Back to Journals » Journal of Inflammation Research » Volume 17
Novel Insights from Comprehensive Bioinformatics Analysis Utilizing Large-Scale Human Transcriptomes and Experimental Validation: The Role of Autophagy in Periodontitis
Authors Liu F , Zhu Z, Zou H, Huang Z, Xiao S, Li Z
Received 1 October 2024
Accepted for publication 21 December 2024
Published 30 December 2024 Volume 2024:17 Pages 11861—11880
DOI https://doi.org/10.2147/JIR.S492048
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Fen Liu,1 Zhipeng Zhu,1 Huaxi Zou,2 Zhen Huang,1 Shengkai Xiao,1 Zhihua Li1
1School of Stomatology, Jiangxi Medical College, Nanchang University, Jiangxi Provincial Key Laboratory of Oral Diseases, Jiangxi Provincial Clinical Research Center for Oral Disease, Nanchang, Jiangxi, People’s Republic of China; 2Department of Cardiovascular Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang, Jiangxi, People’s Republic of China
Correspondence: Zhihua Li, School of Stomatology, Jiangxi Medical College, Nanchang University, Jiangxi Provincial Key Laboratory of Oral Diseases, Jiangxi Provincial Clinical Research Center for Oral Disease, No. 49 FuZhou Road, Nanchang, Jiangxi, People’s Republic of China, Email [email protected]
Objective: Autophagy plays a crucial role in the pathophysiology of periodontitis, yet its precise involvement in the disease process remains elusive. The aim of the present study was thus to investigate the involvement of autophagy in the pathology of periodontitis. This investigation involved transcriptomic analysis of a broad range of human samples and complemented by in vitro experimentation.
Materials and Methods: We analyzed the transcriptomes of human gingival tissues from individuals with periodontitis and health controls to identify the differential expression of autophagy-related genes (DEARGs) and to investigate their potential interactions and functional pathways. Additionally, protein-protein interaction (PPI) networks were constructed to identify key functional modules and hub genes. Experimental validation of autophagy regulation in periodontitis and identification of key autophagy-regulating genes was accomplished through in vitro cellular experiments. Subsequently, a comprehensive analysis of immune cell infiltrate utilizing the CIBERSORT algorithm was performed. Finally, leveraging the DSigDB database, potential candidate drugs for periodontitis treatment targeting autophagy were predicted.
Results: A total of 79 genes have been identified as DEARGs in periodontitis. An intricate interplay among the DEARGs and their impact on the regulatory mechanisms of autophagy within the context of periodontitis was observed. Subsequently, 10 hub genes were discerned through the establishment of a PPI network. Furthermore, dysregulated autophagic activity in periodontitis was validated, and 9 key genes (APP, KDR, IL1B, CXCL12, CXCR4, IL6, FOS, LCK, and SHC1) were identified through in vitro experiments. Our analysis unveiled an association between these genes and altered immune cell infiltration in periodontitis. Additionally, we predicted potential therapeutic agents such as curcumin, 27-hydroxycholesterol, and Trolox, showing promise in the treatment of periodontitis by modulating the autophagic process.
Conclusion: This study identified nine key genes for autophagy regulation and potential therapeutic agents in periodontitis. These findings not only enhance our comprehension of the pathological mechanisms of periodontitis but also provide substantial evidence for the advancement of novel therapeutic strategies.
Keywords: periodontitis, transcriptome, autophagy, key genes, diagnosis, database
Introduction
Periodontitis, a prevalent chronic oral inflammatory disease, is characterized by the pathological loss of tooth-supporting tissues like the gingiva and alveolar bone.1 Its progression can lead to compromised mastication and tooth loss.2 Additionally, numerous studies have indicated periodontitis as a relevant risk factor for a range of non-oral health conditions, encompassing cardiovascular disease, diabetes, and kidney injury, highlighting its systemic health implications.1 Nonetheless, the etiology of periodontitis remains enigmatic, with current diagnostic practices predominantly relying on clinical and radiographic assessments. These methods offer limited therapeutic direction, contributing to treatment delays or excessive medication in a considerable proportion of cases.3–5 Advances in molecular biology have paved the way for tailored and precise management of periodontitis, and the associated precise diagnostic modalities and targeted therapies may play a crucial role in future periodontal care.4,6,7 Therefore, a thorough investigation into the molecular biology aspects of periodontitis pathogenesis is imperative for comprehensive understanding and effective intervention.
Autophagy, a highly conserved and meticulously regulated physiological process, serves as an important regulatory mechanism for maintaining cellular homeostasis and organismal balance.8 In general, autophagy clears detrimental cytosolic components via lysosomal degradation pathways, safeguarding against cellular harm and fostering survival. Conversely, the breakdown of autophagy compromises this protective function.9–11 Although autophagy has been intensively studied, excessive activation of autophagy, as a distinct form of autophagic dysregulation, participates in the advancement of numerous diseases.12 Excessive autophagic activation results in the degradation of crucial cellular components and the dysfunction of organelles, which may ultimately lead to autophagic cell death.11,13–15
Autophagy-related genes (ARGs) are pivotal in governing the autophagic pathway.16,17 The significant regulatory influence of ARGs in inflammatory conditions has been substantiated,18,19 A dual role of autophagy in periodontitis has been observed.20–22 The upregulation of LC3B, serving as a marker for autophagy, has been observed in human periodontitis gingival tissue and experimental models, with contributory factors like dental plaque instigating atypical autophagy in diverse gingival cells, thereby promoting periodontitis development.23–25 Conversely, autophagy may counter apoptosis-induced cell death, preserving cell viability in periodontal diseases, and inadequate autophagic responses could heighten inflammation and susceptibility to periodontitis.26,27 Considering the crucial yet debated implications of autophagy in periodontitis, further investigations are warranted to elucidate the exact functional role of autophagy.28,29
The aim of this study is to examine the function and pathological mechanisms of autophagy in periodontitis using transcriptomic data from a large cohort of human samples and in vitro experiments.
Materials and Methods
Ethics Approval and Consent to Participate
The study protocol for this study was approved by the Ethics Committee of The Affiliated Stomatological Hospital, Jiangxi Medical College, Nanchang University (Approval No. 2022016).
The Collection and Processing of Data
In this study, we downloaded the GSE16134 microarray dataset from the NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/). The study involved transcriptomic analysis of gingival tissues from 120 patients diagnosed with moderate to severe periodontitis. The ID conversion process is based on the platform file provided by the original researchers in the GSE16134 dataset. This involves transforming the original IDs directly into gene symbols. In cases where multiple IDs correspond to the same gene symbol, a strategy of taking the median of the expression levels associated with these IDs is adopted to ensure the accuracy and representativeness of the data.
Identifying Differentially Expressed Genes (DEGs) and Differentially Expressed ARGs (DEARGs)
Utilizing the limma package (version 3.48.0) in the R software (version 4.1), we identified DEGs in the gingiva of periodontitis patients. The moderated t-test from the limma package was employed for statistical assessment, with the results subsequently adjusted using the Benjamini-Hochberg method. Prior to analysis, the data underwent preprocessing via quantile normalisation. The boxplots before and after standardization are presented in Figure S1. Subsequently, DEGs meeting the criteria of |log2FC| ≥ 0.5 and the false discovery rate (FDR) < 0.05 were selected.30,31 Autophagy-related genes (ARGs) were identified utilizing resources such as the Autophagy Database (http://www.tanpaku.org/autophagy/index.html),32 the Human Autophagy Database (http://www.autophagy.lu/index.html),33 and the Human Autophagy Modulator Database (http://hamdb.scbdd.com).34 After removing duplicate genes, a merged gene set comprising 1174 ARGs was established, as shown in Table S1. For the extraction of DEARGs from the DEGs, the Venn diagram package in the R software was employed.
Gene Set Enrichment Analysis (GSEA)
We conducted GSEA utilizing the GSEA software (version 3.0.0) (http://software.broadinstitute.org/gsea/index.jsp), based on phenotypic data, to explore the potential link between ARGs and human gingiva in periodontitis.35 A gene set comprising 1174 ARGs was employed as previously described. Genes from the GSE16134 dataset were quantitatively assessed and ranked according to their respective expression levels, enabling the calculation of an enrichment score (ES). Significance in GSEA was defined by an FDR value of less than 0.05.
Comprehensive Enrichment Analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
The DEARGs were subjected to GO and KEGG enrichment analyses utilizing the ClusterProfiler package (version 3.12.0) in R software. GO (Gene Ontology) functional analysis encompasses three domains: Cellular Component (CC), Molecular Function (MF), and Biological Process (BP). Enrichment results with an FDR below 0.05 were considered statistically significant.
The Construction and Analysis of Protein-Protein Interaction (PPI) Networks
The PPI network of DEARGs was established and visualized utilizing the STRING database (https://string-db.org/) and the Cytoscape software (version 3.8.2).36,37 To identify the functional key module, we employed the Cytoscape plugin MCODE (degree cutoff = 2, node score cutoff = 0.2, K-core = 2, and max depth = 100). Using Cytohubba’s built-in MCC algorithm, hub genes were determined by evaluating each gene within the PPI network and ranking them based on their values. These 10 top-ranked genes were recognized as hub genes.
Culture of Primary Human Gingival Fibroblasts (HGFs)
HGFs were purchased from Wuhan Procell Life Science and Technology Co., Ltd. (Wuhan, China). The cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (H-DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% fresh fetal bovine serum (FBS) (Hyclone, Logan, UT, USA) and 1% penicillin-streptomycin (Solaibao, Beijing, China). The cell culture process occurred in an incubator, where the cells were exposed to 5% CO2, 21% O2, and a temperature of 37°C. Subsequent experiments utilized cells derived from the 4th to 6th generation.
We established an in vitro periodontitis model by exposing HGFs to lipopolysaccharide (LPS) (Sigma-Aldrich, St. Louis, USA). Inhibition of autophagy was achieved through the utilization of 3-methyladenine (3-MA)(MedChemExpress, NJ, USA).24 In the LPS-induced periodontitis model, HGFs were subjected to a 24-hour treatment with 10 μg/mL LPS. Prior to LPS treatment, cells in the LPS + 3-MA group were pre-treated with 3-MA (5 mm) for an additional 24 h. The control group received only the respective solvent.
Cytotoxicity Assays
To assess cytotoxicity, the lactate dehydrogenase (LDH) activity was quantitatively measured employing a specific LDH assay kit (Beyotime, Shanghai, China). HGFs were first seeded into a 96-well plate following different treatments. Subsequently, the supernatant was incubated with the active solution of the LDH assay for 30 min at a temperature of 37°C. The absorbance was then quantified at 490 nm utilizing the Spark R microplate reader. Finally, cytotoxicity was determined using the formula provided in the assay instructions. Cytotoxicity or Mortality (%) = (Absorbance of Treated Sample - Absorbance of Sample Control Well) / (Absorbance of Maximum Cellular Enzyme Activity - Absorbance of Sample Control Well) × 100.
Quantitative Real-Time PCR (qRT-PCR)
After treating the HGFs according to the aforementioned protocol, total RNA extraction was carried out. Subsequently, the RNA concentration was accurately determined, and reverse transcription into cDNA was performed employing the AidMM reverse transcription kit (ThermoFisher Scientific, Massachusetts, USA). qRT-PCR analysis was executed utilizing SYBR Premix EX Taq (Takara, Japan) on the StepOnePlus PCR System (Applied Biosystems, CA, USA). Normalization of the mRNA expression levels was conducted using ACTB as a reference, with calculations performed according to the 2−ΔΔCT method. The sequences of the primers used for the detection of the expression patterns of the genes are detailed in Table S2.
Western Blot
Cells were subjected to lysis using RIPA lysis buffer (Beyotime, Shanghai, China), supplemented with 1% phenylmethylsulfonyl fluoride (PMSF). The lysates were then centrifuged for 15 min. Following centrifugation, the supernatant was divided into two portions. One portion was utilized for protein quantification using a BCA kit, while the other part was prepared for further analysis by mixing with loading buffer and boiling for denaturation.
After being separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were migrated onto a polyvinylidene fluoride (PVDF) membrane for subsequent analysis. The membranes were blocked utilizing 5% skim milk powder for 2 h, followed by an overnight incubation at 4°C with primary antibodies specific to LC3 (Proteintech, 14600-1-AP, Wuhan, China) and β-actin (Biosharp, Hefei, China). Following three washes with Tris-buffered saline containing Tween (TBST), the membranes were incubated for 2 h with the appropriate secondary antibodies (Beyotime, Shanghai, China).
Following additional TBST washes, the blots were developed for detection using an electrochemical luminescence (ECL) kit (Beyotime, Shanghai, China).
Immunofluorescence Analysis
The HGFs were cultured in 24-well plates and treated as described previously. Following treatment, the cells were immobilized by fixing with a 4% paraformaldehyde solution for 10 min. After fixation, the cells underwent a 30-minute blocking with 5% bovine serum albumin (BSA) in PBS at 37°C, followed by an overnight incubation at 4°C with an anti-LC3 primary antibody (Proteintech, 14600-1-AP, Wuhan, China). In the dark, the cells were incubated with a secondary antibody (Beyotime, China) for 1 h, followed by staining of their nuclei with 4ʹ,6-diamidino-2-phenylindole (DAPI) for 10 min under the same conditions. Images were captured employing a fluorescence microscope (Nikon, Tokyo, Japan), with appropriate excitation wavelengths.
Imaging by Transmission Electron Microscopy (TEM)
After centrifugation and culture medium removal, the cells were fixed with 2.5% glutaraldehyde at 4°C. Following the initial fixation, the samples were further fixed with 1% osmic acid at room temperature for 2 hours. The samples then underwent a dehydration process in a series of alcohol solutions, followed by embedding.
The resulting resin blocks were sectioned into ultra-thin slices of 60–80 nanometers in thickness using an ultra-microtome, and these slices were picked up on 150-mesh formvar-coated copper grids. Subsequently, the copper grid-mounted slices were stained with 2% uranyl acetate and 2.6% lead citrate. Finally, autophagosomes and autolysosomes were observed and imaged using a transmission electron microscope (Hitachi 7800, Japan).
Immune Infiltration Analysis
Employing the CIBERSORT algorithm (Version released in 2015), we quantified the proportions of 22 distinct immune cell types present in the sample, thereby enabling a comprehensive analysis.38
Subsequently, the correlation between hub genes and differentially infiltrating immune cells (DIICs) was analyzed employing a linear regression analysis. The data utilized for analysis are identical to the standardized data employed for differential analysis.
Predicting the Potential of Therapeutic Drugs
Interaction data between proteins and drugs were obtained from the DSigDB database (Version released in 2015) (http://dsigdb.tanlab.org/) to predict potential drugs capable of modulating autophagy for the treatment of periodontitis.39 The prediction process involved applying specific criteria, including a stringent cutoff of FDR < 0.001 and a combined score exceeding 1000. A lower FDR value indicated a more robust correlation between the drugs and their target proteins. Identification of the chemical structures of the predicted drugs was achieved through the use of the Drugbank (http://www.drugbank.ca) and PubChem databases (http://pubchem.ncbi.nlm.nih.gov).
Statistical Analysis
The GraphPad Prism software (version 8.0.2) (La Jolla, CA, USA) was employed to carry out the statistical analyses. Group comparisons were conducted utilizing t-tests or signed rank tests, based on the data characteristics. In cases involving multiple groups, differences were assessed using an ordinary one-way analysis of variance (ANOVA). Following the ANOVA results indicating statistical significance, subsequent post-hoc tests were performed to identify specific group differences. Spearman correlation test was utilized for correlation analyses to evaluate relationships between variables. Data are presented as mean values with standard deviation (SD).
Results
Comprehensive Protocol of This Study
Figure 1 illustrates the comprehensive design of the study. Transcriptome data from a total of 310 human gingival samples were included in our study, comprising 241 samples from periodontitis gingival tissues and 69 from healthy controls. Prior to subsequent analysis, all raw expression data underwent normalization following the methodology described above.
 |
Figure 1 Schematic representation of the overall study protocol. |
Identification of DEG and DEARGs
GSEA is a classic and widely used method for assessing the overall correlation between a set of signature genes and the disease transcriptome. GSEA was conducted using dataset GSE16134 to investigate the variation in ARGs within the human periodontitis transcriptome. The results, depicted in Figure 2A, demonstrated a substantial positive correlation of the ARGs set in periodontitis gingiva compared to healthy gingiva (Normalized Enrichment Score (NES) = 1.468, FDR = 0.027). This finding underscores the widespread dysregulation of ARGs as a key characteristic in the altered transcriptome of human periodontitis gingiva. The data derived from the human gingival transcriptome establish a significant association between autophagy and periodontitis.
Our transcriptomic analysis identified 1279 DEGs (782 upregulated, 497 downregulated) in periodontitis samples compared to controls (Figure 2B and C). Notably, 79 ARGs overlapped with these DEGs, signifying their importance in periodontitis (Figure 3A–C and Table S3).
GO and KEGG Analysis of DEARGs
We investigated the functions and associated pathways of DEARGs through GO and KEGG enrichment analyses. These DEARGs are implicated in regulating crucial biological processes (BP), including lymphocyte differentiation, T cell activation, peptidyl-tyrosine phosphorylation, peptidyl-tyrosine modification, positive regulation of cell adhesion, T cell differentiation, and neutrophil homeostasis (Figure 4A). Considering the established connections between autophagy and these specific BPs in a variety of biological contexts,40,41 it is conceivable that autophagy may initiate or influence these processes in the context of periodontitis, indicating a potential interplay between autophagy dysregulation and BP modulation in the pathophysiology of periodontitis. Regarding their cellular components (CCs), the DEARGs were predominantly localized in the membrane region, endocytic vesicle, and membrane raft (Figure 4B). With regard to their molecular functions (MFs), the DEARGs demonstrated enrichment in protein serine/threonine kinase activity, cytokine receptor binding, and growth factor receptor binding. (Figure 4C). The KEGG pathway analysis highlighted the involvement of DEARGs in autophagy, the chemokine signaling pathway, and rheumatoid arthritis. (Figure 4D).
Previous studies have unveiled reciprocal crosstalk between these pathways or functions and the modulation of autophagy in diverse disease processes, with some being recognized as regulators of autophagy in the context of periodontitis.23 Consequently, our subsequent analysis delved into exploring the interplay between these genes and various pathways or functions. The results indicate the presence of intricate interactions between DEARGs and associated functions or pathways, with a significant impact on the biological function of autophagy and, consequently, on the pathological process in periodontitis (Figure 4E–H). This intricate network of interactions emphasizes the multifaceted involvement of autophagy dysregulation in the pathophysiology of periodontitis, suggesting the potential therapeutic targets that could be harnessed to modulate autophagy and ameliorate the disease process.
The Generation and Analysis of a PPI Network
We have used the construction of a PPI network to elucidate the complex relationship between DEARGs and functions or pathways. Utilizing the STRING database, we were able to establish a PPI network and identified functional modules and hub genes through the Cytoscape software (Figure 5A). By applying the MCODE plugin, we revealed one key gene module (Figure 5B). Subsequently, employing the MCC algorithm within the Cytohubba plugin, we ranked the top 10 genes (CXCR4, CD38, FOS, CXCL12, IL1B, KDR, IL6, LCK, APP, SHC1) as hub genes (Figure 5C). The distinct expression patterns of these hub genes in periodontitis gingival are visually represented in Figure 5D, illustrating their differential expression within this pathological framework. Figure 5E shows interactions among hub genes, while Figure 5F demonstrates the large number of connections between hub genes and other DEARGs, shedding light on the complex interplay within the regulatory network associated with autophagy dysregulation in periodontitis.
Validation by in vitro Experiments
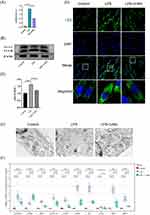
To study the autophagy levels and expression of hub genes in periodontitis, an in vitro periodontitis model using LPS-treated HGFs was implemented. Figure 6A demonstrates a significant increase in cytotoxicity in the LPS-induced periodontitis model, while the 3-MA autophagy inhibition group showed a decrease in cytotoxicity. As shown in Figure 6B and C, LPS treatment led to a substantial elevation in the LC3-II/LC3-I ratio, a pivotal functional marker of autophagic flux, while treatment with 3-MA alleviated this ratio. Furthermore, immunofluorescence analysis showed an augmentation of LC3-positive puncta in the LPS-induced periodontitis model compared to the control group. These autophagic characteristics were markedly suppressed by 3-MA. Notably, Transmission Electron Microscopy (TEM) analysis further confirmed these findings (Figure 6D and E).
The results from both the in vitro experiments and transcriptome analysis of human samples demonstrated dysregulation of autophagy levels in periodontitis. Subsequent studies were conducted to assess the level of expression of the hub genes in vitro experiments. The results of the qRT-PCR experiments showed that there was a significant increase in the expression of these hub genes after treatment with LPS. This trend was notably mitigated by 3-MA pretreatment, except for CD38, indicating that they are involved in autophagy regulation in periodontitis. Therefore, it was determined that the nine genes (CXCR4, FOS, CXCL12, IL1B, KDR, IL6, APP, LCK, and SHC1) from the original ten hub genes were the key genes (Figure 6F).
The results of this study further confirm the extensive presence of autophagic dysregulation in periodontitis (Figure 6B–E), indicating that autophagy may impact the pathophysiological processes of periodontitis by regulating the functions of these key genes (Figure 6F).
Immune Infiltration Analysis
The immune response, which serves as a vital defense mechanism for the body against external pathogens and maintains homeostasis, plays a complex and central role in the pathogenesis of diseases such as periodontitis, and its dynamic fluctuations are closely linked to disease conditions.42,43 Therefore, this study aimed to investigate the interactions between ARGs and immune responses in periodontitis via an immune infiltration analysis.
Through the application of the CIBERSORT algorithm, we quantitatively assessed and subsequently visualized the percentage of DIICs within gingival tissue samples sourced from the GSE16134 (Figure 7A). Clustering heatmaps were generated to illustrate the differences in the percentage of DIICs between the periodontitis gingival group and the healthy gingival group sourced from the GSE16134 dataset (Figure 7B). Furthermore, the correlation heatmap in Figure 7C depicts the relationships among DIICs. Linear regression analysis revealed a noteworthy correlation between hub genes with DIICs in periodontitis (Figure 7D).
In comparison to healthy controls, our analysis revealed a notable rise in the proportions of plasma cells, gamma delta T cells (γδ T cells), activated memory CD4+T cells, and neutrophils were significantly increased in periodontitis tissues. Conversely, periodontitis tissues exhibited a pronounced decline in the proportions of follicular helper T cells (Tfh), resting dendritic cells, CD8+T cells, macrophages M1, and resting mast cells (Figure 7E). Therefore, our results suggest that these autophagy hub genes not only play a key regulatory role in autophagy but also modulate immune cell infiltration.
Predicting Potential Autophagy-Regulating Drugs
The prediction of targeted drugs based on protein-drug interactions may provide direction for the treatment of periodontitis. We performed a predictive drug target analysis using the DSigDB database to identify the top 10 autophagy-regulating drugs associated with key genes, sorted by FDR value. These drugs are deemed particularly promising targets for the treatment of periodontitis through autophagy mediation (Figure 8A). The findings indicated that curcumin (Figure 8B) exhibited the strongest drug-target correlation, followed by 27-hydroxycholesterol (Figure 8C) and Trolox (Figure 8D). A comprehensive depiction of the three aforementioned drugs is presented in Figure 8e. These drugs could potentially play a protective role in periodontitis through autophagy mediation, yet more in-depth studies are needed to verify this.
In this study, we used bioinformatics analysis and preliminary in vitro experimental validation to reveal the prevalence of autophagy dysregulation in gingival tissue from patients with periodontitis and successfully screened nine key autophagy-related genes. Further studies showed that changes in the expression of these genes were closely associated with changes in immune cell infiltration. On this basis, we screened drugs, such as curcumin, that can act on these key genes. These findings suggest that dysregulation of autophagy plays an important role in the pathogenesis of periodontitis and provides potential targets and drug candidates for the development of novel therapeutic strategies against periodontitis in the future.
Discussion
Periodontitis, a chronic inflammatory disease of the oral cavity caused by plaque microorganisms, is the primary etiological factor contributing to tooth loss and chewing dysfunction in adults. This condition arises from the prolonged destruction of periodontal supporting tissues.44,45 As periodontitis progresses, patients may encounter a significant decline in their quality of life due to associated comorbidities such as dyspepsia and facial aging.46 The exact etiology and pathogenesis of periodontitis remain incompletely understood. Previous research suggests that periodontitis may be linked to various pathogenic factors including immunity, inflammation, and environment.47 Recently, autophagy in periodontitis pathogenesis has garnered significant attention, with multiple studies emphasizing its importance. Nevertheless, the exact molecular mechanisms remain elusive, highlighting the need for more intensive research to elucidate them.23
The results from GSEA in this study highlighted significant dysregulation of ARGs in periodontitis gingival tissues, suggesting a crucial role for autophagy in the pathogenesis of periodontitis.23,27 Furthermore, experimental investigations using an LPS-induced periodontitis cell model revealed that LPS, a key bacterial component in periodontitis, can induce autophagic responses in HGFs, leading to heightened autophagy levels. Inhibition of autophagy with 3-MA reduced cytotoxicity and inflammatory responses triggered by LPS, indicating a potential pathological implication of autophagy in periodontitis development.23,24 Additionally, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses unveiled that differentially expressed autophagy-related genes (DEARGs) are enriched in processes associated to lymphocyte differentiation, T-cell activation and differentiation, and autophagy. This suggests that dysregulated autophagy might impact immune cell functions, potentially leading to an imbalance in immune responses.22 Such imbalance could manifest as an exaggerated inflammatory response or immunosuppression, exacerbating the damage to periodontal tissues.
We performed a comprehensive bioinformatics analysis using transcriptomic data from a substantial number of human samples, which was further validated through in vitro experiments to explore the contribution of autophagy to periodontitis progression. The GSEA results revealed widespread abnormal expression of ARGs in periodontitis gingival tissues, suggesting that autophagy may represent a potential mechanistic pathway in the pathogenesis of periodontitis.23,27 Furthermore, GO enrichment analysis was employed to delve deeper into the BP, CC localization, and MF of autophagy-related genes in periodontitis. Accumulating evidence from studies underscores the participation of autophagy in the regulation of T-cell activation and differentiation,48 cell adhesion,49 and neutrophil homeostasis.50 Conversely, T-cell activation and differentiation, as well as cell adhesion can also impact autophagy regulation.51,52 The autophagy process within mesenchymal stem cells may regulate the migration and differentiation of CD4+ T cell cells via the modulation of CXCL8 and TGF-β1 signaling pathways.48 The differentiation of Th1/Th2 regulated by IRF2-INPP4B promotes autophagy while diminishing apoptosis.52 It has been demonstrated that autophagy exerts a modulating influence on the processes of macrophage adhesion and migration in diabetic nephropathy.49 Cell adhesion can hinder autophagy through Src/FAK-mediated phosphorylation and AMPK reduction.51 Our research reveals a significant association between autophagy and protein serine/threonine kinase activity, cytokines, growth factors, heat shock proteins, and misfolded proteins.
Additionally, previous studies have indicated a link between their interactions and various diseases.53–57 Indeed, serine/threonine kinases like AMPK, mTOR, and ULK1 play crucial roles in autophagy regulation.58–61 This mechanism holds the potential to serve as a pathologic pathway in periodontitis, warranting further investigation. Studies have shown that the inhibition of the Akt signaling pathway can effectively block the positive regulation of mTOR activation by Porphyromonas gingivalis (P. gingivalis). This disruption results in the downregulation of mTOR and its downstream target ULK1 phosphorylation, ultimately leading to the activation of ULK1 and the initiation of autophagy.62 Furthermore, in a high-glucose environment, metformin was found to effectively attenuate periodontal tissue damage in db/db mice. This beneficial effect was attributed to the activation of the AMPK/SIRT1/autophagy signaling pathway mechanism.63 Moreover, the chemokine signaling pathway can impact the disease progression by modulating autophagy,64,65 emphasizing the importance of elucidating its regulatory mechanism in periodontitis. Therefore, it is essential to concentrate on unraveling the interactions between these molecules and pathways to comprehensively understand the molecular biological mechanisms governing autophagy regulation in periodontitis.
The construction and subsequent analysis of the PPI network enabled the identification of ten hub autophagy genes associated with periodontitis. Furthermore, our in vitro experimentation corroborated the findings of dysregulated autophagic activity in periodontitis and identified nine key genes (APP, KDR, IL1B, CXCL12, CXCR4, IL6, FOS, LCK, SHC1) from the ten hub genes.
Although autophagy-associated genes have been recognized as crucial regulators of pathological mechanisms in numerous diseases, their specific role in modulating autophagy within the context of periodontitis has been seldom reported. Current research on periodontitis indicates that autophagy may influence the production of inflammatory factors, including IL-6 and IL-1b.23,66,67 Nevertheless, the exact molecular mechanism governing the interaction of IL-6 and IL-1b with autophagy remains unclear, highlighting an intriguing avenue for further exploration. Bacterial virulence factors such as LPS and leukotoxin have been shown to attack oral epithelial cells, triggering the release of pro-inflammatory factors interleukin-1 alpha (IL-1α) and interleukin-1 beta (IL-1β), which play a crucial role in periodontal destruction.68 Autophagy acts to eliminate pathogens and inhibit the production of inflammatory factors, thereby mitigating inflammatory responses.69 It is conceivable that IL1β may intensify inflammation in periodontal tissues by influencing the activity of the autophagy pathway. Previous studies have demonstrated that APP expression levels are significantly higher in the gingival tissues of individuals with periodontitis compared to healthy controls.70 It is established that autophagy processes regulate the intracellular translocation and processing of APP.71 We hypothesize that aberrant autophagy could impact the transport and processing of APP during the pathological process of periodontitis, resulting in the accumulation of aberrant proteins in periodontal tissue cells and the exacerbation of inflammation and damage. Therefore, monitoring the expression levels of APP and autophagy-related marker genes could offer valuable insights into assessing the conditions and prognosis of periodontitis. Furthermore, periodontal lesions have been l associated with pathological angiogenesis, with the VEGF/VEGFR-2 (KDR) axis identified as a potential factor influencing the pathological progression of periodontal disease and angiogenesis.72 Autophagy also plays a regulatory role in the formation of pathological blood vessels.73 Therefore, the interaction between KDR and autophagy may represent a promising target for anti-angiogenic therapy in periodontitis. Furthermore, the interaction between CXCL12 and its receptor CXCR4 has been associated with immune cell migration in periodontitis.74 Autophagy contributes to the immune response within periodontal tissues, influencing the activity and function of immune cells.22 Therefore, the interplay between the CXCL12/CXCR4 axis and autophagy could represent a potential target for immunomodulatory therapy in periodontitis.
This study is the first to reveal an association between CXCL12, KDR, APP, LCK, and SHC1 with the autophagy pathway in periodontitis, relationships that have not been previously documented. This discovery potentially sets the stage for further investigations into the intricate molecular mechanisms underlying periodontitis. Furthermore, while IL1β and IL6 have been implicated in autophagy in periodontitis, these associations lacked substantiation from high-quality evidence obtained from clinical samples. Our study provides the initial conclusive evidence at the transcriptomic level of the correlation of these genes with autophagy by analyzing a substantial number of clinical samples. Through in-depth investigation of the functions of these autophagy-related genes and their mechanisms of action in periodontitis, there is potential for the development of novel therapeutic strategies aimed at alleviating inflammatory responses and tissue damage, thereby improving patients’ oral health status.
The development of periodontitis is characterized by intricate immune and inflammatory responses, with immune dysregulation playing a critical role in its pathogenesis.75 The interplay between autophagy and immunity and inflammation is notably complex, as autophagy can modulate immune and inflammatory responses, while immune and inflammatory signals can reciprocally influence autophagy induction and inhibition.76 Therefore, the interplay between autophagy and immune cell infiltration in periodontitis warrants attention and further investigations. Our current analysis reveals elevated proportions of plasma cells, T cells gamma delta, Neutrophils, and activated memory CD4+ T cells in periodontitis groups compared to that of healthy controls, whereas the percentages of Tfh cells, CD8+ T cells, resting mast cells, macrophages M1, and resting dendritic cells were decreased. Some of these findings align with previous studies. In comparison to healthy human gingiva tissues, inflamed gingival tissues from individuals with periodontitis exhibited a significant elevation in the number of CD4+ T cells, B cells, and plasma cells.77 Additionally, a separate study reported a notable elevation in neutrophil infiltration levels within periodontitis gingival tissues.78
Autophagy, a pivotal regulatory mechanism of cellular metabolism, indeed shows pervasive dysregulation in the gingival tissues during periodontitis. Our research findings indicate an increased proportion of activated memory CD4+ T cells, plasma cells, and neutrophils in periodontitis — cells closely associated with autophagy. Autophagy exerts a profound influence on the biological functions of CD4+ T cells, impacting their proliferation, differentiation, metabolism, and survival, underscoring its crucial role in the regulation of autophagy of periodontal inflammation progression.79 Neutrophils play a pivotal role in defending against pathogen invasion via autophagy mechanisms, emphasizing the significance of autophagy in immune defense.22 Furthermore, plasma cells, as specialized antibody-secreting cells, require autophagy regulation during their differentiation process. Autophagy is involved in maintaining endoplasmic reticulum homeostasis and modulating the equilibrium between plasma cell differentiation and antibody production, indicating its essential role in plasma cell function and adaptive immune responses.80 In summary, autophagy is intricately intertwined with the functions of CD4+ T cells, plasma cells, and neutrophils, collectively governing the immunopathological processes of periodontitis.
Conversely, the infiltration ratio of mast cells, dendritic, and macrophages was decreased in inflamed gingival tissues.81 Our study, utilizing transcriptome analyses, demonstrated a notable reduction in M1 macrophages in periodontitis. In contrast, a previous investigation reported a significant increase in M1 macrophages within the periodontitis microenvironment in human gingival tissues.82 These conflicting findings underscore that the pathogenesis of periodontitis encompasses more than just a spectrum of pathogenic microorganisms and is intricately intertwined with complex immune response mechanisms. As such, as the disease progresses, the quantity and functionality of macrophages are likely subjected to dynamic shifts and alterations.83 Porphyromonas gingivalis (P. gingivalis) has been found to regulate apoptotic and/or autophagic processes within dendritic cells (DCs), contributing to their survival mechanisms and disrupting immune homeostasis.62 Therefore, it is suggested that the restoration of immune homeostasis in patients with periodontitis should involve boosting antibacterial autophagy and apoptosis. This could help prevent peripheral inflammation and augment bacterial clearance.62
Our investigation revealed a significant correlation between key DEARGs and infiltrating immune cells in periodontitis. It is recognized that certain immune cells may possess dual pro-inflammatory and anti-inflammatory functions. Moreover, as periodontitis advances, the composition of the immune infiltration undergoes dynamic changes. Therefore, a comprehensive study of the close and complex relationship between immune infiltration and autophagy-related genes in periodontitis, as well as the specific regulatory mechanisms, is important for the exploration of novel therapeutic strategies for periodontitis.
In light of the pivotal roles played by key genes in periodontitis and their associated mechanisms, numerous researchers are actively investigating effective therapeutic interventions. In this study, we proposed Curcumin as a potential protective agent against periodontitis by modulating autophagy. Curcumin, a natural compound derived from the rhizomes of plants within the Zingiberaceae and Araceae families,84 exhibits diverse biological functions, such as anti-tumor, anti-inflammatory, and analgesic properties.85 Previous studies have demonstrated that Curcumin can effectively alleviate and treat periodontitis through various pathways. It has been shown that Curcumin mitigates periodontal damage by inhibiting ferroptosis.86 Furthermore, curcumin has been demonstrated to have a suppressing impact on the expression of key virulence genes in Porphyromonas gingivalis, resulting in a reduction in its virulence and an improvement in periodontitis.87 Notably, Curcumin’s therapeutic potential extends to diverse conditions such as tumors, inflammation, and heart disease through the regulation of autophagy.88–90
Previous studies have demonstrated that curcumin possesses the ability to inhibit the growth of periodontal pathogens and their biofilm formation, and can alleviate inflammatory responses in periodontal tissues by inhibiting the activation of the MAPK (mitogen-activated protein kinase) inflammatory signaling pathway.91 Recent research suggests that curcumin exerts its protective effects by modulating autophagy processes through the regulation of signaling pathways such as AMPK, MAPK, and mTOR.92,93 In this study, through GO enrichment analysis, we identified a notable association between DEARGS and protein serine/threonine kinase activity. Based on this finding, we propose a hypothesis that curcumin may influence autophagy processes by regulating signaling pathways such as MAPK, AMPK, and mTOR, which are related to protein serine/threonine kinase activity, thereby potentially alleviating inflammation in periodontal tissues. Nevertheless, this hypothesis necessitates further in-depth research for validation. With its ability to regulate autophagy, curcumin presents itself as a promising and innovative therapeutic strategy for addressing periodontitis.
As a transcriptomic analysis of a limited number of human samples, accompanied by preliminary in vitro experimental validation, our study is subject to certain limitations. Firstly, the limitations in the source of transcriptomic samples and sample size may affect the depth of analysis of specific disease stages and complex clinical phenotypes, which in turn reduces the comprehensiveness and accuracy of the findings. It is thought that larger multicentre clinical studies will provide more comprehensive conclusions. Secondly, the environment of in vitro experiments is unable to fully replicate the complex environment present in animals or humans. Consequently, in vitro experiments can only provide initial validation of bioinformatics findings. Further, in vivo experiments and studies with human samples can provide a higher level of evidence for the study, which is the key to further clarifying and translating our findings in the future. It is also important to acknowledge that the various algorithms employed in the bioinformatics analyses of this study may exhibit certain systematic biases. For instance, the use of the CIBERSORT algorithm to determine the proportion of immune cells in human gingival tissues and the prediction of potential therapeutic drugs based on drug-protein relationships are potential sources of bias that, while currently deemed acceptable in the majority of studies, should not be overlooked. Consequently, further in-depth studies are imperative to elucidate the findings.
Conclusion
The present study revealed a general dysregulation of autophagy in periodontitis and identified nine key genes for autophagy regulation (APP, KDR, IL1B, CXCL12, CXCR4, IL6, FOS, LCK, and SHC1) through bioinformatics analysis and in vitro experimental validation. Furthermore, our study identified a correlation between these genes and altered immune cell infiltration in periodontitis. Additionally, we predicted potential therapeutic agents, including curcumin, 27-hydroxycholesterol, and Trolox, which exhibit promise in the treatment of periodontitis by mediating autophagy. Our study provides evidence and novel perspectives to unravel the pathological mechanisms of autophagy regulation in the pathogenesis of periodontitis and to explore new strategies for periodontitis diagnosis and treatment. The conclusions drawn from this study were based on biological information analysis conducted on a relatively limited number of transcriptomes of human gingival tissues, supported by initial in vitro experiments. It is acknowledged that there are several limitations regarding the generalizability and accuracy of these findings, highlighting the critical need for additional validation and refinement through broader and more comprehensive experimental research in the future.
Abbreviations
DEG, differentially expressed genes; ARG, autophagy-related gene; DEARG, differentially expressed ARG; GSEA, Gene set enrichment analysis; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; DIIC, differentially infiltrating immune cell; LPS, lipopolysaccharide.
Author Contributions
All authors contributed to all aspects of this study, from its conception and experimental design to the collection of data, bioinformatics analysis, and in vitro experimental validation. Furthermore, they were actively involved in the writing, revision, and review of the article, ensuring the accuracy and rigor of the research findings. All authors reached a consensus on the submission of the article and assumed full responsibility for the research work.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82360195), the Science and Technology Plan Project of the Health Commission of Jiangxi Province (No. 202210673 and No.202410036), and Jiangxi Province Chinese Medicine Science and Technology Program (No. 2021A385).
Disclosure
The authors declare no competing interests with regard to the paper.
References
1. Deng Y, Xiao J, Ma L, et al. Mitochondrial dysfunction in periodontitis and associated systemic diseases: implications for pathomechanisms and therapeutic strategies. Int J Mol Sci. 2024;25(2):1024. doi:10.3390/ijms25021024
2. Agnese CCD, Schöffer C, Kantorski KZ, Zanatta FB, Susin C, Antoniazzi RP. Periodontitis and oral health-related quality of life: a systematic review and meta-analysis. J Clin Periodontol. 2024. doi:10.1111/jcpe.14074
3. Slots J. Periodontitis: facts, fallacies and the future. Periodontol. 2017;75(1):7–23. doi:10.1111/prd.12221
4. Bartold PM. Lifestyle and periodontitis: the emergence of personalized periodontics. Periodontol. 2018;78(1):7–11. doi:10.1111/prd.12237
5. Rakic M, Pejcic N, Perunovic N, Vojvodic D. A roadmap towards precision periodontics. Medicina. 2021;57(3). doi:10.3390/medicina57030233
6. Jain P, Hassan N, Khatoon K, et al. Periodontitis and systemic disorder-an overview of relation and novel treatment modalities. Pharmaceutics. 2021;13(8):1175. doi:10.3390/pharmaceutics13081175
7. Tonetti MS, Greenwell H, Kornman KS. Staging and grading of periodontitis: framework and proposal of a new classification and case definition. J Periodontol. 2018;89(Suppl 1):S159–s172. doi:10.1002/JPER.18-0006
8. Shang JN, Yu CG, Li R, et al. The nonautophagic functions of autophagy-related proteins. Autophagy. 2024;20(4):720–734. doi:10.1080/15548627.2023.2254664
9. Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. 2020;383(16):1564–1576. doi:10.1056/NEJMra2022774
10. Kim KH, Lee MS. Autophagy--A key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. doi:10.1038/nrendo.2014.35
11. Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J. 2021;40(19):e108863. doi:10.15252/embj.2021108863
12. Martinet W, Agostinis P, Vanhoecke B, Dewaele M, De Meyer GR. Autophagy in disease: a double-edged sword with therapeutic potential. Clin Sci. 2009;116(9):697–712. doi:10.1042/CS20080508
13. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi:10.1126/science.1099993
14. Nah J, Fernández ÁF, Kitsis RN, Levine B, Sadoshima J. Does autophagy mediate cardiac myocyte death during stress? Circ Res. 2016;119(8):893–895. doi:10.1161/CIRCRESAHA.116.309765
15. Nah J, Zablocki D, Sadoshima J. Autosis: a new target to prevent cell death. JACC. 2020;5(8):857–869. doi:10.1016/j.jacbts.2020.04.014
16. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi:10.1016/j.cell.2018.09.048
17. Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22(3):367–376. doi:10.1038/cdd.2014.143
18. Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity. 2021;54(3):437–453. doi:10.1016/j.immuni.2021.01.018
19. Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and Inflammation. Ann Rev Immunol. 2018;36:73–101. doi:10.1146/annurev-immunol-042617-053253
20. Zhuang H, Ali K, Ardu S, Tredwin C, Hu B. Autophagy in dental tissues: a double-edged sword. Cell Death Dis. 2016;7(4):e2192. doi:10.1038/cddis.2016.103
21. Jiang M, Li Z, Zhu G. The role of autophagy in the pathogenesis of periodontal disease. Oral Dis. 2020;26(2):259–269. doi:10.1111/odi.13045
22. Yang Y, Huang Y, Li W. Autophagy and its significance in periodontal disease. J Periodontal Res. 2021;56(1):18–26. doi:10.1111/jre.12810
23. Kim WJ, Park SY, Kim OS, Park HS, Jung JY. Autophagy upregulates inflammatory cytokines in gingival tissue of patients with periodontitis and lipopolysaccharide-stimulated human gingival fibroblasts. J Periodontol. 2022;93(3):380–391. doi:10.1002/JPER.21-0178
24. He S, Zhou Q, Luo B, Chen B, Li L, Yan F. Chloroquine and 3-methyladenine attenuates periodontal inflammation and bone loss in experimental periodontitis. Inflammation. 2020;43(1):220–230. doi:10.1007/s10753-019-01111-0
25. Ebersole JL, Kirakodu SS, Neumann E, Orraca L, Gonzalez Martinez J, Gonzalez OA. Oral microbiome and gingival tissue apoptosis and autophagy transcriptomics. Front Immunol. 2020;11:585414. doi:10.3389/fimmu.2020.585414
26. Ebersole JL, Kirakodu S, Novak MJ, et al. Gingival tissue autophagy pathway gene expression profiles in periodontitis and aging. J Periodontal Res. 2021;56(1):34–45. doi:10.1111/jre.12789
27. Tan YQ, Zhang J, Zhou G. Autophagy and its implication in human oral diseases. Autophagy. 2017;13(2):225–236. doi:10.1080/15548627.2016.1234563
28. Song B, Zhou T, Yang WL, Liu J, Shao LQ. Programmed cell death in periodontitis: recent advances and future perspectives. Oral Dis. 2017;23(5):609–619. doi:10.1111/odi.12574
29. Greabu M, Giampieri F, Imre MM, et al. Autophagy, one of the main steps in periodontitis pathogenesis and evolution. Molecules. 2020;25(18):4338. doi:10.3390/molecules25184338
30. Zou HX, Qiu BQ, Zhang ZY, et al. Dysregulated autophagy-related genes in septic cardiomyopathy: comprehensive bioinformatics analysis based on the human transcriptomes and experimental validation. Front Cardiovasc Med. 2022;9:923066. doi:10.3389/fcvm.2022.923066
31. Yi C, Liu J, Deng W, et al. Macrophage elastase (MMP12) critically contributes to the development of subretinal fibrosis. J Neuroinflammation. 2022;19(1):78. doi:10.1186/s12974-022-02433-x
32. Homma K, Suzuki K, Sugawara H. The autophagy database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011;39(Database issue):D986–990. doi:10.1093/nar/gkq995
33. Moussay E, Kaoma T, Baginska J, et al. The acquisition of resistance to TNFα in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7(7):760–770. doi:10.4161/auto.7.7.15454
34. Wang NN, Dong J, Zhang L, et al. HAMdb: a database of human autophagy modulators with specific pathway and disease information. J Cheminf. 2018;10(1):34. doi:10.1186/s13321-018-0289-4
35. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
36. Zou HX, Hu T, Zhao JY, et al. Exploring dysregulated ferroptosis-related genes in septic myocardial injury based on human heart transcriptomes: evidence and new insights. J Inflamm Res. 2023;16:995–1015. doi:10.2147/JIR.S400107
37. Liao X, Ruan X, Wu X, Deng Z, Qin S, Jiang H. Identification of Timm13 protein translocase of the mitochondrial inner membrane as a potential mediator of liver fibrosis based on bioinformatics and experimental verification. J Transl Med. 2023;21(1):188. doi:10.1186/s12967-023-04037-2
38. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
39. Yoo M, Shin J, Kim J, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31(18):3069–3071. doi:10.1093/bioinformatics/btv313
40. Paul S, Schaefer BC. Selective autophagy regulates T cell activation. Autophagy. 2012;8(11):1690–1692. doi:10.4161/auto.21581
41. Vlahakis A, Debnath J. The interconnections between autophagy and integrin-mediated cell adhesion. J Mol Biol. 2017;429(4):515–530. doi:10.1016/j.jmb.2016.11.027
42. Pan W, Wang Q, Chen Q. The cytokine network involved in the host immune response to periodontitis. Int J Oral Sci. 2019;11(3):30. doi:10.1038/s41368-019-0064-z
43. Liu J, Dan R, Zhou X, Xiang J, Wang J, Liu J. Immune senescence and periodontitis: from mechanism to therapy. J Leukoc Biol. 2022;112(5):1025–1040. doi:10.1002/JLB.3MR0822-645RR
44. Wei W, Ren J, Yin W, et al. Inhibition of Ctsk modulates periodontitis with arthritis via downregulation of TLR9 and autophagy. Cell Proliferation. 2020;53(1):e12722. doi:10.1111/cpr.12722
45. Chapple IL. Time to take periodontitis seriously. BMJ. 2014;348:g2645.
46. Weng Y, Wang H, Li L, Feng Y, Xu S, Wang Z. Trem2 mediated Syk-dependent ROS amplification is essential for osteoclastogenesis in periodontitis microenvironment. Redox Biol. 2021;40:101849. doi:10.1016/j.redox.2020.101849
47. Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. doi:10.1038/nrdp.2017.38
48. Cen S, Wang P, Xie Z, et al. Autophagy enhances mesenchymal stem cell-mediated CD4(+) T cell migration and differentiation through CXCL8 and TGF-β1. Stem Cell Res Ther. 2019;10(1):265. doi:10.1186/s13287-019-1380-0
49. Jiang Y, Zhao Y, Zhu X, et al. Effects of autophagy on macrophage adhesion and migration in diabetic nephropathy. Renal Failure. 2019;41(1):682–690. doi:10.1080/0886022X.2019.1632209
50. Huang Z, Zhang H, Fu X, et al. Autophagy-driven neutrophil extracellular traps: the Dawn of sepsis. Pathol Res Pract. 2022;234:153896. doi:10.1016/j.prp.2022.153896
51. Zhao M, Finlay D, Kwong E, et al. Cell adhesion suppresses autophagy via Src/FAK-mediated phosphorylation and inhibition of AMPK. Cell Signal. 2022;89:110170. doi:10.1016/j.cellsig.2021.110170
52. Zhang F, Zhu K, Liu L, et al. IRF2-INPP4B axis inhibits apoptosis of acute myeloid leukaemia cells via regulating T helper 1/2 cell differentiation. Cell Biochem Funct. 2020;38(5):582–590. doi:10.1002/cbf.3511
53. Hu F, Song D, Yan Y, et al. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat Commun. 2021;12(1):3651. doi:10.1038/s41467-021-23923-1
54. Feng B, Amponsah AE, Guo R, et al. Autophagy-mediated inflammatory cytokine secretion in sporadic ALS patient iPSC-derived astrocytes. Oxid Med Cell Longev. 2022;2022:6483582. doi:10.1155/2022/6483582
55. Xu Y, Propson NE, Du S, Xiong W, Zheng H. Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. Proc Natl Acad Sci U S A. 2021;118(27). doi:10.1073/pnas.2023418118
56. Shan R, Liu N, Yan Y, Liu B. Apoptosis, autophagy and atherosclerosis: relationships and the role of Hsp27. Pharmacol Res. 2021;166:105169. doi:10.1016/j.phrs.2020.105169
57. Cybulsky AV. Endoplasmic reticulum stress, the unfolded protein response and autophagy in kidney diseases. Nat Rev Nephrol. 2017;13(11):681–696. doi:10.1038/nrneph.2017.129
58. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19(2):121–135. doi:10.1038/nrm.2017.95
59. Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125(1):25–32. doi:10.1172/JCI73939
60. Yang Y, Zhu Y, Zhou S, et al. TRIM27 cooperates with STK38L to inhibit ULK1-mediated autophagy and promote tumorigenesis. EMBO J. 2022;41(14):e109777. doi:10.15252/embj.2021109777
61. Li Y, Chen Y. AMPK and autophagy. Adv Exp Med Biol. 2019;1206:85–108.
62. Meghil MM, Tawfik OK, Elashiry M, et al. Disruption of immune homeostasis in human dendritic cells via regulation of autophagy and apoptosis by porphyromonas gingivalis. Front Immunol. 2019;10:2286. doi:10.3389/fimmu.2019.02286
63. Ye X, Wang Y, Tian Y, et al. Metformin alleviates junctional epithelium senescence via the AMPK/SIRT1/autophagy pathway in periodontitis induced by hyperglycemia. Heliyon. 2024;10(6):e27478. doi:10.1016/j.heliyon.2024.e27478
64. Festa BP, Siddiqi FH, Jimenez-Sanchez M, et al. Microglial-to-neuronal CCR5 signaling regulates autophagy in neurodegeneration. Neuron. 2023;111(13):2021–2037.e2012. doi:10.1016/j.neuron.2023.04.006
65. Hu X, Mei S, Meng W, et al. CXCR4-mediated signaling regulates autophagy and influences acute myeloid leukemia cell survival and drug resistance. Cancer Lett. 2018;425:1–12. doi:10.1016/j.canlet.2018.03.024
66. Tang H, Ye Y, Li L, et al. A20 alleviated caspase-1-mediated pyroptosis and inflammation stimulated by Porphyromonas gingivalis lipopolysaccharide and nicotine through autophagy enhancement. Human Cell. 2022;35(3):803–816. doi:10.1007/s13577-022-00678-5
67. Du L, Li Y, Liu W. Maresin 1 regulates autophagy and inflammation in human periodontal ligament cells through glycogen synthase kinase-3β/β-catenin pathway under inflammatory conditions. Arch Oral Biol. 2018;87:242–247. doi:10.1016/j.archoralbio.2017.12.023
68. Papathanasiou E, Conti P, Carinci F, Lauritano D, Theoharides TC. IL-1 superfamily members and periodontal diseases. J Dent Res. 2020;99(13):1425–1434. doi:10.1177/0022034520945209
69. Wang K, Yang C, Tao B, Guo S, Wang H. Editorial: epigenetic regulation of autophagy in inflammatory diseases. Front Immunol. 2024;15:1387459. doi:10.3389/fimmu.2024.1387459
70. Nezu A, Kubota T, Maruyama S, et al. Expression of neprilysin in periodontitis-affected gingival tissues. Arch Oral Biol. 2017;79:35–41. doi:10.1016/j.archoralbio.2017.03.003
71. Mayer J, Boeck D, Werner M, et al. Inhibition of autophagy alters intracellular transport of APP resulting in increased APP processing. Traffic. 2024;25(4):e12934. doi:10.1111/tra.12934
72. Vladau M, Cimpean AM, Balica RA, Jitariu AA, Popovici RA, Raica M. VEGF/VEGFR2 axis in periodontal disease progression and angiogenesis: basic approach for a new therapeutic strategy. Vivo. 2016;30(1):53–60.
73. Schaaf MB, Houbaert D, Meçe O, Agostinis P. Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ. 2019;26(4):665–679. doi:10.1038/s41418-019-0287-8
74. Kim AR, Bak EJ, Yoo YJ. Distribution of neutrophil and monocyte/macrophage populations induced by the CXCR4 inhibitor AMD3100 in blood and periodontal tissue early after periodontitis induction. J Periodontal Res. 2022;57(2):332–340. doi:10.1111/jre.12963
75. Cekici A, Kantarci A, Hasturk H, Van Dyke TE. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol. 2014;64(1):57–80. doi:10.1111/prd.12002
76. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi:10.1038/nature09782
77. Kim YC, Ko Y, Hong SD, et al. Presence of Porphyromonas gingivalis and plasma cell dominance in gingival tissues with periodontitis. Oral Dis. 2010;16(4):375–381. doi:10.1111/j.1601-0825.2009.01649.x
78. Zhang Z, Yuan W, Deng J, et al. Granulocyte colony stimulating factor (G-CSF) regulates neutrophils infiltration and periodontal tissue destruction in an experimental periodontitis. Mol Immunol. 2020;117:110–121. doi:10.1016/j.molimm.2019.11.003
79. Jeong J, Choi YJ, Lee HK. The role of autophagy in the function of CD4(+) T cells and the development of chronic inflammatory diseases. Front Pharmacol. 2022;13:860146. doi:10.3389/fphar.2022.860146
80. Cui B, Lin H, Yu J, Yu J, Hu Z. Autophagy and the immune response. Adv Exp Med Biol. 2019;1206:595–634.
81. Li W, Zhang Z, Wang ZM. Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC Oral Health. 2020;20(1):293. doi:10.1186/s12903-020-01287-0
82. Zhang L, Nie F, Zhao J, et al. PGRN is involved in macrophage M2 polarization regulation through TNFR2 in periodontitis. J Transl Med. 2024;22(1):407. doi:10.1186/s12967-024-05214-7
83. Sun X, Gao J, Meng X, Lu X, Zhang L, Chen R. Polarized macrophages in periodontitis: characteristics, function, and molecular signaling. Front Immunol. 2021;12:763334. doi:10.3389/fimmu.2021.763334
84. Jiang Y, Zong Y, Du Y, et al. Curcumin inhibits the pruritus in mice through mast cell MrgprB2 receptor. Inflamm Res. 2023;72(5):933–945. doi:10.1007/s00011-023-01724-0
85. Shakeri A, Cicero AFG, Panahi Y, Mohajeri M, Sahebkar A. Curcumin: a naturally occurring autophagy modulator. J Cell Physiol. 2019;234(5):5643–5654. doi:10.1002/jcp.27404
86. Wang Y, Lin H, Huang W, et al. Curcumin attenuates periodontal injury via inhibiting ferroptosis of ligature-induced periodontitis in mice. Int J Mol Sci. 2023;24(12):9835.
87. Kumbar VM, Peram MR, Kugaji MS, et al. Effect of curcumin on growth, biofilm formation and virulence factor gene expression of Porphyromonas gingivalis. Odontology. 2021;109(1):18–28. doi:10.1007/s10266-020-00514-y
88. Gong X, Jiang L, Li W, Liang Q, Li Z. Curcumin induces apoptosis and autophagy inhuman renal cell carcinoma cells via Akt/mTOR suppression. Bioengineered. 2021;12(1):5017–5027. doi:10.1080/21655979.2021.1960765
89. Wu X, Liu Z, Yu XY, Xu S, Luo J. Autophagy and cardiac diseases: therapeutic potential of natural products. Med Res Rev. 2021;41(1):314–341. doi:10.1002/med.21733
90. Yin H, Guo Q, Li X, et al. Curcumin suppresses IL-1β secretion and prevents inflammation through inhibition of the NLRP3 inflammasome. J Immun. 2018;200(8):2835–2846. doi:10.4049/jimmunol.1701495
91. Li Y, Jiao J, Qi Y, et al. Curcumin: a review of experimental studies and mechanisms related to periodontitis treatment. J Periodontal Res. 2021;56(5):837–847. doi:10.1111/jre.12914
92. Duan H, Yang S, Yang S, et al. The mechanism of curcumin to protect mouse ovaries from oxidative damage by regulating AMPK/mTOR mediated autophagy. Phytomedicine. 2024;128:155468. doi:10.1016/j.phymed.2024.155468
93. Noor A, Shafi S, Sehar N, et al. Curcuminoids as cell signaling pathway modulators: a potential strategy for cancer prevention. Curr Med Chem. 2024;31(21):3093–3117. doi:10.2174/0929867331666230809100335
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Exploring Dysregulated Ferroptosis-Related Genes in Septic Myocardial Injury Based on Human Heart Transcriptomes: Evidence and New Insights
Zou HX, Hu T, Zhao JY, Qiu BQ, Zou CC, Xu QR, Liu JC, Lai SQ, Huang H
Journal of Inflammation Research 2023, 16:995-1015
Published Date: 9 March 2023
Yunnan Baiyao Might Mitigate Periodontitis Bone Destruction by Inhibiting Autophagy and Promoting Osteoblast Differentiation in vivo, ex vivo and in vitro
Liu W, Li Y, An Y, Zhao R, Wei C, Ren X, He H
Journal of Inflammation Research 2024, 17:2271-2284
Published Date: 15 April 2024
Validation of Senescence of the Role of ATM/P53 Pathway in Myocardial Senescence in Mice with Sepsis
Yan Z, Shi X, Ding R, Xia F, Du Y, Wang X, Peng Q
Infection and Drug Resistance 2025, 18:1961-1974
Published Date: 19 April 2025
The Role of Autophagy in the Regulation of Bidirectional Relationships in Diabetic Periodontitis
Li N, Chen Q, Feng Y, Wang Y
Journal of Inflammation Research 2025, 18:7781-7794
Published Date: 13 June 2025