Back to Journals » Infection and Drug Resistance » Volume 18
Validation of Senescence of the Role of ATM/P53 Pathway in Myocardial Senescence in Mice with Sepsis
Authors Yan Z, Shi X, Ding R, Xia F, Du Y, Wang X, Peng Q
Received 19 November 2024
Accepted for publication 9 April 2025
Published 19 April 2025 Volume 2025:18 Pages 1961—1974
DOI https://doi.org/10.2147/IDR.S505836
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhonghan Yan,1,* Xuemei Shi,1,* Ruilin Ding,2 Fenfen Xia,1 Yan Du,3 Xiaojie Wang,4 Qing Peng1
1Department of Cardiology, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, People’s Republic of China; 2Institute of Drug Clinical Trial/GCP Center, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, People’s Republic of China; 3Department of Cardiology, Deyang People’s Hospital, Deyang, Sichuan, People’s Republic of China; 4Department of Endocrinology, Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaojie Wang, Department of Endocrinology, Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, 646000, People’s Republic of China, Email [email protected] Qing Peng, Department of Cardiology, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, 646000, People’s Republic of China, Email [email protected]
Background: Sepsis induces multi-organ damage, including myocardial dysfunction, which is often reversible. However, the role of cell senescence in sepsis-induced myocardial dysfunction (SIMD) remains understudied. This study aimed to investigate gene expression changes related to myocardial aging in sepsis.
Methods: Transcriptomic datasets (GSE79962 and GSE141864) were analyzed to identify senescence-related genes (SRGs) by intersecting differentially expressed genes (DEGs) with the CellAge database. Functional enrichment and protein-protein interaction (PPI) network analysis were performed to identify key pathways and hub genes. A murine sepsis model was established via intraperitoneal lipopolysaccharide (LPS) injection, and the Ataxia Telangiectasia Mutated Protein (ATM) inhibitor KU60019 was used to assess the effects on cardiac function and cellular aging.
Results: Bioinformatics analysis revealed 15 aging-related genes, including MYC, TP53, CXCL1, and SERPINE1, which were upregulated in septic myocardial tissue. Functional enrichment analysis highlighted pathways related to DNA damage repair, cell senescence, and immune response. In vivo validation using murine LPS-induced sepsis models confirmed significant myocardial damage, which was alleviated by treatment with KU60019, an inhibitor of the DNA damage response pathway.
Conclusion: Cellular senescence and immune dysregulation play critical roles in SIMD. Targeting DDR pathways, as demonstrated by KU60019 treatment, provides novel insights into the role of cellular senescence in severe sepsis and its potential therapeutic implications for improving cardiovascular prognosis in septic patients.
Keywords: sepsis-induced myocardial dysfunction, cell senescence, transcriptome, key genes, database, ATM/P53 pathway
Introduction
Sepsis is a life-threatening condition characterized by an uncontrolled inflammatory response to infection, which leads to compromised systemic arterial function.1 Although the survival rates of sepsis have significantly increased in high-income nations, the long-term consequences of sepsis are accompanied by the emergence of new health complications,2 which impose significant burdens on both patients and society.3 Post-sepsis syndrome encompasses a wide range of long-lasting complications affecting multiple systems, including the immune, cognitive, mental, renal, and cardiovascular systems.4 Among these, cardiovascular diseases are particularly prominent, with a heightened long-term risk of myocardial dysfunction. Despite being reversible in some cases, sepsis-induced myocardial dysfunction (SIMD) contributes to the long-term morbidity of survivors and remains a critical factor in their prognosis.5
Recent studies6,7 have identified cell senescence as an important mechanism driving the long-term consequences of sepsis and clinical observational evidence suggests a link between the cell senescence component and worse cardiac outcomes.8 Cell senescence refers to the irreversible replication arrest in typically proliferating cells, which was initially observed by Hayflick et al in human fibroblasts cultured in vitro.9 Cells undergoing senescence secrete a variety of proinflammatory cytokines, growth factors, and extracellular matrix proteins, known as the senescence-associated secretory phenotype (SASP),10 which can further exacerbate the inflammatory environment and drive pathogenesis in multiple organs, including the heart.11
Oxidative stress, a hallmark of sepsis, plays a central role in the induction of senescence.12 Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are elevated in sepsis, causing cellular damage, particularly to DNA.13 This DNA damage triggers the DNA damage response (DDR),14 a critical mechanism that helps maintain genome integrity. The DDR involves several key signaling pathways, including activation of proteins such as Ataxia Telangiectasia Mutated Protein (ATM) and p53,6,15,16 which coordinate cellular repair, cell cycle arrest, and apoptosis. In sepsis, sustained oxidative stress leads to prolonged activation of the DDR, particularly in myocardial cells, and promotes premature senescence in these cells. Myocardial senescence exacerbates cardiac dysfunction by impairing contractility, promoting fibrosis, and enhancing inflammation.17 These mechanisms collectively contribute to the progression of SIMD.
Although previous studies have identified oxidative stress and DNA damage as key contributors to sepsis-induced myocardial injury, the role of cellular senescence and the DDR pathway in sepsis-related myocardial dysfunction remains unclear. Furthermore, while glucocorticoids have demonstrated potential in modulating cellular senescence18 and glucocorticoid receptor antagonism has been implicated in cardiomyocyte regeneration,19,20 their effects on myocardial senescence and regeneration in sepsis have not been fully explored.
In this study, we integrated transcriptomic analysis and experimental validation to systematically investigate the role of oxidative stress, DNA damage, and the DDR pathway in myocardial senescence during sepsis. Additionally, we assessed the therapeutic potential of ATM inhibition and glucocorticoids in regulating myocardial senescence and improving cardiac function, providing novel insights into the prevention and treatment of long-term cardiovascular complications in sepsis survivors.
Materials and Methods
Data Collection
The microarray datasets GSE79962 and GSE141864, both containing human heart tissue samples, were obtained from the NCBI GEO (Gene expression omnibus) database (https://www.ncbi.nlm.nih.gov/geo/), an international public repository that archives and distributes high-throughput gene expression data and other functional genomics data. The data were anonymized and de-identified to ensure patient privacy and comply with relevant ethical standards. Specifically, GSE79962 dataset on the GPL6244 platform included transcriptomic data from 20 post-mortem heart samples of sepsis patients and 11 non-heart failure controls. Conversely, the GSE141864 dataset on the GPL17586 platform provided transcriptomic data from heart samples of 5 patients with meningococcal septic shock and 2 controls.
Differentially Expressed Genes (DEGs) and Senescence-Related Genes (SRGs)
Differentially expressed genes (DEGs) were identified in a suppurative heart using the limma package (version 3.40.6) in R software, with a significance threshold of P < 0.05 and |log2FC (Fold change) | ≥ 0.5. Senescence-Related Genes (SRGs) were obtained from the Cellage database (https://www.cellage.org/). Venn diagrams of Weisheng were used to identify DEGs linked to senescence.
Gene Set Enrichment Analysis (GSEA)
To assess aging-related gene enrichment in heart tissue from sepsis patients, we conducted Gene Set Enrichment Analysis (GSEA) on the Sangerbox platform (https://www.sangerbox.com/), with nominal P < 0.05 and False discovery rate (FDR) < 0.25 as thresholds.
GO and KEGG Enrichment Analysis
To investigate the biological functions of aging-related genes, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using the clusterProfiler package (version 3.12.0) in R software. Gene sets significantly enriched for senescence were defined by an FDR < 0.1 and P < 0.05.
Protein-Protein Interaction (PPI) Network and Hub Genes Identification
A PPI network for DEGs associated with aging was constructed using the STRING database (https://string-db.org/) and Cytoscape software (version 3.9.1). Essential modules were identified through MCODE based on specific parameters such as degree cutoff = 2, node fraction cutoff = 0.2, k-core = 2, and maximum depth = 100. Hub genes were identified using the Cytohubba plugin, where genes were ranked based on their Maximal Clique Centrality (MCC) scores, with the top 7 genes as important hub genes.
Animal Grouping and Modeling
All animal procedures were carried out according to the ARRIVE guidelines and approved by the Animal Experiment Center of Southwest Medical University (GB/T35892-2018), following the Regulations on the Management of Laboratory Animals and Guidelines for Ethical Review of Laboratory Animal Welfare. Specifically, fifteen C57BL/6J mice (6–8 weeks old) were housed individually under controlled conditions (22±2°C, 55±10% humidity) with a 12-h light/dark cycle. After one week of acclimatization with ad libitum access to food and water,21,22 the mice were randomly assigned to five experimental groups (n=3 per group): (i) control (saline, 10 mg/kg, i.p).; (ii) DMSO (saline + DMSO); (iii) LPS (lipopolysaccharides, 10 mg/kg, i.p).; (iv) LPS + DMSO and (v) LPS + DMSO + KU60019 (LPS + DMSO + 5 mg/kg KU60019, i.p). In the LPS+DMSO+KU60019 group, the ATM inhibitor KU60019 was administered 1 hour after LPS injection.23 Echocardiography was performed 24h later, followed by blood collection via cardiac puncture. Mice were then euthanized by cervical dislocation, and myocardial tissues were immediately harvested for subsequent analysis.
Echocardiography Analysis of Cardiac Function
Mice were anesthetized by inhaling 1.5–2% isoflurane and evaluated for heart function with echocardiography (30 MHz, VisualSonics Vevo 3100), Parameters including ejection fraction (EF%) and fractional shortening (FS%) were measure.
Collection and Treatment of Heart and Serum Samples
Blood samples were obtained by retro-orbital puncture, centrifuged at 3000 rpm and 4°C for 20 minutes, and stored at −80°C for future analysis. Mouse hearts were collected, washed, and divided into two portions. One part was stored at −80°C for Quantitative-Realtime fluorescence quantitative (qRT-PCR) and Western blot analysis, and the other was preserved in 4% paraformaldehyde for histopathological studies.
Hematoxylin and Eosin (H&E) Staining
Following treatment with 10% paraformaldehyde, mouse heart tissues were encased in paraffin wax and cut into 4-mm-thick sections. The deparaffinization process was performed at a temperature of 60°C, then followed by two rounds of incubation in xylene and dehydration using various ethanol solutions. Thereafter, the segments were dyed using Harris hematoxylin staining solution for 10 min and then immersed in 1% HCl in ethanol for 30s. The slices were washed with tap water for 15 min, followed by staining with 1% eosin iron-red and incubated in 90% ethanol. Then, the parts were rinsed with 95% ethanol for 1 min and then three times with xylene. Next, the H&E-stained sections were left to dry for 20 min before examination under light microscopy at a magnification of 400 × to evaluate the structural alterations in the myocardium.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA was conducted to determine the levels of sepsis biomarkers in serum, including IL-8, IL-6, CK-MB, 8-OhdG, and CTNT. In this study, the ELISA kits used were obtained from Hengyi Sai Biotechnology Co., LTD in Wuhan, China. The tests were performed following the guidelines provided by the manufacturer, using IL-8 (Cat ml063162, ml063162), 8-OhdG (Cat ELK7861, ELK Biotechnology), IL-6 (Cat ELK1157, ELK Biotechnology), CK-MB (Cat ELK1286, ELK Biotechnology), and CTNT (Cat ELK6207, ELK Biotechnology) ELISA kits. Results were documented in ng/pg per total protein. Detailed methods can be found in the Supplementary File 1.
Western Blot Analysis
After collecting, washing, and pre-cooling the remaining cardiac tissue, it was grounded and proteins were lysed to extract total protein, which were then quantified for concentration. Thereafter, the altered protein was isolated using gel electrophoresis according to its size, moved to a nitrocellulose membrane, obstructed with milk at ambient temperature, and subsequently left to sit overnight at a cold temperature with primary antibodies such as p-ATM (abcam, ab315019, 1:500), ATM (CST, #2873, 1:1000), P53 (Wuhan Sanying, 60283-2-Ig, 1:2000), P21 (CST, #37543, 1:1000), P16 (CST, #29271, 1:1000), and γH2Ax (abcam, ab81299, 1:500). The next day, the membrane was rinsed and then exposed to a secondary antibody linked to HRP (Horseradish peroxidase) (ASPEN, AS1058, 1:10000) at room temperature, followed by another wash. The protein bands were captured and examined to determine their expression levels by utilizing a gel electrophoresis machine (DYY-6C, BEIJING LIUYI BIOTECHNOLOGY CO, LTD). The optical density values of the target bands were analyzed by the AlphaEaseFC software processing system. Detailed methods can be found in the Supplementary File 2.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from heart tissue following the guidelines provided by the manufacturer, utilizing TRIzol reagent from ELK Biotechnology. Thereafter, a 2-μg RNA sample was converted into cDNA via reverse transcription with EntiLink 1st Strand cDNA Synthesis Super Mix from ELK Biotechnology (EQ031). The cDNA samples were subjected to qRT-PCR using the EnTurbo SYBR Green PCR SuperMix (ELK Biotechnology, EQ001). The QuantStudio 6 Flex system from Life Technologies was utilized, with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal control. The qPCR primers in Table 1 were created and produced by HY Cell Biotechnology in Wuhan, China. Gene expression levels were analyzed using the 2−ΔΔCt technique to determine their relative values. Detailed methods can be found in the Supplementary File 3.
 |
Table 1 Primers Designed for Use in Quantitative Real-Time PCR |
Immune Infiltration Analysis
Immunoinfiltration analysis on Sangerbox platform was used to evaluate the existence of immune cells in septic heart and control heart. Gene expression data from GSE79962 and GSE141864 were standardized, and the proportion of 22 immune cells in each sample was determined by using Sangerbox’s immune infiltration calculation tool and CIBERSORT algorithm.
Receiver Operating Characteristic (ROC) Curve
Receiver operating characteristic (ROC) curves were employed to assess the diagnostic capacity of genes. To measure this ability, the Area Under Curve (AUC) was computed. Genes with AUC > 0.6 were classified as diagnostic, with those exceeding 0.8 recognized for their exceptional diagnostic potential.
Statistical Analysis
GraphPad Prism and the SangerBox platform were utilized for statistical analyses. The choice between t-tests or the signed rank test for comparing two groups depended on the data’s characteristics, whereas ANOVA (Ordinary one-way analysis of variance) was utilized for comparing multiple groups. Spearman correlation tests were utilized for correlation analysis. The data is displayed as the mean ± standard deviation (SD).
Results
Overview of Key Findings
This study aimed to investigate the molecular mechanisms underlying SIMD, with a particular focus on the role of cellular senescence and immune dysregulation in septic cardiomyocytes. Through an integrated approach combining bioinformatics analysis, immune infiltration profiling, and experimental validation, we identified key SRDEGs and explored their functional pathways. Additionally, we assessed the impact of immune cell infiltration and validated the therapeutic potential of targeting the DDR pathway in a murine sepsis model.
Identification of DEGs and SRDEGs
To identify SRDEGs in septic cardiomyocytes, we conducted gene set enrichment analysis and differential expression analysis using the merged GSE79962 + GSE141864 dataset, which comprised 41 samples, including 13 controls and 28 sepsis cases (Figure 1A). Our findings showed a significant activation of aging-related gene sets in septic myocardial tissue. Furthermore, differential analysis and screening of genes within the dataset identified 739 differentially expressed genes (Figure 1B) with |log2FC| > 0.5 and P < 0.05. The remaining aging-related genes were obtained via an intersection from the cell database, resulting in the identification of 15 common differentially expressed genes (ZFP36, BHLHE40, BCL6, MYC, TP53, PRPF19, SERPINE1, NOX4, ETS1, SMG1, CXCL8, CXCL1, CDKN1A, and FOS were upregulated, and PROX1 was downregulated) (Figure 1C and D).
Functional Enrichment Analysis of SRDEGs
GO and KEGG analysis unveiled the potential biological roles and enriched pathways of SRDEGs. The GO analysis was divided into three categories: biological processes, cellular components, and molecular functions. Enrichment analysis of GO terms revealed that biological processes of genes with differential expression were enriched in regulating metabolism of nuclear-containing base compounds, responding to stress, promoting cell proliferation, inhibiting cell proliferation, responding to cytokines, aging, undergoing cell senescence, and reacting to gamma radiation (Figure 2A). The cell components were mainly enriched in the nuclear cavity, nuclear part, nucleoplasm, nucleus, chromosome part, replication fork, transcription factor complex, double-strand break sites, DNA damage sites, and transcription factor AP-1 complex (Figure 2B). The primary molecular functions included binding to specific DNA sequences, binding to DNA in regions that regulate transcription, binding to nucleic acids in the regulatory regions, binding to transcription factors, binding to specific DNA sequences in regions that regulate RNA polymerase II activity, binding to DNA in regions that regulate RNA polymerase II activity, binding to specific DNA sequences in regions that regulate RNA polymerase II activity, binding to specific sequences of double-stranded DNA, binding to core promoters, and binding to activating transcription factors (Figure 2C). KEGG enrichment analysis revealed Kaposi sarcoma-associated herpes virus infection, cell senescence, human T-cell leukemia virus infection, hepatitis B infection, and transcriptional dysregulation (Figure 2D).
PPI Network Analysis and Hub Gene Identification and Identification and Validation of Diagnostic Characteristic Biomarkers
A PPI network consisting of 15 gene-related proteins was built using the STRING online tool (https://string-db.org/) to detect interactions among differentially expressed genes. Following the network construction, hub genes were identified using the Mcode plug-in, resulting in a gene cluster with seven nodes and 40 edges (Figure 3A). The seven hub genes—CXCL8, MYC, TP53, CXCL1, CDKN1A, FOS, and SERPINE1—were identified as central to the network (Figure 3B). To evaluate the diagnostic value of these hub genes in SIMD, ROC curve analysis was performed, revealing that CXCL8, MYC, TP53, CXCL1, and SERPINE1 exhibited high diagnostic accuracy with AUC values greater than 0.800 (Figure 3C).
Immune Infiltration Analysis
Confirming the significant role of the immune response in the pathogenesis of SIMD, we conducted an analysis of immune infiltration to uncover the interplay between the immune response in SRG and SIMD. We utilized the CIBERSORT algorithm to calculate the percentage of immune cells in heart samples GSE79962+GSE141864, as shown in Figure 4B. The heatmap, organized by clusters, illustrates the differences in the ratio of immune cells infiltrating the septic and control heart samples. Notable differences were identified in the plasma cells (eg P < 0.05); follicular helper T cells (eg P < 0.05); quiescent NK cells (eg P < 0.05); M0, M1 and M2 macrophages (eg P < 0.05); and neutrophils between the control and septic myocardial samples (Figure 4C). Correlation analysis did not reveal any significant association with the key genes (Figure 4A). These findings suggested a significant alteration in immune cell profiles in septic hearts, which may play a crucial role in the progression of SIMD.
KU60019 Treatment Alleviated LPS-Induced Heart Injury in Mice
The LPS-induced septic mouse model is a common animal model utilized in SIMD research. Echocardiography, CK-MB level (eg P < 0.05), cTnT level (eg P < 0.05), and H&E staining all indicated a notable decrease in cardiac function in the LPS group, accompanied by inflammatory cell infiltration, myocardial tissue swelling, damage, and structural irregularities. In the LPS + DMSO + KU60019 group, these conditions were alleviated except for the FS% level (Figure 5A–D). To validate the experimental findings, DMSO and DMSO + LPS groups were established based on multiple studies indicating the cardiotoxic effects of DMSO.24 Based on these results, DMSO did not cause any significant differences.
Hub Gene Expression Validation
In the LPS-induced SIMD mouse model, we confirmed the expression levels of hub genes that are diagnostically valuable (the mouse model was unable to detect CXCL8). The qRT-PCR findings indicated that the expression levels of the four central genes followed a pattern similar to that of the cardiac tissue in patients with sepsis. Treatment with KU60019 notably reduced these levels (eg P < 0.05), indicating its role in controlling SIMD cells (Figure 6A–D). Thus, we identified four key genes (MYC, TP53, CXCL1, and SERPINE1) that regulate SIMD cell senescence.
 |
Figure 6 The mRNA expression level of the hub gene detected by qRT-PCR (A–D). ****: p < 0.0001. |
Sepsis May Induce Premature Aging of Cardiac Cells
We measured the P53, P21, and P16 levels in the mouse cardiomyocytes (Figure 7A). These findings indicated that LPS caused a notable increase in the levels of P53, P21, and P16 proteins in the cardiac tissue of mice, whereas treatment with KU60019 led to a reduction in the levels of these proteins (eg P < 0.05) in the cardiac tissue of mice. The proteins’ expression was not significantly affected by DMSO (Figure 7B).
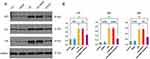 |
Figure 7 Western blotting was utilized to detect the presence of aging-related markers P21, P16, and P53 (A), with the findings displayed in a bar chart (B). Significance levels are denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (Full-length gels and blots are visible in the Supplementary File 4). |
Inhibition of DNA Damage Response Can Reduce Myocardial Senescence
The levels of SASP (IL-6 and IL-8) and DDR markers (γH2AX, 8-OhdG, and pATM/ATM) were evaluated in mouse heart tissue through Western blotting (Figure 8B) and ELISA. Compared to the control group, we observed a notable increase in DDR markers 8-OhdG (Figure 8A), γH2AX, and pATM/ATM (eg P < 0.05) (Figure 8C) in the LPS group. These findings indicate a notable rise in DDR levels within the heart muscle of individuals with sepsis, along with a marked increase in the expression levels of SASP factors IL-6 and IL-8 (eg P < 0.05), providing additional evidence of myocardial senescence. In mice treated with KU60019, we observed markedly reduced expression levels of all these substances compared to the LPS group (eg P < 0.05). Furthermore, the presence or absence of DMSO did not significantly affect the detection results, further validating the accuracy (Figure 8A–C).
 |
Figure 8 The levels of IL-6, IL-8, and 8-OHdG in mice from each group were analyzed using the ELISA method and presented in a bar chart format. The sample size for each group was n=3 (A). The expression of aging-related markers p-ATM, ATM, and γH2Ax was detected via Western blotting. The results are presented in a bar chart (B–C). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Full-length gels and blots are visible in the Supplementary File 4). |
Discussion
Summary of Our Work
This study revealed the intricate relationship between cellular senescence and SIMD. Using bioinformatics methods, we discovered an unusual expression of genes associated with aging in cardiac tissue of sepsis patients, analyzed and hypothesized the key genes and pathways regulating this process, validated cellular senescence occurrence in the myocardial tissue of SIMD in animal models. We demonstrated that inhibiting ATM activation can reduce the synthesis of P53, thereby improving the aging of cardiomyocytes caused by sepsis, which provides theoretical support for potential therapeutic strategies to improve the prognosis of sepsis patients, especially in enhancing survival rates and quality of life.
SIMD-Induced Premature Aging in Cardiomyocytes
The mechanism and occurrence of SIMD-induced premature aging in cardiac myocytes remains unclear. Our study first utilized GSEA analysis to identify aging-related gene sets significantly upregulated in cardiomyocytes from sepsis patients. A total of 15 genes related to aging were identified by comparing gene expression between sepsis and healthy hearts. KEGG analysis indicated notable enrichment in aging and cellular senescence pathways, and GO enrichment analysis revealed information regarding the biological processes like cellular senescence, cell proliferation, nucleotide metabolism, and DNA repair. These findings suggest that sepsis may induce premature aging of cardiomyocytes, potentially through DNA damage repair mechanisms, which was in line with those of prior studies.25,26
Key Genes Regulating Cellular Aging in SIMD
Five key genes — MYC, TP53, CXCL1, CXCL8, and SERPINE1 — were identified and validated their expression in animal experiments. Specifically, CXCL8 and CXCL1, as chemokines,27 are generally secreted by aging cells (mainly monocytes) to maintain chronic inflammation during the aging process and are classic molecules of the aging-related secretory phenotype.28,29 Research has shown that CXCL1 recruits neutrophils to the liver of old mice, inducing tissue aging and inflammation.30 MYC plays a crucial role in the regeneration of cardiomyocytes and may also promote aging by inhibiting PARKIN, a protein necessary for mitochondrial autophagy.31 Conversely, impaired mitochondrial autophagy is a key factor in cellular aging.32 The upregulation of MYC expression could exacerbate sepsis-induced aging. SERPINE1, which encodes PAI-1,33 regulates blood clotting and is implicated in aging and fibrosis. Studies have shown that PAI-1 plays a crucial role in both stress-induced and replicative aging in cells and can be readily detected in accelerated and physiological aging models.34 Although the precise process remains unknown, numerous research studies have showed that PAI-1 production can be triggered by different inflammatory agents, such as IL-6 and TNF-α.35 The TP53 gene produces the P53 protein, essential for regulating cell growth and apoptosis. It activates in response to stressors like radiation, toxins, low oxygen, reactive oxygen species, and abnormal cell division. In sepsis, mitochondrial dysfunction involves oxidative phosphorylation issues, reactive oxygen species production, altered energy metabolism, and mitophagy.36 These results indicate that sepsis can cause premature aging of the cardiac tissue, leading to abnormal expression of these genes.
The Role of the P53 Pathway in Myocardial Aging
In the PPI interaction network, we noted that P53 occupies the most central position, combined with our enrichment analysis results, aligning with some prior findings.37,38 Thus, we hypothesized that myocardial cell senescence may be caused by ROS accumulation leading to DNA damage in myocardial tissue, triggering DDR, and ultimately resulting in a series of cascading reactions. We found that inhibiting the ATM/P53 pathway in LPS mice led to a significant decrease in the DDR response level, cell aging markers (P53, P16, and P21), and SASP expression, including a minimal improvement in cardiac function and tissue damage. This indicates that improving DDR in the early stage of sepsis can alleviate myocardial injury by modulating myocardial aging.
Immune Microenvironment in Sepsis
We also explored the relationship between early myocardial injury in sepsis and the immune microenvironment and analyzed the correlation between relevant immune cells and aging-related hub genes. We noted abnormal immune cell infiltration and significant differences in numerous immune cell types between sepsis and control myocardial samples. Despite previous studies indicating that aging can alter tissue immune microenvironments,39 our study yielded similar results, but did not find a significant correlation between changes in numerous immune cells and these key genes. We may need more samples and experiments for analysis and validation.
Implications for Therapeutic Strategies
Our study demonstrated that LPS-induced sepsis accelerated myocardial aging through DNA damage repair mechanisms, primarily involving the ATM/P53 pathway. This finding offers important theoretical insight into one of the key mechanisms underlying myocardial dysfunction in sepsis. Although our results suggested that targeting the ATM/P53 pathway and other anti-aging strategies could help alleviate sepsis-associated deficits (including immune, cognitive, psychological, renal, and cardiovascular impairments), it is crucial to emphasize that these findings were based on animal models. Therefore, the therapeutic potential of these interventions needs to be validated in human populations.
Study Limitations and Future Directions
This study has some limitations. First, the mouse model of sepsis used may not accurately represent the myocardial dysfunction seen in humans; therefore, it is crucial to conduct additional preclinical and clinical research, along with in vitro tests, to gain a deeper insight into the mechanisms that cause myocardial cell aging due to sepsis. Second, although this study confirmed cell aging in sepsis animal models, the sample size was relatively small; therefore, larger datasets and more samples are needed to validate our findings. Finally, it is important to mention that cellular aging can be reversed as a biological process, and conducting animal experiments with longer time points would be valuable in investigating whether sepsis-induced cell aging contributes to poor long-term cardiac outcomes.
Conclusion
This study highlights the role of the ATM/P53 pathway in sepsis-induced myocardial aging. Our findings suggest that targeting DNA damage repair and modulating cellular aging could offer potential therapeutic strategies to improve the prognosis of sepsis patients. Further studies with larger sample sizes and longer follow-up are needed to validate these results and explore clinical applications.
Accessibility of Information and Resources
The research used information obtained from publicly available databases, which is thoroughly explained in the paper. Information can be accessed in the GEO data repository (https://www.ncbi.nlm.nih.gov/geo/) using the accession numbers GSE79962 and GSE141864. Furthermore, the CellAge database can be found at https://genomics.senescence.info/cells/.
Approval of Ethics and Agreement to Participate
The transcriptomic datasets used in this study, GSE79962 and GSE141864, are obtained from the NCBI GEO database and are legally acquired public data. All data are anonymized and do not involve any sensitive personal information or commercial interests. In accordance with item 1 and 2 of Article 32 of the Measures for Ethical Review of Life Science and Medical Research Involving Human Subjects issued on February 18, 2023, China, the use of human data in this study is exempt from ethical review. Animal experiments were evaluated and authorized by the Ethics Committee of the Affiliated Hospital of Southwest Medical University (reference number 20231115-003).
Consent for Publication
All authors consent to the publication of this study.
Acknowledgments
We appreciate the online resources of online analysis sites Weisheng (bioinformatics.com.cn) and sangerbox3.0 (SangerBox.con/home/. html). Zhonghan Yan and Xuemei Shi share the first authorship.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by the Science Foundation of the Sichuan Medical Association Project (grant number: S23007), the Scientific Project of the Southwest Medical University (grant number: 2020ZRQNA004), the Medical Research of the Sichuan Medical Association Project (grant number S2024010), and the Doctoral Research Foundation of the Affiliated Hospital of Southwest Medical University (grant number: 2018099). The sponsors were not involved in the study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors claim that they have no conflicting financial interests or personal relationships that could have affected the results discussed in this article.
References
1. Clere-Jehl R, Mariotte A, Meziani F, Bahram S, Georgel P, Helms J. JAK-STAT targeting offers novel therapeutic opportunities in sepsis. Trends Mol Med. 2020;26(11):987–1002. doi:10.1016/j.molmed.2020.06.007
2. Shankar-Hari M, Rubenfeld GD. Understanding long-term outcomes following sepsis: implications and challenges. Curr Infect Dis Rep. 2016;18(11):37. doi:10.1007/s11908-016-0544-7
3. Dupuis C, Bouadma L, Ruckly S, et al. Sepsis and septic shock in France: incidences, outcomes and costs of care. Ann Intens Care. 2020;10(1):145. doi:10.1186/s13613-020-00760-x
4. van der Slikke EC, An AY, Hancock REW, Bouma HR. Exploring the pathophysiology of post-sepsis syndrome to identify therapeutic opportunities. EBioMedicine. 2020;61:103044. doi:10.1016/j.ebiom.2020.103044
5. Prescott HC, Osterholzer JJ, Langa KM, Angus DC, Iwashyna TJ. Late mortality after sepsis: propensity matched cohort study. BMJ. 2016;353:i2375. doi:10.1136/bmj.i2375
6. Chen J, Chen XY, Cong XX, et al. Cellular senescence implicated in sepsis-induced muscle weakness and ameliorated with metformin. Shock. 2023;59(4):646–656. doi:10.1097/SHK.0000000000002086
7. Merdji H, Kassem M, Chomel L, et al. Septic shock as a trigger of arterial stress-induced premature senescence: a new pathway involved in the post sepsis long-term cardiovascular complications. Vascular Pharmacol. 2021;141:106922. doi:10.1016/j.vph.2021.106922
8. Mehdizadeh M, Aguilar M, Thorin E, Ferbeyre G, Nattel S. The role of cellular senescence in cardiac disease: basic biology and clinical relevance. Nat Rev Cardiol. 2022;19(4):250–264. doi:10.1038/s41569-021-00624-2
9. Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi:10.1016/0014-4827(61)90192-6
10. Coppé JP, Patil CK, Rodier F, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi:10.1371/journal.pbio.0060301
11. Mohamad Kamal NS, Safuan S, Shamsuddin S, Foroozandeh P. Aging of the cells: insight into cellular senescence and detection methods. Eur J Cell Biol. 2020;99(6):151108. doi:10.1016/j.ejcb.2020.151108
12. Joffre J, Hellman J. Oxidative stress and endothelial dysfunction in sepsis and acute inflammation. Antioxid Redox Signaling. 2021;35(15):1291–1307. doi:10.1089/ars.2021.0027
13. Van Houten B, Santa-Gonzalez GA, Camargo M. DNA repair after oxidative stress: current challenges. Curr Opin Toxicol. 2018;7:9–16. doi:10.1016/j.cotox.2017.10.009
14. Huff LA, Yan S, Clemens MG. Mechanisms of Ataxia Telangiectasia Mutated (ATM) control in the DNA damage response to oxidative stress, epigenetic regulation, and persistent innate immune suppression following sepsis. Antioxidants. 2021;10(7):1146. doi:10.3390/antiox10071146
15. Hwang SY, Kuk MU, Kim JW, et al. ATM mediated-p53 signaling pathway forms a novel axis for senescence control. Mitochondrion. 2020;55:54–63. doi:10.1016/j.mito.2020.09.002
16. Weintz G, Olsen JV, Frühauf K, et al. The phosphoproteome of toll-like receptor-activated macrophages. mol Syst Biol. 2010;6:371. doi:10.1038/msb.2010.29
17. Mankowski RT, Yende S, Angus DC. Long-term impact of sepsis on cardiovascular health. Intensive Care Med. 2019;45(1):78–81. doi:10.1007/s00134-018-5173-1
18. Laberge RM, Zhou L, Sarantos MR, et al. Glucocorticoids suppress selected components of the senescence-associated secretory phenotype. Aging Cell. 2012;11(4):569–578. doi:10.1111/j.1474-9726.2012.00818.x
19. Sethi Y, Padda I, Sebastian SA, et al. Glucocorticoid receptor antagonism and cardiomyocyte regeneration following myocardial infarction: a systematic review. Curr Prob Cardiol. 2023;48(12):101986. doi:10.1016/j.cpcardiol.2023.101986
20. Pianca N, Sacchi F, Umansky KB, et al. Glucocorticoid receptor antagonization propels endogenous cardiomyocyte proliferation and cardiac regeneration. Nature Cardiovasc Res. 2022;1(7):617–633. doi:10.1038/s44161-022-00090-0
21. Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 2018;18(12):733–744. doi:10.1038/s41577-018-0065-8
22. Sun Y, Yao X, Zhang QJ, et al. Beclin-1-dependent autophagy protects the heart during sepsis. Circulation. 2018;138(20):2247–2262. doi:10.1161/CIRCULATIONAHA.117.032821
23. Yan P, Li Z, Xiong J, et al. LARP7 ameliorates cellular senescence and aging by allosterically enhancing SIRT1 deacetylase activity. Cell Rep. 2021;37(8):110038. doi:10.1016/j.celrep.2021.110038
24. Jacob SW, de la Torre JC. Pharmacology of dimethyl sulfoxide in cardiac and CNS damage. Pharmacol Rep. 2009;61(2):225–235. doi:10.1016/S1734-1140(09)70026-X
25. Merdji H, Schini-Kerth V, Meziani F, Toti F. Long-term cardiovascular complications following sepsis: is senescence the missing link? Ann Intens Care. 2021;11(1):166. doi:10.1186/s13613-021-00937-y
26. Yang H, Jiang Z, Feng L, et al. Nppb contributes to sepsis-induced myocardial injury by regulating Senescence-Related genes. Int Immunopharmacol. 2024;143(Pt 2):113461. doi:10.1016/j.intimp.2024.113461
27. Sawant KV, Poluri KM, Dutta AK, et al. Chemokine CXCL1 mediated neutrophil recruitment: role of glycosaminoglycan interactions. Sci Rep. 2016;6:33123. doi:10.1038/srep33123
28. Kumar M, Seeger W, Voswinckel R. Senescence-associated secretory phenotype and its possible role in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2014;51(3):323–333. doi:10.1165/rcmb.2013-0382PS
29. Ravalet N, Guermouche H, Hirsch P, et al. Modulation of bone marrow and peripheral blood cytokine levels by age and clonal hematopoiesis in healthy individuals. Clin Immunol. 2023;255:109730. doi:10.1016/j.clim.2023.109730
30. Su L, Li N, Tang H, et al. Kupffer cell-derived TNF-α promotes hepatocytes to produce CXCL1 and mobilize neutrophils in response to necrotic cells. Cell Death Dis. 2018;9(3):323. doi:10.1038/s41419-018-0377-4
31. Soh JEC, Shimizu A, Molla MR, et al. RhoA rescues cardiac senescence by regulating Parkin-mediated mitophagy. J Biol Chem. 2023;299(3):102993. doi:10.1016/j.jbc.2023.102993
32. Broda M, Millar AH, Van Aken O. Mitophagy: a mechanism for plant growth and survival. Trends Plant Sci. 2018;23(5):434–450. doi:10.1016/j.tplants.2018.02.010
33. Aihemaiti A, Yamamoto N, Piao J, et al. A novel PAI-1 inhibitor prevents ageing-related muscle fiber atrophy. Biochem Biophys Res Commun 2021;534:849–856. doi:10.1016/j.bbrc.2020.10.089
34. Vaughan DE, Rai R, Khan SS, Eren M, Ghosh AK. Plasminogen activator inhibitor-1 is a marker and a mediator of senescence. Arteriosclerosis Thrombosis Vasc Biol. 2017;37(8):1446–1452. doi:10.1161/ATVBAHA.117.309451
35. Kang S, Tanaka T, Inoue H, et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci USA. 2020;117(36):22351–22356. doi:10.1073/pnas.2010229117
36. Beyfuss K, Hood DA. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018;23(1):100–117. doi:10.1080/13510002.2017.1416773
37. Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules. 2020;10(3):420. doi:10.3390/biom10030420
38. Pawge G, Khatik GL. p53 regulated senescence mechanism and role of its modulators in age-related disorders. Biochem Pharmacol. 2021;190:114651. doi:10.1016/j.bcp.2021.114651
39. Gu J, Wang S, Guo H, et al. Inhibition of p53 prevents diabetic cardiomyopathy by preventing early-stage apoptosis and cell senescence, reduced glycolysis, and impaired angiogenesis. Cell Death Dis. 2018;9(2):82. doi:10.1038/s41419-017-0093-5
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Exploring Dysregulated Ferroptosis-Related Genes in Septic Myocardial Injury Based on Human Heart Transcriptomes: Evidence and New Insights
Zou HX, Hu T, Zhao JY, Qiu BQ, Zou CC, Xu QR, Liu JC, Lai SQ, Huang H
Journal of Inflammation Research 2023, 16:995-1015
Published Date: 9 March 2023
Novel Insights from Comprehensive Bioinformatics Analysis Utilizing Large-Scale Human Transcriptomes and Experimental Validation: The Role of Autophagy in Periodontitis
Liu F, Zhu Z, Zou H, Huang Z, Xiao S, Li Z
Journal of Inflammation Research 2024, 17:11861-11880
Published Date: 30 December 2024






