods are often used to achieve these operations. Wang, Xinghao et al used LIFEx software and ITK-SNAP tool to extract features, Pearson correlation coefficient (PCC), principal component analysis (PCA), Lasso regression and other methods for feature selection, and used common classifiers. Including support vector machine (SVM), logistic regression (LAD), decision tree, random forest and naive Bayes algorithm to build the model. Models with high AUC values and strong generalization ability are selected from different models to be included in the final model selection. Kaplan-Meier survival curve and K-M test were used to evaluate the model’s ability to predict progression-free survival. Decision curve analysis (DCA) was performed to evaluate the classification and predictive power of different models, as well as the predictive power of patient outcomes. These steps develop and validate a PET/CT image-based radiomic model for predicting Ki-67 status in high-grade serous ovarian cancer (HGSOC) and explore the stratification effect of this model on progression-free survival (PFS) in ovarian cancer patients.43 In this mini-review, we summarize the key findings, challenges, and future directions of habitat analysis by analyzing multiple articles.
Evaluation of Tumor Heterogeneity, Classification of Tumor Subtypes and Tumor Staging
Habitat analysis can be used to assess intra-tumor heterogeneity, including features such as metabolism, perfusion, and tissue diffusion, to reveal the complexity and diversity of the tumor. Park, Ji Eun et al Electrical characteristic Tomography (EPT), diffusion-weighted imaging (DWI) and perfusion weighted imaging (PWI) were used to identify and analyze different microenvironments inside tumors, namely the so-called “tumor habitat”.34 Xie, Peiyi et al Explored the subregional histogram features of APTw MRI and compared them with those of diffusion-weighted imaging (DWI), proposed a new method to predict the tumor bud (TB) grade of rectal cancer (RC). This may have important implications for clinical decision-making and treatment planning.44 Xu, Run et al, used the K-means clustering algorithm to categorize voxels from dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) and diffusion-weighted imaging (DWI), and assigned them to three distinct spatial habitats defined based on perfusion and diffusion patterns. Studies have shown that the ability to distinguish triple-negative breast cancer from non-triple-negative breast cancer can be improved by analyzing the multi-parameter MRI habitat and fractal characteristics of tumors, providing potential imaging biomarkers for clinical diagnosis and treatment.45 Some studies have found that the Metabolic Tumor Volume (MTV) and Total Lesion Glycolysis (TLG) in FDG PET/CT imaging are closely related to the staging of lung cancer,46 laryngeal cancer,47 and ovarian cancer,48 thereby providing a more comprehensive basis for the assessment of treatment response and survival prognosis.
Tumor Prognosis Assessment, Treatment Response Monitoring and Treatment Resistance Analysis
Habitat analysis can also be used to predict progression-free survival (PFS) and overall survival (OS) of patients, providing quantitative indicators for clinical prognosis assessment. Mu, Wei et al Study to predict the prognosis of locally advanced cervical cancer patients receiving chemotherapy and radiotherapy. Habitat analysis is used to monitor tumor response to treatment, including early response assessment and post-treatment changes.49 Lee, Da Hyun et al Studied whether tumor habitat on structural and physiological magnetic resonance imaging (MRI) could distinguish between surviving tumors and radionecrosis in brain metastases after stereotactic radiation therapy (SRS), involving treatment response monitoring.29 Longitudinal physiological magnetic resonance imaging (MRI) was used to analyze tumor habitats of brain metastases to predict tumor recurrence after stereotactic radiation therapy (SRS), involving treatment response monitoring and treatment resistance analysis.50
Radiogenomics Research and Non-Invasive Diagnosis
Combined with habitat analysis and genomic data, genetic and molecular characteristics of the tumor were explored to provide basis for personalized treatment. Beig, Niha et al Studied the Radiomic Risk Score (RRS) based on radiomic characteristics. Used to predict Progression-Free Survival (PFS) of Glioblastoma (GBM) patients from the tumor microenvironment on conventional magnetic resonance imaging (MRI). The association between these prognostic radiomic features and molecular signaling pathways was explored.37 Zhang, Yunfei et al, studied habitat imaging to predict the recurrence risk of hepatocellular carcinoma (HCC) and its surrounding microenvironment, involving non-invasive diagnosis.51 Wu, Jingran et al, developed a radiomic nomogram combining deep learning, radiomics and clinical variables to predict EGFR mutation status in patients with stage I non-small cell lung cancer (NSCLC), which is a non-invasive diagnostic method.52 These involve radiometric genomics studies that use image data to identify tumor features associated with genetic information and explore the use of non-invasive diagnostic methods in tumor evaluation. A study53 developed a predictive model for the KRAS/NRAS/BRAF mutation status in colorectal cancer patients through habitat analysis, integrating radiological features and genomic data. By analyzing the metabolic activity and structural characteristics within the tumor, it is possible to identify tumor subregions with specific gene mutations, thereby providing a basis for personalized treatment.
The Emerging Role of Habitat Analysis in the Context of Artificial Intelligence and Machine Learning
With the rapid development of Artificial Intelligence (AI) and Machine Learning (ML) technologies, Habitat Analysis can be combined with them to achieve automated feature extraction: The study by Sachpekidis, Christos et al54 demonstrates how to use deep learning algorithms to automatically extract heterogeneous features within tumors. Through Convolutional Neural Networks (CNNs), researchers are able to extract high-dimensional features from multimodal imaging data (such as PET/CT and MRI), which can more accurately reflect the biological characteristics of tumors. Li, Hebei et al55 explored in their study how to use Generative Adversarial Networks (GANs) to generate high-quality synthetic imaging data for model training and validation. This method not only improves the generalization ability of the model but also reduces the need for large amounts of real data. In addition to this, it can also enhance the predictive power of the model. Liu, J et al56 and others have developed models for predicting tumor prognosis and treatment response by combining habitat analysis features with machine learning algorithms (such as random forests and support vector machines). These models, validated on multiple independent datasets, have shown high accuracy and reproducibility.
Challenges and Limitations of Habitat Analysis
Although habitat analysis has shown great potential in multiple studies, there are still some challenges and limitations in the field of oncology imaging. First, the accuracy of habitat analysis is highly dependent on image quality and imaging techniques. Different equipment and scanning parameters may lead to variation of habitat characteristics and affect the consistency and repeatability of analysis results. Second, the automation and standardization of habitat analysis needs to be improved. Currently, many habitat analysis methods require manual intervention by experts, which limits their widespread application in clinical practice. In addition, the biological interpretation of habitat analysis remains a challenge. The association between habitat features and tumor biology is not fully understood, and further studies are needed to elucidate the biological significance of these features. Finally, the clinical application of habitat analysis requires more prospective studies to verify its predictive performance and clinical value.57,58
Future Development Directions of Habitat Analysis
In the future, the development of habitat technology will be committed to integrating various imaging techniques (such as MRI, PET, CT, etc) to achieve multimodal data fusion, and combining clinical data (such as pathological information, genomic data, etc) to deeply explore the correlation between habitat features and tumor biology, thereby providing a more solid basis for personalized treatment. Transitioning habitat analysis technology from the research phase to clinical application is a key objective for the future, which requires large-scale, diversified clinical data validation to assess its practical effects in tumor diagnosis, treatment response monitoring, and prognosis evaluation. Meanwhile, to realize the widespread application of habitat analysis, it is essential to enhance its level of automation and standardization, develop standardized imaging protocols and analysis processes, reduce manual intervention, and improve the reproducibility and consistency of results.
Conclusion and Prospect
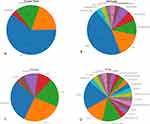
The application of habitat analysis in medical imaging has a broad prospect, which can not only provide the microstructure information of diseases, but also reveal the biological characteristics of diseases, and provide an important basis for the diagnosis, treatment and prognosis evaluation of diseases. Through the analysis of this review, we summarize the common methods used in habitat analysis, and see the key role of habitat analysis in improving diagnostic accuracy, optimizing treatment plans, and driving the development of precision medicine. The proportion of tumor types, image types, clustering methods, research purposes and other data in the literatures summarized are roughly summarized in Table 1 and Figure 1. The word cloud map showing the frequency of keywords in the table is shown in Figure 2. Future research should focus on addressing existing challenges and driving innovation and clinical application of habitat analysis techniques to enable more precise disease management and patient care.
 |
Figure 2 Keyword occurrence frequency word cloud map. |
 |
Table 1 Summary Table of Main Information of Literature |
Through the above summary, we recognize the important role of habitat analysis in medical imaging diagnosis and treatment. It provides a new perspective for the diagnosis, prognosis assessment, and monitoring of treatment responses of tumors by accurately identifying intratumoral heterogeneity. It can not only reveal the complexity and diversity of tumors but also combine multimodal imaging techniques and genomic data to provide a solid basis for the development of personalized treatment strategies. In the future, with the optimization of algorithms, integration of multimodal data, and the advancement of automation and standardization, habitat analysis is expected to play a greater role in clinical practice, promote the development of precision medicine, and bring more precise and effective treatment plans to patients.
Future research should aim to increase the level of automation and standardization of habitat analysis to reduce reliance on expert intervention and improve its applicability in different clinical Settings. Combining multimodal image data with artificial intelligence techniques such as deep learning is expected to further improve the accuracy and efficiency of habitat analysis.At the same time, the integration of habitat analysis with clinical data, such as pathological information and genomic data, will contribute to a more comprehensive understanding of the biology of diseases and promote the development of precision medicine. In addition, the application of habitat analysis in other disease areas, such as cardiovascular disease and inflammatory diseases, also deserves further exploration.
Funding
The study was supported by Science and technology project of Suzhou Health and Family Planning Commission /Suzhou science and technology plan project (No.KJXW2022077).
Disclosure
The authors have no conflicts of interest to declare in this work.
References
1. Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi:10.1016/j.ccr.2012.02.022
2. Nia HT, Munn LL, Jain RK. Physical traits of cancer. Science. 2020;370:eaaz0868. doi:10.1126/science.aaz0868
3. Park JE, Kim HS, Kim N, Park SY, Kim YH, Kim JH. Spatiotemporal heterogeneity in multiparametric physiologic MRI is associated with patient outcomes in IDH-wildtype glioblastoma. Clin Cancer Res. 2021;27:237–245. doi:10.1158/1078-0432.CCR-20-2156
4. Wu J, Cao G, Sun X, et al. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 2018;288:26–35. doi:10.1148/radiol.2018172462
5. Kim J, Ryu SY, Lee SH, Lee HY, Park H. Clustering approach to identify intratumour heterogeneity combining FDG PET and diffusion-weighted MRI in lung adenocarcinoma. Eur Radiol. 2019;29:468–475.
6. Gillies RJ, Balagurunathan Y. Perfusion MR imaging of breast cancer: insights using “habitat imaging”. Radiol Soc N Am. 2018;288(1):36–37.
7. Juan-Albarracín J, Fuster-Garcia E, Pérez-Girbés A, et al. Glioblastoma: vascular habitats detected at preoperative dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging predict survival. Radiology. 2018;287:944–954. doi:10.1148/radiol.2017170845
8. Jeong SY, Park JE, Kim N, et al. Hypovascular cellular tumor in primary central nervous system lymphoma is associated with treatment resistance: tumor habitat analysis using physiologic MRI. Am J Neuroradiol. 2022;43(1):40–47. doi:10.3174/ajnr.A7351
9. Lin F, Zhu L-X, Ye Z-M, et al. Computed tomography-based intratumor heterogeneity predicts response to immunotherapy plus chemotherapy in esophageal squamous cell carcinoma. Acad Radiol. 2024;31(12):4886–4899. doi:10.1016/j.acra.2024.06.032
10. Zhao H, Su Y, Wang Y, et al. Using tumor habitat-derived radiomic analysis during pretreatment 18F-FDG PET for predicting KRAS/NRAS/BRAF mutations in colorectal cancer. Cancer Imaging. 2024;24(1):26. doi:10.1186/s40644-024-00670-2
11. Prior O, Macarro C, Navarro V, et al. Identification of precise 3D CT radiomics for habitat computation by machine learning in cancer. Radiol. 2024;6(2):e230118. doi:10.1148/ryai.230118
12. Fang M, Kan Y, Dong D, et al. Multi-habitat based radiomics for the prediction of treatment response to concurrent chemotherapy and radiation therapy in locally advanced cervical cancer. Front Oncol. 2020;10:563. doi:10.3389/fonc.2020.00563
13. Li J, Qiu Z, Zhang C, et al. ITHscore: comprehensive quantification of intra-tumor heterogeneity in NSCLC by multi-scale radiomic features. Eur Radiol. 2023;33(2):893–903. doi:10.1007/s00330-022-09055-0
14. Wu H, Tong H, Du X, et al. Vascular habitat analysis based on dynamic susceptibility contrast perfusion MRI predicts IDH mutation status and prognosis in high-grade gliomas. Eur Radiol. 2020;30(6):3254–3265. doi:10.1007/s00330-020-06702-2
15. Jardim-Perassi BV, Mu W, Huang S, et al. Deep-learning and MR images to target hypoxic habitats with evofosfamide in preclinical models of sarcoma. Theranostics. 2021;11(11):5313–5329. doi:10.7150/thno.56595
16. Wang S, Liu X, Wu Y, et al. Habitat-based radiomics enhances the ability to predict lymphovascular space invasion in cervical cancer: a multi-center study. Front Oncol. 2023;13:1252074. doi:10.3389/fonc.2023.1252074
17. Liu D, Chen J, Hu X, et al. Imaging-genomics in glioblastoma: combining molecular and imaging signatures. Front Oncol. 2021;11:699265. doi:10.3389/fonc.2021.699265
18. Zhang W, Liang F, Zhao Y, et al. Multiparametric MR-based feature fusion radiomics combined with ADC maps-based tumor proliferative burden in distinguishing TNBC versus non-TNBC. Phys Med Biol. 2024;69(5). doi:10.1088/1361-6560/ad25c0
19. Choi SW, Cho -H-H, Koo H, et al. Multi-habitat radiomics unravels distinct phenotypic subtypes of glioblastoma with clinical and genomic significance. Cancers. 2020;12(7):1707. doi:10.3390/cancers12071707
20. Parra NA, Lu H, Li Q, et al. Predicting clinically significant prostate cancer using DCE-MRI habitat descriptors. Oncotarget. 2018;9(98):37125–37136. doi:10.18632/oncotarget.26437
21. Wu J, Gensheimer MF, Zhang N, et al. Tumor subregion evolution-based imaging features to assess early response and predict prognosis in oropharyngeal cancer. J Nucl Med. 2020;61(3):327–336. doi:10.2967/jnumed.119.230037
22. Bailo M, Pecco N, Callea M, et al. Decoding the heterogeneity of malignant gliomas by PET and MRI for spatial habitat analysis of hypoxia, perfusion, and diffusion imaging: a preliminary study. Front Neurosci. 2022;16:885291. doi:10.3389/fnins.2022.885291
23. Cho H-H, Kim H, Nam SY, et al. Measurement of perfusion heterogeneity within tumor habitats on magnetic resonance imaging and its association with prognosis in breast cancer patients. Cancers. 2022;14(8):1858. doi:10.3390/cancers14081858
24. Sagreiya H. Finding the pieces to treat the whole: using radiomics to identify tumor habitats. Radiol. 2024;6(2):e230547. doi:10.1148/ryai.230547
25. Chang Y-CC, Ackerstaff E, Tschudi Y, et al. Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep. 2017;7(1):9746. doi:10.1038/s41598-017-09932-5
26. Verma R, Hill VB, Statsevych V, et al. Stable and discriminatory radiomic features from the tumor and its habitat associated with progression-free survival in glioblastoma: a multi-institutional study. Am J Neuroradiol. 2022;43(8):1115–1123. doi:10.3174/ajnr.A7591
27. Verma R, Correa R, Hill VB, et al. Tumor habitat-derived radiomic features at pretreatment MRI that are prognostic for progression-free survival in glioblastoma are associated with key morphologic attributes at histopathologic examination: a feasibility study. Radiology. 2020;2(6):e190168. doi:10.1148/ryai.2020190168
28. Chen L, Liu K, Zhao X, et al. Habitat imaging-based 18F-FDG PET/CT radiomics for the preoperative discrimination of non-small cell lung cancer and benign inflammatory diseases. Front Oncol. 2021;11:759897. doi:10.3389/fonc.2021.759897
29. Lee DH, Park JE, Kim N, et al. Tumor habitat analysis by magnetic resonance imaging distinguishes tumor progression from radiation necrosis in brain metastases after stereotactic radiosurgery. Eur Radiol. 2022;32(1):497–507. doi:10.1007/s00330-021-08204-1
30. Wei R, Lu S, Lai S, et al. A subregion-based RadioFusionOmics model discriminates between grade 4 astrocytoma and glioblastoma on multisequence MRI. J Cancer Res Clin Oncol. 2024;150(2):73. doi:10.1007/s00432-023-05603-3
31. Huang H, Chen H, Zheng D, et al. Habitat-based radiomics analysis for evaluating immediate response in colorectal cancer lung metastases treated by radiofrequency ablation. Cancer Imaging. 2024;24(1):44. doi:10.1186/s40644-024-00692-w
32. Weinfurtner RJ, Abdalah M, Stringfield O, et al. Quantitative changes in intratumoral habitats on MRI correlate with pathologic response in early-stage ER/PR+ HER2- breast cancer treated with preoperative stereotactic ablative body radiotherapy. J Breast Imaging. 2022;4(3):273–284. doi:10.1093/jbi/wbac013
33. Bernatowicz K, Grussu F, Ligero M, et al. Robust imaging habitat computation using voxel-wise radiomics features. Sci Rep. 2021;11(1):20133. doi:10.1038/s41598-021-99701-2
34. Park JE, Kim HS, Kim N, et al. Low conductivity on electrical properties tomography demonstrates unique tumor habitats indicating progression in glioblastoma. Eur Radiol. 2021;31(9):6655–6665. doi:10.1007/s00330-021-07976-w
35. Lee J, Narang S, Martinez J, et al. Spatial habitat features derived from multiparametric magnetic resonance imaging data are associated with molecular subtype and 12-month survival status in glioblastoma multiforme. PLoS One. 2015;10(9):e0136557. doi:10.1371/journal.pone.0136557
36. Stringfield O, Arrington JA, Johnston SK, et al. Multiparameter MRI predictors of long-term survival in glioblastoma multiforme. Tomography. 2019;5(1):135–144. doi:10.18383/j.tom.2018.00052
37. Beig N, Bera K, Prasanna P, et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin Cancer Res. 2020;26(8):1866–1876. doi:10.1158/1078-0432.CCR-19-2556
38. Sala E, Mema E, Himoto Y, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72(1):3–10. doi:10.1016/j.crad.2016.09.013
39. Tabassum M, Suman AA, Suero Molina E, et al. Radiomics and machine learning in brain tumors and their habitat: a systematic review. Cancers. 2023;15(15):3845. doi:10.3390/cancers15153845
40. Kazerouni AS, Hormuth DA, Davis T, et al. Quantifying tumor heterogeneity via MRI habitats to characterize microenvironmental alterations in HER2+ breast cancer. Cancers. 2022;14(7):1837. doi:10.3390/cancers14071837
41. Yu S, Yang Y, Wang Z, et al. CT-based conventional radiomics and quantification of intratumoral heterogeneity for predicting benign and malignant renal lesions. Cancer Imaging. 2024;24(1):130. doi:10.1186/s40644-024-00775-8
42. Su G-H, Xiao Y, You C, et al. Radiogenomic-based multiomic analysis reveals imaging intratumor heterogeneity phenotypes and therapeutic targets. Sci Adv. 2023;9(40):eadf0837. doi:10.1126/sciadv.adf0837
43. Wang X, Xu C, Grzegorzek M, et al. Habitat radiomics analysis of pet/ct imaging in high-grade serous ovarian cancer: application to Ki-67 status and progression-free survival. Front Physiol. 2022;13(948767). doi:10.3389/fphys.2022.948767
44. Xie P, Huang Q, Zheng L, et al. Sub-region based histogram analysis of amide proton transfer-weighted MRI for predicting tumor budding grade in rectal adenocarcinoma: a prospective study. Eur Radiol. 2024;35:1382–1393. doi:10.1007/s00330-024-11172-x
45. Xu R, Yu D, Luo P, et al. Do habitat MRI and fractal analysis help distinguish triple-negative breast cancer from non-triple-negative breast carcinoma. Canad Associat Radiolog J. 2024;75(3):584–592. doi:10.1177/08465371241231573
46. Huang Y, Jiang X, Xu H, et al. Preoperative prediction of mediastinal lymph node metastasis in non-small cell lung cancer based on 18F-FDG PET/CT radiomics. Clin Radiol. 2023;78(1):8–17. doi:10.1016/j.crad.2022.08.140
47. Al-Ibraheem A, Abdlkadir AS, Shagera QA, et al. The diagnostic and predictive value of 18F-fluorodeoxyglucose positron emission tomography/computed tomography in laryngeal squamous cell carcinoma. Cancers. 2023;15(22):5461. doi:10.3390/cancers15225461
48. Yusufaly TI, Zou J, Nelson TJ, et al. Improved prognosis of treatment failure in cervical cancer with nontumor PET/CT radiomics. J Nucl Med. 2022;63(7):1087–1093. doi:10.2967/jnumed.121.262618
49. Mu W, Liang Y, Hall LO, et al. 18 F-FDG PET/CT habitat radiomics predicts outcome of patients with cervical cancer treated with chemoradiotherapy. Radiol. 2020;2(6):e190218. doi:10.1148/ryai.2020190218
50. Lee DH, Park JE, Kim N, et al. Tumor habitat analysis using longitudinal physiological MRI to predict tumor recurrence after stereotactic radiosurgery for brain metastasis. Korean J Radiol. 2023;24(3):235–246. doi:10.3348/kjr.2022.0492
51. Zhang Y, Yang C, Sheng R, et al. Predicting the recurrence of hepatocellular carcinoma (≤ 5 cm) after resection surgery with promising risk factors: habitat fraction of tumor and its peritumoral micro-environment. La Radiologia medica. 2023;128(10):1181–1191. doi:10.1007/s11547-023-01695-6
52. Wu J, Meng H, Zhou L, et al. Habitat radiomics and deep learning fusion nomogram to predict EGFR mutation status in stage I non-small cell lung cancer: a multicenter study. Sci Rep. 2024;14(1):15877. doi:10.1038/s41598-024-66751-1
53. Chen S-W, Lin C-Y, Ho C-M, et al. Genetic alterations in colorectal cancer have different patterns on 18F-FDG PET/CT. Clin Nucl Med. 2015;40(8):621–626. doi:10.1097/RLU.0000000000000830
54. Sachpekidis C, Enqvist O, Ulén J, et al. Application of an artificial intelligence-based tool in [18F]FDG PET/CT for the assessment of bone marrow involvement in multiple myeloma. Eur J Nucl Med Mol Imaging. 2023;50(12):3697–3708. doi:10.1007/s00259-023-06339-5
55. Li H, Xu C, Xin B, et al. 18 F-FDG PET/CT radiomic analysis with machine learning for identifying bone marrow involvement in the patients with suspected relapsed acute leukemia. Theranostics. 2019;9(16):4730–4739. doi:10.7150/thno.33841
56. Liu J, Tu J, Yao L, et al. MRI-based radiomics virtual biopsy for BCL6 in primary central nervous system lymphoma. Clin Radiol. 2025;80:106746. doi:10.1016/j.crad.2024.106746
57. Mar Álvarez-Torres M, Juan‐Albarracín J, Fuster‐Garcia E, et al. Robust association between vascular habitats and patient prognosis in glioblastoma: an international multicenter study. J Magn Reson Imaging. 2020;51(5):1478–1486. doi:10.1002/jmri.26958
58. Caii W, Wu X, Guo K, Chen Y, Shi Y, Chen J Integration of deep learning and habitat radiomics for predicting the response to immunotherapy in NSCLC patients. Cancer Immunol Immunother. 2024;73(8):153. PMID: 38833187; PMCID: PMC11150226. doi:10.1007/s00262-024-03724-3
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.