Back to Journals » International Journal of Nanomedicine » Volume 19
The Genetic and Epigenetic Toxicity of Silica Nanoparticles: An Updated Review
Authors Zheng M, Chen Z, Xie J, Yang Q, Mo M, Liu J, Chen L
Received 15 July 2024
Accepted for publication 15 November 2024
Published 24 December 2024 Volume 2024:19 Pages 13901—13923
DOI https://doi.org/10.2147/IJN.S486858
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sachin Mali
Manjia Zheng,1 Ziwei Chen,1 Jiling Xie,1 Qiyuan Yang,1 Minhua Mo,1 Jia Liu,2 Liangjiao Chen1
1Department of Orthodontics, School and Hospital of Stomatology, Guangdong Engineering Research Center of Oral Restoration and Reconstruction & Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou Medical University, Guangzhou, People’s Republic of China; 2Stomatological Hospital, Southern Medical University, Guangzhou, People’s Republic of China
Correspondence: Jia Liu, Stomatological Hospital, Southern Medical University, No. 366 jiangnan Road South, Guangzhou, Guangdong, 510280, People’s Republic of China, Email [email protected] Liangjiao Chen, Department of Orthodontics, School and Hospital of Stomatology, Guangdong Engineering Research Center of Oral Restoration and Reconstruction & Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou Medical University, No. 31 huangsha Road, Guangzhou, Guangdong, 510145, People’s Republic of China, Email [email protected]
Abstract: Silica nanoparticles (SiNPs) are widely used in biomedical fields, such as drug delivery, disease diagnosis, and molecular imaging. An increasing number of consumer products containing SiNPs are being used without supervision, and the toxicity of SiNPs to the human body is becoming a major problem. SiNPs contact the human body in various ways and cause damage to the structure and function of genetic material, potentially leading to carcinogenesis, teratogenicity and infertility. This review summarizes SiNPs-induced genetic and epigenetic toxicity, especially to germ cells, and explore their potential mechanisms. SiNPs cause genetic material damage mainly by inducing oxidative stress. Furtherly, the molecular mechanisms of epigenetic toxicity are discussed in detail for the first time. SiNPs alter DNA methylation, miRNA expression, histone modification and inhibit chromatin remodeling by regulating epigenetic-related enzymes and transcription factors. This review is beneficial for investigating potential solutions to avoid toxicity and provide guidance for better application of SiNPs in the biomedical field.
Keywords: silica nanoparticles, genotoxicity, epigenetic, DNA damage, germ cells
Introduction
Silica nanoparticles (SiNPs) are among the three most commonly used nanomaterials in the world. Owing to their controllable particle size, large specific surface area, and presence of silanol groups, SiNPs exhibit unique physicochemical properties and excellent biocompatibility.1 In the biomedical field, SiNPs have been introduced into the human body for drug delivery, disease diagnosis and molecular imaging.2–4 SiNPs can contact the human body through intravenous injection, lung inhalation, skin contact and gastrointestinal routes and have toxic effects on many organs, including the lungs, liver, heart, brain, spleen, and kidneys.5–10 As an increasing number of consumer products containing nanomaterials are used without supervision, the toxicity of nanomaterials to the human body is becoming a major problem.
SiNPs are mainly divided into two types: crystalline SiNPs and amorphous SiNPs. Crystalline SiNPs are released from the natural environment and from construction and industrial processes. Amorphous SiNPs are divided into mesoporous SiNPs and nonporous SiNPs on the basis of the presence or absence of pores.3 Amorphous SiNPs are widely used in food, cosmetics, the automotive industry, and construction, among other industries.11 The commonly reported toxicities of SiNPs are respiratory toxicity, immunotoxicity, cardiovascular toxicity, etc.12–14 Therefore, it has become necessary to understand the effects and hazards of SiNPs upon exposure to the human body. Recently, the genetic and epigenetic toxicity of SiNPs has attracted widespread attention and are important parts of evaluating SiNPs’ safety. Any abnormal changes in gene information can affect gene expression and pose a threat to human health.
The DNA sequence is the basis of inheritance and the skeleton of genes.15 Genotoxicity refers to genetic material damage at the base, molecular, and chromosome levels, including gene mutations, DNA damage and chromosomal damage.16 Epigenetics refers to heritable changes in gene expression that occur without altering the DNA structural sequence, including modifications of DNA or chromatin structures or proteins related to them.17 Therefore, genetic and epigenetic abnormalities may affect gene expression and induce toxicity. Genetic and epigenetic toxicity can cause harm, such as cancer, teratogenicity, and infertility, by affecting biological genetic information, resulting in serious health consequences. In this review, we summarize the genetic and epigenetic toxicity induced by SiNPs, elucidate the potential adverse effects of such toxicity (Figure 1), and further discuss the potential underlying molecular mechanisms involved. Considering the genetic effects of reproductive system toxicity on offspring, we paid special attention to genetic and epigenetic toxicity in the reproductive system. This review provides guidance for evaluating the safety of SiNPs and preventing their potential adverse effects on the human body so that they can be better applied in the biomedical field in the future.
Genetic and Epigenetic Toxicity of SiNPs Causes Potential Risks
SiNPs Increase the Risk of Cancer Occurrence
The occurrence of cancer is closely related to genetic mutations. Gene mutations affect the steady-state development of key cell functions, causing uncontrolled cell growth and driving the occurrence of cancer.18 These cancer-driving genes include oncogenes and tumor suppressor genes, which are affected by point mutations, translocations or copy number changes.19 Previous studies have shown that NPs are harmful components of pollution particles in the air.20 Inhaling tungsten carbide cobalt NPs can lead to “hard metal lung disease”, doubling the risk of lung cancer.21 Long-term exposure to inhalable crystalline silica can induce silicosis and may even lead to lung tumors.22 Amorphous SiNPs increase the frequency of gene mutations and induce the malignant transformation of some somatic cells, leading to the loss of growth inhibition signals or cell dysfunction.23–25 Therefore, the genotoxicity of SiNPs causes precancerous transformation and increases the risk of cancer. However, whether genetic material damage caused by SiNPs leads to cancer is related to the dose, exposure time, cell sensitivity, etc.
SiNPs Cause Embryonic Malformation and Behavioral Changes
Embryonic development is tightly regulated by genes, and DNA damage is considered the main mechanism leading to abnormalities in embryonic development.26 SiNPs may penetrate the placental barrier, accumulate in fetal tissue, induce DNA damage, and lead to embryonic developmental abnormalities, including pericardial and yolk sac edema, blood clots and delays in embryonic development.27,28
A low concentration of SiNPs does not cause changes in the developmental morphology of zebrafish embryos, but it causes behavioral changes in embryonic light movement and the larval motor response.29 However, SiNPs can enrich pollutants. When absorbing environmental pollutants such as cadmium and tetrabromobisphenol A, SiNPs amplify the teratogenic toxicity of these pollutants and cause abnormal embryonic development.30–32 In addition, large SiNPs are blocked by placental chorionic villi and do not cause embryonic development abnormalities, whereas small SiNPs, such as those that are ≤ 70 nm, can penetrate the placental barrier and accumulate in the placenta, fetal liver, and brain, causing malformations of the embryo.33 Therefore, exposure to small SiNPs should be avoided during pregnancy.
SiNPs Increase the Risk of Infertility
DNA damage and histone modification abnormalities in germ cells induced by SiNPs can lead to infertility.34,35 Many studies have reported the negative impact of SiNPs on fertility.36,37 In the male reproductive system, SiNPs can penetrate the blood‒testis barrier and accumulate in the testes and epididymis, leading to abnormal sperm morphology, decreased sperm count, and spermatogenesis dysfunction.34,36 In the female reproductive system, SiNPs are internalized and accumulate in the ovaries, leading to granulosa cell apoptosis and follicular atresia, impairing the survival and developmental ability of ovarian cells.32,37 One study revealed that damage caused by Stöber SiNPs to the reproductive system could be reversed after exposure was stopped, suggesting that the adverse effects of SiNPs may be temporary.35
Other Diseases
The genotoxicity induced by SiNPs may also promote the occurrence of autoimmune diseases, endocrine diseases, etc. For example, SiNPs damage macrophage DNA and chromosomes, induce immune system toxicity, and may cause immune system diseases.13 DNA hypermethylation leads to abnormal insulin secretion and promotes the occurrence of type 2 diabetes mellitus.38
The Genetic and Epigenetic Toxicity of SiNPs
Genetic Material Damage
SiNPs cause genetic material damage, which has been reported to occur mainly in human lung epithelial cells, fibroblasts, and germ cells.35,39 SiNPs can cause damage to genetic material at the base, DNA molecular and chromosomal levels, leading to genotoxicity. DNA damage induced by SiNPs includes gene mutation, oxidative modification of bases, or DNA strand breaks.40–44 For example, exposure to spherical amorphous-SiNPs causes gene mutation and comet-like nuclear changes (Figure 2A and D).41,45 Comet-like nuclear changes are caused by DNA breakage and migration away from the nucleus. In addition, SiNPs induce the phosphorylation of histone family member X (γ-H2AX), which is a specific biomarker of DNA double-strand breaks (Figure 2B and C).43 The type of DNA damage is influenced by the size of the NPs. Smaller NPs are prone to insertion into DNA molecules, causing base mismatch, whereas larger NPs tend to bind to the DNA strand, causing DNA strand breakage.46,47
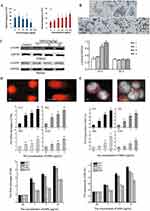 |
Figure 2 Genetic material damage induced by SiNPs. (A) Mutant frequencies in MEF-LacZ cells exposed to (a) 10-nm and (b) 30-nm spherical A-SiNPs. The 30-nm SiNPs increased the mutation frequency in a dose-dependent manner. *P < 0.05. Source: Park MVDZ, Verharen HW, Zwart E, et al. Genotoxicity evaluation of amorphous silica nanoparticles of different sizes using the micronucleus and the plasmid lacZ gene mutation assay. Nanotoxicology. 2011;5(2):168–181. reprinted by permission of the publisher (Taylor&FrancisLtd, http://www.tandfonline.com).41 (B) Effect of SiNPs on the expression of γ-H2AX in GC-2 cells after 24 h of exposure. γ-H2AX (marked with red arrows) is a biomarker of DNA strand breaks. Obvious γ-H2AX-positive staining was observed in the 25 and 50 μg/mL SiNPs groups. Source: Reprinted from Zhang J, Liu J, Ren L, et al. Silica nanoparticles induce abnormal mitosis and apoptosis via PKC-delta mediated negative signaling pathway in GC-2 cells of mice. Chemosphere. 2018;208:942–950. With permission from Elsevier.43 (C) Western blot analyses of γ-H2AX proteins in the ovaries of Balb/c female mice after exposure to SiNPs by intratracheal instillation. On the 15th day after the first dose, the expression of γ-H2AX significantly increased. On the 30th day after the first dose, the γ-H2AX level did not obviously differ among the groups. C, control group; L, 7 mg/kg SiNPs group; M, 21 mg/kg SiNPs group; H, 35 mg/kg SiNPs group. *P < 0.05. Source: Reprinted with permission from Liu J, Yang M, Jing L, et al. Silica nanoparticle exposure inducing granulosa cell apoptosis and follicular atresia in female Balb/c mice. Environ Sci Pollut Res Int. 2018;25(4):3423–3434. Springer Nature.48 (D) Changes in the nuclear shape of HUVECs, as determined by the alkaline comet assay. The typical “comet” shapes of the cell nuclei were observed in the A-SiNPs group (exposed for 4 h). DNA damage was interpreted by the OTM. The OTM values revealed the dose- and size-dependent effects on DNA damage induced by SiNPs. *P < 0.05, **P < 0.01. Source: Reprinted with permission from Zhou F, Liao F, Chen L, Liu Y, Wang W, Feng S. The size-dependent genotoxicity and oxidative stress of silica nanoparticles on endothelial cells. Environ Sci Pollut Res Int. 2019;26(2):1911–1920. Springer Nature.45 (E) Chromosomal damage in HUVECs was detected via a cytokinesis-block MN assay. MNs in binucleated cells (marked with white arrows) exposed to A-SiNPs. A-SiNPs (exposed for 24 h) had dose-dependent effects on MN%. The results are presented as the percentage of micronucleated cells (MN%) per 1000 binucleated cells. *P < 0.05, **P < 0.01. Source: Reprinted with permission from Zhou F, Liao F, Chen L, Liu Y, Wang W, Feng S. The size-dependent genotoxicity and oxidative stress of silica nanoparticles on endothelial cells. Environ Sci Pollut Res Int. 2019;26(2):1911–1920. Springer Nature.45 MEF-LacZ, containing lacZ as a reporter gene; A-SiNPs, amorphous silica nanoparticles; γ-H2AX, histone family member X phosphorylation; OTM, olive tail moment; GC-2 cells, spermatocyte lines; MN, micronucleus. |
In addition, DNA damage caused by NPs also includes abasic sites and DNA adducts. Graphite oxide induces abasic sites in DNA, ie, sites with neither a purine base nor a pyrimidine base.49 NPs with electrophilic groups, such as magnetic nanoparticles, combine covalently with DNA to form DNA adducts.50 However, whether SiNPs induce abasic sites and DNA adducts is unclear.
At the chromosome level, SiNPs induce chromosomal aberrations, including structural and numerical aberrations.51 Amorphous SiNPs of four sizes (100 nm, 50 nm, 25 nm, and 10 nm) induce dose-dependent micronucleus frequency changes (Figure 2E).45 SiNPs may also cause chromosomal numerical aberration by inhibiting chromosome segregation or inducing whole chromosome loss in the cell division stage.52–54 In conclusion, SiNPs induce DNA and chromosomal damage, posing potential risks to the human body, such as cancer, deformities, and infertility.
Abnormal Epigenetic Changes
Changes in DNA Methylation
SiNPs can induce abnormal DNA methylation, regulate gene expression and cause toxicity. DNA methylation is currently the most widely studied epigenetic mechanism.55 DNA methylation inhibits gene expression by altering transcription factor binding and chromatin conformation.56 It regulates many cellular processes, including cell differentiation, embryonic development, and gene transcription.57 Abnormal (high or low) methylation affects genomic stability and has many adverse consequences. Hypermethylation of specific promoter regions and global DNA hypomethylation are closely related to the occurrence of cancer. In addition, abnormal methylation can lead to autoimmune diseases, respiratory diseases, skin diseases, etc.38
SiNPs induce extensive DNA methylation changes in GC-2 cells, including 51.0% hyperdifferentially methylated regions and 49.0% hypodifferentially methylated regions.58 This abnormal DNA methylation leads to abnormal transcription and translation, mitochondrial damage, and sperm apoptosis. SiNPs also induce abnormal DNA methylation changes in specific genes. SiNPs inhibit outer dense fiber 1 (Odf1) and Bcl-xl in spermatocytes by inducing Crem hypermethylation, leading to sperm bundle structure and spermatocyte apoptosis, thereby disrupting spermatogenesis.59 Notably, abnormal DNA methylation induced by SiNPs can affect offspring. Prenatal exposure to high-dose food-grade SiNPs E 551 can lead to hypermethylation in the maternal and fetal liver, causing liver metabolic disorders and teratogenicity (Figure 3).60 Long interspersed element-1 (LINE-1) and Alu elements are important noncoding elements in the human genome, and changes in their methylation levels are closely related to the development of cancer and autoimmune diseases. SiNPs have been shown to induce DNA hypomethylation of Alu elements in HaCaT cells but have no effect on the DNA methylation levels of LINE-1.61 These findings suggest that the impact of SiNPs on DNA methylation is DNA sequence specific. However, the current research on the impact of SiNPs on DNA methylation is mostly based on in vitro experimental models, and further verification is needed to determine whether abnormal methylation occurs in vivo and causes corresponding adverse consequences.
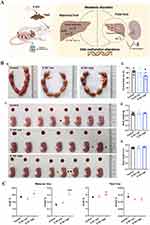 |
Figure 3 Food-grade SiNPs (E 551) induce genome-wide DNA methylation changes in mothers and fetuses. (A) E 551 accumulated in maternal and fetal liver tissues, causing genome-wide DNA methylation changes in liver tissues after high-dose prenatal exposure. The methylation and altered expression of genes are related mainly to glycolipid metabolism, which impairs glucose tolerance in pregnant mice. E 551 has a risk of inducing metabolic disorders in both the maternal and fetal liver, leading to fetal resorption. (B) (a) Representative uterine morphology after E 551 exposure at GD19. The red arrows indicate that low-dose and high-dose E551 resulted in fetal resorption. (b) Normal fetal rates after E551 exposure. High-dose exposure decreased normal fetal rates. (c) Representative fetal and placental morphology at GD19. (d and e) Weights (d) and body lengths (e) of all fetuses in the control group (n = 14), low-dose E 551 group (n = 7), and high-dose E 551 group (n = 7). (C) Genomic 5-mC and 5-hmC levels in the maternal liver and fetal liver, n=7 for each group. 5-mC levels increased in high-dose E 551-exposed livers. The 5-hmC levels in the maternal liver increased only in the high-dose E 551 group. *P < 0.05, ***P < 0.001, compared with the control group. Source: Reprinted from Zhan Y, Lou H, Shou R, et al. Maternal exposure to E 551 during pregnancy leads to genome-wide DNA methylation changes and metabolic disorders in the livers of pregnant mice and their fetuses. J Hazard Mater. 2024;465:133233. Copyright 2024, with permissions from Elsevier.60 GD19, gestational day 19; 5-methylcytosine; 5-hmC, 5-hydroxymethylcytosine. |
Changes in microRNA Expression
MicroRNAs (miRNAs) can affect gene expression by binding to the 3ʹ untranslated region (3ʹ-UTR) of mRNAs to increase the degradation of posttranscriptional mRNAs or inhibit gene translation.62 miRNAs play important roles in cell differentiation, proliferation, apoptosis, and angiogenesis. SiNPs induce abnormal expression of miRNAs. The abnormal expression of these miRNAs leads to reproductive dysfunction, endothelial dysfunction, and even tumorigenesis.25,63–66
SiNPs induce abnormal miRNA expression in germ cells. These abnormal changes in miRNA expression interfere with DNA replication, DNA repair, fatty acid metabolism, and autophagy, resulting in damage to reproductive system function. SiNPs increase the expression of 10 types of miRNAs and decrease the expression of 5 types of miRNAs, which are functionally enriched in DNA replication and fatty acid metabolism, in GC-2 spd cells (Figure 4). Among them, miRNA-450b-3p inhibits cell cycle progression, and miRNA-138-1-3p may inhibit fatty acid metabolism.67 Zhao et al reported that SiNPs inhibit the expression of the DNA repair-related protein zinc finger CW-type and PWWP domain containing 1 (ZCWPW1) by increasing the expression of miRNA-5622-3p, leading to DNA repair failure in GC-2 spd cells.65 SiNPs have also been shown to upregulate miRNA-494, which ultimately causes autophagy dysfunction and spermatocyte death.68 Moreover, another study revealed that SiNPs downregulated miRNA-450b-3p expression to increase cytoskeletal protein expression, thus disrupting the mitochondrial structure and inducing cell apoptosis, ultimately inhibiting sperm development.64 Notably, the reproductive dysfunction caused by miRNA deregulation is caused by short-term exposure (less than 3 months) to SiNPs, and long-term effects have not yet been reported. This may be because long-term tracking and observation require a significant amount of time and manpower.
 |
Figure 4 SiNPs-induced changes in the miRNA expression profile of GC-2 spd cells. (A) The expression levels of 15 miRNAs changed in GC-2 spd cells after SiNPs (5 μg/mL, 24 h) exposure. The relative up- and downregulation of miRNAs are indicated by yellow and blue, respectively. (B) Percentages of differentially expressed miRNAs in GC-2 spd cells. Fifteen miRNAs (0.08%) were differentially expressed. Among them, 5 were upregulated (33.3%), and 10 were downregulated (66.7%). (C) Pathways associated with significantly up- and downregulated miRNAs according to the GO enrichment database. Top 30 significant GO terms for the 15 miRNAs. (a) Biological processes of upregulated miRNA target genes. Biological processes primarily involve small-molecule metabolic processes. (b) Cellular components of upregulated miRNA target genes. Cellular components primarily include intracellular components. (c) Molecular functions of upregulated miRNA target genes, which involve mainly nucleoside phosphate binding. (d) Biological processes of downregulated miRNA target genes, which primarily involve the regulation of tyrosine phosphorylation of the Stat1 protein. (e) Cellular components of downregulated miRNA target genes, which primarily involve the membrane. (f) Molecular functions of downregulated miRNA target genes, which involve mainly core promoter sequence-specific DNA binding. X-axis, negative logarithm of the P value (-LgP); the larger the number is, the smaller the P value. Source: Reprinted from Zhou G, Ren L, Yin H, et al. The alterations of miRNA and mRNA expression profile and their integration analysis induced by silica nanoparticles in spermatocyte cells. NanoImpact. 2021;23:100348. Copyright 2021, with permission from Elsevier.67 GC-2 spd cells, spermatocyte lines. |
SiNPs activate the IL6R/STAT/TF signaling pathway by downregulating miRNA-451a, leading to endothelial dysfunction and thrombosis.66 In addition to causing dysfunction, abnormal miRNA expression may lead to severe consequences of tumorigenesis. Amorphous SiNPs affect ATP5H/SOD1 and EIF4G2/PAPPC1 gene expression through the upregulation of miRNA-3648/572/661 and downregulation of miRNA-4521, thereby promoting the occurrence of cancer.25 These findings suggest that these four miRNAs may be potential biomarkers of cancer and can serve as potential therapeutic targets for preventing the toxicity of SiNPs. However, the above studies are based on microarray data predictions and require wet experiments to demonstrate the interaction between identified genes and miRNA expression, as well as their impact on cell proliferation and apoptosis.
Changes in Histone Modifications
Chromosomes are composed of DNA and histones, which bind together. Histones are classified into five types: H1, H2A, H2B, H3, and H4. The N-terminal and C-terminal tails of histones can undergo posttranslational modifications, including acetylation, ubiquitination, phosphorylation, and methylation. The different histone modification methods determine the structural state of chromatin condensation or loosening and play a role in transcriptional regulation, replication, repair, and recombination.56 The abnormal modification of histones can lead to abnormal gene expression, which is closely related to the pathogenesis of cancer, neurodevelopmental disorders, and autoimmune diseases.38
SiNPs can induce abnormal states of ubiquitination, phosphorylation and acetylation of histones. SiNPs inhibit the ubiquitination of H2A and H2B in the sperm nucleus, leading to sperm production disorders (Figure 5A and B).35,69 Histone phosphorylation is a key phenomenon in DNA damage and response at different stages of the cell cycle. SiNPs increase the phosphorylation of H2AX, causing the apoptosis of spermatocytes.35 In addition, SiNPs increase the acetylation levels of histones H3K9 and H3K56, leading to human lung epithelial cell apoptosis.70 Histone methylation is the most stable form of modification. It is involved in the activation and inhibition of transcription as well as in the compaction of chromatin. Nanoparticles, such as selenium nanoparticles, can induce abnormal histone methylation, which increases the methylation of histones H3K9 and H3K27.71 However, the impact of SiNPs on histone methylation has not yet been reported. This requires further exploration.
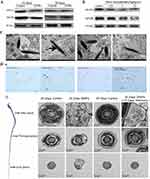 |
Figure 5 SiNPs inhibit histone ubiquitination and chromatin remodeling. (A) SiNPs (20 mg/kg.bw) inhibited ubH2A/ubH2B protein expression in nuclear extracts of elongating spermatids from male ICR mice after intratracheal instillation for 35 days. After the 15-day withdrawal period, the ubH2A/ubH2B levels recovered. Source: Reprinted from Liu J, Li X, Zhou G, et al. Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ Pollut. 2021;284:117446. Copyright 2021, with permission from Elsevier.69 (B) Western blot analyses of ubH2A/ubH2B in the nuclear extracts of germ cells from male ICR mice after exposure to SiNPs by intratracheal instillation every 3 days for 15 days. SiNPs inhibited the expression of ubH2A/ubH2B in a dose-dependent manner. The internal control protein was PCNA. Source: Reprinted from Liu J, Li X, Zhou G, et al. Silica nanoparticles induce spermatogenesis disorders via L3MBTL2-DNA damage-p53 apoptosis and RNF8-ubH2A/ubH2B pathway in mice. Environ Pollut. 2020;265(Pt A):114974. Copyright 2020, with permission from Elsevier.35 (C) TEM images showing a defect in DNA condensation in sperm heads after exposure to SiNPs (20 mg/kg.bw) for 35 days. After the 15-day withdrawal period, there was no significant difference in the amount of sperm nuclear chromatin. (a) Control group after 35 days. (b) SiNPs group after 35 days. (c) Control group after the 15-day withdrawal period. (d) SiNPs group after the 15-day withdrawal period. The black thick arrow indicates the less condensed chromatin in the sperm. Source: Reprinted from Liu J, Li X, Zhou G, et al. Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ Pollut. 2021;284:117446. Copyright 2021, with permission from Elsevier.69 (D) Effects of SiNPs on sperm quality. (a) Epididymal sperm morphology detected via sperm smears. The black arrows point to sperm folded at the neck, and the white arrow points to sperm with its head falling off. More sperm with abnormal morphology, including neck folding and head shedding, were observed in the 35 days SiNPs group than in the 35 days SiNPs +15 days recovery group. (b) Electron microscope image of the structure of each segment of the sperm flagella. The cross section of the middle piece includes the CP, OD, ODF and MS. The cross section of the principal piece contains the CP, OD, ODFs, and FS. The solid arrows represent abnormal structures. The sperm flagella were significantly damaged in the 35 days SiNPs group. Source: Reprinted with permission from Sang Y, Liu J, Dong X, et al. Silica nanoparticles induce male reproductive toxicity via Crem hypermethylation mediated spermatocyte apoptosis and sperm flagella damage. Environ Sci Pollut Res. 2024;31(9):13856–13866. Springer Nature.59 ubH2A, ubiquitinated H2A; ubH2B, ubiquitinated H2A; PCNA, proliferating cell nuclear antigen; bw, body weight; ICR mice, Institute of Cancer Research mice; CP, central pair; OD, outer doublet microtubules; ODF, outer dense fiber; MS, mitochondrial sheath; FS, fibrous sheath. |
Inhibiting Chromatin Remodeling
Chromatin remodeling refers to the dynamic process of changing chromatin between concentrated and relaxed states and plays an important role in optimizing cell adaptation and body development.72 Chromatin remodeling regulates the accessibility of chromatin to transcription factors.73 This process is achieved through changes in the position and structure of nucleosomes. When remodeling is inhibited, it leads to abnormal gene expression and causes genotoxicity.
Chromatin remodeling occurs during spermatogenesis, in which more than 90% of the core histones in the nucleosome are successively replaced by testicle-specific histone variants, transition proteins (TNPs), and protamine (PRM).74 Studies have indicated that nanoparticles may inhibit chromatin remodeling. Titanium NPs inhibit chromatin remodeling by reducing the expression of PRM1 and TNP2 in male mice.75 Gold NPs inhibit chromatin remodeling, resulting in increased chromatin instability.76
SiNPs inhibit chromatin remodeling in the haploid phase of sperm cells, hindering sperm cell differentiation and ultimately leading to a decrease in sperm quantity and quality (Figure 5C and D).69 Notably, after a 15-day withdrawal period, this damage disappeared, indicating that the sperm toxicity induced by SiNPs may be reversible. Whether this reversible damage is cell-specific or environment-specific and when irreversible damage can be caused are unknown. SiNPs affect the maturation of oocytes and interfere with the meiotic division of ova.32 However, it is not yet clear whether the interference of SiNPs in ovum formation is caused by chromatin remodeling.
Genetic and Epigenetic Toxicity Mechanisms of SiNPs
As mentioned above, the genotoxicity of SiNPs mainly manifests as damage to genetic material and abnormal genetic information caused by abnormal epigenetic changes, affecting body development and causing related diseases. The possible molecular mechanisms are discussed below.
Genetic Material Damage
The oxidative stress damage caused by reactive oxygen species (ROS) in genetic materials is the main mechanism of SiNP genotoxicity.45,77 ROS may attack bases, deoxyribose or DNA main chains, resulting in nonbulky (nonhelix distortion) and bulky (helix distortion) damage.78,79 ROS may also attack chromosomes, causing chromosome breakage and fragmentation.
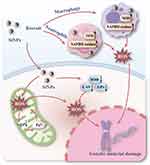
Mitochondrial dysfunction is one of the mechanisms of SiNP-induced oxidative stress damage in genetic material. On the one hand, owing to the silicon-bonded hydroxyl groups on the particle surface and the unsaturated bonds, SiNPs have a high oxidation capacity, which can induce the electron respiratory transmission chain on the inner membrane of mitochondria to produce ROS (Figure 6A).80,81 On the other hand, exposure to SiNPs may lead to the depletion of intracellular antioxidant defense systems, including antioxidant enzymes and reduced glutathione (GSH), resulting in the accumulation of ROS (Figure 6B).66,82 In vivo studies have shown that SiNPs reduce the activities of superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx) and induce excessive ROS production.83 The excessive production of ROS results in mitochondrial swelling and crista rupture (Figure 6C).84 Moreover, ROS also damage mitochondrial DNA, leading to a reduction in the mitochondrial membrane potential (MMP) and mitochondrial dysfunction.85 These changes cause defects in the transport of mitochondrial iron sulfur cluster proteins and the accumulation of Fe2+ in mitochondria, further aggravating oxidative stress damage.86 Therefore, a vicious cycle caused by SiNPs occurs between mitochondrial dysfunction and mitochondrial oxidative stress, further promoting the accumulation of ROS in cells and damaging genetic material.
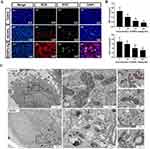 |
Figure 6 SiNPs induce ROS production and disrupt mitochondrial structure. (A) ROS assay of hCECs exposed to 100 μg/mL MSiNPs or MSiNP-Ag+ for 24 h. Immunofluorescence images of specific markers in hCECs, including intracellular ROS (red), DAPI (blue) and FITC (green), are shown. The ROS levels of hCECs increased in the MSiNP and MSiNP-Ag+ groups. Source: Reprinted with permission from Royal Society of Chemistry, Chen X, Zhu S, Hu X, et al. Toxicity and mechanism of mesoporous silica nanoparticles in eyes. Nanoscale. 2020;12(25):13637–13653. permission conveyed through Copyright Clearance Center, Inc.80 (B) SiNPs decreased the activities of SOD and GSH-Px in a dose-dependent manner in the aortic arch of Sprague‒Dawley rats after 30 days of exposure via intratracheal instillation. **P < 0.01. Source: Reprinted with permission from Feng L, Yang X, Liang S, et al. Silica nanoparticles trigger the vascular endothelial dysfunction and prethrombotic state via miR-451 directly regulating the IL6R signaling pathway. Part Fibre Toxicol. 2019;16(1):16. (http://creativecommons.org/licenses/by/4.0/).66 (C) TEM image of mitochondrial morphology after exposure to SiNPs (50 μg/mL, 24 h). More aberrantly shaped mitochondria were observed (red arrow) in SiNPs group. The mitochondria in the SiNPs group were mainly short rod-shaped. Scale bars: 2 μm or 500 nm. Source: Reprinted with permission from Royal Society of Chemistry, Qi Y, Ma R, Li X, et al. Disturbed mitochondrial quality control involved in hepatocytotoxicity induced by silica nanoparticles. Nanoscale. 2020;12(24):13034–13045. permission conveyed through Copyright Clearance Center, Inc.84 MSiNPs, mesoporous SiNPs; MSiNPs-Ag+, silver ion-adsorbed mesoporous SiNPs; SOD, superoxide dismutase; GSH-Px, glutathione peroxidase; hCECs, primary human corneal epithelial cells; Mt, mitochondrion; ER, endoplasmic reticulum; Av, autophagic vacuole. |
In addition to directly affecting mitochondrial function, amorphous SiNPs promote macrophage or neutrophil aggregation through inflammation and produce reactive oxygen species (ROS) and reactive nitrogen species (RNS) by upregulating NADPH oxidase and nitric oxide synthase (NOS), which cause genetic material damage to cells at the site of inflammation (Figure 7).5,87
Regulatory Mechanism of DNA Methylation
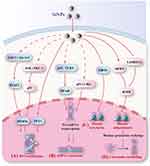
DNA methylation refers to the transfer of a methyl group from S-adenosylmethionine to the fifth carbon of a cytosine residue to form 5-methylcytosine (5-meC) under the catalysis of DNA methyltransferases (DNMTs).55 Multiple mechanisms are involved in DNA methylation induced by SiNPs, including the regulation of DNMTs, ten-eleven translocation (TET) and DNA methylation substrates (Figure 8).
In mammals, there are three types of DNMTs: DNMT1, DNMT3a, and DNMT3b. DNMT1 is a DNA methylation maintenance factor that maintains DNA methylation patterns during replication. DNMT3a and DNMT3b are de novo synthesized DNMTs that transfer methyl groups to DNA. Generally, hypermethylation is related to the overexpression of DNMTs, whereas hypomethylation is related to a lack of DNMTs.88 SiNPs induce whole-genome hypomethylation by decreasing the expression of DNMT1 and DNMT3a in HaCaT cells (Figure 9A).89 Further investigation revealed that knockdown of DNMT1 restored the normal expression and promoter methylation levels of PARP-1, indicating that hypermethylation of the PARP-1 promoter induced by SiNPs is mediated by DNMT1.90 The food-grade SiNPs E 551 slightly increased DNMT3a expression, leading to hypermethylation, which was subsequently inherited by the offspring.60 SiNPs may promote the transcription of DNMT3a and DNMT3b by activating the JAK2/STAT3 or Akt/Src/STAT3 pathway, which can induce hypermethylation.91–95 SiNPs decrease DNMT1 and DNMT3a expression by activating JNK or PKC-δ to phosphorylate p53 or cause the cytoplasmic localization of p53, leading to hypomethylation.43,96–98
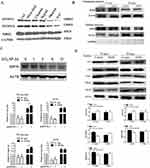 |
Figure 9 SiNPs induce epigenetic changes by regulating enzymes. (A) 15-nm SiNPs (10μg/mL, 24 h) exposure decreased DNMT1 and DNMT3a protein levels in HaCaT cells. Source: Reprinted from Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397(3):397–400. Copyright 2010, with permission from Elsevier.89 (B) SiNPs (20 mg/kg.bw) exposure for 35 days inhibited RNF8 expression in nuclear extracts of testes. After the 15-day withdrawal period, the RNF8 levels recovered. Source: Reprinted from Liu J, Li X, Zhou G, et al. Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ Pollut. 2021;284:117446. Copyright 2021, with permission from Elsevier.69 (C) SiNPs (50 μg/mL) decreased SIRT6 protein expression in A549 cells. A549 cells were infected with a virus expressing shRNA targeting SIRT6 or control shRNA. ChIP analysis was performed with antibodies against Ac-H3K9, Ac-H3K56 or control IgG and analyzed by qPCR. SIRT6 reduced Ac-H3K9 and Ac-H3K56 levels at the FST promoter region. The ACTB gene was used as a control. *P < 0.05, **P < 0.01. Source: Reprinted from Zhang L, Han B, Xiang J, Liu K, Dong H, Gao X. Silica nanoparticle releases SIRT6-induced epigenetic silencing of follistatin. Int J Biochem Cell Biol. 2018;95:27–34. Copyright 2018, with permission from Elsevier.70 (D) SiNPs (20 mg/kg.bw) altered the levels of histones and protamine. On day 35 after the first dose, histones (H2A, H2B, H3, H4) were dramatically upregulated, and TNP1, PRM1 and PRM2 were downregulated in the SiNP group. After the 15-day withdrawal period, the injury was reversed. n=5 for each group. *P< 0.05. Source: Reprinted from Liu J, Li X, Zhou G, et al. Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ Pollut. 2021;284:117446. Copyright 2021, with permission from Elsevier.69 Micro-SiO2, microsized SiNPs; DAC, 5-aza-deoxycytidine, a DNA methyltransferase inhibitor; DNMT, DNA methyltransferase; MBD2, methyl-CpG binding protein 2; RNF8, ring finger protein 8; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SIRT6, sirtuin 6; ACTB, beta-actin; FST, follistatin; A549 cells, lung epithelial cells; ChIP, chromatin immunoprecipitation; Ac-H3K9, acetylated H3K9; Ac-H3K56, acetylated H3K56. |
DNA demethylation is mediated by DNA demethylases, which are referred mainly to as the TET protein family.99 The TET family includes TET1, TET2, and TET3. Among them, TET1 can promote the oxidation of 5mC to 5-hydroxymethylcytosine (5-hmC). Prenatal high-dose exposure to food-grade SiNPs resulted in a 1.8-fold upregulation of TET1 expression in the maternal liver, whereas low-dose and high-dose exposure significantly downregulated TET1 expression levels in fetal liver tissue.60 At present, the specific mechanism by which SiNPs regulate TET1 is still unclear. Furthermore, no studies have reported the effects of SiNPs on the expression of TET2 and TET3, and future research on the mechanism of SiNP-induced DNA methylation is needed.
In addition to regulating related enzymes, SiNPs may block DNA as a substrate for DNMTs, resulting in abnormal DNA methylation. The ROS generated by SiNPs attack DNA to form 8-hydroxy-2ʹ-deoxyguanosine (8-OHdG), which inhibits DNA methylation of nearby cytosine bases to induce DNA hypomethylation. Another type of DNA oxidative damage product, 5hmC, can promote DNA demethylation, leading to DNA hypomethylation.100 Inflammation caused by SiNPs damages DNA,101–103 and damage products such as 5-chlorocytosine and 5-bromocytidine stimulate 5-meC to increase the binding of methyl-binding proteins, thus promoting abnormal hypermethylation.104
Regulatory Mechanism of microRNA Expression
miRNAs cause gene silencing by inhibiting mRNA translation or degrading mRNAs.105 Posttranscriptional regulation of gene expression by miRNAs is one of the key mechanisms of the epigenetic toxicity of SiNPs. In the nucleus, miRNA genes are transcribed into pri-miRNAs. Pri-miRNAs are processed in the nucleus and cytoplasm to ultimately produce mature miRNAs.106 SiNPs may affect the transcription of miRNA genes by regulating transcription factors and subsequently altering the expression of miRNAs (Figure 8). These transcription factors may include NF-κB, p53 and c-Myc.
NF-κB regulates the expression of various miRNAs, such as by promoting the transcription of miRNA-146 and miRNA-21 and inhibiting the transcription of miRNA-29 and miRNA let-7.107 SiNPs activate NF-κB through p62 or TLR4. In BEAS-2B cells, SiNPs not only damage lysosomes and block autophagic flux but also stimulate Nrf2 translocation to the nucleus, where it binds to the p62 promoter and activates p62 transcription. These two mechanisms lead to the accumulation of p62, activating the p62/TRAF6/NF-κB pathway.108,109 Moreover, amorphous SiNPs promote the translocation and release of HMGB1 from the nucleus to the cytoplasm and the expression of TLR4 in HUVECs. Subsequently, TLR4 binds to HMGB1, increasing the expression of MyD88 and ultimately activating the NF-κB signaling pathway.102
Exposure to SiNPs can induce p53 activation.98 p53 regulates the transcription of miRNAs, such as those in the miRNA-34 family, and promotes Drosha-mediated editing of pri-miRNAs into precursor miRNAs.110 c-Myc regulates the transcription of miRNAs, including miRNA-17 and miRNA-34.111 Amorphous SiNP NM-203 has been shown to increase the mRNA and protein levels of c-Myc, possibly by triggering the transcription of the P1 promoter of the c-Myc gene. However, low concentrations of NM-203 increased c-Myc protein levels but did not affect mRNA expression levels.112 This finding indicates that other mechanisms may be involved. SiNPs may also promote c-Myc expression by activating p38 MAPK or ERK1/2.36,113,114
Regulatory Mechanisms of Histone Modification and the Inhibition of Chromatin Remodeling
As mentioned earlier, the histone modifications caused by SiNPs include ubiquitination, phosphorylation, and acetylation. Ring finger protein 8 (RNF8) is a ubiquitin ligase, E3, that regulates the ubiquitination of H2A and H2B and promotes the exchange of histones with protamines.115 SiNPs led to a reduction in lethal (3) malignant brain tumor-like 2 (L3MBTL2) levels, which inhibited RNF8-ubH2A/ubH2B and affected spermatogenesis.35 Moreover, SiNPs can reduce RNF8 in the sperm nucleus by increasing PIWI-like protein 1 (MIWI) expression or inhibiting the degradation of MIWI (Figure 9B).69 SiNPs may also activate S-phase kinase associated protein (Skp2). Skp2 is also an important E3 ubiquitin ligase. Akt and AMPK activated by SiNPs are upstream regulatory molecules of Skp2.116–119 These findings suggest that Skp2 may be one of the mechanisms by which SiNPs regulate histone ubiquitination.
Histone acetylation is mediated by histone acetyltransferase (HAT) and histone deacetylase (HDAC). Sirtuin 6 (SIRT6) is an H3K9 and H3K56 hDAC that can regulate the deacetylation of histones. Research has shown that the overexpression of SIRT6 leads to a decrease in H3K9 and H3K56 acetylation levels.120 Exposure to SiNPs has been shown to inhibit SIRT6 expression and increase the acetylation levels of H3K9 and H3K56 at the follistatin (FST) gene promoter region (Figure 9C). Further research revealed that SiNPs shorten the half-life of SIRT6 mRNA without altering the activity of the SIRT6 promoter.70 These findings suggest that SiNPs may reduce the expression of SIRT6 through a posttranscriptional regulatory mechanism. SiNPs may play a posttranscriptional regulatory role in SIRT6 by activating PI3K. A previous study reported that SiNPs activate the PI3K pathway.121 PI3K activation promotes miRNA-34a expression. miRNA-34a binds to the 3ʹ-UTR of SIRT6 mRNA, reducing its stability and leading to its degradation.122
SiNPs inhibit chromatin remodeling by inhibiting histone ubiquitination (Figure 8). The ubiquitination of H2A and H2B promotes the removal of histones, which is beneficial for replacing histones with histone–protamines. SiNPs inhibited histone‒protamine exchange by decreasing H2A and H2B ubiquitination (Figure 9B and D), resulting in the inhibition of chromatin remodeling.35,69 Histone acetylation also regulates chromatin remodeling. Histone variant hyperacetylation is beneficial for chromatin remodeling. During the process of histone‒protamine exchange, histones are first replaced by histone variants. Histone variant hyperacetylation leads to the unwinding of nucleosome DNA, resulting in a loose chromatin structure. Subsequently, topoisomerase II beta causes DNA breaks, promotes the removal of histone variants, and is replaced by transition proteins.74 NPs can reduce the acetylation level of histones. For example, zinc oxide nanoparticles reduce the acetylation level of histone H4K5.123 Nickel nanoparticles reduce acetylation levels of histone H3 in human bronchial epithelial cells.124 A previous study reported that SiNPs alter histone H3K9 and H3K56 acetylation levels, but their effects on the acetylation levels of histone variants are still unclear.70 These findings indicate that SiNPs may inhibit chromatin remodeling by inhibiting histone variant acetylation, but further research is needed for verification.
Factors Influencing the Genetic and Epigenetic Toxicity of SiNPs
Particle Size
Particle size is one of the key factors affecting the genetic and epigenetic toxicity of SiNPs. Microsized SiNPs (1–5 μm) do not induce changes in DNA methylation levels in HaCaT cells, whereas 15-nm SiNPs reduce global DNA methylation levels.89 The genotoxicity of SiNPs increases with decreasing particle size. Amorphous SiNPs of four sizes (10, 25, 50, and 100 nm) were used to study the genotoxicity of SiNPs in human umbilical vein endothelial cells. The results indicated that the smaller the particle size was, the greater the induced genotoxicity.45 This may be because smaller particles have a larger surface area per unit mass, and their reactivity increases accordingly, which can produce more ROS at a given mass.125 The cellular uptake efficiency of SiNPs of different sizes is different. Smaller NPs are more likely to penetrate the cell membrane and cause cell damage.126,127 Larger than 100 nm SiNP have been proven to have good biocompatibility and not induce genotoxicity.128 In addition, small SiNPs may aggregate into larger agglomerated particles and cause severe genotoxicity.129 Therefore, SiNPs induce genetic and epigenetic toxicity in a size-dependent manner.
Dose
The genetic and epigenetic toxicity induced by SiNPs is dose dependent.130 With increasing doses of SiNPs, the genetic and epigenetic toxicity effects increase. SiNPs inhibited the ubiquitination of histones H2A and H2B in a dose-dependent manner in the sperm of mice.35 SiNPs induce DNA breakage in rat lymphocytes in a linear dose-dependent manner.131 Similarly, the highest concentration of SiNPs (100 μg/mL) induced the most significant DNA damage in onion tissue cells.42 This occurs because the higher the concentration of NPs is, the greater the degree of cellular uptake. After passing through the cell membrane, NPs are localized mainly in lysosomes. When excessive NPs accumulate, cells eliminate excess NPs through extracellular secretion and lysosomal escape mechanisms.132,133 However, when the amount of NPs exceeds a certain threshold, the NPs accumulate in the extracellular matrix and damage the cell membrane, leading to lysosomal rupture due to lysosomal overload. This damage ultimately leads to cytotoxicity and cell death.132
Geometric Shape
SiNPs are spherical, rod shaped, plate shaped, etc. The geometric shape of SiNPs affects cell internalization. Compared with spherical mesoporous SiNPs, rod-shaped mesoporous SiNPs induce more severe genotoxicity;52 this may be due to the high aspect ratio of rod NPs, which are taken up by cells faster and in greater quantities.134 The use of mesoporous SiNPs with different aspect ratios also revealed that particle shape affected the development of actin tissue and filopodia in HeLa cells.135 In addition, at the same volume, rod-shaped NPs have a larger surface area than spherical NPs do, and the surface area available for interaction and contact with the cell membrane is larger, making them easier to absorb by cells and causing more severe genotoxicity.136 The shape of SiNPs also affects their biological distribution and clearance rate. Spherical mesoporous SiNPs are easily retained in the liver, whereas long rod-shaped mesoporous SiNPs are easily distributed in the spleen.137 The clearance rate of short rod-shaped mesoporous SiNPs is faster than that of long rod-shaped mesoporous SiNPs.138 This finding indicates that SiNPs with a longer aspect ratio have a longer residence time in the body and greater genotoxic effects. At present, it is not clear whether the shape of SiNPs affects the magnitude of epigenetic toxicity.
Structure Type
Crystalline SiNPs are arranged in a regular manner, whereas amorphous SiNPs are arranged irregularly. Different types of SiNPs have different parameters, such as morphology, porosity and crystallinity, which affect the biological distribution and genetic and epigenetic toxicity of SiNPs.139 Generally, the genetic and epigenetic toxicity effects of crystalline SiNPs are greater than those of amorphous SiNPs, possibly because the physical and chemical properties are affected by the crystal structure.112,140 In addition, the toxicity of mesoporous SiNPs has been shown to be significantly greater than that of nonporous SiNPs. This occurs because the density of mesoporous SiNPs is lower, and therefore, the weight of each particle is lower, resulting in a greater number of particles per unit mass.141
Administration Route
SiNPs can enter the human body through intravenous injection, airway inhalation, skin contact, etc. Different administration routes affect the biological distribution and local tissue concentration of SiNPs, leading to different genetic and epigenetic toxicity effects. After oral or intravenous injection, SiNPs are distributed mainly in the liver or spleen. Through tracheal instillation, they are distributed mainly in lung tissue.139 Different biological distributions result in different types of cells being exposed to SiNPs, thus affecting genotoxicity. Cabellos et al studied the genotoxicity of the oral administration of nonporous and mesoporous SiNPs and reported that DNA in several organs and tissues of rats was not damaged.142 Similarly, studies on intratracheal instillation have not shown genotoxic effects of amorphous SiNPs (15 nm and 55 nm in size).143 However, in a study by Downs et al, SiNPs (15 nm and 55 nm in size) caused DNA damage in the liver and micronucleus when injected intravenously.144 This may be because the concentration of SiNPs that reach local tissues via the intravenous route is greater than that via tracheal drip or the oral route.
Surface Charge
Compared with negatively charged SiNPs, SiNPs with a positive surface charge cause greater genotoxicity.145 On the one hand, positively charged NPs are more easily internalized by cells. The cell membrane is negatively charged because of the phospholipid layer on the outer surface. NPs with positive charges are more likely to enter cells through the cell membrane due to electrostatic interactions.146 On the other hand, positively charged SiNPs are more likely to interact with negatively charged DNA, causing damage to genetic material.138 Therefore, SiNPs with positive charges are more likely to penetrate the cell membrane, bind to negatively charged DNA, and damage DNA. However, whether surface charge of SiNPs cause epigenetic toxicity is unclear.
Other Factors
The genetic and epigenetic toxicity effects induced by SiNPs may also be related to the exposure time of the particles and the type of toxicity tested. The longer the exposure time is, the greater the dose of NPs reaching the local tissues of the organism, and the corresponding prolongation of contact time with cells results in greater genetic and epigenetic toxicity. In addition, the type of experiment can affect the experimental results. In the Ames test, spherical amorphous SiNPs were shown not to induce gene mutations.147 This is because not all particles can pass through bacterial membranes; thus, the Ames test may not be suitable for evaluating the genotoxicity of NPs. Therefore, evaluating the genotoxicity of SiNPs also requires consideration of the type of experiment.
Limitations and Future Outlook
The genetic and epigenetic toxicity of SiNPs has received increasing attention, especially their adverse effects on the reproductive system. The experimental data on the genetic and epigenetic toxicity of SiNPs to the reproductive system have been mostly derived from animal models, with a lack of actual human exposure and epidemiological data. Currently, deriving impacts on human health from animal data is not possible. In the future, emphasis should be placed on transforming basic toxicology research results into medical indicators for measuring human risk. In addition, most studies are based on the results of short-term exposure (less than 3 months) to SiNPs. Long-term in vivo exposure experiments are needed to accurately predict harm to the human body and adverse consequences for offspring.
There are inconsistent or even contradictory results regarding the genetic and epigenetic toxicity of SiNPs, which may depend on differences in particle properties (such as size, structure, and shape), exposure status (such as exposure pathway, dose, and time), and cell type. Therefore, when evaluating genetic and epigenetic toxicity, a thorough physical and chemical characterization of SiNPs, including size, surface area, shape, crystallinity, porosity, surface charge, agglomeration, etc., is needed. Second, differences in evaluation methods should be considered to avoid false-negative results. It is necessary to establish a standard guideline for evaluation. In addition, a comprehensive and complete evaluation of the epigenetic changes induced by SiNPs is needed. Owing to the ability of SiNPs to induce multiple abnormal epigenetic changes, an aim of future research should be to simultaneously detect multiple types of epigenetic changes, including at least DNA methylation, miRNA expression changes, and histone modifications.
Exploring the potential mechanisms of SiNPs-induced genetic and epigenetic toxicity will be beneficial for developing new treatment strategies and reducing the adverse effects of SiNPs genetic and epigenetic toxicity on human health. However, there are still many gaps in our understanding of how SiNPs induce epigenetic toxicity. In the future, it will be necessary to further explore the potential molecular mechanisms by which SiNPs induce changes in miRNA expression, such as how SiNPs affect the maturation and processing of miRNAs. The effects of SiNPs on enzymes related to histone modification, such as E1, E2 and HAT, as well as how to regulate chromatin remodeling processes through histone modification, are all worthy of attention.
Avoiding and reducing toxicity risk are urgent tasks. First, an assessment standard for SiNPs should be established. The best testing and testing conditions should be selected to increase the likelihood of extrapolating genetic and epigenetic toxicity results to human risk. Second, safe and nontoxic SiNPs should be designed and developed to reduce genetic and epigenetic toxicity: (1) the size, shape, and structure of SiNPs should be optimized; (2) by doping, metal loading should be used to reduce the oxidative stress damage caused by SiNPs; and (3) the surface chemical properties and in vivo clearance rate of SiNPs should be modified through functional group surface modification. Third, researchers should focus on the key targets of genetic and epigenetic toxicity mechanisms. For example, key mechanistic targets for regulating the expression of miRNAs, such as NF-κB and p53, have been identified. The development of drugs that inhibit these pathways in clinical practice and block key components of these pathways, such as upstream stimuli or downstream targets, could be used to treat SiNP genetic and epigenetic toxicity. These methods provide broader application prospects for SiNPs and can improve their application in the biomedical field.
Conclusion
The genetic and epigenetic toxicity induced by SiNPs, especially damage to germ cells, cause a risk of infertility and teratogenesis. SiNPs induce genetic material damage, which manifests as gene mutations, DNA strand breaks, and chromosomal aberrations. The main mechanism is oxidative stress damage mediated by mitochondrial dysfunction and inflammation. Furthermore, the molecular mechanisms of epigenetic toxicity are discussed in detail for the first time, SiNPs alter DNA methylation, miRNA expression, histone modification and inhibit chromatin remodeling by regulating epigenetic-related enzymes and transcription factors. The potential solutions to avoid toxicity were prospected and provide a guidance for better application of SiNPs in the biomedical field. In the future, it will be necessary to focus on the key targets of genetic and epigenetic toxicity mechanisms. How to reduce the genetic and epigenetic risk or toxicity of SiNPs is still challenging, and material modification or the synthesis of safe SiNPs is urgently needed.
Acknowledgments
This work was supported by the National Natural Science Fund (grant numbers: 81600904) and Guangzhou Science and Technology Plan Project jointly funded by City and Universities (grant numbers: 2023A03J0328).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Li Z, Mu Y, Peng C, Lavin MF, Shao H, Du Z. Understanding the mechanisms of silica nanoparticles for nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2021;13(1):e1658. doi:10.1002/wnan.1658
2. Ghaferi M, Koohi Moftakhari Esfahani M, Raza A, Al Harthi S, Ebrahimi Shahmabadi H, Alavi SE. Mesoporous silica nanoparticles: synthesis methods and their therapeutic use-recent advances. J Drug Target. 2021;29(2):131–154. doi:10.1080/1061186x.2020.1812614
3. Huang Y, Li P, Zhao R, et al. Silica nanoparticles: biomedical applications and toxicity. Biomed Pharmacother. 2022;151:113053. doi:10.1016/j.biopha.2022.113053
4. Ding R, Li Y, Yu Y, Sun Z, Duan J. Prospects and hazards of silica nanoparticles: biological impacts and implicated mechanisms. Biotechnol Adv. 2023;69:108277. doi:10.1016/j.biotechadv.2023.108277
5. Inoue M, Sakamoto K, Suzuki A, et al. Size and surface modification of silica nanoparticles affect the severity of lung toxicity by modulating endosomal ROS generation in macrophages. Part Fibre Toxicol. 2021;18(1):21. doi:10.1186/s12989-021-00415-0
6. Abulikemu A, Zhao X, Xu H, et al. Silica nanoparticles aggravated the metabolic associated fatty liver disease through disturbed amino acid and lipid metabolisms-mediated oxidative stress. Redox Biol. 2023;59:102569. doi:10.1016/j.redox.2022.102569
7. Wang F, Liang Q, Ma Y, et al. Silica nanoparticles induce pyroptosis and cardiac hypertrophy via ROS/NLRP3/Caspase-1 pathway. Free Radic Biol Med. 2022;182:171–181. doi:10.1016/j.freeradbiomed.2022.02.027
8. Ning X, Li X, Ma K, et al. VDAC1 protein regulation of oxidative damage and mitochondrial dysfunction-mediated cytotoxicity by silica nanoparticles in SH-SY5Y cells. Mol Neurobiol. 2023;60(11):6542–6555. doi:10.1007/s12035-023-03491-9
9. Tassinari R, Martinelli A, Valeri M, Maranghi F. Amorphous silica nanoparticles induced spleen and liver toxicity after acute intravenous exposure in male and female rats. Toxicol Ind Health. 2021;37(6):328–335. doi:10.1177/07482337211010579
10. Sasai F, Rogers KL, Orlicky DJ, et al. Inhaled silica nanoparticles cause chronic kidney disease in rats. Am J Physiol Renal Physiol. 2022;323(1):F48–F58. doi:10.1152/ajprenal.00021.2022
11. Marques Da Silva V, Benjdir M, Montagne P, Pairon J-C, Lanone S, Andujar P. Pulmonary toxicity of silica linked to its micro- or nanometric particle size and crystal structure: a review. Nanomaterials. 2022;12(14):2392. doi:10.3390/nano12142392
12. Ao LH, Wei YG, Tian HR, Zhao H, Li J, Ban JQ. Advances in the study of silica nanoparticles in lung diseases. Sci Total Environ. 2024;912:169352. doi:10.1016/j.scitotenv.2023.169352
13. Chen L, Liu J, Zhang Y, et al. The toxicity of silica nanoparticles to the immune system. Nanomedicine. 2018;13(15):1939–1962. doi:10.2217/nnm-2018-0076
14. Yu J, Dan N, Eslami SM, Lu X. State of the art of silica nanoparticles: an overview on biodistribution and preclinical toxicity studies. Aaps J. 2024;26(3):35. doi:10.1208/s12248-024-00906-w
15. Ravegnini G, Sammarini G, Hrelia P, Angelini S. Key genetic and epigenetic mechanisms in chemical carcinogenesis. Toxicol Sci. 2015;148(1):2–13. doi:10.1093/toxsci/kfv165
16. Choudhuri S, Kaur T, Jain S, Sharma C, Asthana S. A review on genotoxicity in connection to infertility and cancer. Chem Biol Interact. 2021;345:109531. doi:10.1016/j.cbi.2021.109531
17. Chakraborty A, Ghosh S, Biswas B, Pramanik S, Nriagu J, Bhowmick S. Epigenetic modifications from arsenic exposure: a comprehensive review. Sci Total Environ. 2022;810:151218. doi:10.1016/j.scitotenv.2021.151218
18. Paul P, Malakar AK, Chakraborty S. The significance of gene mutations across eight major cancer types. Mutat Res Rev Mutat Res. 2019;781:88–99. doi:10.1016/j.mrrev.2019.04.004
19. Martinez-Jimenez F, Muinos F, Sentis I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi:10.1038/s41568-020-0290-x
20. Yin XH, Xu YM, Lau ATY. Nanoparticles: excellent materials yet dangerous when they become airborne. Toxics. 2022;10(2):50. doi:10.3390/toxics10020050
21. Armstead AL, Li B. Nanotoxicity: emerging concerns regarding nanomaterial safety and occupational hard metal (WC-Co) nanoparticle exposure. Int J Nanomed. 2016;11:6421–6433. doi:10.2147/IJN.S121238
22. Nowak-Pasternak J, Lipińska-Ojrzanowska A, Świątkowska B. Silicosis after short-term exposure. Occup Med. 2023;73(1):33–35. doi:10.1093/occmed/kqac113
23. Guo C, Wang J, Yang M, et al. Amorphous silica nanoparticles induce malignant transformation and tumorigenesis of human lung epithelial cells via P53 signaling. Nanotoxicology. 2017;11(9–10):1176–1194. doi:10.1080/17435390.2017.1403658
24. Fontana C, Kirsch A, Seidel C, et al. In vitro cell transformation induced by synthetic amorphous silica nanoparticles. Mutat Res Genet Toxicol Environ Mutagen. 2017;823:22–27. doi:10.1016/j.mrgentox.2017.08.002
25. Xie D, Zhou Y, Luo X. Amorphous silica nanoparticles induce tumorigenesis via regulating ATP5H/SOD1-related oxidative stress, oxidative phosphorylation and EIF4G2/PABPC1-associated translational initiation. PeerJ. 2019;7:e6455. doi:10.7717/peerj.6455
26. Baran V, Pisko J. Cleavage of early mouse embryo with damaged DNA. Int J Mol Sci. 2022;23(7):3516. doi:10.3390/ijms23073516
27. Valadares LPDA, Lima LCO, Saboia-Morais SMTD, et al. Embryotoxicity of silica nanoparticles in the drug delivery of domperidone in zebrafish. Aquat Toxicol. 2023;258:106454. doi:10.1016/j.aquatox.2023.106454
28. Bai Y, Li FF, Zhang Y, Ding YB. Silicon dioxide nanoparticles compromise decidualization via autophagy impairment to possibly cause embryo resorption. Toxicol Lett. 2023;381:72–82. doi:10.1016/j.toxlet.2023.05.003
29. Li X, Dang J, Li Y, et al. Developmental neurotoxicity fingerprint of silica nanoparticles at environmentally relevant level on larval zebrafish using a neurobehavioral-phenomics-based biological warning method. Sci Total Environ. 2021;752:141878. doi:10.1016/j.scitotenv.2020.141878
30. Zhu B, Lei L, Fu K, et al. Neurotoxicity of tetrabromobisphenol A and SiO2 nanoparticle co-exposure in zebrafish and barrier function of the embryonic chorion. Sci Total Environ. 2022;845:157364. doi:10.1016/j.scitotenv.2022.157364
31. Liu P, Wang S, Chang Z, Li L, Xing H, Dong WF. Combined toxicity of silica nanoparticles and cadmium chloride on the cardiovascular system of zebrafish (Danio rerio) larvae. Comp Biochem Physiol C Toxicol Pharmacol. 2021;239:108895. doi:10.1016/j.cbpc.2020.108895
32. Liu P, Zhao Y, Wang S, Xing H, Dong WF. Effect of combined exposure to silica nanoparticles and cadmium chloride on female zebrafish ovaries. Environ Toxicol Pharmacol. 2021;87:103720. doi:10.1016/j.etap.2021.103720
33. Yamashita K, Yoshioka Y, Higashisaka K, et al. Silica and titanium dioxide nanoparticles cause pregnancy complications in mice. Nat Nanotechnol. 2011;6(5):321–328. doi:10.1038/nnano.2011.41
34. Guo Z, Wang X, Zhang P, et al. Silica nanoparticles cause spermatogenesis dysfunction in mice via inducing cell cycle arrest and apoptosis. Ecotoxicol Environ Saf. 2022;231:113210. doi:10.1016/j.ecoenv.2022.113210
35. Liu J, Li X, Zhou G, et al. Silica nanoparticles induce spermatogenesis disorders via L3MBTL2-DNA damage-p53 apoptosis and RNF8-ubH2A/ubH2B pathway in mice. Environ Pollut. 2020;265(Pt A):114974. doi:10.1016/j.envpol.2020.114974
36. Meng F, Hao H, Guo Z, et al. Silica nanoparticles induces sperm granuloma formation and blood-epididymal barrier disruption via the p38 MAPK pathway in mice. Food Chem Toxicol. 2023;182:114113. doi:10.1016/j.fct.2023.114113
37. Chen F, Sun J, Wang Y, et al. Silica nanoparticles induce ovarian granulosa cell apoptosis via activation of the PERK-ATF4-CHOP-ERO1α pathway-mediated IP3R1-dependent calcium mobilization. Cell Biol Toxicol. 2023;39(4):1715–1734. doi:10.1007/s10565-022-09776-4
38. Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. doi:10.1007/978-981-15-3449-2_1
39. Yazdimamaghani M, Moos PJ, Dobrovolskaia MA, Ghandehari H. Genotoxicity of amorphous silica nanoparticles: status and prospects. Nanomedicine. 2019;16:106–125. doi:10.1016/j.nano.2018.11.013
40. Demir E, Castranova VJTR. Genotoxic effects of synthetic amorphous silica nanoparticles in the mouse lymphoma assay. Toxicol Rep. 2016;3:807–815. doi:10.1016/j.toxrep.2016.10.006
41. Park MVDZ, Verharen HW, Zwart E, et al. Genotoxicity evaluation of amorphous silica nanoparticles of different sizes using the micronucleus and the plasmid lacZ gene mutation assay. Nanotoxicology. 2011;5(2):168–181. doi:10.3109/17435390.2010.506016
42. Liman R, Acikbas Y, Cigerci IH, Ali MM, Kars MD. Cytotoxic and genotoxic assessment of silicon dioxide nanoparticles by allium and comet tests. Bull Environ Contam Toxicol. 2020;104(2):215–221. doi:10.1007/s00128-020-02783-3
43. Zhang J, Liu J, Ren L, et al. Silica nanoparticles induce abnormal mitosis and apoptosis via PKC-delta mediated negative signaling pathway in GC-2 cells of mice. Chemosphere. 2018;208:942–950. doi:10.1016/j.chemosphere.2018.05.178
44. Villani P, Eleuteri P, Pacchierotti F, et al. Pyrogenic synthetic amorphous silica (NM-203): genotoxicity in rats following sub-chronic oral exposure. Mutat Res Genet Toxicol Environ Mutagen. 2022;876–877:503458. doi:10.1016/j.mrgentox.2022.503458
45. Zhou F, Liao F, Chen L, Liu Y, Wang W, Feng S. The size-dependent genotoxicity and oxidative stress of silica nanoparticles on endothelial cells. Environ Sci Pollut Res Int. 2019;26(2):1911–1920. doi:10.1007/s11356-018-3695-2
46. Wu L, Zhang P, Zhou H, et al. Molecular dynamics simulation of the interaction between graphene oxide quantum dots and DNA fragment. Materials. 2022;15(23):8506. doi:10.3390/ma15238506
47. Kong Z, Hu W, Jiao F, et al. Theoretical evaluation of DNA genotoxicity of graphene quantum dots: a combination of density functional theory and molecular dynamics simulations. J Phys Chem B. 2020;124(42):9335–9342. doi:10.1021/acs.jpcb.0c05882
48. Liu J, Yang M, Jing L, et al. Silica nanoparticle exposure inducing granulosa cell apoptosis and follicular atresia in female Balb/c mice. Environ Sci Pollut Res Int. 2018;25(4):3423–3434. doi:10.1007/s11356-017-0724-5
49. Flasz B, Dziewiecka M, Ajay AK, et al. Age- and lifespan-dependent differences in GO caused DNA damage in acheta domesticus. Int J Mol Sci. 2022;24(1):290. doi:10.3390/ijms24010290
50. Ishino K, Kato T, Kato M, et al. Comprehensive DNA adduct analysis reveals pulmonary inflammatory response contributes to genotoxic action of magnetite nanoparticles. Int J Mol Sci. 2015;16(2):3474–3492. doi:10.3390/ijms16023474
51. Li L, Deng Y, Meng X, et al. Genotoxicity evaluation of silica nanoparticles in murine: a systematic review and meta-analysis. Toxicol Mech Methods. 2022;32(1):1–17. doi:10.1080/15376516.2021.1965277
52. Niu M, Zhong H, Shao H, et al. Shape-dependent genotoxicity of mesoporous silica nanoparticles and cellular mechanisms. J Nanosci Nanotechnol. 2016;16(3):2313–2318. doi:10.1166/jnn.2016.10928
53. Nallathamby PD, Xu XH. Study of cytotoxic and therapeutic effects of stable and purified silver nanoparticles on tumor cells. Nanoscale. 2010;2(6):942–952. doi:10.1039/c0nr00080a
54. Ivask A, Voelcker NH, Seabrook SA, et al. DNA melting and genotoxicity induced by silver nanoparticles and graphene. Chem Res Toxicol. 2015;28(5):1023–1035. doi:10.1021/acs.chemrestox.5b00052
55. Mattei AL, Bailly N, Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38(7):676–707. doi:10.1016/j.tig.2022.03.010
56. Zhang W, Liu S, Han D, He Z. Engineered nanoparticle-induced epigenetic changes: an important consideration in nanomedicine. Acta Biomater. 2020;117:93–107. doi:10.1016/j.actbio.2020.09.034
57. Angeloni A, Bogdanovic O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019;63(6):707–715. doi:10.1042/ebc20190030
58. Sang Y, Liu J, Li X, et al. The effect of SiNPs on DNA methylation of genome in mouse spermatocytes. Environ Sci Pollut Res Int. 2021;28(32):43684–43697. doi:10.1007/s11356-021-13459-8
59. Sang Y, Liu J, Dong X, et al. Silica nanoparticles induce male reproductive toxicity via Crem hypermethylation mediated spermatocyte apoptosis and sperm flagella damage. Environ Sci Pollut Res. 2024;31(9):13856–13866. doi:10.1007/s11356-024-32046-1
60. Zhan Y, Lou H, Shou R, et al. Maternal exposure to E 551 during pregnancy leads to genome-wide DNA methylation changes and metabolic disorders in the livers of pregnant mice and their fetuses. J Hazard Mater. 2024;465:133233. doi:10.1016/j.jhazmat.2023.133233
61. Sooklert K, Nilyai S, Rojanathanes R, et al. N-acetylcysteine reverses the decrease of DNA methylation status caused by engineered gold, silicon, and chitosan nanoparticles. Int J Nanomed. 2019;14:4573–4587. doi:10.2147/ijn.S204372
62. Ferragut Cardoso AP, Banerjee M, Nail AN, Lykoudi A, States JC. miRNA dysregulation is an emerging modulator of genomic instability. Semin Cancer Biol. 2021;76:120–131. doi:10.1016/j.semcancer.2021.05.004
63. Zhou G, Liu J, Li X, et al. Silica nanoparticles inducing the apoptosis via microRNA-450b-3p targeting MTCH2 in mice and spermatocyte cell. Environ Pollut. 2021;277:116771. doi:10.1016/j.envpol.2021.116771
64. Zhou G, Wang J, Ren L, et al. Silica nanoparticles suppressed the spermatogenesis via downregulation of miR-450b-3p by targeting Layilin in spermatocyte of mouse. Environ Pollut. 2023;318:120864. doi:10.1016/j.envpol.2022.120864
65. Zhao M, Zhou G, Wang J, et al. MiR-5622-3p inhibits ZCWPW1 to induce apoptosis in silica-exposed mice and spermatocyte cells. Nanotoxicology. 2023;17(4):372–384. doi:10.1080/17435390.2023.2223632
66. Feng L, Yang X, Liang S, et al. Silica nanoparticles trigger the vascular endothelial dysfunction and prethrombotic state via miR-451 directly regulating the IL6R signaling pathway. Part Fibre Toxicol. 2019;16(1):16. doi:10.1186/s12989-019-0300-x
67. Zhou G, Ren L, Yin H, et al. The alterations of miRNA and mRNA expression profile and their integration analysis induced by silica nanoparticles in spermatocyte cells. NanoImpact. 2021;23:100348. doi:10.1016/j.impact.2021.100348
68. Ren L, Liu J, Zhang J, et al. Silica nanoparticles induce spermatocyte cell autophagy through microRNA-494 targeting AKT in GC-2spd cells. Environ Pollut. 2019;255(Pt 1):113172. doi:10.1016/j.envpol.2019.113172
69. Liu J, Li X, Zhou G, et al. Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ Pollut. 2021;284:117446. doi:10.1016/j.envpol.2021.117446
70. Zhang L, Han B, Xiang J, Liu K, Dong H, Gao X. Silica nanoparticle releases SIRT6-induced epigenetic silencing of follistatin. Int J Biochem Cell Biol. 2018;95:27–34. doi:10.1016/j.biocel.2017.12.011
71. Toubhans B, Alkafri N, Quintela M, et al. Selenium nanoparticles modulate histone methylation via lysine methyltransferase activity and S-adenosylhomocysteine depletion. Redox Biol. 2023;61:102641. doi:10.1016/j.redox.2023.102641
72. Morrison AJ. Chromatin-remodeling links metabolic signaling to gene expression. Mol Metab. 2020;38:100973. doi:10.1016/j.molmet.2020.100973
73. Xu R, Li C, Liu X, Gao S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell. 2021;12(1):7–28. doi:10.1007/s13238-020-00757-z
74. Hao SL, Ni FD, Yang WX. The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene. 2019;706:201–210. doi:10.1016/j.gene.2019.05.027
75. Gao G, Ze Y, Zhao X, et al. Titanium dioxide nanoparticle-induced testicular damage, spermatogenesis suppression, and gene expression alterations in male mice. J Hazard Mater. 2013;258–259:133–143. doi:10.1016/j.jhazmat.2013.04.046
76. Nazar M, Talebi AR, Hosseini Sharifabad M, Abbasi A, Khoradmehr A, Danafar AH. Acute and chronic effects of gold nanoparticles on sperm parameters and chromatin structure in Mice. Int J Reprod Biomed. 2016;14(10):637–642.
77. Tarantini A, Lanceleur R, Mourot A, et al. Toxicity, genotoxicity and proinflammatory effects of amorphous nanosilica in the human intestinal Caco-2 cell line. Toxicol In Vitro. 2015;29(2):398–407. doi:10.1016/j.tiv.2014.10.023
78. Juan CA, Perez de la Lastra JM, Plou FJ, Perez-Lebena E. The chemistry of Reactive Oxygen Species (ROS) revisited: outlining their role in biological macromolecules (DNA, Lipids and Proteins) and induced pathologies. Int J Mol Sci. 2021;22(9):4642. doi:10.3390/ijms22094642
79. Lee TH, Kang TH. DNA oxidation and excision repair pathways. Int J Mol Sci. 2019;20(23):6092. doi:10.3390/ijms20236092
80. Chen X, Zhu S, Hu X, et al. Toxicity and mechanism of mesoporous silica nanoparticles in eyes. Nanoscale. 2020;12(25):13637–13653. doi:10.1039/d0nr03208e
81. Guo C, Zhao X, Ma R, et al. Silica nanoparticles promoted pro-inflammatory macrophage and foam cell transformation via ROS/PPARγ/NF-κB signaling. Sci Total Environ. 2023;881:163430. doi:10.1016/j.scitotenv.2023.163430
82. Jiang X, Gao H, Cao Y, et al. SiNPs induce ferroptosis in HUVECs through p38 inhibiting NrF2 pathway. Front Public Health. 2023;11:1024130. doi:10.3389/fpubh.2023.1024130
83. Aouey B, Boukholda K, Gargouri B, et al. Silica nanoparticles induce hepatotoxicity by triggering oxidative damage, apoptosis, and Bax-Bcl2 signaling pathway. Biol Trace Elem Res. 2022;200(4):1688–1698. doi:10.1007/s12011-021-02774-3
84. Qi Y, Ma R, Li X, et al. Disturbed mitochondrial quality control involved in hepatocytotoxicity induced by silica nanoparticles. Nanoscale. 2020;12(24):13034–13045. doi:10.1039/d0nr01893g
85. Wu R, Högberg J, Adner M, Ramos-Ramírez P, Stenius U, Zheng H. Crystalline silica particles cause rapid NLRP3-dependent mitochondrial depolarization and DNA damage in airway epithelial cells. Part Fibre Toxicol. 2020;17(1):39. doi:10.1186/s12989-020-00370-2
86. Quan Y, Xin Y, Tian G, Zhou J, Liu X. Mitochondrial ROS-Modulated mtDNA: a Potential Target for Cardiac Aging. Oxid Med Cell Longev. 2020;2020:9423593. doi:10.1155/2020/9423593
87. Guo C, Xia Y, Niu P, et al. Silica nanoparticles induce oxidative stress, inflammation, and endothelial dysfunction in vitro via activation of the MAPK/Nrf2 pathway and nuclear factor-kappaB signaling. Int J Nanomed. 2015;10:1463–1477. doi:10.2147/IJN.S76114
88. Hu C, Liu X, Zeng Y, Liu J, Wu F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin Clin Epigenet. 2021;13(1):166. doi:10.1186/s13148-021-01154-x
89. Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397(3):397–400. doi:10.1016/j.bbrc.2010.05.076
90. Gong C, Tao G, Yang L, et al. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2012;209(3):264–269. doi:10.1016/j.toxlet.2012.01.007
91. Smith AD, Lu C, Payne D, et al. Autocrine IL6-mediated activation of the STAT3-DNMT axis silences the TNFα-RIP1 necroptosis pathway to sustain survival and accumulation of myeloid-derived suppressor cells. Cancer Res. 2020;80(15):3145–3156. doi:10.1158/0008-5472.Can-19-3670
92. Luo JF, Zhou H, Lio CK. Akebia Saponin D inhibits the inflammatory reaction by inhibiting the IL-6-STAT3-DNMT3b axis and activating the Nrf2 pathway. Molecules. 2022;27(19):6236. doi:10.3390/molecules27196236
93. Mahmoud AM, Desouky EM, Hozayen WG, et al. Mesoporous Silica nanoparticles trigger liver and kidney injury and fibrosis via altering TLR4/NF-kappaB, JAK2/STAT3 and Nrf2/HO-1 signaling in rats. Biomolecules. 2019;9(10):528. doi:10.3390/biom9100528
94. Yu Y, Zhu T, Li Y, et al. Repeated intravenous administration of silica nanoparticles induces pulmonary inflammation and collagen accumulation via JAK2/STAT3 and TGF-β/Smad3 pathways in vivo. Int J Nanomed. 2019;14:7237–7247. doi:10.2147/ijn.S209458
95. Kundu J, Kim DH, Chae IG, et al. Silicon dioxide nanoparticles induce COX-2 expression through activation of STAT3 signaling pathway in HaCaT cells. Toxicol In Vitro. 2018;52:235–242. doi:10.1016/j.tiv.2018.06.008
96. Huang DY, Lu ST, Chen YS, Cheng CY, Lin WW. Epigenetic upregulation of spleen tyrosine kinase in cancer cells through p53-dependent downregulation of DNA methyltransferase. Exp Cell Res. 2023;425(2):113540. doi:10.1016/j.yexcr.2023.113540
97. Sandoval JE, Reich NO. p53 and TDG are dominant in regulating the activity of the human de novo DNA methyltransferase DNMT3A on nucleosomes. J Biol Chem. 2021;296:100058. doi:10.1074/jbc.RA120.016125
98. Zhao X, Wu Y, Li J, et al. JNK activation-mediated nuclear SIRT1 protein suppression contributes to silica nanoparticle-induced pulmonary damage via p53 acetylation and cytoplasmic localisation. Toxicology. 2019;423:42–53. doi:10.1016/j.tox.2019.05.003
99. López-Moyado IF, Ko M, Hogan PG, Rao A. TET enzymes in the immune system: from DNA demethylation to immunotherapy, inflammation, and cancer. Annu Rev Immunol. 2024;42(1):455–488. doi:10.1146/annurev-immunol-080223-044610
100. Wu Q, Ni X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr Drug Targets. 2015;16(1):13–19. doi:10.2174/1389450116666150113121054
101. Refsnes M, Skuland T, Lilleaas E, Øvrevik J, Låg M. Concentration-dependent cytokine responses of silica nanoparticles and role of ROS in human lung epithelial cells. Basic Clin Pharmacol Toxicol. 2019;125(3):304–314. doi:10.1111/bcpt.13221
102. Liu X, Lu B, Fu J, Zhu X, Song E, Song Y. Amorphous silica nanoparticles induce inflammation via activation of NLRP3 inflammasome and HMGB1/TLR4/MYD88/NF-kb signaling pathway in HUVEC cells. J Hazard Mater. 2021;404(Pt B):124050. doi:10.1016/j.jhazmat.2020.124050
103. Feng L, Ning R, Liu J, et al. Silica nanoparticles induce JNK-mediated inflammation and myocardial contractile dysfunction. J Hazard Mater. 2020;391:122206. doi:10.1016/j.jhazmat.2020.122206
104. Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: a mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67(12):5583–5586. doi:10.1158/0008-5472.Can-07-0846
105. Balasubramanian S, Gunasekaran K, Sasidharan S, Jeyamanickavel Mathan V, Perumal E. MicroRNAs and xenobiotic toxicity: an overview. Toxicol Rep. 2020;7:583–595. doi:10.1016/j.toxrep.2020.04.010
106. Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25(1):1–16. doi:10.1261/rna.068692.118
107. Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-B signaling. J Mol Cell Biol. 2011;3(3):159–166. doi:10.1093/jmcb/mjr007
108. Wu Y, Jin Y, Sun T, et al. p62/SQSTM1 accumulation due to degradation inhibition and transcriptional activation plays a critical role in silica nanoparticle-induced airway inflammation via NF-κB activation. J Nanobiotechnol. 2020;18(1):77. doi:10.1186/s12951-020-00634-1
109. Ma Y, Liang Q, Wang F, et al. Silica nanoparticles induce pulmonary autophagy dysfunction and epithelial-to-mesenchymal transition via p62/NF-κB signaling pathway. Ecotoxicol Environ Saf. 2022;232:113303. doi:10.1016/j.ecoenv.2022.113303
110. Capaccia C, Diverio S, Zampini D, Guelfi G. The complex interaction between P53 and miRNAs joins new awareness in physiological stress responses. Cells. 2022;11(10):1631. doi:10.3390/cells11101631
111. Psathas JN, Thomas-Tikhonenko A. MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med. 2014;4(8):a014175. doi:10.1101/cshperspect.a014175
112. Seidel C, Kirsch A, Fontana C, et al. Epigenetic changes in the early stage of silica-induced cell transformation. Nanotoxicology. 2017;11(7):923–935. doi:10.1080/17435390.2017.1382599
113. Arnst J, Jing Z, Cohen C, Ha SW, Viggeswarapu M, Beck GR Jr. Bioactive silica nanoparticles target autophagy, NF-κB, and MAPK pathways to inhibit osteoclastogenesis. Biomaterials. 2023;301:122238. doi:10.1016/j.biomaterials.2023.122238
114. Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi:10.1016/j.cell.2012.03.003
115. Xue J, Li X, Chi Y, et al. Decabromodiphenyl ether induces the chromosome association disorders of spermatocytes and deformation failures of spermatids in mice. J Environ Sci. 2024;138:531–542. doi:10.1016/j.jes.2023.03.043
116. Lin YJ, Yang CC, Lee IT, et al. Reactive oxygen species-dependent activation of EGFR/Akt/p38 mitogen-activated protein kinase and JNK1/2/FoxO1 and AP-1 pathways in human pulmonary alveolar epithelial cells leads to up-regulation of COX-2/PGE(2) induced by silica nanoparticles. Biomedicines. 2023;11(10):2628. doi:10.3390/biomedicines11102628
117. Tian J, Li J, Yin H, et al. In vitro and in vivo uterine metabolic disorders induced by silica nanoparticle through the AMPK signaling pathway. Sci Total Environ. 2021;762:143152. doi:10.1016/j.scitotenv.2020.143152
118. Fan X, Wang Y, Fan J, Chen R. Deletion of SMURF 1 represses ovarian cancer invasion and EMT by modulating the DAB2IP/AKT/Skp2 feedback loop. J Cell Biochem. 2019;120(6):10643–10651. doi:10.1002/jcb.28354
119. Hu R, Wang MQ, Liu LY, et al. Calycosin inhibited autophagy and oxidative stress in chronic kidney disease skeletal muscle atrophy by regulating AMPK/SKP2/CARM1 signalling pathway. J Cell Mol Med. 2020;24(19):11084–11099. doi:10.1111/jcmm.15514
120. Fan Y, Yang Q, Yang Y, et al. Sirt6 suppresses high glucose-induced mitochondrial dysfunction and apoptosis in podocytes through AMPK activation. Int J Biol Sci. 2019;15(3):701–713. doi:10.7150/ijbs.29323
121. Lee KI, Su CC, Fang KM, Wu CC, Wu CT, Chen YW. Ultrafine silicon dioxide nanoparticles cause lung epithelial cells apoptosis via oxidative stress-activated PI3K/Akt-mediated mitochondria- and endoplasmic reticulum stress-dependent signaling pathways. Sci Rep. 2020;10(1):9928. doi:10.1038/s41598-020-66644-z
122. Baker JR, Vuppusetty C, Colley T, et al. Oxidative stress dependent microRNA-34a activation via PI3Kα reduces the expression of sirtuin-1 and sirtuin-6 in epithelial cells. Sci Rep. 2016;6:35871. doi:10.1038/srep35871
123. Gao F, Ma N, Zhou H, et al. Zinc oxide nanoparticles-induced epigenetic change and G2/M arrest are associated with apoptosis in human epidermal keratinocytes. Int J Nanomed. 2016;11:3859–3874. doi:10.2147/ijn.S107021
124. Yuan J, Mo Y, Zhang Y, Zhang Y, Zhang Q. Nickel nanoparticles induce epithelial-mesenchymal transition in human bronchial epithelial cells via the HIF-1α/HDAC3 pathway. Nanotoxicology. 2022;16(6–8):695–712. doi:10.1080/17435390.2022.2142169
125. Tanaka Y-K, Ogra Y. Quantitative determination of the intracellular uptake of silica nanoparticles using asymmetric flow field flow fractionation coupled with ICP mass spectrometry and their cytotoxicity in HepG2 cells. Arch Toxicol. 2024;98(3):769–777. doi:10.1007/s00204-023-03672-4
126. Dong X, Wu Z, Li X, et al. The size-dependent cytotoxicity of amorphous silica nanoparticles: a systematic review of in vitro studies. Int J Nanomed. 2020;15:9089–9113. doi:10.2147/IJN.S276105
127. Miao C, Jia P, Luo C, et al. The size-dependent in vivo toxicity of amorphous silica nanoparticles: a systematic review. Ecotoxicol Environ Saf. 2024;271:115910. doi:10.1016/j.ecoenv.2023.115910
128. Chan WT, Liu CC, Chiang Chiau JS, et al. In vivo toxicologic study of larger silica nanoparticles in mice. Int J Nanomed. 2017;12:3421–3432. doi:10.2147/IJN.S126823
129. Ferreira LF, Picco AS, Galdino FE, Albuquerque LJC, Berret JF, Cardoso MB. Nanoparticle-protein interaction: demystifying the correlation between protein corona and aggregation phenomena. ACS Appl Mater Interfaces. 2022;14(25):28559–28569. doi:10.1021/acsami.2c05362
130. Yang Y, Du X, Wang Q, et al. Mechanism of cell death induced by silica nanoparticles in hepatocyte cells is by apoptosis. Int J Mol Med. 2019;44(3):903–912. doi:10.3892/ijmm.2019.4265
131. Jimenez-Villarreal J, Rivas-Armendariz DI, Arellano Perez-Vertti RD, et al. Relationship between lymphocyte DNA fragmentation and dose of iron oxide (Fe2O3) and silicon oxide (SiO2) nanoparticles. Genet Mol Res. 2017;16(1). doi:10.4238/gmr16019206
132. Liu N, Tang M. Toxic effects and involved molecular pathways of nanoparticles on cells and subcellular organelles. J Appl Toxicol. 2020;40(1):16–36. doi:10.1002/jat.3817
133. Parodi A, Evangelopoulos M, Arrighetti N, et al. Endosomal escape of polymer‐coated silica nanoparticles in endothelial cells. Small. 2020;16(36). doi:10.1002/smll.201907693
134. Hadji H, Bouchemal K. Effect of micro- and nanoparticle shape on biological processes. J Control Release. 2022;342:93–110. doi:10.1016/j.jconrel.2021.12.032
135. Meng H, Yang S, Li Z, et al. Aspect ratio determines the quantity of mesoporous silica nanoparticle uptake by a small GTPase-dependent macropinocytosis mechanism. ACS Nano. 2011;5(6):4434–4447. doi:10.1021/nn103344k
136. Banerjee A, Qi J, Gogoi R, Wong J, Mitragotri S. Role of nanoparticle size, shape and surface chemistry in oral drug delivery. J Control Release. 2016;238:176–185. doi:10.1016/j.jconrel.2016.07.051
137. Shao D, Lu M-M, Zhao Y-W, et al. The shape effect of magnetic mesoporous silica nanoparticles on endocytosis, biocompatibility and biodistribution. Acta Biomater. 2017;49:531–540. doi:10.1016/j.actbio.2016.11.007
138. Tan Y, Yu D, Feng J, et al. Toxicity evaluation of silica nanoparticles for delivery applications. Drug Deliv Transl Res. 2023;13(9):2213–2238. doi:10.1007/s13346-023-01312-z
139. Murugadoss S, Lison D, Godderis L, et al. Toxicology of silica nanoparticles: an update. Arch Toxicol. 2017;91(9):2967–3010. doi:10.1007/s00204-017-1993-y
140. Rafieepour A, Azari RM, Khodagholi F. Cytotoxic effects of crystalline silica in form of micro and nanoparticles on the human lung cell line A549. Toxicol Ind Health. 2023;39(1):23–35. doi:10.1177/07482337221140644
141. Hadrup N, Aimonen K, Ilves M, et al. Pulmonary toxicity of synthetic amorphous silica-effects of porosity and copper oxide doping. Nanotoxicology. 2021;15(1):96–113. doi:10.1080/17435390.2020.1842932
142. Cabellos J, Gimeno-Benito I, Catalan J, et al. Short-term oral administration of non-porous and mesoporous silica did not induce local or systemic toxicity in mice. Nanotoxicology. 2020;14(10):1324–1341. doi:10.1080/17435390.2020.1818325
143. Maser E, Schulz M, Sauer UG, et al. In vitro and in vivo genotoxicity investigations of differently sized amorphous SiO2 nanomaterials. Mutat Res Genet Toxicol Environ Mutagen. 2015;794:57–74. doi:10.1016/j.mrgentox.2015.10.005
144. Downs TR, Crosby ME, Hu T, et al. Silica nanoparticles administered at the maximum tolerated dose induce genotoxic effects through an inflammatory reaction while gold nanoparticles do not. Mutat Res. 2012;745(1–2):38–50. doi:10.1016/j.mrgentox.2012.03.012
145. Vecchio G, Fenech M, Pompa PP, Voelcker NH. Lab‐on‐a‐chip‐based high‐throughput screening of the genotoxicity of engineered nanomaterials. Small. 2014;10(13):2721–2734. doi:10.1002/smll.201303359
146. Kwon JY, Koedrith P, Seo YR. Current investigations into the genotoxicity of zinc oxide and silica nanoparticles in mammalian models in vitro and in vivo: carcinogenic/genotoxic potential, relevant mechanisms and biomarkers, artifacts, and limitations. Int J Nanomed. 2014;9:271–286. doi:10.2147/ijn.S57918
147. Lee GH, Kim YS, Kwon E, Yun JW, Kang BC. Toxicologic evaluation for amorphous silica nanoparticles: genotoxic and non-genotoxic tumor-promoting potential. Pharmaceutics. 2020;12(9):826. doi:10.3390/pharmaceutics12090826
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.